Abstract
One of the major concerns in the production of dairy concentrates is the risk of contamination by heat-resistant spores from thermophilic bacteria. In order to acquire more insight in the composition of microbial communities occurring in the dairy concentrate industry, a bar-coded 16S amplicon sequencing analysis was carried out on milk, final products, and fouling samples taken from dairy concentrate production lines. The analysis of these samples revealed the presence of DNA from a broad range of bacterial taxa, including a majority of mesophiles and a minority of (thermophilic) spore-forming bacteria. Enrichments of fouling samples at 55°C showed the accumulation of predominantly Brevibacillus and Bacillus, whereas enrichments at 65°C led to the accumulation of Anoxybacillus and Geobacillus species. Bacterial population analysis of biofilms grown using fouling samples as an inoculum indicated that both Anoxybacillus and Geobacillus preferentially form biofilms on surfaces at air-liquid interfaces rather than on submerged surfaces. Three of the most potent biofilm-forming strains isolated from the dairy factory industrial samples, including Geobacillus thermoglucosidans, Geobacillus stearothermophilus, and Anoxybacillus flavithermus, have been characterized in detail with respect to their growth conditions and spore resistance. Strikingly, Geobacillus thermoglucosidans, which forms the most thermostable spores of these three species, is not able to grow in dairy intermediates as a pure culture but appears to be dependent for growth on other spoilage organisms present, probably as a result of their proteolytic activity. These results underscore the importance of abiotic and microbiotic factors in niche colonization in dairy factories, where the presence of thermophilic sporeformers can affect the quality of end products.
INTRODUCTION
Contamination by spore-forming bacteria is an important concern in the production of dairy concentrates. Besides mesophilic bacteria, thermophiles are problematic in food-producing industrial facilities operating from 40°C to 65°C, as these temperatures support growth and biofilm formation of thermophilic sporeformers (1). The growth of these thermophiles in biofilms in factories can result in numbers of up to 106 CFU/g of bacteria and spores released in the final products, including whey and milk concentrates (2). These spores could germinate when the conditions are favorable, finally resulting in high numbers of bacteria and off-flavor in end products (2, 3). In order to prevent the presence and outgrowth of the accumulated spores, costly precautions such as frequent cleaning, short production runs, and intensive microbial product control are required.
Most thermophilic sporeformers which have been identified so far in dairy processing lines and products belong to the genera of Bacillus, Geobacillus, and Anoxybacillus (2, 4, 5). Geobacillus spp. and A. flavithermus are the most frequently reported species in thermophilic dairy biofilms (1). The presence of spores from these thermophilic bacilli in the final products most likely results from the detachment of spores from biofilms on stainless steel surfaces found within a milk powder plant (2). However, it is not evident that these organisms are the only organisms important for biofilm formation in dairy processing environments. Insight into the species diversity and the contribution of both thermophilic and mesophilic species in microbial populations at the different sites in dairy concentrate production lines is currently lacking. In this study, we applied a bar-coded 16S amplicon sequencing approach (6) to get insight into the microbial composition of fouling samples in dairy concentrate-processing plants and evaluated the effect of enrichments at high temperatures, at air-liquid interfaces, or on different surfaces. We isolated three thermophilic species on the basis of their ability to grow at high temperatures and efficiency in forming biofilms under laboratory conditions. We provide evidence that suggests that growth in milk-based media of G. thermoglucosidans is dependent on proteolytic activity of other species present in dairy concentrate-processing environments.
MATERIALS AND METHODS
Sampling, culturing, and enrichment.
A number of fouling sites were selected along dairy concentrate production lines for the bar-coded 16S amplicon sequencing analysis of the microbial flora (see Table S1 in the supplemental material). Samples of standard milk, fouling material isolated from the processing line, and final products were collected. Standard milk was flash frozen by dripping in liquid nitrogen. The frozen milk pellets were stored at −80°C. Fouling samples were scraped from pipelines, dispersed at a 1:1 (wt/vol) dilution in sterile antifreeze Microbank medium (Pro-Lab Diagnostics, Canada), and stored at −80°C. Final products were dissolved in sterile water (2% to 10% [wt/vol]) and stored at −80°C.
