Abstract
Single-cell methods are a powerful application in microbial research to study the molecular mechanism underlying phenotypic heterogeneity and cell-to-cell variability. Here, we describe the optimization and application of single-cell time-lapse fluorescence microscopy for the food spoilage bacterium Bacillus cereus specifically. This technique is useful to study cellular development and adaptation, gene expression, protein localization, protein mobility, and cell-to-cell communication over time at the single-cell level. By adjusting existing protocols, we have enabled the visualization of growth and development of single B. cereus cells within a microcolony over time. Additionally, several different fluorescent reporter proteins were tested in order to select the most suitable green fluorescent protein (GFP) and red fluorescent protein (RFP) candidates for visualization of growth stage- and cell compartment-specific gene expression in B. cereus. With a case study concerning cotD expression during sporulation, we demonstrate the applicability of time-lapse fluorescence microscopy. It enables the assessment of gene expression levels, dynamics, and heterogeneity at the single-cell level. We show that cotD is not heterogeneously expressed among cells of a subpopulation. Furthermore, we discourage using plasmid-based reporter fusions for such studies, due to an introduced heterogeneity through copy number differences. This stresses the importance of using single-copy integrated reporter fusions for single-cell studies.
INTRODUCTION
The Gram-positive soil bacterium Bacillus cereus is a known cause of food spoilage and food-borne illnesses of both diarrheal and emetic types. Over the last decades, research concerning this problematic microorganism has focused on various different research disciplines, including toxin production and virulence (1, 2), resistance mechanisms against applied stresses (3, 4), cell structure, metabolism (5, 6), and cellular developments, such as cell division, biofilm formation (7), sporulation (8), and spore germination (9). In this way, B. cereus is becoming an increasingly studied model organism for Gram-positive pathogenic bacteria next to its well-known but fairly distant and nonpathogenic relative Bacillus subtilis. In contrast to such studies of the latter, in-depth studies on B. cereus are still hampered by the lack of suitable and effective molecular biological tools and are often restricted due to poor genetic accessibility of the strains. Importantly, there has been an increasing number of reports that mention phenotypic heterogeneity in the above-described processes (10–13), and phenotypic heterogeneity can be defined as the emergence of subpopulations within an isogenic culture. Such phenotypic heterogeneity increases the chances of survival of a given species during frequent and random environmental changes (14). In industrial application, however, it complicates the predictability of microbial behavior and, as a result, the eradication of food spoilage Bacillus spp. from foodstuffs by conventional food preservation techniques (15, 16).
Using the sporulation and spore germination processes as an example, we previously discussed the molecular basis of such heterogeneity in B. cereus populations (16). We suggested that application of single-cell techniques would achieve further insights into this phenotypic variation. Combinations of such techniques with fluorescent reporters can further couple the observed phenotypic heterogeneity to underlying molecular mechanisms.
Here, we describe the applicability of time-lapse fluorescence microscopy for B. cereus to investigate levels and dynamics of and heterogeneity in gene expression and furthermore demonstrate important technical constraints of the technique. Studies with B. subtilis have shown that heterogeneity in sporulation and in spore properties can be attributed to differences in gene expression and protein levels between individual cells (17–19). In order to study these phenomena in B. cereus, we have optimized time-lapse fluorescence microscopy specifically for this organism. Even though usability of this technique has been described for several species of bacteria, including B. subtilis, Escherichia coli, and Streptococcus pneumoniae (20, 21), application of the described methods for B. cereus cells was unsuccessful. By adjusting medium composition, cell culture preparation, and slide preparation, we demonstrated growth and sporulation of B. cereus cells under the microscope in a single layer from single cell to microcolony. In this way, we have studied the dynamics of expression of the sporulation gene cotD of B. cereus ATCC 14579 in single cells. CotD is produced during late-stage sporulation in the mother cell under the control of sporulation-specific sigma factor σK (23). It is localized in the inner coat of the spore, which is important for the spores' resistance properties (23, 24).
For decades, fluorescent proteins (FPs) have been successfully employed as tools for biological imaging (25–27). Continued development of a wide range of FP variants has decreased the biological and spectral limitations of the ones initially available (25). On the other hand, choosing an FP for any experimental question or organism at study has become more complicated. The selection of the most suitable candidate for a specific experiment greatly depends on influencing factors such as the pH of the environment, the presence of ions, multimerization and toxicity, cultivation temperature, the availability of oxygen, photostability, and spectral overlap (28). Species-specific codon-optimized or mutated variants of FPs are increasingly utilized to maximize the transcription, translation, and fluorescent capacities of the proteins for optimal fluorescence in specific species or experimental setups (29–31). To enable tracking of gene expression during all growth stages of B. cereus and to improve the available molecular tool box, we have also tested a few readily available green fluorescent proteins (GFPs) and red fluorescent proteins (RFPs) in order to select the most suitable candidates for this organism.
MATERIALS AND METHODS
Strains and plasmids.
All strains and plasmids used in this study are listed in Table 1. As a reference strain, Bacillus cereus ATCC 14579 was used. Foreign DNA was introduced in this strain either by using multicopy plasmids or by single-crossover DNA integration using the nonreplicative pSG1151 vector, introduced via electroporation (32).
Table 1.
