Abstract
Salmonella enterica serovar Typhimurium DT104 is a recognized food-borne pathogen that displays a multidrug-resistant phenotype and that is associated with systemic infections. At one extreme of the food chain, this bacterium can infect humans, limiting the treatment options available and thereby contributing to increased morbidity and mortality. Although the antibiotic resistance profile is well defined, little is known about other phenotypes that may be expressed by this pathogen at key points across the pork production food chain. In this study, 172 Salmonella enterica serovar Typhimurium DT104/DT104b isolated from an extensive “farm-to-fork” surveillance study, focusing on the pork food chain, were characterized in detail. Isolates were cultured from environmental, processing, retail, and clinical sources, and the study focused on phenotypes that may have contributed to persistence/survival in these different niches. Molecular subtypes, along with antibiotic resistance profiles, tolerance to biocides, motility, and biofilm formation, were determined. As a basis for human infection, acid survival and the ability to utilize a range of energy sources and to adhere to and/or invade Caco-2 cells were also studied. Comparative alterations to biocide tolerance were observed in isolates from retail. l-Tartaric acid and d-mannose-1-phosphate induced the formation of biofilms in a preselected subset of strains, independent of their origin. All clinical isolates were motile and demonstrated an enhanced ability to survive in acidic conditions. Our data report on a diverse phenotype, expressed by S. Typhimurium isolates cultured from the pork production food chain. Extending our understanding of the means by which this pathogen adapts to environmental niches along the “farm-to-fork” continuum will facilitate the protection of vulnerable consumers through targeted improvements in food safety measures.
INTRODUCTION
Salmonella enterica serovar Typhimurium is a well-known food-borne zoonotic pathogen that causes gastrointestinal disease in humans and is of importance to public health (1). Increased morbidity and mortality is associated with the multidrug-resistant (MDR) phenotype, which is expressed by variants of this bacterium (2). MDR-Salmonella Typhimurium definitive phage type 104 (DT104) is associated with food-producing animals, a feature that has focused attention on the development of strategies designed to reduce the selective pressure following the use of antimicrobial agents for livestock production (3). The majority of the studies report on the use of subtyping methods to follow the clonal dissemination of Salmonella (4). However, only very few studies, if any, consider this dissemination in the context of the bacterial phenotype, i.e., how changes in Salmonella phenotype could support bacterial adaptation, as the nature of the selective pressures change across the food chain.
Strategies used to develop microbial risk assessments consider the bacterial load, the frequency of contamination, the potential for bacterial growth to occur, and the amount consumed (5). Whether or not Salmonella species and other bacteria of public health importance can adapt as they cross the food chain is not well understood. Such adaptation could increase the virulence of the bacterium arising from cross-resistance to other stresses applied to eliminate the pathogen (including the use of biocides to control the production environment and acid produced by the animal and human stomach). Salmonella species capable of adapting to these stresses would likely increase the risk to public health (6).
A previous study, conducted in the Republic of Ireland, investigated selected points of Salmonella contamination from the time pigs entered the lairage to the time the carcass was processed in the boning hall, to determine the importance of different sources of Salmonella along the Irish pork production chain (7). Lairage was found to be the major source of cross-contamination with Salmonella, followed by the hands of evisceration operatives, conveyor belts, and equipment in the boning hall. This study, as well as others (8, 9), highlighted not only the major source of contamination along the food chain but also the critical points that should be assessed. However, there are to date very few studies that assess the change in the bacterial phenotype in response to the stresses encountered along the food chain. With that in mind, we designed the present study to assess the phenotypic variation of Salmonella Typhimurium DT104 isolates previously subtyped and determined to be largely indistinguishable, based on a computational analysis of their DNA macrorestriction profiles. All were cultured from the pork production chain and were composed of definitive phage types DT104 (n = 85) and DT104b (n = 87). Each isolate was extensively characterized, as shown in Fig. 1, and its phenotype was determined using tests relevant to stresses encountered at the various culturing points across the food chain. The relationship(s) of all phenotypes observed was thoroughly assessed by using suitable statistical algorithms to explore associations between these phenotypes and to determine any critical features that might be useful for inclusion as a future food safety measure.
Fig 1.
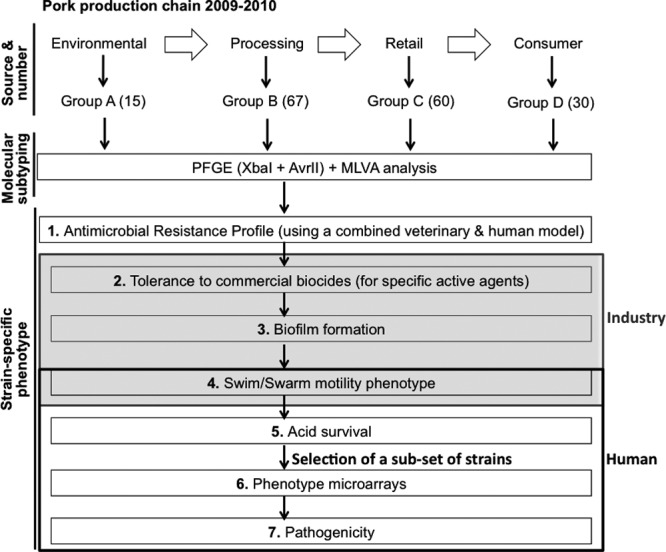
Study design. A total of 172 S. Typhimurium DT104 strains (collected from 2002 to 2010) originating from the pork food chain in Ireland were studied. Group A, isolates obtained from the environment (n = 15); group B, isolates obtained from the processing chain (n = 67); group C, isolates obtained from retail outlets (n = 60); group D, clinical isolates (n = 30).
MATERIALS AND METHODS
Bacterial isolates and culture methods.
A total of 172 Salmonella Typhimurium DT104/DT104b strains originating from the pork food chain, in Ireland, were included for study. These strains were cultured as part of an extensive surveillance study of the Irish pork food chain (7, 10, 11). The collection included bacteria cultured from the farm environment (11), the abattoir environment (7), retail pork products (10), and from clinical specimens (gently given by Martin Cormican, from the National Salmonella Reference Laboratory, NUI Galway, Ireland). In the present study a subset of these isolates was chosen for study and divided into groups according to their origin: group A, isolates obtained from the environment (n = 15); group B, isolates obtained from the processing chain (n = 67); group C, isolates obtained from retail outlets (n = 60); and group D, clinical isolates (n = 30) (see Dataset S1 in the supplemental material). All were collected between 2002 and 2010. Two phage types are represented and these consisted of DT104 (n = 85) and DT104b (n = 87). Salmonella Typhimurium DT104 NCTC 13348 was included as a reference strain in all of the assays performed.
All Salmonella isolates were stored on beads in cryopreservation fluid at −80°C (Technical Service Consultants, Ltd., Lancashire, England). When needed, isolates were streaked onto Mueller-Hinton (MH) agar (Oxoid, Cambridge, United Kingdom), and a few isolated colonies were taken and used to inoculate 5 ml of MH broth (Oxoid) and then grown overnight at 37°C, with shaking at ∼200 rpm. The overnight culture was then used to perform the tests described in more detail below.
PFGE.
Molecular subtyping was carried out on all isolates. Pulsed-field gel electrophoresis (PFGE) was performed using the restriction endonucleases XbaI and AvrII (New England BioLabs) by following the CDC PulseNet protocol (http://www.cdc.gov/pulsenet/protocols.htm). Salmonella enterica serovar Braenderup H9812 (ATCC BAA-664) was used as the reference strain for the molecular size standard. Gels were stained in a 1-μg/ml solution of ethidium bromide and visualized under UV light transillumination with Gel Doc (Bio-Rad, Munich, Germany). Macrorestriction patterns were compared using the BioNumerics fingerprinting software (version 6.5; Applied Math, Austin, TX). The similarity index of the isolates was calculated using the Dice correlation coefficient with a band position tolerance of 1% and an optimization of 1%. The unweighted-pair group method using average linkages (UPGMA) was used to construct a combined-restriction profile dendrogram. Generation of the dendrogram was based on a single-experiment analysis (PFGE only) using the two restriction enzymes XbaI and AvrII.
MLVA subtyping.