Viable counts were carried out for all samples analyzed with bar-coded 16S amplicon sequencing. Growth analysis of strains and CFU determination were performed on tryptone soy broth (TSB) or tryptone soy agar (TSA) (Tritium Microbiologie, The Netherlands). All CFU determinations in this study were performed by plating 80 μl on TSA plates followed by overnight (O/N) incubation at 30°C (for nonthermophilic CFU determination) or at 55°C (for thermophilic CFU determination). Dilution series were made in PPS (0.1% peptone, 0.9% NaCl). Thermophilic aerobic spore counts at 55°C were similarly determined after pretreatment of the samples at 100°C for 30 min to eliminate vegetative cells and to activate thermoresistant spores (2). The CFU determinations of samples after enrichment at 55 or 65°C were obtained by plating on TSA and incubating at the respective enrichment temperatures.
Thermophilic enrichment was carried out by O/N culturing of 50 μl of a sample in 2 ml TSB at 55 or 65°C followed by inspection for growth according to increases of optical density (OD) (no growth [−], little growth [±], or outgrowth [+]). Initially, enrichment was performed at 55°C in TSB, since it is a classical method to determine dairy thermophilic bacterial loads (2). However, to prevent extensive overgrowth of mesophilic species at 55°C, enrichment at 65°C was included as well, in order to facilitate selection of the thermophilic species.
Biofilm model systems.
In order to study biofilm formation by thermophilic spore-forming dairy isolates on a laboratory scale, a standing steel biofilm model system was developed. This biofilm system included a sterile, vertically standing, 14-by-14-mm stainless steel coupon (P. 316 grade) in a well of a sterile 24-well plate (Corning, The Netherlands). The plate was incubated in a tight plastic bag containing a wetted paper towel to limit evaporation of the culture media (see Fig. S1 in the supplemental material). In addition, a submerged steel biofilm system was developed, consisting of a steel coupon lying horizontally on the well bottom of a 24-well plate.
For enrichment in the static biofilm models, 2-ml industrial milk samples (standardized milk with a standardized composition) and 50 μl of a fouling sample in 2 ml of heat-sterilized milk (120°C, 20 min) were cultured O/N at 55 or 65°C (nonshaken) in separate wells. After O/N incubation, the various fractions (including culture medium, polystyrene well wall, and coupon surfaces) were harvested and directly subjected to CFU determinations or stored at −80°C until DNA isolation (see below). The medium fractions were directly harvested from the culture wells. The metal coupons and empty wells were gently rinsed with sterile PPS (three times with 3 ml each time) and separately swabbed (coupons were first transferred to clean sterile wells) with sterile cotton swabs, each sampled twice in 150 μl sterile PPS.
Air-liquid interface biofilms of industrial isolates were studied as well by the use of vertical, sterile 15-by-15-mm glass coupons (cut from standard microscopy object glass) in 12-well plates. After O/N cultivation, the glass coupons were gently washed with demineralized water (demiwater) and fixed by drying for 10 min at 60°C. Culture wells were washed with sterile water (three times with 3 ml/well) and fixed by incubation for 10 min at 60°C. Water-washed and air-dried coupons or culture wells were used for crystal violet (CV) staining (performed for 5 min with 1% [wt/vol] CV followed by three washes with water). CV-stained coupons were analyzed by light microscopy. CV-stained culture wells were destained for 5 min at room temperature with 33% acetic acid (at 1.1× the volume originally cultured in the well), and the OD between 580 and 600 nm was measured with a plate well reader (Tecan, Switzerland) to determine the amount of CV-stainable biofilm.
Fluorescence microscopy.
Coupons were incubated for 2 min with 0.1% Auramine (Merck, The Netherlands) for visualizing the attached cells (7). Spores were stained in the water-washed and air-dried coupons by the Auramine-Safranine method (7). Briefly, stainless steel or glass coupons were incubated for 2 min with 0.1% Auramine (Merck, The Netherlands), water washed, incubated for 1 min with 0.25% Safranin (BD Biosciences), water washed, and air dried for 10 min at 60°C, and bright-field microscopy (glass coupons) and fluorescence microscopy (Zeiss Axio Observer Z1; filter set, Endow green fluorescent protein [GFP]; excitation bandpass [Ex BP], 470 to 40; beam splitter frustrated total [BS FT], 495; emission [EM] BP, 525 to 55) were performed directly on the stained, dried, and covered glass coupons.
DNA isolation.
Genomic DNA (gDNA) was isolated from the (enriched) fouling samples and fractions from the static biofilm model. The bacterial samples (50 to 200 μl) were added to a 1.5-ml screw-cap Eppendorf tube with 0.3 g zirconium-silica beads (0.1-mm-diameter bead size), 800 μl phenol (pH 8.0), and 400 μl Agowa buffer without detergent. Next, the samples were homogenized with a Bio101 BeadBeater (Biospec Products) twice for 45 s each time with a 30-s interval of cooling on ice and spun down for 10 min at 10,000 × g. The upper, aqueous phase was taken and extracted with an Agowa mag Mini DNA isolation kit (Agowa, Germany) eluted in 45 μl Agowa BL buffer. The quality and quantity of gDNA were determined using an agarose gel and a Nanodrop ND-1000 spectrophotometer (NanoDrop Technologies).