Plasmids and strains used in this study
| Strain or plasmid | Description | Source(s) |
|---|---|---|
| Strains | ||
| B. cereus ATCC 14579 | Enterotoxic strain of B. cereus wild-type isolate | ATCC, BGSC ID6A5 |
| B. cereus ATCC 14579 PcotD-gfpmut1 | PcotD-gfpmut1 | This study |
| Plasmids | ||
| pSG1151 | Vector for integrative P-gfpmut1 fusions in B. subtilis | 33 |
| pSG14cotD | pSG1151 with gfpmut1 driven by B. cereus cotD promoter | This study |
| pAD123 | E. coli Gram-positive shuttle vector containing gfpmut3a | 34 |
| pAD123-A | gfpmut3a driven by B. cereus secA promoter | This study |
| pAD123-D | gfpmut3a driven by B. cereus cotD promoter | This study |
| pAD123-G | gfpmut3a driven by B. cereus sigG promoter | This study |
| pAD43-25 | gfpmut3a driven by 1.4-kb insert of B. cereus UW85 DNA | 34 |
| pAD641 | pAD123 derivative with gfpmut3a replaced by sfgfp(Bs) | This study |
| pAD641-A | sfgfp(Bs) driven by B. cereus secA promoter | This study |
| pAD641-D | sfgfp(Bs) driven by B. cereus cotD promoter | This study |
| pAD641-G | sfgfp(Bs) driven by B. cereus sigG promoter | This study |
| pAD641-25 | sfgfp(Bs) driven by 1.4-kb insert of B. cereus UW85 DNA | This study |
| pAD642 | pAD123 derivative with gfpmut3a replaced by gfp(Sp) | This study |
| pAD642-A | gfp(Sp) driven by B. cereus secA promoter | This study |
| pAD642-D | gfp(Sp) driven by B. cereus cotD promoter | This study |
| pAD642-G | gfp(Sp) driven by B. cereus sigG promoter | This study |
| pAD642-25 | gfp(Sp) driven by 1.4-kb insert of B. cereus UW85 DNA | This study |
| pAD650 | pAD123 derivative with gfp replaced by mCherry | This study |
| pAD650-A | mCherry driven by B. cereus secA promoter | This study |
| pAD650-D | mCherry driven by B. cereus cotD promoter | This study |
| pAD650-G | mCherry driven by B. cereus sigG promoter | This study |
| pAD650-25 | mCherry driven by 1.4-kb insert of B. cereus UW85 DNA | This study |
| pAD651 | pAD123 derivative with gfp replaced by mKate2 | This study |
| pAD651-A | mKate2 driven by B. cereus secA promoter | This study |
| pAD651-D | mKate2 driven by B. cereus cotD promoter | This study |
| pAD651-G | mKate2 driven by B. cereus sigG promoter | This study |
| pAD651-25 | mKate2 driven by 1.4-kb insert of B. cereus UW85 DNA | This study |
For the construction of the pSG14cotD vector, an 800-bp fragment of the region upstream of the B. cereus ATCC 14579 cotD gene (BC1560) was amplified using primers Bce14_cD800HindIII-F and Bce14_cDpstI-R (see Table S1 in the supplemental material). The fragment was cleaved with HindIII and PstI and ligated into the corresponding sites of the pSG1151 vector (33), enabling a transcriptional fusion to the gfpmut1 gene. Electroporation of the resulting pSG14cotD vector into competent cells of B. cereus ATCC 14579 and selection on LB agar plates (containing 4 μg/ml chloramphenicol) resulted in a single-crossover event of the vector on the region of homology with the region upstream of cotD. Correct integration of the fusion into the chromosome was verified using PCR and sequencing (Macrogen).
To study optimal fluorescence performance in B. cereus, we selected several readily available GFPs and RFPs. For green fluorescence, we tested GFPmut3a (34), sfGFP(Bs) (superfolder GFP [35], which was B. subtilis codon optimized using the dual-codon method (48), and GFP(Sp) (GFP codon optimized for S. pneumoniae) (36). For red fluorescence, we selected mCherry (37) and mKate2 (38, 39). The gfpmut3a gene on pAD123 (34) was replaced by genes encoding these variants, using the XbaI and HindIII restriction sites. The resulting vectors (pAD641, pAD642, pAD650, and pAD651) are listed in Table 1. The expression of the FP-encoding genes in B. cereus was driven by different promoters, as listed in Table 2. For this purpose, promoter-containing fragments (approximately 250 bp each) were amplified by PCR from B. cereus ATCC 14579 chromosomal DNA using primers TIFN73 and TIFN74, TIFN75 and Bce14-upctD-XbaI-R, and TIFN77 and TIFN78 (see Table S1 in the supplemental material). Alternatively, the 1,400-bp upp fragment was cleaved from pAD43-25 (34) using KpnI and XbaI restriction enzymes (Fermentas FastDigest). These fragments were subsequently ligated into the KpnI/XbaI sites of the obtained pAD vectors. The vectors were introduced into B. cereus ATCC 14579 via electroporation and selection on LB agar plates with the addition of chloramphenicol (4 μg/ml).
Table 2.
Selected promoters for transcription of various gfp and rfp genes
| Promoter | Gene locus tag | Properties |
|---|---|---|
| PsecA | BC5189 | Strong promoter for the essential secA gene during vegetative growth (22) |
| PcotD | BC1560 | Sporulation-specific promoter activated during late-stage sporulation in the mother cell (23) |
| PsigG | BC3903 | Sporulation-specific promoter activated during late-stage sporulation in the forespore (41) |
| Pupp | The ′glyA fragment plus the upstream region of the upp gene from B. cereus UW85, which has been described as a strong constitutive promoter during vegetative growth (34) |
Culture preparation.
All B. cereus cultures were grown at 30°C with shaking at 220 rpm. For the preparation of cells for time-lapse microscopy, cells were diluted to an optical density at 600 nm (OD600) of 0.1 from overnight cultures in hydrolyzed casein (CH) medium (40), with the addition of 4 μg/ml chloramphenicol when appropriate. To allow for sporulation, cells were harvested at early exponential phase (OD600, ∼0.7) and resuspended in the same volume of chemically defined medium (CDM) (20). Cells were allowed to adapt to this medium for 15 min at 30°C with shaking at 220 rpm prior to a 1:4 dilution in CDM. Of this dilution, 1.5 μl was spread on a strip of time-lapse polyacrylamide (PAA) (see below).
Slide preparation.
To support growth and development under the microscope, we spread cells on specially prepared time-lapse microscopy slides as previously described by de Jong et al. in 2011 (20), with a few adjustments. Most importantly, instead of low-melting-point agarose, polyacrylamide was used. A detailed description of the slide preparation can be found in the text in the supplemental material. In short, we performed the following steps:
Prepare a gene frame glass slide as is described in steps 2.1 to 2.3 in the protocol description by de Jong et al. (20).
Mix 1.5 ml of supplemented time-lapse medium (see the text in the supplemental material) with 500 μl 40% PAA solution (acrylamine-BIS [N,N-methylenebisacrylamide], 37.5:1; Serva). Add 20 μl 10% ammonium persulfate (APS) and 2 μl TEMED (N,N,N′,N′-tetramethylethylenediamine) and pour 500 μl of this liquid within the gene frame. Apply pressure with another glass slide. Leave at room temperature for 30 min to allow for polymerization.
Cut the PAA patch in six equal-sized strips using a sterilized scalpel. Wash the strips in sterile deionized water and supplemented time-lapse medium.