Multilocus variable number of tandem repeats analysis (MLVA) was performed by a modified method based on a previously described protocol (12) using an Agilent 2100 Bioanalyzer (Agilent Technologies Ireland, Ltd.). Briefly, MLVA was performed using primers designed to amplify repeats for the five loci detailed. These primers were denoted as ST7, ST6, ST5, ST2, and ST8 (Centers for Disease Control and Prevention [CDC], PulseNet) for the five of the loci detailed (12, 13). These loci were located on the main chromosome of S. Typhimurium LT2 (AE 006468). Details of the loci, repeat sizes, and PCR and thermal cycling conditions were as previously published (14). Amplicon size estimates were obtained by using the Agilent 2100 Expert Software Version B.02.03.SI307 firmware C.01.055 (Agilent Technologies). Unique alleles at each locus were assigned numbers, which were used for strain comparison purposes. Allele numbers were assigned based on fragment size offset (the base pair length of the left and right flanking regions of the tandem repeat loci) divided by the copy number in base pairs. The loci in the allele strings were previously described (12; CDC, PulseNet) and imported into BioNumerics. Population modeling using the minimum-spanning tree method was carried out on all of the isolates typed. A minimum spanning tree was constructed using the single-locus variants as a priority rule with the creation of hypothetical (or missing) types and based on the isolate's source.
Antibiotic susceptibility profiling.
Resistance profiles to various antimicrobial agents—ampicillin (A), chloramphenicol (C), gentamicin (Gm), kanamycin (Kan), nalidixic acid (NAL), streptomycin (S), sulfonamides (Su), tetracycline (T), and trimethoprim (Tm)—were determined for all isolates. This was accomplished by using the Kirby-Bauer method and susceptibility or resistance classified according to Clinical and Laboratory Standards Institute (CLSI) guidelines (15). S. Typhimurium DT104 NCTC 13348 (reference strain) and Escherichia coli ATCC 25922 (control strain) were also included.
Tolerance to commercially available biocidal actives.
All isolates were tested for their susceptibility to four biocidal actives (compounds normally used alone or as a component in biocides formulations) used in food grade sanitizer formulations. The biocidal actives tested were benzalkonium chloride, chlorhexidine, hydrogen peroxide, and triclosan. All of the compounds were prepared fresh on the day of the experiment. Triclosan was prepared in absolute ethanol to a final concentration of 0.1 mg/ml; chlorhexidine, benzalkonium chloride, and hydrogen peroxide were prepared in sterile water to final concentrations of 1 mg/ml, 5 mg/ml, and 100 mM, respectively. From these stocks, working solutions were prepared for triclosan (0.02 mg/ml), chlorhexidine (0.5 mg/ml), benzalkonium chloride (0.5 mg/ml), and hydrogen peroxide (40 mM). The pH of the chlorhexidine solution was adjusted to ∼7 using sodium carbonate solution (1 M) and 10 μl of acetic acid. The tolerance to the biocidal actives was evaluated by determining the MICs in 96-well plates by the broth microdilution method, as described previously (16). All assays were performed in triplicate on three separate occasions.
Biofilm formation.
The ability to form biofilms was investigated, and the method used was adapted from a previously described study using crystal violet (CV) dye binding (17). The quantitative analysis of biofilm production was performed by adding 150 μl of 95% (vol/vol) ethanol to destain the wells, and the optical density at 595 nm (OD595) was measured using a microtiter plate reader (Tecan, Männedorf, Switzerland). The average OD of the negative control (denoted as ODc) was subtracted from the OD595 for tested wells. The experiment was performed on three separate occasions for all S. Typhimurium isolates. Averages and standard deviations were calculated for all experiments. The ability of the isolates to form biofilm was classified using the following algorithm: OD595 ≤ ODc indicates no biofilm formation, ODc < OD595 ≤ (2 × ODc) indicates weak biofilm formation, (2 × ODc) < OD595 ≤ (4 × ODc) indicates moderate biofilm formation, and (4 × ODc) < OD595 indicates strong biofilm formation (18).
Bacterial motility assays.
To assess bacterial motility, swim and swarm assays were performed on all isolates. For the swim assay, culture plates were previously prepared (Luria-Bertani [LB] broth with 0.3% [wt/vol] agar; Sigma) and stab inoculated (19). These plates were then incubated for 8 h at 37°C. The region of visible colony spread on the agar was measured (in mm) with Vernier calipers. Swarm motility assays were done and bacterial cells were grown overnight in LB broth. One microliter of this cell suspension was spotted directly onto a motility plate (LB broth with 0.6% [wt/vol] agar with 0.5% glucose; Sigma) and incubated at 37°C overnight for 24 h (20). The region of visible colony spread on the agar was recorded.
Acid resistance.
The ability of all bacterial isolates to survive a low-pH environment or culture medium was evaluated according to the method described by Berk et al. (21) with minor modifications. Briefly, bacterial cultures were grown overnight (18 h) without shaking in 3 ml of buffered LB (pH 5) containing 0.4% (wt/vol) glucose (LBG) at 37°C. The overnight cultures should yield ∼108 CFU/ml. The pH of the culture medium after overnight incubation was ∼4.6. On the next day, an aliquot of 333.4 μl of the overnight culture (diluted 1:6) was taken and centrifuged at 16,060 × g for 3 min. The supernatant was discarded, and the cells were resuspended in 2 ml of prewarmed LBG at pH 2.5 (prepared fresh on the day of the experiment) and incubated at 37°C in a prewarmed 24-well plate for a defined period of time. Time points selected for the present study included: t = 0 h, before acidification; t = 1 h, and t = 2 h. At the defined time points, a 10-μl aliquot of the bacterial culture was removed and diluted in 1,990 μl of maximum recovery diluent (MRD) medium (Oxoid) (1:200 dilution gives a pH of ∼7.0) distributed into a 24-well plate. Bacteria were allowed to recover for 30 min at room temperature. Samples were then serially diluted in MRD, and 100-μl aliquots of the relevant dilutions were plated on Luria-Bertani agar plates for CFU counting. The plates were then incubated for 18 h at 37°C. On the following day, CFU were counted, and the survival rates calculated according to the following formula: %AR = (CFUtx/CFUt0) × 100, where AR denotes acid resistance, t is time, x is the selected time point (1 or 2 h), and t0 denotes the initial time point.
Selection of strains for phenotypic microarray and ex vivo studies.
Based on their acid survival ability, two isolates from each of the defined groups (A through D) were selected for further studies. Isolates that showed a marked ability to survive in acidic conditions compared to other members from their group, were studied in order to assess the growth in specific substrate conditions, biofilm formation under those same conditions, and the ability to adhere/invade epithelial cells.
Phenotypic microarrays.
Metabolic profiling of all isolates was performed using a phenotypic microarray, wherein respiration output was monitored in real time. The ability of the bacterial strains to respond to different carbon/energy sources was examined using the Biolog phenotype microarray platform (Biolog, Inc., Hayward, CA). Briefly, bacteria were cultured on blood agar overnight at 37°C. Distinct colonies were picked with a sterile cotton swab and suspended in 10 ml of IF-0a (Biolog), and the cell density adjusted to an OD600 of 0.035 with a spectrophotometer (Biomate 5; Thermospectronic, Cambridge, United Kingdom). A 750-μl aliquot of this cell suspension was added to 150 ml of IF-10 (Biolog). Microtiter plates (PM plates; PM1 through PM4, together with PM9 and PM10) were inoculated with 100 μl of the cell suspension per well and then incubated at 37°C for 48 h in an OmniLog incubator (Biolog) and monitored continuously for color changes in the wells. Kinetic data were analyzed with OmniLog PM software (Biolog, Inc., Hayward, CA). All of these assays were conducted in duplicate.
Biofilm formation under defined substrate-growth conditions.
As an addendum to the PM analysis, the ability to form biofilms under these conditions was also tested, using the CV assay (17). Briefly, plates used for the phenotypic array were incubated for further 24 h at 37°C. After this incubation period, the supernatants were discarded. Each well was washed three times with phosphate-buffered saline (PBS). Plates were air dried for 15 min, after which 100 μl of 0.4% (vol/vol) crystal violet solution was added to each well. Plates were then incubated for 15 min at room temperature. A further wash was performed (three times with PBS), and plates were left to air dry for a further 15 min. Finally, 100 μl of acetic acid (33% [vol/vol]) were added to each well, and the OD measured using a microplate reader (Multiskan FC Microplate Photometer with incubator; Thermo Scientific) at 570 nm.
Ex vivo studies: adhesion and invasion of Caco-2 cells.