Bar-coded 16S amplicon sequencing.
Mass sequencing was performed as described earlier (6). Briefly, barcoded 16S rRNA fragments were amplified with forward primer 785F (5′-GCCTCCCTCGCGCCATCAGGGATTAGATACCCBRGTAGTC-3′) and reverse primer 1175R (5′-GCCTTGCCAGCCCGCTCAGNNNNACGTCRTCCCCDCCTTCCTC-3′). Pyrosequencing of equimolar mixes of 24 amplicon pools was performed by Keygene N.V. (The Netherlands) using a Roche Genome Sequencer-20 (GS-20) and FLX 454 pyrosequencing technology, yielding on average 1,145 reads per amplicon pool (standard deviation, 456; minimum, 277; maximum, 2,583). The FASTA format sequences and corresponding quality scores were extracted from the .sff data files generated by the GS-FLX system using the GS Amplicon software package (Roche, Branford, CT). Sequence data were processed using modules implemented in the mothur v. 1.25.0 software platform (8). Sequences were binned by sample of origin by the unique barcode sequences in each amplicon pool. For further downstream analyses, barcodes and primer sequences were trimmed and low-quality reads were excluded from the analyses. The data set was simplified by using the “unique.seqs” command to generate a nonredundant (unique) set of sequences. Unique sequences were aligned using the “align.seqs” command and an adaptation of the Bacterial SILVA SEED database as a template (available at http://www.mothur.org/wiki/Alignment_database). In order to ensure that we were analyzing comparable regions of the 16S rRNA gene across all reads, sequences that started before the 2.5 percentile or ended after the 97.5 percentile in the alignment were filtered. Sequences were denoised using the “pre.cluster” command. This command applies a pseudo-single-linkage algorithm with the aim of removing sequences that are likely due to pyrosequencing errors (9). Potentially chimeric sequences were detected and removed using the “chimera.slayer” command (10). High-quality aligned sequences were classified using the RDP-II naive Bayesian Classifier implemented into the mothur platform. Aligned sequences were clustered into operational taxonomic units (OTUs) (defined by 97% similarity) using the average linkage clustering method. Typing to the level of Anoxybacillus and Geobacillus species was performed using the most abundant unique sequence of these OTUs in the Seqmatch tool of RDP. Relative abundances of genera and species were calculated as fractions of the total reads per sample.
Typing of industrial isolates.
A set of around 100 bacterial isolates (single colonies) were obtained from raw and enriched samples. These isolates were cultured to determine growth and biofilm formation at temperatures of 30, 60, 65, and 70°C in TSB medium. Of all 100 isolates tested, 20 isolates were able to grow (OD > 0.08) and form biofilms (OD > 0.11) at 60°C and 70°C. DNA of 20 industrial isolates was isolated as described above. For typing of industrial isolates, the 16S rRNA gene region 8 to 1408 was PCR amplified from gDNA using forward (F) and reverse (R) primers 8F (5′-AGAGTTTGATCHTGGYTCAG-3′) and 1408R (5′-TGACGGGCGGTGTGTACAA-3′). PCR amplicons were purified and bidirectionally sequenced by GATC-biotech AG, Germany, using primers 8F, 27F (5′-AGAGTTTGATCMTGGCTCAG-3′), 1408R, and 1392R (5′-ACGGGCGGTGTGTGTRC-3′). The sequences were typed at the species level with the RDP SeqMatch tool (http://rdp.cme.msu.edu/) (11) and by selection of the best hit reported from the RDP database (type strains, non-type strains, unculturable strains, and isolates with a size of >1,200 bp and of good quality). Growth curves of selected model strains from thermophilic spore-forming species A. flavithermus TNO-09.006, G. stearothermophilus TNO-09.008, and G. thermoglucosidans TNO-09.020 were determined in TSB medium at various temperatures by the use of a temperature gradient in a PCR machine (model DNA Engine Tetrad, PTC-225) (100 μl of culture per well; range, 38 to 74°C) and multiple OD measurements during cultivation (20 min each) (50 μl/well of a 384-well plate, using a Tecan F500 plate reader at 600 nm). Exponential-growth rates (μ) and doubling times (tD) were calculated with the equations μ = ln[d(OD)]/dt and tD = ln2/μ, respectively. The Tmin and Tmax are defined as the minimum and maximum temperatures at which growth could be detected under the conditions used. The Topt is defined as the temperature with the highest growth rate, as expressed in doubling time (tD).