Place up to three strips between a new gene frame, making sure there are gaps on either side of the slice.
Add 1.5 μl of prepared cells (see “Culture preparation”) onto a PAA strip as described in steps 2.9 and 2.10 in the protocol description in reference 20.
Place a clean microscope slide coverslip (24 by 50 mm) on the gene frame from one side to the other. Immediately search for single cells by using an inverted microscope with a prewarmed climate chamber and time-lapse microscopy software.
Time-lapse microscopy.
For the visualization of growth and development of B. cereus cells, we used an IX71 microscope (Olympus) with a CoolSNAP HQ2 camera (Princeton Instruments) and DeltaVision softWoRx 3.6.0 (Applied Precision) software as previously described in point 3 of the protocol section of reference 20, with the following adjustments. A 60× phase-contrast objective was used with a GFP filter set (Chroma) (excitation at 470/40 nm and emission at 525/50 nm) for visualization of green fluorescence. Images were taken every 15 or 20 min using 32% APLLC white-light-emitting diode (LED) light and 0.05-s exposure for bright-field pictures and 10% xenon light with 0.5 s exposure for GFP detection.
Fluorescence microscopy.
B. cereus cells containing pAD vectors (Table 1) were grown in maltose sporulation medium (MSM) with the addition of chloramphenicol (4 μg/ml) at 30°C and shaking at 220 rpm. Fluorescence at various time points was analyzed using the Nikon TI-E microscope with a CoolSNAP H2Q camera. The fluorescein isothiocyanate (FITC) filter (excitation at 465 to 495 nm, dichroic mirror [DM] at 505 nm, barrier filter [BA] at 515 to 555 nm) was used for visualization of green fluorescence and the tetramethyl rhodamine isothiocyanate (TRITC) filter (excitation at 528 to 553 nm, DM at 565 nm, BA at 590 to 650 nm) for red fluorescence. A ×100 magnification lens was used. By use of IS Elements software, all pictures were taken using the following settings: pixel size, 1,280 by 1,024 (no binning); phase-contrast exposure at 150 ms, gain of 8; and FITC/TRITC exposure at 60 ms, gain of 2.
Image analysis.
Pictures were adjusted and analyzed in ImageJ software (http://rsb.info.nih.gov/ij/). Equal adjustment settings were applied to images taken for each FP variant but were optimized per promoter. This allowed for signal comparison between GFPs or RFPs but not between promoters.
GFP intensities per cell per frame were determined in arbitrary units by measuring the pixel intensity of 16-bit images by using the region of interest (ROI) tool. For the time-lapse data analysis, the GFP intensity of strains containing PcotD-gfp as an integrated single copy or on the pAD vector was determined for 180 cells or 131 cells, respectively. For quantification of the fluorescence signal derived from several FP variants expressed from the pAD vector, a minimum of 300 cells per strain was counted. Measurements were exported to Excel and normalized by subtracting background fluorescence levels of the microscopy slide (agarose) and autofluorescence of the cells or spores (highest fluorescence value measured for promoterless vectors). Normalized fluorescence levels were determined and set to absolute values (<0 = 0). Mean fluorescence values and the intensity distributions were calculated using Excel, and after this, the data were plotted.
Flow cytometry analysis.
To verify fluorescence distributions of cells containing GFP variants controlled by different promoters, flow cytometry analysis was applied. Overnight cultures were diluted to an OD600 of 0.1 in fresh MSM with the addition of chloramphenicol (4 μg/ml) and grown at 30°C with shaking at 220 rpm. Multiple samples were analyzed using a BD FACS Canto flow cytometer at specific time points. The best signals were observed at T3 for Pupp, T4 for PsecA, and T23 for PcotD and PsigG, where the variable represents the number of hours after initial dilution. The voltage settings were set to forward scatter (FSC) 10, side scatter (SSC) 300, and FL-1 600 (FITC/GFP). FSC and SSC threshold levels were set to 200, measuring 50,000 events per sample. Once the cells had reached late exponential growth phase, they were briefly treated in a bead beater (Mini-Bead-Beater-8; Biospec Products) without glass beads prior to being measured for homogeneity. Data were captured using FACSDiva software (BD Biosciences) and further analyzed using WinMDI 2.8 software (http://facs.scripps.edu/software.html).
RESULTS
Optimization of culture and slide preparation results in visualization of growth and development of B. cereus cells using time-lapse microscopy.
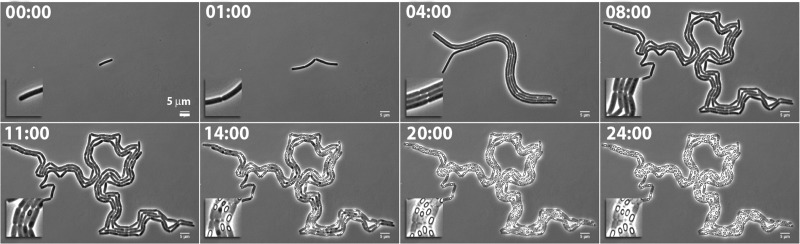
Studying cellular development (as well as gene expression and protein localization) at the single-cell level provides valuable insights that are generally missed by using techniques focused on or limited to bacterial populations. Time-lapse microscopy is a powerful technique which allows for tracking and visualization of growth and development from a single cell into a microcolony over time. Application of this tool for B. cereus cells by using previously described techniques and protocols for B. subtilis (20, 21) was unsuccessful (data not shown). Cells grew in multiple layers in the z direction, grew out of the field of vision, showed a deviant morphology from cells grown in liquid culture, showed increased lysis, and failed to sporulate. For this reason, the protocol was optimized and adjusted specifically for B. cereus. The adjustments included a two-step cultivation method in rich CH medium and a poor chemically defined medium (20). Moreover, instead of agarose, polyacrylamide (PAA) was used for the solid medium on the microscopy slide. Use of agarose caused the cells to grow in multiple layers (causing focus problems) and in long strings that grew out of the field of vision. With PAA, the growth was more controlled and remained in a monolayer. The PAA was mixed with time-lapse CDM supplemented with a 1:50 dilution of maltose sporulation medium (MSM). This defined proportion of available nutrients resulted in the most suitable growth and development of B. cereus cells on the PAA solid medium, with adequate sporulation events (Fig. 1; see Movie S1 in the supplemental material). For a detailed description of the optimized protocol, see Materials and Methods and also the text in the supplemental material.