Caco-2 cells were cultured in Dulbecco modified Eagle medium (DMEM)-high glucose, with l-glutamine and sodium pyruvate (Thermo Fisher Scientific, Ireland) supplemented with 10% (vol/vol) fetal bovine serum, in a humidified atmosphere of 5% (vol/vol) CO2 at 37°C. Cells were then seeded (105 cells/ml) into 24-well plates and cultured for ∼15 days. Cell medium was changed three times during each 7-day period. An overnight culture of bacteria was diluted, 1 in 100 into freshly prepared LB broth and allowed to grow to mid-log phase. Bacterial cells were collected by centrifugation (1,968 × g for 10 min) and resuspended in PBS at a concentration of ∼108 bacteria per ml. Caco-2 cells previously cultured in the 24-well plates were washed twice in PBS, serum-free medium was then added, and cells were allowed to equilibrate for 2 h in 5% (vol/vol) CO2 at 37°C. The DMEM serum-free medium was removed, and the Caco-2 cells were incubated with the bacterial inoculum for 1 h in 5% (vol/vol) CO2 at 37°C.
For the adhesion assay, after infection, the Caco-2 cells were washed four times with PBS before disruption with a 1-ml volume of PBS containing 1% (vol/vol) Triton X-100 (Sigma-Aldrich). Cells where then incubated at room temperature for 5 min. The number of bacteria per well was determined by plating appropriate dilutions of this final suspension directly onto LB agar. Numbers of adherent bacteria were reported as the number of total bacteria (adhered and invaded) less the number of invaded bacteria.
For the invasion assay, after the bacteria were allowed to adhere to the monolayers, wells were washed twice with Hanks' balanced salt solution (HBSS) (Sigma-Aldrich), which was then replaced with DMEM containing gentamicin sulfate (50 μg/ml; Sigma-Aldrich), followed by incubation for 30 min to kill all external bacteria. The culture medium was then removed, and the cells were washed twice with HBSS to remove any residual gentamicin sulfate before lysis with 1% (vol/vol) Triton X-100. The number of bacteria invaded was estimated by plating serial dilutions. Adhesion and invasion assays were performed on three separate occasions, with four wells per assay used for each isolate.
Statistical analysis.
All data were analyzed using Microsoft Excel and PASW Statistics 20.0 software. The average MIC values from the biocides were examined using descriptive statistics. The Student t test and null hypothesis were performed to access significance in the different groups of isolates. Possible correlations were analyzed by performing a Spearman's rho correlation test. Significance was defined at the 0.01 and 0.05 levels.
RESULTS
Bacterial isolates, their sources, and molecular subtypes.
A total of 172 Salmonella Typhimurium DT104 and DT104b strains were included for study. These isolates were divided into categories according to their origin, namely: group A, isolates from environmental sources (n = 15); group B, isolates obtained in the pig processing environment (n = 67); group C, isolates from retail outlets (n = 60); and group D, clinical sources (n = 30) (see Dataset S1 in the supplemental material). All were collected from 2002 to 2010 and consisted of Salmonella strains DT104 (n = 85) and DT104b (n = 87). Salmonella Typhimurium DT104 NCTC 13348 was included as a reference strain, for comparative purposes, in all of the assays performed.
PFGE analysis of S. Typhimurium DT104 was carried out using the restriction enzymes, XbaI and AvrII. We observed ca. 24 different PFGE patterns among the 172 isolates. To analyze the clonal relationships between the isolates according to the PFGE patterns, a dendrogram was generated using UPGMA algorithms with Dice coefficients (Fig. 2A). Four distinct clusters (denoted I to IV) with >85% similarity were designated, three of which were major clonal groups (I, III, and IV) (Fig. 2A). Based on the macrorestriction profiles, isolates could be clustered as follows: those cultured from processing samples, clustered almost exclusively together (clusters I and III) or with isolates cultured from the environment and/or retail outlets (cluster II). Clinical isolates clustered almost exclusively together (cluster IV).
Fig 2.
(A) PFGE analysis of S. Typhimurium DT104 using XbaI and AvrII. Isolate groups were color identified for simplicity as follows, and the color codes were maintained in all of the assays: reference strain (white); group A, environmental (green); group B, processing (red); group C, retail (blue); and group D, clinical (yellow). Clonal relationships between the isolates were analyzed according to the PFGE patterns. A dendrogram was generated using UPGMA algorithms with Dice coefficients. Clusters (denoted I to IV) with >85% similarity were designated, three of which were major clonal groups (I, III, and IV). (B) MLVA analysis of S. Typhimurium DT104 isolates. MLVA analysis was based on five VNTR loci. The MLVA profiles were used for categorical clustering in BioNumerics, and a minimum-spanning tree was constructed. MLVA clustering was considered if neighbors differed in no more than one of the five VNTR loci. Groups were additionally colored based on their source: reference strain (white); group A, environmental (green); group B, processing (red); group C, Retail (blue); and group D, clinical (yellow). These color groups were maintained in all of the assays (PFGE dendrogram clustering and phenotypic microarrays). Based on the isolates distribution, four main clusters depicted in panel B were labeled A, B, C, and D.
All isolates were also subtyped using MLVA based on five variable-number tandem repeat (VNTR) loci and were divided into different MLVA groups (Fig. 2B). The MLVA profiles were used for categorical clustering in BioNumerics, and a minimum-spanning tree was constructed (Fig. 2B). MLVA clustering was achieved if neighbors differed in no more than one of the five VNTR loci. As shown in Fig. 2B, the MLVA profiles were classified into four main clusters (A, B, C, and D). In general, the clustering patterns obtained from the MLVA subtyping, performed in all of the isolates, were similar to those obtained by PFGE. Environmental strains clustered with isolates obtained from the processing environment along with some from retail sources. The majority of clinical isolates clustered together in two main clusters (clusters A and C). Some clinical isolates also clustered with strains originated from processing and retail (clusters B and D) (Fig. 2B).
Susceptibility to antimicrobial compounds: antibiotics and biocidal actives.
The studied isolates showed a total of 14 different antibiotic resistance profiles, with the majority of the isolates exhibiting the classical ACSSuT resistance (n = 100), common among Salmonella Typhimurium DT104 strains (Fig. 3). Additional antibiotic resistances to gentamicin (Gm), kanamycin (Kan), nalidixic acid (NAL), and trimethoprim (Tm) were observed in processing (group B), retail (group C), and clinical (group D) isolates but not in environmental isolates (group A). When compared using their resistance profiles, bacteria cultured from retail outlets (group C), demonstrated the broadest resistance profile, being resistant to the greatest number of compounds. Notably, resistance to NAL was found only in the clinical group (group D; Fig. 3). Resistance to NAL, in patients infected with Salmonella, has been reported in cases of treatment failure with fluoroquinolones. Mutations in the gyrA gene and overexpression of efflux pumps are known to contribute to this resistance and to the decreased susceptibility to fluoroquinolones, such as ciprofloxacin (20).
Fig 3.

Susceptibility profile of the isolates to antimicrobial compounds. A, ampicillin; C, chloramphenicol; Gm, gentamicin; Kan, kanamycin; NAL, nalidixic acid; S, streptomycin; Su, sulfonamides; T, tetracycline; Tm, trimethoprim.
The ability of the isolates within this collection to tolerate biocidal actives, i.e., active components of certain biocides, was also investigated. Tolerance to a biocidal active component was defined as a >4-fold increase in the MIC compared to the reference strain. This measure was converted to a percent difference (as shown in Table 1). Among the environmental strains (group A), the majority showed an increased tolerance (IT) phenotype to hydrogen peroxide (60%, n = 9) compared to the reference strain. Isolates from retail sources (group C) showed an IT to three of the four active components tested (Table 1), namely, benzalkonium chloride (71.7%, n = 43), hydrogen peroxide (80%, n = 48), and triclosan (81.7%, n = 49). Isolates from processing points (group B) along the food chain, along with clinical isolates (group D), either showed no IT or had an IT value of <50%.
Table 1.