DNA-DNA hybridizations.
Genomic DNA was extracted from pure cultures according to a modification of the procedure by Gevers et al. (12). Hybridizations were performed in the presence of 50% formamide at 39°C according to a method adapted from Ezaki et al. (13). The DNA-DNA hybridization percentages reported are the means of at least 6 hybridizations.
Assessment of casein-degrading activity.
The selected model strains from thermophilic spore-forming species A. flavithermus TNO-09.006, G. stearothermophilus TNO-09.008, and G. thermoglucosidans TNO-09.020 were examined for their capability to utilize milk protein. Media included Casitone plates (25 g Casitone, 5 g NaCl, 2.5 g K2HPO4, 1.5% [wt/vol] agar), tryptone plates (10 g tryptone, 5 g NaCl, 2.5g K2HPO4, 1.5% [wt/vol] agar), Ca-caseinate plates (1.25% [wt/vol] containing Ca-caseinate [Friesland-Campina, The Netherlands]) and 0.8% agarose, and pancreatin-digested Ca-caseinate plates. The latter plates were prepared by digestion of Ca-caseinate (1.25% [wt/vol]) with 10 mg/ml pancreatin (Sigma P3292) for 3 h at 37°C followed by heat inactivation at 100°C for 10 min. Plates were inoculated by transfer of bacterial cells taken from a TSA plate.
Heat resistance of spores.
A suspension prepared from an plate culture grown overnight was spread on NA++ plates (nutrient agar with supplementation of analysis-grade 1.13 mM CaCl2 and 0.99 mM MnSO4) and incubated for 2 days at 55°C. Bacterial lawns containing spores were harvested and washed with sterile demiwater as described previously (14). This water-washing procedure was repeated three times in order to obtain pure spore suspensions. The spore suspensions were stored at −20°C. The heat inactivation kinetics of the spores were determined as follows. Micropipettes of 100 μl were filled with spore suspensions, and both ends of the micropipettes were sealed by heating. Micropipettes were incubated within a time window at serial temperatures above 100°C in an oil bath filled with glycerol. The spore suspension was diluted 100 times in PPS, a series of dilutions were made, and the dilutions were plated on to TSA plates. The D values of the spore batches were derived from plots with log CFU versus incubation time by fitting a log-linear model with a tail to the data, and the z values were calculated by plotting the logD value against the temperature and performing a linear regression (see Fig. S2 in the supplemental material).
Compartmentalized growth experiments.
The determination of growth dependencies in ultra-heat-treated (UHT) skim milk was performed using a BD Falcon cell culture insertion system. This system allows growth of strains in two compartments separated by a permeable membrane that permits diffusion of medium components (pore size, 0.4 μm). Both the well and the cell culture insertion were filled with 3 ml UHT skim milk and inoculated with approximately 4 × 103 CFU A. flavithermus TNO-09.006 and 3 × 103 CFU G. thermoglucosidans TNO-09.020, respectively. As controls, wells were filled with 3 ml UHT skim milk and inoculated with either A. flavithermus TNO-09.006 or G. thermoglucosidans TNO-09.020 with the same amounts of CFU. All measurements were performed in triplicate. Following inoculation, the 6-well plates were wrapped in a plastic bag and sealed in order to prevent evaporation and incubated at 65°C at 50 rpm. Sampling was performed at 3, 6, 9, 12, and 24 h. CFU counts of each fraction were determined by serial dilutions poured in TSA.
Formation of biofilms on stainless steel coupons in cocultures.
Stainless steel coupons were placed in the wells of a polystyrene 12-well plate (Falcon; Becton, Dickinson, France). The wells were half filled with 3 ml of UHT skim milk, which was inoculated with a 1% (vol/vol) overnight culture of either a mixture or single strain of A. flavithermus strain TNO-09.006 and G. thermoglucosidans TNO-09.020. The plates were wrapped with plastic bags and wet tissues and incubated for 12 h, 24 h, and 48 h at 65°C. The total number of bacterial cells, present in the milk or attached to the surface of the stainless steel coupon, was determined by CFU counting. The coupons were washed in sterile UHT skim milk 3 times. Then they were placed in 50-ml tubes filled with 3 ml of UHT skim milk and 0.5 g of glass beads (100-μm diameter). Tubes were mixed by a vortex procedure for 1 min to detach the cells from the stainless steel coupon. Serial dilutions were made and plated on TSA–5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (TSA–X-Gal) plates for counting after 24 h of incubation at 55°C. Biofilm formation was assessed in triplicate in two independent experiments.