Fig 1.
Time-lapse phase-contrast microscopy of Bacillus cereus ATCC 14579. Snapshots were taken from Movie S1. Time is indicated in hours.
Differences in oxygen distribution on the time-lapse microscopy slide affect growth and development of B. cereus cells.
While executing time-lapse experiments, we observed distinct differences in the phenotypic development of the cells that could be correlated to the position of the cells on the PAA slice (Fig. 2). We speculate that oxygen distribution is the main reason for this effect, as we assume that the oxygen availability on the periphery of the polyacrylamide slice is higher than that in the middle. To illustrate this effect, the polyacrylamide slice was divided into seven equal strips, which were defined by x and y coordinates relative to the periphery. Pictures of cells were taken in each of these locations. Cells closest to the periphery of the PAA slice (Fig. 2, locations 1 and 2) showed the best growth and most efficient sporulation behavior, whereas cells that were located more in the middle of the slice (Fig. 2, locations 3 and 4) grew slower, produced generally smaller microcolonies, and showed very little or no sporulation.
Fig 2.
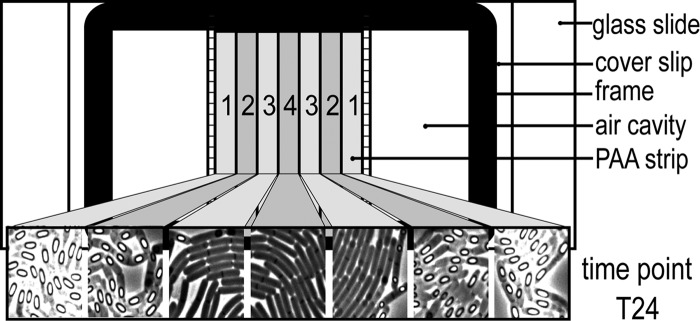
Cellular development depending on the position of the cells on the time-lapse microscopy slide. A schematic representation of the polyacrylamide (PAA) strip is divided into seven equal sections (given numbers 1 through 4). Pictures of cells taken from each of these sections show clear differences in sporulation efficiency depending on the distance of the cells relative to the periphery of the PAA slice (pictures were taken 24 h after the start of the time-lapse experiment).
The cotD promoter of B. cereus shows a monomodal distribution of activity during sporulation.
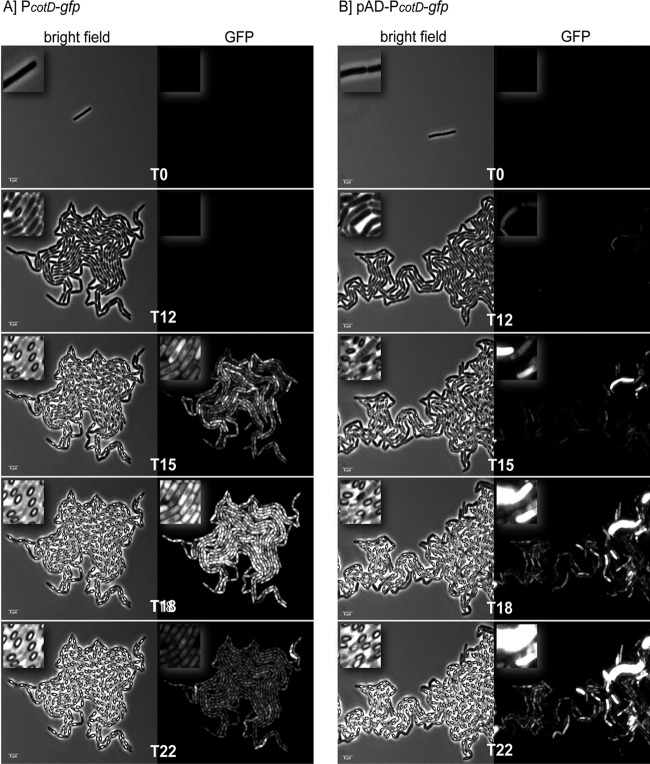
Various studies in B. subtilis have shown heterogeneous expression of some key sporulation and/or germination genes during sporulation. This contributes to differences in the sporulation process or resulting spore properties (17–19). In this study, we investigated the expression of spore coat protein CotD in B. cereus using time-lapse fluorescence microscopy. To investigate whether cotD is expressed heterogeneously in B. cereus during sporulation, its promoter was fused to gfp on the pSG1151 vector. This created pSG14cotD, which was introduced into B. cereus ATCC 14579 via electroporation and integrated as a single copy in the region of homology on the chromosome (PcotD-gfp). Alternatively, the same promoter was fused to gfp(Sp) on the pAD642 vector to create pAD42-D. This vector was also introduced into B. cereus ATCC 14579 cells via electroporation and persists as multiple copies under chloramphenicol selection (pADPcotD-gfp). Both strains were prepared for time-lapse microscopy as described in Materials and Methods. For comparison purposes, the bright-field channels and GFP channels were adjusted in brightness, contrast, and levels using the exact same values for both strains. Results are shown in Fig. 3 and Movies S2 and S3 in the supplemental material.
Fig 3.
PcotD promoter activity in B. cereus. The activity of the PcotD promoter was tested by fusing the cotD upstream region to gfp as described in Materials and Methods. A single-copy integrated version (PcotD-gfp; panel A) and a multicopy plasmid-based version (pAD-PcotD-gfp, panel B) were studied using time-lapse fluorescence microscopy. Snapshots of the bright field and the GFP channel at different time points (indicated in hours) were taken from Movie S2 (panel A) and Movie S3 (panel B). The time-lapse experiment for both strains was performed for cells located on the periphery of the PAA slice (location 1 in Fig. 2).