Tolerance to active components of biocides of Salmonella Typhimurium DT104/DT104b isolates
| Isolate group | No. | Tolerance to biocides (active components)a |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Benzalkonium chloride |
Chlorhexidine |
Hydrogen peroxide |
Triclosan |
||||||
| No. | % IT | No. | % IT | No. | % IT | No. | % IT | ||
| Environmental | 15 | 0 | 0 | 6 | 40 | 9 | 60.0 | 2 | 13.3 |
| Processing | 67 | 2 | 3.0 | 25 | 37.3 | 17 | 25.4 | 8 | 11.9 |
| Retail | 60 | 43 | 71.7 | 14 | 23.3 | 48 | 80.0 | 49 | 81.7 |
| Clinical | 30 | 1 | 3.3 | 5 | 16.7 | 1 | 3.3 | 0 | 0 |
No., number of isolates; IT, increased tolerance. Data in boldface indicate isolate groups that showed >50% IT to the active components of the biocides compared to the reference strain.
Since biocides are widely used in the modern food industry, concerns have been expressed about potential cross-resistance between biocides and other antimicrobial compounds. Based on this, we assessed possible correlations between increased cotolerance to two or more biocidal actives or antibiotics, namely, the possibility that strains that showed IT to one of the biocidal actives might also have an IT to other biocidal actives or antibiotics.
In the processing group (group B), significant cotolerance to triclosan and chlorhexidine (r = 0.336; P = 0.005) was observed in samples cultured from lairage swabs taken after the animals were slaughtered, from rectal contents, and from samples taken from the truck after transport at the 0.01 level. Isolates from this same group showed a significant cotolerance between benzalkonium chloride and hydrogen peroxide (r = 0.301; P = 0.013) and between hydrogen peroxide and chlorhexidine (r = 0.529; P = 0.000) at the 0.05 and 0.01 levels, respectively. Isolates from retail sources (group C) showed a significant correlation between the tolerance to benzalkonium chloride and chlorhexidine (r = 0.255; P = 0.049) at the 0.05 level. Within this same group, we also noted a significant correlation at the 0.01 level between the tolerance to benzalkonium chloride and hydrogen peroxide (r = 0.561; P = 0.000) and similarly for triclosan and hydrogen peroxide (r = 0.596; P = 0.000) and for benzalkonium chloride and triclosan (r = 0.551; P = 0.000). The latter possibly reflects the selection pressure imposed following biocide use. In the clinical group (group D), cross-tolerance was observed only for two of the biocides, namely, benzalkonium chloride and chlorhexidine (r = 0.398; P = 0.030), at the 0.05 level. These values were examined according to the nature of the sample taken and the isolate recovered. This was particularly relevant in the case of isolates obtained from retail sources (group C), since it is arguable that a greater dependence on biocidal formulations occurs at this point in the pig food chain (P = 0.000). When proposing a null hypothesis between the antibiotic resistance profile of the strains and the ability to tolerate biocides such as benzalkonium chloride, chlorhexidine, and hydrogen peroxide, this hypothesis was rejected, showing significance at the 0.05 level. In this way, significance was obtained between the antibiotic resistance profiles of the strains (i.e., ACSSuTTm and ACSSuT) and the tolerance to benzalkonium chloride (P = 0.000) and chlorhexidine (P = 0.009). In the case of the antibiotic resistance profile ACSSuTTm, there was also significance between this profile and the ability to tolerate hydrogen peroxide (P = 0.000).
Biofilm formation.
The ability of all S. Typhimurium isolates to form biofilms on PVC plastic surfaces indicated that all bacteria could attach to this abiotic surface type when grown in M9 minimal medium; however, none were determined to adhere strongly (Table 2) (23). Of the isolates recovered from the environment (group A), 33.3% (5/15) were classified as moderate biofilm formers with an average OD595 of 0.63. Of the remaining 10 isolates, 66.7% (10/15) were defined as weak biofilm formers, as measured by this method with an average OD595 of 0.45. In the processing group (group B; n = 67), 62.7% (42/67) were defined as moderate biofilm formers, while 37.3% (25/67) were weak. From the 60 retail samples (group C), equal numbers (50%) were moderate and weak biofilm formers. Among the clinical strains in group D, 73.3% (22/30) were moderate biofilm formers with an average OD595 of 0.8 and, of the remaining isolates, 26.7% (8/30) were weakly adherent, with an average OD595 of 0.57 (Table 2). By proposing a null hypothesis, in each case, possible significant links between biofilm formation and other phenotypes were tested. A link between the antibiotic resistance profile of the strains and the ability to form biofilms was rejected under this hypothesis, showing significance at the 0.05 level. Significance was obtained between the antibiotic resistance profile of the strains (ACSSuTNAL) and the ability to produce biofilms (P = 0.004). Interestingly, in the processing group (group B) there was a significant negative correlation between the antibiotic resistance profile of the isolates and their ability to form biofilms (r = −0.263; P = 0.031). The isolates were also assessed for possible correlations between the ability to tolerate biocides and form biofilms. When taking these parameters into account, only isolates obtained from the environment (group A) showed a significant negative correlation between the tolerance to benzalkonium chloride and the ability to form biofilms (r = −0.724; P = 0.002) (P < 0.01).
Table 2.
Biofilm formation ability of the Salmonella isolates assessed by the crystal violet-binding methoda
| Isolate group (no.) | Biofilm formationa |
|||
|---|---|---|---|---|
| Moderate |
Weak |
|||
| No. | % | No. | % | |
| Reference (1) | 1 | 100.0 | 0 | 0.0 |
| Environmental (15) | 5 | 33.3 | 10 | 66.7 |
| Processing (67) | 42 | 62.7 | 25 | 37.3 |
| Retail (60) | 30 | 50.0 | 30 | 50.0 |
| Clinical (30) | 22 | 73.3 | 8 | 26.7 |
Attachment to PVC plastic microplates was measured using a crystal violet assay after the cultures were incubated at 28°C for 72 h. Data in boldface indicate instances where >50% of the isolates formed biofilms in a moderate or weak manner. No., number of isolates.
Bacterial motility assays.
The swim activity phenotype of the strains in this collection was diverse (Fig. 4A; see also Table S1 in the supplemental material). Among the environmental isolates (group A), 3 isolates exhibited a reduced swim activity compared to the reference strain, while 12 isolates had a higher swim activity. In group B, 20 processing isolates had lower swim activity, one had an equivalent activity to the reference strain and 46 exhibited a higher swim activity compared to the reference strain. In this group, the swim phenotype was significantly different (P = 0.000) from the other groups. When the isolates from retail sources (group C) were considered, 31 showed a reduced swim activity compared to the reference, two isolates had an equivalent value, and 27 had higher swim activities. In the clinical group (group D), 3 isolates had a lower swim activity than the reference strain, and 27 isolates had a higher swim activity (see Table S1 in the supplemental material). This ability was significantly different (P = 0.000) from the other groups. In total, 57 isolates had a reduced ability to swim compared to the reference strain; three isolates showed an equivalent ability to swim compared to the reference strain, and 112 isolates had a higher ability to swim compared to the reference strain.
Fig 4.

Swim (A) and swarm (B) ability of the S. Typhimurium isolates. The reference strain showed an average swim ability of 65 mm. For plotting purposes, swarm motility was considered in the cases that confluence was achieved after 24 h of incubation.
The swarm activity phenotype also appeared to be variable. A majority of the isolates in the four groups exhibited swarm activity (being classified as motile in Fig. 4B). In the environmental group, 13 isolates were motile, and 2 were defined as being nonswarming. Among the processing group (group B), 62 isolates exhibited a swarm activity, and only 5 isolates were considered nonswarming. In the retail group, 59 isolates exhibited swarm activity, and only 1 isolate was negative (Fig. 4B). All of the clinical isolates (group D) demonstrated swarm activity (n = 30 isolates; 100%), and none were considered nonmotile.
When the swim and swarm abilities of the isolates of the collection were compared, statistical significance (P = 0.002) was observed between these two parameters. When the swim ability was analyzed for possible correlations with other phenotypes, a significant correlation was obtained with the antibiotic-resistant profile (r = 0.564; P = 0.028), the ability to survive in acidic conditions (r = 0.591; P = 0.020) and to tolerate triclosan (r = −0.595; P = 0.019) in the isolates obtained from the environment (group A). Isolates obtained from processing (group B) showed a significant correlations between the swim ability and the antibiotic resistance profile (r = −0.397; P = 0.001), the ability to survive in acidic conditions (r = 0.253; P = 0.039) and to tolerate triclosan (r = 0.325; P = 0.007). In groups C (retail) and D (clinical), no correlations were observed between the ability to swim and the other phenotypes.
Acid resistance among the bacterial collection.