RESULTS
Enrichment of Geobacillus and Anoxybacillus at high temperatures.
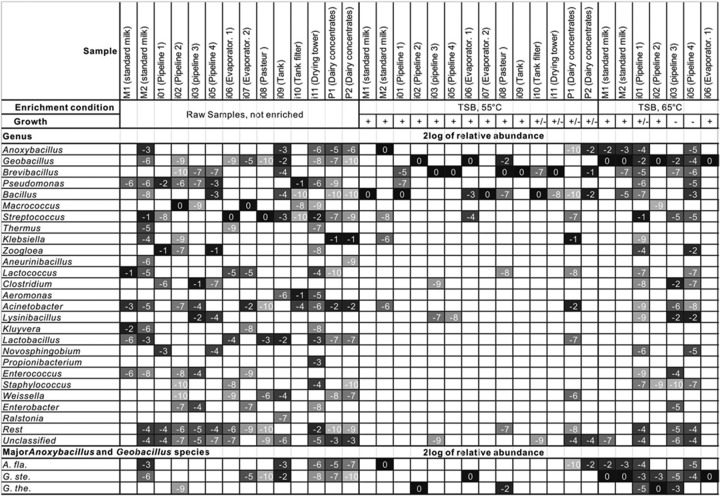
The contribution of thermophilic sporeformers to the contamination of the dairy processing lines and end products was evaluated by an analysis of the microbial composition of dairy fouling samples by bar-coded 16S rRNA amplicon sequencing up to the genus level. At the phylum level, the dairy fouling samples were dominated by Firmicutes and Proteobacteria (55% and 42%, respectively). The majority of the 16S rRNA sequences in each sample represented a wide variety of mesophilic genera (Fig. 1), covering many genera of the classical milk microbial flora (3, 15–18); only a minor fraction of 16S rRNA sequences were associated with the thermophilic genera Anoxybacillus and Geobacillus. It should be noted that the standard enumeration method for thermophilic species at 55°C also provides conditions for some mesophilic species to grow. Therefore, the composition analysis of the 14 dairy samples was also carried out after enrichment at 65°C (Fig. 1). Overnight (O/N) incubation of the 14 dairy samples at 55°C resulted in the enrichment of the spore-forming genus Bacillus (four samples) or Brevibacillus (six samples) and in some cases in the enrichment of thermophilic spore-forming genera Geobacillus (two samples) and Anoxybacillus (one sample). An increase of the enrichment temperature to 65°C resulted in a higher predominance of thermophilic genera, including Geobacillus (seven samples) and Anoxybacillus (three samples). In eight samples, little or no growth occurred (− or ±), showing a composition similar to that present in the samples prior to enrichment at 65°C.
Fig 1.
Microbial inventory of raw and enriched dairy samples. Genomic DNA isolated from industrial samples (raw and enriched overnight at 55°C or 65°C in TSB medium) was analyzed by mass-sequencing 16S genotyping (500 to 2,000 sequences per sample). Gray levels of cells represent relative abundances of microbial composition at the genus level or species level; black to white shading indicates high to low abundance levels. Numbers represent 2log values (relative abundance). From the top to the bottom of the figure, each row presents the abundance of one genus or species in each of 14 locations selected from a dairy processing plant. Abbreviations: A. fla, Anoxybacillus flavithermus; G. ste, Geobacillus stearothermophilus; G. the., Geobacillus thermoglucosidans.
Preference of thermophiles for air-liquid interface or submerged biofilms.
The next experiment was aimed at the identification of thermophilic genera in different types of biofilms formed at high temperatures. Static biofilm systems were inoculated with three of our previously isolated dairy samples (two standard milk samples and one whey evaporator sample). The incubations were carried out at 55°C and 65°C in the submerged steel biofilm system and the standing steel biofilm model system, which includes an air-liquid interface, as described in Materials and Methods and displayed in Fig. S1 in the supplemental material. The total viable counts of the different fractions in the standing steel biofilm model (medium, standing steel coupon, and plastic well) were determined (see Table S2). The counts in the planktonic fraction at 55°C and 65°C were approximately 1,000-fold higher than the initial counts of the dairy samples at 55°C, indicating that enrichment of thermophiles occurred at 55°C and 65°C in milk.