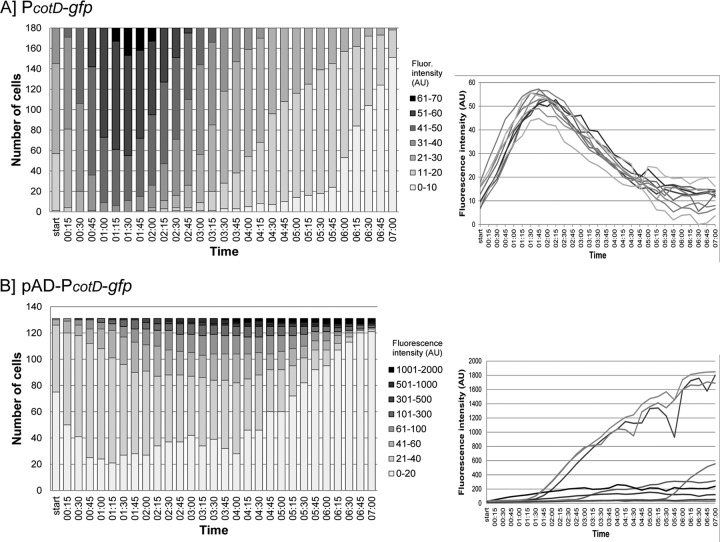
To quantify the signal distribution between cells in the microcolony, the pixel intensity in the GFP channel for 180 cells or 131 cells was measured for the PcotD-gfp fusion (Movie S2) or the pADPcotD-gfp fusion (Movie S3), respectively. The obtained values (in arbitrary units [AU]) were categorized, and the number of cells per category was calculated. As can be seen in Fig. 4A, the single-copy PcotD-gfp fusion generated a consistent monomodal fluorescence pattern in all cells (only 10 individual cells are shown for clarity). Importantly, the variations in signal strength between all cells at a given time are limited (<40 AU), indicating a homogeneous expression of cotD in the population during sporulation. In contrast, gfp expression from the same promoter on the pAD vector generated an inconsistent fluorescence intensity among cells of the microcolony (Fig. 3B and 4B). In some cells, the intensity of the signal was of such strength that it overpowered the signal in neighboring cells. Furthermore, the fluorescence signal remained detectable for an extended time compared to the single-copy fusion. Quantification of the signal showed strong variations in signal strength between individual cells (ranging from 0 to 2,000 AU) (Fig. 4B).
Fig 4.
Quantitative analysis of cotD expression using time-lapse fluorescence microscopy. The fluorescence intensity from GFP expressed from the cotD promoter in single cells was measured in 29 frames from Movies S2 (single copy integrated fusion [PcotD-gfp]) (A) and S3 (plasmid-based fusion [pAD-PcotD-gfp]) (B) in the supplemental material. The distribution of the signal intensity among all cells per strain is represented by the gray bars of different shades (left graphs). The legend shows the categorization of the fluorescence value (in arbitrary units [AU]). Ten cells were selected to show the dynamics of the signal intensity per cell (right graphs [time on the x axis, fluorescence intensity {AU} on the y axis]). Each line represents one individual cell.
Optimization of the usage of fluorescent reporters to study gene expression in B. cereus at the single-cell level.
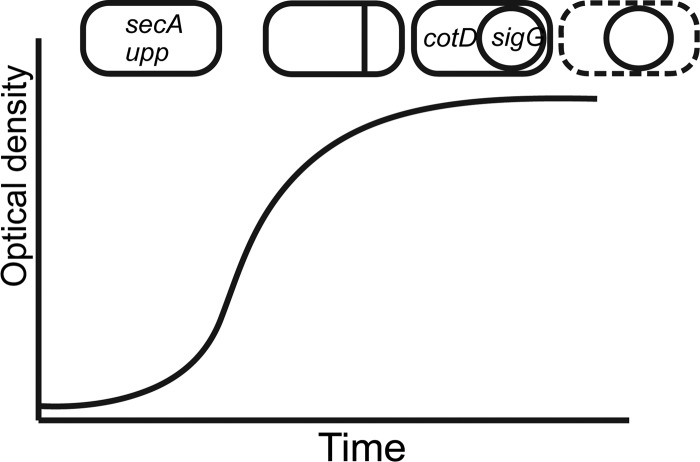
Fluorescent proteins are useful tools for studying gene expression, protein production, and localization events within cells. Observed differences in gene expression (and regulation of such) often provide vital clues for understanding heterogeneity in phenotypes within a microbial population. To optimize usage of fluorescent reporters in B. cereus in conjunction with the time-lapse microscopy technique, we studied the signal intensities and distributions of several green and red fluorescent reporters expressed by various promoters. Especially when dealing with a relatively weak promoter, a strong and stable signal by the fluorescent protein is crucial, as it minimizes the time and intensity of exposure of the cells to the phototoxic excitation light during the course of the time-lapse experiment. For this purpose, the gfpmut3a gene on the pAD123 vector (34) was replaced by genes encoding two different GFP variants and two different RFP variants as described in Materials and Methods. For green fluorescence, the sfGFP(Bs) protein (35) and GFP(Sp) (36) were selected, as these were found to yield the strongest and most stable signals in Streptococcus pneumoniae and Bacillus subtilis, respectively (48). For red fluorescence, we selected two widely used RFP variants, namely, mCherry (37) and mKate2 (38, 39). To test the usability of these proteins as reporters during various stages of the cell's life cycle, four different promoters were selected to drive the expression of these genes (Fig. 5 and Table 2). The first of these was PsecA, which is a strong promoter driving the expression of the essential secA gene during exponential growth phase (22). The second of these was PcotD, which is activated during the late stages of sporulation in the mother cell compartment only (23). In contrast, PsigG is activated in the forespore compartment somewhat earlier during sporulation (41). Lastly, we selected the previously described Pupp for strong and constitutive expression of the FP genes (34). The optimal time point of activity of these promoters as well as the signal intensity deriving from each FP was analyzed using flow cytometry (GFP only) and fluorescence microscopy, as described in Materials and Methods (see Fig. S1 in the supplemental material).
Fig 5.
Timing and location of promoter activity for four selected promoters in this study. The secA and upp genes are expressed constitutively during exponential growth, whereas cotD and sigG are expressed during late-stage sporulation in two different compartments of the cell (mother cell and forespore, respectively).