The strains that showed a reduced ability to survive in acidic conditions were obtained from the environment (Fig. 5) (group A; n = 15; 13.3%), whereas in the group representative of processing points of the food chain (group B; n = 67) 95.5% could survive at pH 2.5 under the test conditions. The latter finding was similar to the strains from retail sources (group C; n = 60), 96.7% of which survived at this low pH. The highest resistance to acidic conditions was noted among the clinical isolates, with 100% survival (Fig. 5). This difference was statistically significant when the isolates from the environment (group A) were compared to isolates from processing (group B; P = 0.000), retail (group C; P = 0.000) or clinical (group D; P = 0.000) sources. When analyzing a possible correlation for the four groups between the ability to form biofilms and to survive in acidic conditions, the clinical isolates showed a significant correlation (r = 0.398; P = 0.029) between these two parameters at the 0.05 level. This correlation was only significant in this group of isolates. In the isolates obtained from the environment (group A), a significant correlation was obtained between the acid survival ability of these isolates and their antibiotic resistance profile (r = 0.782; P = 0.001) and the ability to swim (r = 0.591; P = 0.020) at the 0.01 and 0.05 levels, respectively. There was also a significant correlation (P < 0.05) between acid survival (after 2 h) and tolerance to triclosan (r = 0.254; P = 0.038) and the ability to swim (r = 0.253; P = 0.039) in the strains obtained from processing (group B). In the retail group (group C), no significant correlations were obtained. In the clinical group (group D), the ability to survive in acidic conditions and to form biofilms was significantly correlated (r = 0.398; P = 0.029) at the 0.05 level.
Fig 5.
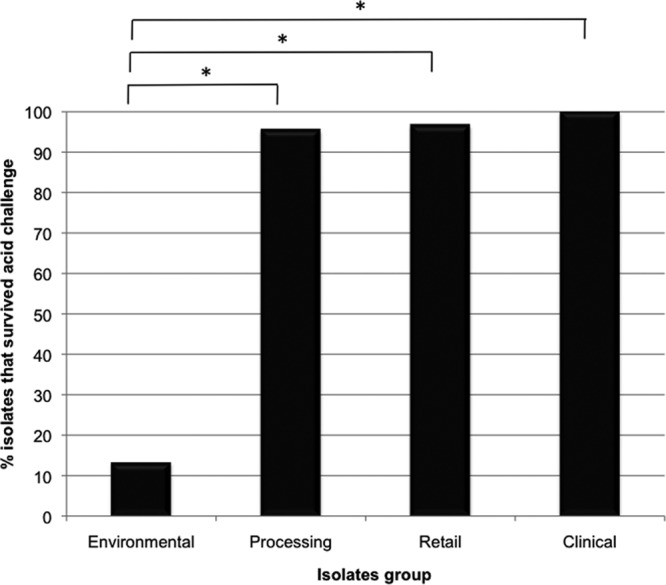
Acid survival ability of S. Typhimurium strains. Statistical difference was obtained in the resistance to acid compared to the reference strain. The reference strain did not survived when exposed to the acidic conditions (2 h after exposure at pH 2.5).
Phenotypic microarrays.
In order to assess in more detail possible changes in the isolates phenotype when grown in the presence of different substrates, carbon sources or other chemical substances, a partial phenotypic microarray (PM1 to -4 and PM9 and -10) was performed on two isolates from each of the defined groups (A through D) that showed a marked ability to survive in acidic conditions. Differences observed in terms of slower growths were recorded when the isolates were grown in different carbon sources, osmolytes and at various pHs (Fig. 6A and see Table S2 in the supplemental material). The environmental isolates (group A) exhibited a slower growth rate in plates containing l-tartaric acid or uridine-2′-monophosphate. One of the isolates in this group also showed decreased growth when in the presence of l-glutamic acid at pH 4.5. In the presence of substrates such as l-tartaric acid, sodium lactate (at 3, 4, 5, and 10%), sodium nitrate (40 mM), and pH 9.5 and in the presence of l-tryptophan, the isolates from the different groups showed slower growth/increased sensitivity compared to the reference strain (see Table S2 in the supplemental material).
Fig 6.
(A) Phenotypic microarrays results showing the number of phenotypes lost (slower growth) by the tested isolates. The isolates (color coded according to their source) were grown in the presence of different conditions/substrates. (I) Carbon, phosphorus, and sulfur sources; (II) osmolytes; (III) pH. The plotted results are shown as difference (arbitrary units) in the growth of the isolates compared to the reference strain. (B) Phenotypic microarrays results showing the number of phenotypes gained (faster growth) by the tested isolates. The isolates were grown in the presence of different substrates, identified on the outside of the figure (clockwise orientation: pH, osmolytes and phosphorus and sulfur sources). All isolates were color coded (according to their source) for simplicity. Arrows represent the quantitative increase in growth units observed for the isolates when these were compared to the reference strain.
When in the presence of d-mannose-1-phosphate, isolates J3 (environmental, group A), O2 (processing, group B), ST1333 (retail, group C), and MS100185 (clinical, group D) exhibited faster growth compared to the reference strain. In the presence of l-tyrosine at pH 9.5, faster growth was also recorded for isolates A15 (environmental) and ST1164 (retail) (Fig. 6B and see Table S3 in the supplemental material). Two other substrates that induce faster growth for some isolates were agmatine at pH 9.5 (for isolate A15; environmental, group A) and 8% sodium lactate (for isolate A43; processing, group B). Of the clinical isolates, MS100185 showed faster growth in the presence of d-mannose-1-phosphate (Fig. 6B and see Table S3 in the supplemental material).
The ability of the isolates to form biofilms when in the presence of specific substrates was evaluated by processing the PM plates and using the CV-binding assay. Isolate A15 (environmental, group A) showed decreased growth in the presence of uridine-2′-monophosphate or l-glutamic acid at pH 4.5 and also a reduced ability to form biofilms in these conditions compared to the reference strain (Fig. 7A). Similarly, for isolate O2 (processing, group B) the ability to form biofilms was reduced compared to the reference strain in the presence of 3, 4, 5, and 10% sodium lactate and also at pH 9.5 in the presence of l-tryptophan (Fig. 7A).
Fig 7.
(A) Biofilm formation from Biolog plates (phenotypes lost). The ability of the isolates to form biofilms when in the presence of specific substrates was evaluated by processing the PM plates and using the CV-binding assay. (B) Biofilm formation from Biolog plates (phenotypes gained). The biofilm production was assessed after processing the PM plates and using the CV-binding assay.
When analyzed in more detail, the retail isolate ST1333 exhibited a slight decrease in the ability to form biofilms in the presence of 3 and 4% sodium lactate compared to the reference strain (Fig. 7A). The same phenotype was also noted for isolate ST1164 when grown at pH 9.5 in the presence of l-tryptophan. For the clinical isolate MS100410, the ability to form biofilms was decreased in the presence of 3, 4, 5, and 10% of sodium lactate compared to the reference strain (Fig. 7A). In the case of the clinical isolate MS100185, there was only a slight difference in the ability to produce biofilms at pH 9.5 and in the presence of phenylethylamine compared to the reference strain (Fig. 7A). In all of the cases mentioned previously the reference strain exhibited a greater ability to form biofilms in the presence of specific substrates. Isolates that exhibited a reduced growth rate compared to the reference strain were able to produce more biofilms, as measured by CV dye binding, when in the presence of l-tartaric acid (Fig. 7A).
In the presence of substrates such as d-mannose-1-phosphate, isolates that exhibited a greater growth (see Table S3 in the supplemental material) were also able to form stronger biofilm structures (Fig. 7B). In the presence of the latter substrate, the test strains J3 (group A, environment), O2 (group B, processing), ST1333 (group C, retail), and MS100185 (group D, clinical) exhibited an enhanced ability to form biofilms compared to the reference strain. At pH 9.5 and in the presence of l-tyrosine, isolates A15 (group A, environment) and ST1164 (group C, retail) also showed increased biofilm structures (Fig. 7B).
Ex vivo studies: adhesion and invasion of Caco-2 cells.
The ability of a subset of isolates to adhere and invade Caco-2 epithelial cells was evaluated (Fig. 8). Based on these data, the adherence properties were similar, since representative isolates from all four groups could adhere to Caco-2 cells to a similar level (Fig. 8). Isolates A15 (group A, environment) and A43 (group B, processing) exhibited a slightly greater ability to adhere to the epithelial cells, followed by the two clinical isolates (group D, MS100185 and MS100410). For some isolates, the adherence to Caco-2 cells did not always correlate with the ability to invade these cells (Fig. 8).