Subsequently, the different fractions in the biofilm model system were analyzed for their microbiological composition by bar-coded 16S amplicon sequencing. The thermophilic genera Anoxybacillus and Geobacillus dominated in most of the samples (Fig. 2). Relatively high numbers of Anoxybacillus were found after enrichment at both 55°C and 65°C, whereas 16S rRNA gene sequences affiliated with Geobacillus dominated the population when samples were incubated at 65°C. Besides, the mesophilic spore-forming genus Aneurinibacillus was enriched at 55°C. We observed that the contribution of the thermophilic genera Geobacillus and Anoxybacillus in biofilms was higher in the air-liquid-interface biofilms (standing steel) than in the submerged biofilms, where the genus Pseudomonas dominated at 55°C and 65°C. Although the latter genus is not a known thermophilic biofilm former, it should be noted that a thermophilic Pseudomonas species growing at 55°C has been previously described (19). Thermophilic populations which adhere to steel and plastic surfaces were found to be nearly identical in our model system (Fig. 2). The presence of the species A. flavithermus, G. stearothermophilus, and G. thermoglucosidans is shown in the three bottom rows of Fig. 2. While A. flavithermus and G. stearothermophilus were frequently enriched in the standing steel biofilm system, G. thermoglucosidans was not found in any of the samples enriched in milk medium. Apparently, this species does not readily accumulate in milk medium, possibly as a result of a growth dependence, as described below.
Fig 2.
Microbial inventory of thermophilic bacteria in a static biofilm model. Standard milk (M1 and M2) or sterile milk inoculated with industrial fouling from a whey evaporator (i7) was cultured O/N at 55 or 65°C in plastic culture wells containing steel coupons (static biofilm model) in order to study thermophile enrichment in the medium (planktonic) and on steel and plastic surfaces (biofilm). Genomic DNA isolated from raw milk and fouling (raw) milk, steel or submerged steel, and plastic well wall fractions were analyzed by mass-sequencing 16S genotyping (500 to 2,000 sequences per sample). Gray values, numbers, and abbreviations are the same as described for Fig. 1. M, media; subm.S, submerged-stainless-steel-surface-attached biofilm; St.S, standing-stainless-steel-surface-attached biofilm; P, plastic-surface-attached biofilm.
Isolation and characterization of novel thermophilic biofilm and sporeformers.
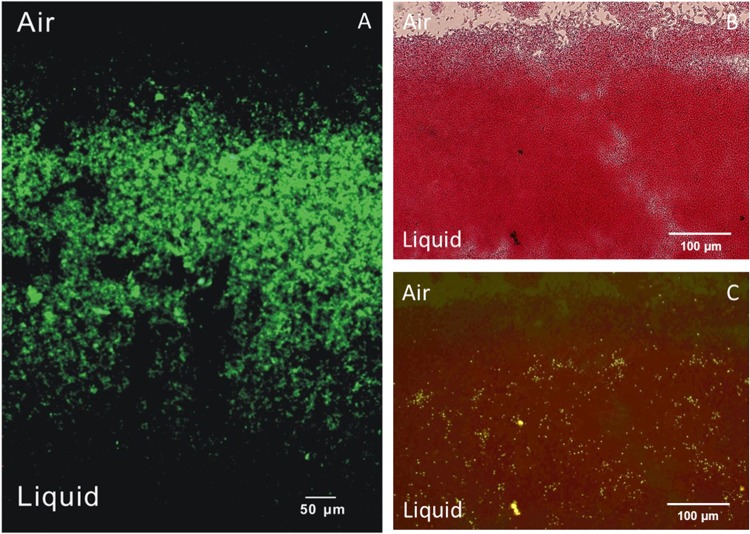
In this study, approximately 200 strains were isolated by selection of colonies from TSA plates incubated at 55°C after inoculation with fouling samples from the dairy concentrate production line. Twenty strains with morphologically different colonies were characterized with respect to their 16S rRNA genotypes, growth rates, and biofilm-forming performance (data not shown). All culturable isolates from the standard milk sources (M1 and M2) were typed as A. flavithermus, except for two isolates of Bacillus licheniformis; the isolate from the dairy concentrate end product was typed as G. stearothermophilus, and those from the fouling samples showed a larger variety of species, including A. flavithermus, G. stearothermophilus, and G. thermoglucosidans. The occurrence of the thermophilic isolates A. flavithermus, G. stearothermophilus, and G. thermoglucosidans in raw and enriched samples was confirmed by an exact match to the 16S rRNA sequences of these species (see the three bottom rows in Fig. 1). Most isolates showed significant biofilm formation at 60°C and 70°C, as derived from the OD values from crystal violet staining of surface-attached biomass after growth. On the basis of their ability to efficiently form biofilms in a laboratory model system, the isolates A. flavithermus TNO-09.006, G. stearothermophilus TNO-09.008, and G. thermoglucosidans TNO-09.020 were selected and their species identity was confirmed by DNA-DNA hybridizations with genomic DNA isolated from the three corresponding type strains. The percentage of relatedness to the type strain matched the >70% criterion for the assignment of all three bacterial species (Table 1). The sequences of the full genomes