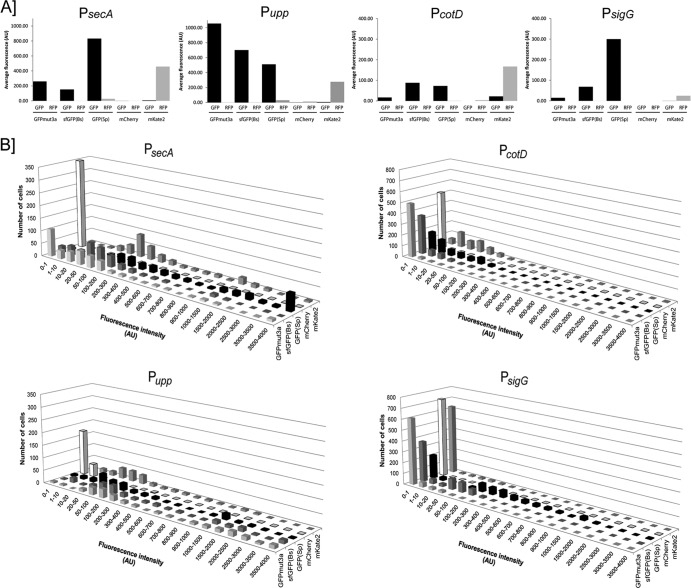
Significant differences in fluorescence intensity and fluorescence distribution in the strains were observed, and these differences seem to be both FP and promoter dependent (Fig. 6). Interestingly, all three GFP variants performed very well when placed under the control of the constitutive Pupp promoter (see Fig. S1 in the supplemental material and Fig. 6). Furthermore, the GFPmut3a protein yielded the strongest average signal when expressed by the Pupp promoter (34), whereas it showed the weakest average signal when placed under the control of the three other selected promoters (Fig. 6A). Generally, the GFP(Sp) protein yielded an overall strong average fluorescent signal in a larger number of cells (Fig. 6B). For red fluorescence, the mKate2 protein clearly performed better in B. cereus than mCherry did. The latter barely yielded a signal for all tested promoters, whereas mKate2 yielded a strong signal in a widespread distribution of cells (Fig. 6B). On the other hand, GFP(Sp) and mKate2 also yielded the highest levels of background fluorescence in the red and green channels, respectively [TRITC for GFP(Sp) and FITC for mKate2] (Fig. 6A). This is important to take into account when dual-labeling experiments are foreseen.
Fig 6.
Fluorescence intensities and distributions of GFP and RFP variants expressed by different promoters on multicopy plasmids. A minimum of 300 cells per strain was counted and the fluorescence intensity per cell measured as described in Materials and Methods. In panel A, the average fluorescence intensity per FP variant is shown in arbitrary units in the FITC channel (GFP; black bars) and the TRITC channel (RFP; gray bars) for each studied promoter (from left to right, PsecA, Pupp, PcotD, and PsigG). Error bars are not included, due to a wide distribution of signal intensity for some promoters (the error bars would then exceed 50%). This variation of fluorescence for all cells is visualized in panel B. Signal intensities (in arbitrary units [AU]) were grouped (x axis), and the number of fluorescent cells per group was calculated (y axis). The distribution of fluorescence intensity is shown for GFPmut3a (light gray shading), sfGFP(Bs) (dark gray shading), GFP(Sp) (black shading), mCherry (white shading), and mKate2 (patterned shading).
DISCUSSION
Time-lapse microscopy is a powerful single-cell technique for the visualization of growth and development of single cells into a microcolony. By use of specific tracking software, such as ImageJ (http://rsbweb.nih.gov/ij/) or Microbetracker (Mathworks Matlab), the exact history of each cell in the microcolony can be tracked and analyzed, enabling detailed studies on cell lineages and heritable traits of observed phenotypes. Even though this technique is already widely used for studying a number of bacterial species, no such reports on its application for Bacillus cereus cells have yet emerged. To enable such an application, we have optimized both cultivation and slide preparation protocols for B. cereus cells specifically.
With time-lapse microscopy, cells are spotted on a slice of solid medium within a chamber on a glass microscopy slide. The chamber is encased by a frame several millimeters thick (20). This allows for complete sealing of the chamber to prevent desiccation of the solid medium, while oxygen is still provided to support growth of the cells. Several conditions have to be met before cells can be successfully studied using the time-lapse microscopy technique. For instance, growth in a monolayer is crucial, especially when working with fluorescent reporters for which signal registration artifacts may occur when cells overlap in the z direction. Also, cells must grow in a patchwise fashion to prevent growth out of the field of vision, which renders cell division tracking and lineage studies impossible. Finally, when studying cellular developmental processes such as sporulation, proper timing and development of such processes under the conditions used are crucial.
When the previously described protocol for B. subtilis time-lapse microscopy studies (20) was applied to B. cereus ATCC 14579 cells, the conditions described above were not met (data not shown). Cells rather grew in complex multidimensional structures and did not sporulate. Both cell preparation and time-lapse conditions were optimized to allow for single-cell studies of growing and developing B. cereus cells. These conditions were optimized for B. cereus ATCC 14579 but are also appropriate for other strains of B. cereus (E. Frenzel, R. Eijlander, O. Kuipers, and M. Ehling-Schulz, personal communication, unpublished data). Importantly, the location of the selected cells on the microscope slide is crucial, as differences in cellular development were observed depending on the position of the cells relative to the periphery of the PAA slice (Fig. 2). This is probably due to differences in oxygen availability. We speculate that this affects not only cellular development but also the fluorescence signal, as oxygen availability is required for proper activity of GFP (42). Selecting cells as close to the periphery as possible is therefore required for more reliable analysis of the results obtained (locations 1 and 2 in Fig. 2). For a slice 4 mm in width, this corresponds to no more than 1 mm away from the edge of the slice. All movies described in this study were obtained with cells within this parameter.
To minimize exposure of the cells to the toxicity of the excitation light during time-lapse microscopy, a strong and relatively stable fluorescence signal is required. For this reason, we tested several readily available GFPs and RFPs in our lab to determine the best-performing variant in B. cereus ATCC 14579 during various growth stages. Under the conditions tested, the overall best performers for green and red fluorescence were GFP(Sp) and mKate2, respectively. Both yielded a well-detectable signal for all four promoters (Fig. 6; see also Fig. S1 in the supplemental material). However, there may be specific conditions in which other (nontested) variants work better.
Fluorescent reporter proteins are useful tools for, e.g., studying promoter activity, heterogeneity in gene expression, protein localization, and cellular dynamics. Nevertheless, visualization of these reporters more often than not merely provides clues to possible mechanisms and often does not reflect the true nature of the process studied. For instance, translational fusions of native proteins to homo-oligomeric FPs can cause mislocalization and/or aggregation of the protein (43). The maturation time of one fluorescent protein may differ from the maturation time of another, causing a delay in signal between two different fluorophores (K. Beilharz, personal communication, unpublished data). Furthermore, the growth stage of the cell can affect the signal captured depending on the intrinsic properties of the FP used (44). In this study, we have shown that the choice of the most optimal FP may also be promoter specific. This calls for optimization and the necessity of important controls when performing new experiments. Fluorescent proteins are continuously being adapted and optimized for improved signal strength, limited spectral overlap, decreased phototoxicity, and increased protein stability. These adaptations include mutations and species-specific codon optimization for improved translation. There is, however, no guarantee that this will yield the best FP variant for that species (48).