Fig 8.
Adhesion and invasion of the Salmonella isolates to Caco-2 cells. In this assay, the adherent bacteria are reported as the number of total bacteria (adhered and invading) minus the number of invading bacteria. The number of invading bacteria was estimated by plating serial dilutions. Adhesion and invasion assays were performed on three separate occasions, with four wells per assay used for each isolate.
Isolates A15 (group A, environment) and A43 (group B, processing) showed a greater ability to invade the Caco-2 cells compared to all of the other strains. These were the same isolates that showed greater adherence abilities (Fig. 8). Isolates O2 (group B, processing), ST1164 and ST1333 (group C, retail), and MS100410 (group D, clinical) exhibited a reduced ability to invade Caco-2 cells compared to the reference strain. The clinical isolate MS100410 (group D) could invade Caco-2 cells, but at a reduced level, despite its greater ability to previously adhere (Fig. 8) to these cells.
DISCUSSION
Molecular subtyping of strains is the gold standard for tracking Salmonella contamination along the food chain (9, 13). To date, there have been few, if any, studies examining the corresponding phenome(s) of these Salmonella isolates. The present study sought to examine the phenotypic characteristics of S. Typhimurium DT104 and DT104b isolates collected between 2002 and 2010 from across the pork “farm-to-fork” food chain, as well as from clinical samples (see Dataset S1 in the supplemental material), and to assess their ability to survive and adapt to various physiological challenges, including nutrient depletion, acidic conditions, and tolerance to biocides.
All bacterial isolates were initially subtyped by PFGE and MLVA in order to reduce any bias, based on the macrorestriction profiles. Unsurprisingly, for phage types DT104 and DT104b, the isolates were found to be clonal, based on the degree of genetic identity, as determined by analyses of their macrorestriction banding and MLVA allelic profiles. Significantly, isolates cultured from the environment (group A) clustered almost exclusively with those recovered from processing sites (group B), along with fewer isolates from retail sources (group C). In addition, clinical isolates (group D) clustered almost exclusively with those of processing and retail origin (groups B and C; Fig. 2A). The same broad clustering was noted when MLVA was performed (Fig. 2B). These findings are consistent with the view that most contamination of meat and meat products originates during the processing phase of the food chain and the fact that there is limited transfer from environmental sources (5, 7, 8).
In terms of resistance to antimicrobial compounds associated with this serovar, the majority exhibited the classical ACSSuT resistance profile. Resistance to NAL, in patients infected with Salmonella, has been reported in cases of treatment failure with fluoroquinolones. Mutations in the gyrA gene and overexpression of efflux pumps are known to contribute to this resistance and to the decreased susceptibility to fluoroquinolones, such as ciprofloxacin (22). In the isolates of our study, resistance to NAL was only recorded for the clinical isolates (group D). This may be due to the fact that Salmonella-infected patients can be treated with fluoroquinolone drugs, and thus had already been exposed to this antibiotic. Resistance to Gm was found in isolates (n = 2) from processing and retail (groups B and C, respectively) sources, resistance to Tm (n = 40) and Kan (n = 2) was found in isolates from processing, retail, and clinical (groups B, C and D, respectively) sources, and resistance to Kan (n = 2) was found in isolates originating from retail sources (Group C). Isolates from the environment (group A) were not found to be resistant to any of these antibiotics.
Biocides are widely used in the food industry to control hygiene and limit bacterial spread. However, these organisms have developed mechanisms to overcome the action of these compounds, enabling their adaptation and increasing their ability to survive in the food production environment (16). Not surprisingly, arising from this bacterial characteristic, the future efficacy of biocides may be compromised. In the present study, we assessed the ability of all isolates to tolerate active components found in biocide formulations, including benzalkonium chloride, chlorhexidine, hydrogen peroxide, and triclosan. Isolates obtained from retail sources (denoted as group C) exhibited an increased tolerance to these compounds compared to the reference strain and strains from other groups (groups A, B, and D). It is difficult to explain this phenotype. This could be due to the exposure of these isolates to certain packaging materials or even to the packaging environment. In this group of isolates, there was also a significant correlation between the cotolerance to benzalkonium chloride and chlorhexidine (r = 0.255; P = 0.049), hydrogen peroxide (r = 0.561; P = 0.000), and triclosan (r = 0.551; P = 0.000).
The emergence of bacteria displaying an increased tolerance to biocides, together with the potential for the development of a cross-resistant phenotype to antibiotics, can pose an obstacle to the use of these compounds in the future (24). Based on this premise, we statistically assessed the collection to ascertain any possible associations between these stresses. When proposing a null hypothesis between the antibiotic resistance profile of the isolates and their ability to tolerate biocidal actives such as benzalkonium chloride, chlorhexidine, and hydrogen peroxide, this was rejected, showing significance at the 0.05 level. Bacterial isolates cultured from processing (Group B) and retail (group C) sources and resistant to ACSSuTTm (n = 33) exhibited a significant increased tolerance to benzalkonium chloride (P = 0.000), chlorhexidine (P = 0.009), and hydrogen peroxide (P = 0.000). This finding supports the data provided in a very recent study reporting that when Salmonella was exposed to sublethal concentrations of individual active biocide agents, tolerant isolates may emerge, and this is associated with changes in the antimicrobial susceptibility profile of the given strain (16). It may be that the mechanisms associated with the tolerance to active components of the biocides are broad spectrum, imparting a reduced susceptibility to other compounds that present a diverse chemical structure (such as antibiotics). Isolates from processing and retail sources encounter different stresses during the food processing process that may trigger modifications of the cell wall, such as the decrease of porins or increased efflux pump activity. Antibiotics such as ampicillin, chloramphenicol, nalidixic acid, tetracycline, etc., are known efflux pump substrates. Therefore, if the isolates are exposed to biocides/biocidal actives during the food production chain, these may prompt the bacteria to change their cell wall in order to overcome the action of the biocides, and this can consequently also be applied to the antibiotics.
Bacteria can experience various stress conditions, and these can cause organisms to alter their phenotype accordingly. As an example, this change can lead to an increase in bacterial adherence to surfaces, culminating in the production of a biofilm (25). Biofilms are a recognized source of contamination, whose eradication at points along the food chain continues to pose a food safety challenge. Furthermore, it has also been reported that bacterial cells contained in a biofilm can survive disinfection, becoming more resistant. This compromises food safety and increases the risk to public health (23, 25). Therefore, the ability to produce biofilms was evaluated in here. As published in one of our earlier studies, all of the isolates tested could attach to plastic, but none adhered strongly to this surface type (23). No major differences were recorded among the groups tested. Overall, 62.7% (n = 42) of the study isolates cultured from the processing environment (group B) were classified as moderate biofilm formers, while those from retail sources (group C) contained equal numbers of moderate and weak biofilm formers. Interestingly, 22 (73%) of the bacterial isolates cultured from clinical sources (group D) could form moderately strong biofilms. These data support the observations of others, which suggested that S. Typhimurium is not a strong biofilm former under standard conditions (26). When the nature of the biofilm formed by these strains was compared to the corresponding antimicrobial resistance profile, a biofilm phenotype significance of P = 0.004 was determined among the clinical isolates, particularly those exhibiting an ACSSuTNAL resistance profile.
The motility of bacteria is known to contribute to virulence in Salmonella. S. Typhimurium strains use their flagella to aid with their orientation and ability to translocate (27). In cooperation with chemosensing systems, flagellum-aided movement facilitates bacterial migration toward nutrient sources (28). Therefore, motility would have a direct impact on the interaction established between the pathogen and its host. In the laboratory, the latter would be important for invasion in tissue culture systems and, in the host, for the induction of inflammation of the gut in vitro (29, 30) and in vivo (31, 32). In the present study, the swim activity reported for the collection of bacteria studied was diverse, with little evidence of any major differences among the groups. A similar observation was recorded for swarm activity, with some isolates classified as being nonmotile. Notably, the clinical isolates (group D) exhibited a greater swarm activity, with none being classified as nonmotile.