of the three strains were determined (20, 21), and the strains were characterized regarding their temperature growth range and optimum, sporulation efficiency, and spore heat resistance (Tables 1 and 2). In addition, their ability to sporulate was confirmed by microscopic examination, showing the formation of bright-phase endospores at the poles (see Fig. S3 in the supplemental material). Heat-resistant spores were enumerated in culture medium and stainless steel biofilm fractions during a cultivation experiment of 30 h, indicating an increase in the number of spores over time in both fractions of up to 105 CFU per ml (see Fig. S4 in the supplemental material). Interestingly, the growth at high temperatures was observed over a temperature window of 19°C for all three thermophilic species, including 43 to 62°C, 48 to 67°C, and 50 to 69°C for A. flavithermus, G. stearothermophilus, and G. thermoglucosidans, respectively. The preference for Geobacillus to grow at relatively high temperatures is reflected in the enrichment experiments, showing accumulation at 65°C rather than 55°C (Fig. 1 and 2). The geobacilli produce heat-resistant spores with decimal reduction values ranging from 18 to 20 min at 110°C, whereas the D value of Anoxybacillus is only 2 min at this temperature (Table 2). The ability to efficiently form biofilms and generate highly heat-resistant spores with high efficiency under laboratory conditions renders G. thermoglucosidans an interesting model organism. The biofilm-forming behavior of the TNO-09.020 isolate on a stainless steel coupon in the static biofilm model system with tryptone-based medium was analyzed microscopically. Examination of biofilms stained with Auramine indicated the presence of the multicellular structures that predominantly formed at the air-liquid interface (Fig. 3A). The bacterial spores formed within these biofilms appeared more or less randomly distributed (Fig. 3B and C).
Table 1.
Typing and growth characteristics of selected model strainsa
| Strain | DNA-DNA hybridization (% homology) | Growth temp range |
tD (min) | ||
|---|---|---|---|---|---|
| Tmin (°C) | Tmax (°C) | Topt (°C) | |||
| TNO-09.006 | Anoxybacillus flavithermus LMG 18397T (75 ± 8) | 43 | 62 | 57 | 52 |
| TNO-09.008 | Geobacillus stearothermophilus LMG 6939T (86 ± 9) | 48 | 67 | 61 | 35 |
| TNO-09.020 | Geobacillus thermoglucosidans LMG 7137T (88 ± 13) | 50 | 69 | 60 | 32 |
Species assignment of model strains was confirmed by DNA-DNA hybridizations with reference strains from the LMG culture collection. The Tmin (°C) and Tmax (°C) are defined as the maximum and minimum temperatures at which growth could still be detected under the conditions used (see Materials and Methods). The Topt (°C) is the temperature at the highest growth rate, which is expressed in minutes of doubling time [tD (min)].
Table 2.
Sporulation efficiency of thermophilic sporeformers and heat resistance of their sporesa
| Strain | Avg sporulation efficiency (%) (on NA++ agar plates) | Heat resistance of spores |
|
|---|---|---|---|
| D110 (min) | z value (°C) | ||
| Anoxybacillus flavithermus TNO-09.006 | 77 ± 40 | 2 | 13 |
| Geobacillus stearothermophilus TNO-09.008 | 38 ± 31 | 18 | 11 |
| Geobacillus thermoglucosidans TNO-09.020 | 91 ± 3 | 20 | 8 |
The sporulation efficiency is expressed as the number of spores (CFU after heat inactivation) divided by the total number of bacterial cells and spores (CFU before heat inactivation). The D values are expressed in minutes of treatment at the indicated temperature for a 10-fold CFU reduction; the z values are expressed in °C temperature increase required for a 10-fold reduction of the D value. The calculations are described in detail in Materials and Methods and Fig. S2 in the supplemental material.
Fig 3.
Geobacillus thermoglucosidans TNO-09.020 biofilms at the air-liquid interface. (A) Fluorescence microscopy image of Auramine-stained biofilm on a standing stainless steel coupon after 10 h of batch cultivation at 65°C. (B and C) Bright-field (B) and fluorescence (C) microscopy images of Auramine- and Safranine-stained biofilm on a standing glass coupon after 16 h of batch cultivation at 65°C.
Growth dependence of Geobacillus thermoglucosidans.