Optimization of the time-lapse fluorescence microscopy technique for B. cereus now allows us to investigate the expression level and dynamics of genes of interest in single cells for this organism specifically. In this study, we have focused on the cotD sporulation gene. The heterogeneous character of spores in food products complicates their eradication (15, 16, 45). To obtain further insights into the origin of spore variability, time-lapse (fluorescence) microscopy is a powerful tool to analyze heterogeneity in single-spore development (17, 20, 45, 46). When studying the dynamics of cotD expression during sporulation in B. cereus ATCC 14579, we did not observe a heterogeneous or bimodal signal pattern (Fig. 3A and 4A). This indicates that cotD is expressed homogeneously and consistently among single cells of a subpopulation. Importantly, when studying the same promoter activity expressed from a multicopy plasmid, a wide distribution in GFP intensity could be observed among single cells (Fig. 3B and 4B). The plasmid copy number can vary per cell, per growth stage, and during sporulation (47), which can cause a significant level of heterogeneity that does not reflect true promoter activity over time. Taken together, these results show that plasmid-based fusions are an unsuitable tool for single-cell and heterogeneity studies.
Unfortunately, the molecular toolbox for genetic manipulation via single or double crossover in the B. cereus genome is not as elaborate as those for other model organisms, such as B. subtilis. The development of an integrative system allowing for single-copy transcriptional and translational fluorescence reporter fusions in B. cereus should be considered for future research efforts, as this will be a valuable addition to the entire B. cereus research field. Together with optimized techniques such as time-lapse fluorescence microscopy, various developmental and molecular processes (such as heterogeneous gene expression during sporulation) can be studied and visualized at the single-cell level. These tools will subsequently increase our understanding of such processes in B. cereus and related species as more specific mechanistic insights can be obtained.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Frenzel for testing the time-lapse microscopy method using other strains of B. cereus and for useful comments on the manuscript. We also thank J. W. Veening for useful discussions and suggestions.
Footnotes
Published ahead of print 12 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01347-13.
REFERENCES
- 1.Bottone EJ. 2010. Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 23:382–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stenfors Arnesen LP, Fagerlund A, Granum PE. 2008. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32:579–606 [DOI] [PubMed] [Google Scholar]
- 3.Mols M, Abee T. 2011. Bacillus cereus responses to acid stress. Environ. Microbiol. 13:2835–2843 [DOI] [PubMed] [Google Scholar]
- 4.Mols M, Abee T. 2011. Primary and secondary oxidative stress in Bacillus. Environ. Microbiol. 13:1387–1394 [DOI] [PubMed] [Google Scholar]
- 5.Abee T, Wels M, de Been M, den Besten H. 2011. From transcriptional landscapes to the identification of biomarkers for robustness. Microb. Cell Fact. 10(Suppl 1):S9. 10.1186/1475-2859-10-S1-S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindbäck T, Mols M, Basset C, Granum PE, Kuipers OP, Kovacs AT. 2012. CodY, a pleiotropic regulator, influences multicellular behaviour and efficient production of virulence factors in Bacillus cereus. Environ. Microbiol. 14:2233–2246 [DOI] [PubMed] [Google Scholar]
- 7.Wijman JG, de Leeuw PP, Moezelaar R, Zwietering MH, Abee T. 2007. Air-liquid interface biofilms of Bacillus cereus: formation, sporulation, and dispersion. Appl. Environ. Microbiol. 73:1481–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Voort M, Abee T. 2013. Sporulation environment of emetic toxin-producing Bacillus cereus strains determines spore size, heat resistance and germination capacity. J. Appl. Microbiol. 114:1201–1210 [DOI] [PubMed] [Google Scholar]
- 9.Abee T, Groot MN, Tempelaars M, Zwietering M, Moezelaar R, van der Voort M. 2011. Germination and outgrowth of spores of Bacillus cereus group members: diversity and role of germinant receptors. Food Microbiol. 28:199–208 [DOI] [PubMed] [Google Scholar]
- 10.den Besten HM, van Melis CC, Sanders JW, Nierop Groot MN, Abee T. 2012. Impact of sorbic acid on germination and outgrowth heterogeneity of Bacillus cereus ATCC 14579 spores. Appl. Environ. Microbiol. 78:8477–8480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yi X, Setlow P. 2010. Studies of the commitment step in the germination of spores of Bacillus species. J. Bacteriol. 192:3424–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Want A, Hancocks H, Thomas CR, Stocks SM, Nebe-von-Caron G, Hewitt CJ. 2011. Multi-parameter flow cytometry and cell sorting reveal extensive physiological heterogeneity in Bacillus cereus batch cultures. Biotechnol. Lett. 33:1395–1405 [DOI] [PubMed] [Google Scholar]
- 13.Cronin UP, Wilkinson MG. 2008. Bacillus cereus endospores exhibit a heterogeneous response to heat treatment and low-temperature storage. Food Microbiol. 25:235–243 [DOI] [PubMed] [Google Scholar]
- 14.Veening JW, Smits WK, Kuipers OP. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62:193–210 [DOI] [PubMed] [Google Scholar]
- 15.Hornstra LM, Ter Beek A, Smelt JP, Kallemeijn WW, Brul S. 2009. On the origin of heterogeneity in (preservation) resistance of Bacillus spores: input for a ‘systems' analysis approach of bacterial spore outgrowth. Int. J. Food Microbiol. 134:9–15 [DOI] [PubMed] [Google Scholar]
- 16.Eijlander RT, Abee T, Kuipers OP. 2011. Bacterial spores in food: how phenotypic variability complicates prediction of spore properties and bacterial behavior. Curr. Opin. Biotechnol. 22:180–186 [DOI] [PubMed] [Google Scholar]