If Salmonella isolates are able to escape the food processing environment and enter the human food chain, then additional risk factors could be predicted for the consumer. When Salmonella infects the human host, it passes into the stomach, a site often considered to be the first line of defense against food-borne pathogens due to its acidic nature. Pathogens that survive this stress could be considered more virulent. Indeed, S. Typhimurium DT104 is reported to be more acid tolerant compared to other Salmonella isolates (21). Acid resistance could thus be regarded as an indicator of virulence, since resistant strains are better able to survive passage from the human stomach into the lower intestine. This phenotype would also support their survival inside macrophages (33). In the present study, none of the isolates cultured from the environment (group A) could survive acidic conditions as tested. In contrast, those from processing (group B; 95.5% survival) and retail (group C; 96.7% survival) sources, when tested, survived conditions at pH 2.5 for up to 2 h. This may indicate an adaptational switch in the phenotype as bacteria transfer from the environment to the processing or retail settings. This adaptation appears to be maintained, since clinical isolates (group D) were completely resistant when exposed to the same acidic conditions. Furthermore, it is not possible to discount the fact that latter phenotypes could be due to preexposure to the acidic conditions of the stomach (in animals and humans) and consequently are more fit to survive when reexposed to this same pH stress. A significant correlation (r = 0.398; P = 0.029) among the clinical isolates (group D) was noted, at the 0.05 level, between their ability to produce biofilms and their ability to survive in acidic conditions.
A few studies have reported adaptive phenotypes that enteric bacterial pathogens demonstrate in response to in vivo-mimicking stress conditions, such as exposure to propionate (34), short chain fatty acids (35), or antibiotics (such as NAL) (36). However, in the majority of these studies, only one condition is considered at a given time point, nor do these studies investigate the isolates along the food production chain, in contrast to what was currently reported by us. Consequently, if these isolates demonstrated an adaptational switch when subjected to different pressures, that type of change would be missed. In these type of studies, it is important to examine bacteria that are passaged through the food chain, as well as testing the stress responses that they demonstrate. Some stresses encountered through the food chain can trigger bacteria to activate specific mechanisms or signals that contribute to them being more adapted to the “new environmental conditions.”
Within the host, Salmonella can adopt an intracellular lifestyle, targeting epithelial cells, macrophages, dendritic cells, and others. It is well known that once internalized by host cells, S. Typhimurium can reside within a Salmonella-containing vacuole (SCV). Depending on the nature of the infected host cell, S. Typhimurium can persist and/or proliferate inside this organelle. An SCV is a hostile environment, being nutrient limiting, and one which imposes a series of stresses on the intracellular bacteria (37). In order to determine the isolates' ability to survive, when challenged with a range of adverse conditions, we used the phenotypic microarray platform to evaluate the phenotypes of selected isolates from the pork production food chain. The parameters measured included utilization of carbon, phosphorus, and sulfur sources, as well as survival under different osmotic conditions, osmolytes, and different nutrients/substrates. Environmental isolates (group A) exhibited a difference in phenotype, when respiring in the presence of various phosphorus and sulfur compounds. Carbon sources, including l-tartaric acid, were not utilized efficiently, as shown by the reduction in growth compared to the reference strain. Tartaric acid is contained in candy, cold drinks, and wines, among other foods, and it can also be used as a preservative, stabilizer, and additive in processed foods. Tartaric acid is metabolized in the absence of oxygen and is an analog of malic acid, which is an intermediate of the Krebs cycle. It is reasonable to postulate that the metabolism of l-tartaric acid could interfere with the Krebs cycle, thereby altering bacterial cell energetics, slowing the growth rate and providing for a dormant mode required to support survival. The fact that strains exhibited a decreased growth rate in the presence of l-tartaric acid was not unexpected. Interestingly, when we compared the ability to produce biofilms among this selection of isolates many were capable of increasing their biofilm formation. Additives used in the food industry to reduce water activity, and consequently bacterial growth may induce the formation of biofilms, as a survival response. Therefore, metabolic switching could contribute to increased bacterial survival.
Sodium lactate metabolism resulted in the opposite effect. When the formation of biofilms was studied using this carbon source, not all of the test isolates showed an increased ability to form biofilms compared to the reference strain. In the case of isolates from processing (group B, isolate O2), retail (group C, isolate ST1333) and clinical (group D, MS100410) sources, their ability to form biofilms was reduced, based on measurements of crystal violet staining. Despite the fact that the isolates were slower growing (see Table S2 in the supplemental material), this effect was not translated into the improved production of biofilm structures (Fig. 7A). Sodium lactate is an antimicrobial widely used in meat products to extend the shelf life and ensure the safety of these foods (38). It can also be used as an antioxidant agent and flavor potentiator to enhance both taste and aroma of the products (39). Sodium lactate can prevent the growth of several bacteria by reducing the water content in the food matrix (38, 39). This may explain why under growth-limiting conditions in the presence of this substrate, these bacteria were unable to form biofilms (36, 37).
In the pathogenesis of Salmonella infections, the interaction between the bacteria and the host is critical to the final outcome. In the case of epithelial cells, S. Typhimurium has been described as having a bimodal lifestyle, being able to replicate in the SCV (40), as well as in the cytosol (41, 42). The ability of the strains to adhere to and invade Caco-2 epithelial cells (Fig. 8) is therefore one important aspect that we considered. From a subset of isolates taken from all four groups (A through D) studied, all could adhere to Caco-2 cells at a higher rate compared to the reference strain. Interestingly, adherence did not always correlate with the subsequent invasion of these epithelial cells. For example, it could be expected that since the strains obtained from clinical sources (group D, isolates MS100185 and MS100410) were already preexposed to the human gut, this would confer them an additional advantage when in contact with the epithelial cells. This feature was not observed in these isolates. The results obtained for the entire collection were plotted on a heat map, provided as supplementary material (see Dataset S1 in the supplemental material), that can be used as a guiding tool to easily assess and analyze the phenome(s) of the isolates.
In conclusion, our study highlights the importance of studying the phenome(s) of Salmonella isolates across the food processing chain and identifying associated risk factors. The most relevant features noted in this collection included an increased tolerance phenotype to biocides, particularly among bacteria associated with processing and retail sources (groups B and C, respectively). Furthermore, increased acid survival exhibited by isolates that ultimately reach the human host should be considered. The ability to form a biofilm in the presence of l-tartaric acid may be an avenue for further study. Taken together, these data suggest that certain additives or processing conditions in the food industry could elicit a phenotypic switch, altering the phenotype toward improved adaptation and increased survival. In order to implement effective measures to control the spread and dissemination of MDR Salmonella, we should focus on practical approaches designed to interrupt dissemination (4). Detailed phenotypic studies such as the one presented here help to establish markers that can contribute to identify isolates with increased virulence, which can pose an increased risk to the food industry and, ultimately, to the consumer.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Department of Agriculture, Fisheries, and Food of Ireland (project 06/TNI-UCD10).
The S. Typhimurium DT104 and DT104b clinical isolates were kindly provided by Martin Cormican of the National Salmonella Reference Laboratory, Department of Medical Microbiology, Galway University Hospitals, Galway, Ireland.
Footnotes
Published ahead of print 14 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01041-13.