- 17.de Jong IG, Veening JW, Kuipers OP. 2010. Heterochronic phosphorelay gene expression as a source of heterogeneity in Bacillus subtilis spore formation. J. Bacteriol. 192:2053–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh S, Scotland M, Setlow P. 2012. Levels of germination proteins in dormant and superdormant spores of Bacillus subtilis. J. Bacteriol. 194:2221–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramirez-Peralta A, Stewart KA, Thomas SK, Setlow B, Chen Z, Li YQ, Setlow P. 2012. Effects of the SpoVT regulatory protein on the germination and germination protein levels of spores of Bacillus subtilis. J. Bacteriol. 194:3417–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Jong IG, Beilharz K, Kuipers OP, Veening JW. 2011. Live cell imaging of Bacillus subtilis and Streptococcus pneumoniae using automated time-lapse microscopy. J. Vis. Exp. 2011(53):e3145. 10.3791/3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young JW, Locke JC, Altinok A, Rosenfeld N, Bacarian T, Swain PS, Mjolsness E, Elowitz MB. 2012. Measuring single-cell gene expression dynamics in bacteria using fluorescence time-lapse microscopy. Nat. Protoc. 7:80–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbort M, Klein M, Manting EH, Driessen AJ, Freudl R. 1999. Temporal expression of the Bacillus subtilis secA gene, encoding a central component of the preprotein translocase. J. Bacteriol. 181:493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng LB, Losick R. 1990. Cascade regulation of spore coat gene expression in Bacillus subtilis. J. Mol. Biol. 212:645–660 [DOI] [PubMed] [Google Scholar]
- 24.Imamura D, Kuwana R, Takamatsu H, Watabe K. 2010. Localization of proteins to different layers and regions of Bacillus subtilis spore coats. J. Bacteriol. 192:518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. 2010. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 90:1103–1163 [DOI] [PubMed] [Google Scholar]
- 26.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. 2006. The fluorescent toolbox for assessing protein location and function. Science 312:217–224 [DOI] [PubMed] [Google Scholar]
- 27.Luo W, He K, Xia T, Fang X. 2013. Single-molecule monitoring in living cells by use of fluorescence microscopy. Anal. Bioanal. Chem. 405:43–49 [DOI] [PubMed] [Google Scholar]
- 28.Shaner NC, Steinbach PA, Tsien RY. 2005. A guide to choosing fluorescent proteins. Nat. Methods 2:905–909 [DOI] [PubMed] [Google Scholar]
- 29.Leroch M, Mernke D, Koppenhoefer D, Schneider P, Mosbach A, Doehlemann G, Hahn M. 2011. Living colors in the gray mold pathogen Botrytis cinerea: codon-optimized genes encoding green fluorescent protein and mCherry, which exhibit bright fluorescence. Appl. Environ. Microbiol. 77:2887–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henriques MX, Catalao MJ, Figueiredo J, Gomes JP, Filipe SR. 2013. Construction of improved tools for protein localization studies in Streptococcus pneumoniae. PLoS One 8:e55049. 10.1371/journal.pone.0055049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sastalla I, Chim K, Cheung GY, Pomerantsev AP, Leppla SH. 2009. Codon-optimized fluorescent proteins designed for expression in low-GC gram-positive bacteria. Appl. Environ. Microbiol. 75:2099–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masson L, Prefontaine G, Brousseau R. 1989. Transformation of Bacillus thuringiensis vegetative cells by electroporation. FEMS Microbiol. Lett. 51:273–277 [DOI] [PubMed] [Google Scholar]
- 33.Lewis PJ, Marston AL. 1999. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227:101–110 [DOI] [PubMed] [Google Scholar]
- 34.Dunn AK, Handelsman J. 1999. A vector for promoter trapping in Bacillus cereus. Gene 226:297–305 [DOI] [PubMed] [Google Scholar]
- 35.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24:79–88 [DOI] [PubMed] [Google Scholar]
- 36.Martin B, Granadel C, Campo N, Henard V, Prudhomme M, Claverys JP. 2010. Expression and maintenance of ComD-ComE, the two-component signal-transduction system that controls competence of Streptococcus pneumoniae. Mol. Microbiol. 75:1513–1528 [DOI] [PubMed] [Google Scholar]
- 37.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 [DOI] [PubMed] [Google Scholar]
- 38.Beilharz K, Novakova L, Fadda D, Branny P, Massidda O, Veening JW. 2012. Control of cell division in Streptococcus pneumoniae by the conserved Ser/Thr protein kinase StkP. Proc. Natl. Acad. Sci. U. S. A. 109:E905–E913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, Lukyanov S, Zaraisky AG, Davidson MW, Chudakov DM. 2009. Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 418:567–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sterlini JM, Mandelstam J. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Vries YP, Hornstra LM, de Vos WM, Abee T. 2004. Growth and sporulation of Bacillus cereus ATCC 14579 under defined conditions: temporal expression of genes for key sigma factors. Appl. Environ. Microbiol. 70:2514–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heim R, Prasher DC, Tsien RY. 1994. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 91:12501–12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J. 2012. Segregation of molecules at cell division reveals native protein localization. Nat. Methods 9:480–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doherty GP, Bailey K, Lewis PJ. 2010. Stage-specific fluorescence intensity of GFP and mCherry during sporulation in Bacillus subtilis. BMC Res. Notes 3:303. 10.1186/1756-0500-3-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pandey R, Ter Beek A, Vischer NO, Smelt JP, Brul S, Manders EM. 2013. Live cell imaging of germination and outgrowth of individual Bacillus subtilis spores; the effect of heat stress quantitatively analyzed with SporeTracker. PLoS One 8:e58972. 10.1371/journal.pone.0058972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veening JW, Stewart EJ, Berngruber TW, Taddei F, Kuipers OP, Hamoen LW. 2008. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc. Natl. Acad. Sci. U. S. A. 105:4393–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turgeon N, Laflamme C, Ho J, Duchaine C. 2008. Evaluation of the plasmid copy number in B. cereus spores, during germination, bacterial growth and sporulation using real-time PCR. Plasmid 60:118–124 [DOI] [PubMed] [Google Scholar]
- 48.Overkamp W, Beilharz K, Weme RDO, Solopova A, Karsens H, Kovács AT, Kok J, Kuipers OP, Veening J-W. Benchmarking various GFP variants in Bacillus subtilis, Streptococcus pneumoniae and Lactococcus lactis for live cell imaging. Appl. Environ. Microbiol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.