REFERENCES
- 1.Newell DG, Koopmans M, Verhoef L, Duizer E, Aidara-Kane A, Sprong H, Opsteegh M, Langelaar M, Threfall J, Scheutz F, van der Giessen J, Kruse H. 2010. Food-borne diseases: the challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 139:S3–S15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaidi MB, Leon V, Canche C, Perez C, Zhao S, Hubert SK, Abbott J, Blickenstaff K, McDermott PF. 2007. Rapid and widespread dissemination of multidrug-resistant blaCMY-2 Salmonella Typhimurium in Mexico. J. Antimicrob. Chemother. 60:398–401 [DOI] [PubMed] [Google Scholar]
- 3.Alcaine SD, Warnick LD, Wiedmann M. 2007. Antimicrobial resistance in nontyphoidal Salmonella. J. Food Prot. 70:780–790 [DOI] [PubMed] [Google Scholar]
- 4.Davis MA, Hancock DD, Besser TE. 2002. Multiresistant clones of Salmonella enterica: the importance of dissemination. J. Lab. Clin. Med. 140:135–141 [DOI] [PubMed] [Google Scholar]
- 5.Soumpasis I, Butler F. 2009. Development and application of a stochastic epidemic model for the transmission of Salmonella Typhimurium at the farm level of the pork production chain. Risk Anal. 29:1521–1533 [DOI] [PubMed] [Google Scholar]
- 6.Oliveira M, Wijnands L, Abadias M, Aarts H, Franz E. 2011. Pathogenic potential of Salmonella Typhimurium DT104 following sequential passage through soil, packaged fresh-cut lettuce and a model gastrointestinal tract. Int. J. Food Microbiol. 148:149–155 [DOI] [PubMed] [Google Scholar]
- 7.Duggan SJ, Mannion C, Prendergast DM, Leonard N, Fanning S, Gonzales-Barron U, Egan J, Butler F, Duffy G. 2010. Tracking the Salmonella status of pigs and pork from lairage through the slaughter process in the Republic of Ireland. J. Food Prot. 73:2148–2160 [DOI] [PubMed] [Google Scholar]
- 8.Hernández M, Gómez-Laguna J, Luque I, Herrera-León S, Maldonado A, Reguillo L, Astorga RJ. 2013. Salmonella prevalence and characterization in a free-range pig processing plant: tracking in trucks, lairage, slaughter line and quartering. Int. J. Food Microbiol. 162:48–54 [DOI] [PubMed] [Google Scholar]
- 9.Hyeon JY, Chon JW, Hwang IG, Kwak HS, Kim MS, Kim SK, Choi IS, Song CS, Park C, Seo KH. 2011. Prevalence, antibiotic resistance, and molecular characterization of Salmonella serovars in retail meat products. J. Food Prot. 74:161–166 [DOI] [PubMed] [Google Scholar]
- 10.Boughton C, Leonard FC, Egan J, Kelly G, O'Mahony P, Markey BK, Griffin M. 2004. Prevalence and number of Salmonella in Irish retail pork sausages. J. Food Prot. 67:1834–1839 [DOI] [PubMed] [Google Scholar]
- 11.Rowe TA, Leonard FC, Kelly G, Lynch PB, Egan J, Quirke AM, Quinn PJ. 2003. Salmonella serotypes present on a sample of Irish pig farms. Vet. Rec. 153:453–456 [DOI] [PubMed] [Google Scholar]
- 12.Lindstedt BA, Heir E, Gjernes E, Vardund T, Kapperud G. 2003. DNA fingerprinting of Shiga toxin producing Escherichia coli O157 based on multiple-locus variable-number tandem-repeats analysis (MLVA). Ann. Clin. Microbiol. Antimicrob. 10:2–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindstedt BA, Vardund T, Aas L, Kapperud G. 2004. Multiple-locus variable-number tandem-repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolor capillary electrophoresis. J. Microbiol. Methods 59:163–172 [DOI] [PubMed] [Google Scholar]
- 14.McCabe E. 2010. The development and evaluation of a real-time PCR-based assay using a two-step enrichment for the sensitive and specific detection of Salmonella enterica in fresh meat. Ph.D. thesis University College Dublin, Dublin, Ireland [Google Scholar]
- 15.CLSI 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. Document M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 16.Condell O, Iversen C, Cooney S, Power KA, Walsh C, Burgess C, Fanning S. 2012. Efficacy of biocides used in the modern food industry to control Salmonella enterica, and links between biocide tolerance and resistance to clinically relevant antimicrobial compounds. Appl. Environ. Microbiol. 78:3087–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Djordjevic D, Wiedmann M, McLandsborough LA. 2002. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 68:2950–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. 2000. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 40:175–179 [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Zhao Y, McClelland M, Harshey RM. 2007. The RcsCDB signaling system and swarming motility in Salmonella enterica serovar Typhimurium: dual regulation of flagellar and SPI-2 virulence genes. J. Bacteriol. 189:8447–8457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Frye JG, McClelland M, Harshey RM. 2004. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol. Microbiol. 52:169–187 [DOI] [PubMed] [Google Scholar]
- 21.Berk PA, Jonge R, Zwietering MH, Abee T, Kieboom J. 2005. Acid resistance variability among isolates of Salmonella enterica serovar Typhimurium DT104. J. Appl. Microbiol. 99:859–866 [DOI] [PubMed] [Google Scholar]
- 22.Lunn AD, Fàbrega A, Sánchez-Céspedes J, Vila J. 2010. Prevalence of mechanisms decreasing quinolone-susceptibility among Salmonella sp. clinical isolates. Int. Microbiol. 13:15–20 [DOI] [PubMed] [Google Scholar]
- 23.O'Leary D, Cabe EM, McCusker MP, Martins M, Fanning S, Duffy G. 2013. Microbiological study of biofilm formation in isolates of Salmonella enterica Typhimurium DT104 and DT104b cultured from the modern pork chain. Int. J. Food Microbiol. 161:36–43 [DOI] [PubMed] [Google Scholar]
- 24.Davin-Regli A, Pagès JM. 2012. Cross-resistance between biocides and antimicrobials: an emerging question. Rev. Sci. Tech. 31:89–104 [PubMed] [Google Scholar]
- 25.Rodrigues D, Cerca N, Teixeira P, Oliveira R, Ceri H, Azeredo J. 2011. Listeria monocytogenes and Salmonella enterica Enteritidis biofilms susceptibility to different disinfectants and stress-response and virulence gene expression of surviving cells. Microb. Drug Resist. 17:181–189 [DOI] [PubMed] [Google Scholar]
- 26.Díez-García M, Capita R, Alonso-Calleja C. 2012. Influence of serotype on the growth kinetics and the ability to form biofilms of Salmonella isolates from poultry. Food Microbiol. 31:173–180 [DOI] [PubMed] [Google Scholar]
- 27.Josenhans C, Suerbaum S. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291:605–614 [DOI] [PubMed] [Google Scholar]
- 28.Porter SL, Wadhams GH, Armitage JP. 2011. Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. 9:153–165 [DOI] [PubMed] [Google Scholar]
- 29.Dibb-Fuller MP, Allen-Vercoe E, Thorns CJ, Woodward MJ. 1999. Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology 145:1023–1031 [DOI] [PubMed] [Google Scholar]
- 30.Jones BD, Lee CA, Falkow S. 1992. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect. Immun. 60:2475–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stecher B, Barthel M, Schlumberger MC, Haberli L, Rabsch W, Kremer M, Hardt WD. 2008. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell Microbiol. 10:1166–1180 [DOI] [PubMed] [Google Scholar]
- 32.Stecher B, Hapfelmeier S, Müller C, Kremer M, Stallmach T, Hardt WD. 2004. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 72:4138–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Jonge R, Ritmeester WS, van Leusden FM. 2003. Adaptive responses of Salmonella enterica serovar Typhimurium DT104 and other S. Typhimurium strains and Escherichia coli O157 to low pH environments. J. Appl. Microbiol. 94:625–632 [DOI] [PubMed] [Google Scholar]
- 34.Calhoun LN, Kwon YM. 2010. The effect of long-term propionate adaptation on the stress resistance of Salmonella Enteritidis. J. Appl. Microbiol. 109:1294–1300 [DOI] [PubMed] [Google Scholar]
- 35.Rishi P, Ricke S. 2007. hilA gene expression in SCFAs adapted and inorganic acid challenged Salmonella enterica serovar typhimurium. Nepal Med. Coll. J. 9:162–165 [PubMed] [Google Scholar]
- 36.Dowd SE, Killinger-Mann K, Blanton J, San Francisco M, Brashears M. 2007. Positive adaptive state: microarray evaluation of gene expression in Salmonella enterica Typhimurium exposed to nalidixic acid. Foodborne Pathog. Dis. 4:187–200 [DOI] [PubMed] [Google Scholar]
- 37.Ibarra JA, Steele-Mortimer O. 2009. Salmonella, the ultimate insider: Salmonella virulence factors that modulate intracellular survival. Cell Microbiol. 11:1579–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tovunac I, Galic K, Prpic T, Juric S. 2011. Effect of packaging conditions on the shelf-life of chicken frankfurters with and without lactate addition. Food Sci. Technol. Int. 17:167–175 [DOI] [PubMed] [Google Scholar]
- 39.Yuan W, Ágoston R, Lee D, Lee SC, Yuk HG. 2012. Influence of lactate and acetate salt adaptation on Salmonella Typhimurium acid and heat resistance. Food Microbiol. 30:448–452 [DOI] [PubMed] [Google Scholar]
- 40.Steele-Mortimer O. 2008. The Salmonella-containing vacuole: moving with the times. Curr. Opin. Microbiol. 11:38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, Montero M, Steele-Mortimer O. 2010. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc. Natl. Acad. Sci. U. S. A. 107:17733–17738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steele-Mortimer O, Brumell JH, Knodler LA, Méresse S, Lopez A, Finlay BB. 2002. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell Microbiol. 4:43–54 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