The selected species A. flavithermus, G. steathermophilus, and G. thermoglucosidans were further characterized for their ability to grow on different nutrient plates. Interestingly, the G. thermoglucosidans strains TNO-09.020 and TNO-09.023 were not capable of growing on milk plates. However, they were capable of growing on plates containing casein, the major protein component of milk, if the casein was proteolytically digested (data not shown). Therefore, we hypothesized that G. thermoglucosidans is dependent on the proteolytic activity of other bacteria for growth in milk. To test this, we analyzed growth of G. thermoglucosidans TNO-09.020 and A. flavithermus TNO-09.006 in a cell culture insertion setup that enables cultivation of the two strains separated by a permeable membrane. This membrane allows the diffusion of enzymes and small organic molecules between the two compartments. A. flavithermus TNO-09.006 readily started growth after 3 h and continued growing until approximately 12 h in the presence and absence of TNO-09.020, after which the CFU number started to decrease (Fig. 4). As expected, G. thermoglucosidans TNO-09.020 inoculated in milk did not show any growth, with the CFU number remaining below 3 log units per ml. However, when G. thermoglucosidans TNO-09.020 was inoculated in the presence of A. flavithermus TNO-09.006, growth started after a long lag time of ≥12 h, reaching a CFU value of approximately 5 log units after 24 h (Fig. 4). Clearly, G. thermoglucosidans TNO-09.020 is dependent on the presence of A. flavithermus TNO-09.006 for growth in the milk medium. The second G. thermoglucosidans strain isolated in this study, TNO-09.023, was also tested in this cell culture insertion setup and showed similar behavior (data not shown).
Fig 4.
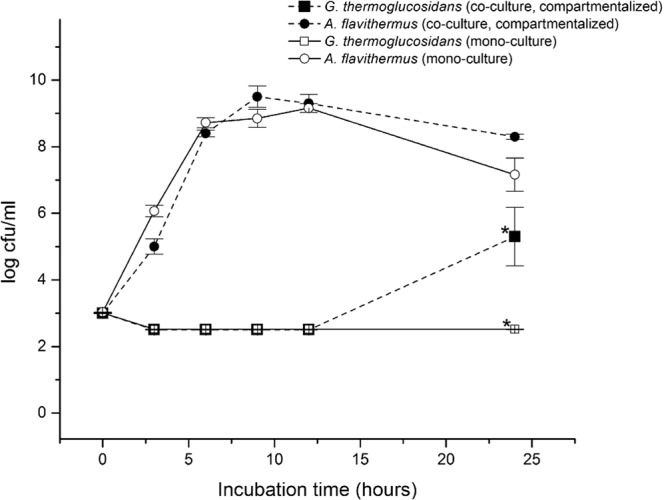
Compartmentalized growth of Geobacillus thermoglucosidans TNO-09.020 and Anoxybacillus flavithermus TNO-09.006. A graphical representation is shown of bacterial counts from two strains in a compartmentalized growth experiment in UHT skim milk with the BD Falcon cell culture insertion system, allowing growth of strains in two compartments separated by a permeable membrane that permits diffusion of medium components (pore size, 0.4 μm). ■, cell counts of TNO-09.020 (with TNO-09.006 in the other compartment); ●, cell counts of TNO-09.006 (with TNO-09.020 in the other compartment); □, cell counts of TNO-09.020 in the absence of TNO-09.006; ○, cell counts of TNO-09.006 in the absence of TNO-09.20. The bacterial cultures were enumerated at 6 different time points; each point represents the mean and standard deviation of triplicate measurements. *, significant difference for growth (log CFU) of G. thermoglucosidans TNO-09.020 in the presence or absence of A. flavithermus TNO-09.006 in the other compartment (t test; P < 0.02).
Next, a coculture experiment in milk was conducted with G. thermoglucosidans TNO-09.020 and A. flavithermus TNO-09.006. Biofilm development was monitored by determining the total number of viable cells in the biofilm attached to stainless steel coupons, and the number of CFU of G. thermoglucosidans TNO-09.020 was selectively determined, as they appear as white colonies on TSA–X-Gal plates at 55°C, in contrast to colonies of the A. flavithermus TNO-09.006 strain, which appear blue on TSA X-gal plates as a result of its galactosidase activity (Fig. 5). In agreement with the results of the compartmentalized growth experiment, the strain TNO-09.020 is able to form biofilms only when TNO-09.006 is present, and the number of CFU of TNO-09.020 in the biofilm reached a level of approximately 105 CFU/biofilm after 24 h and approximately 107 CFU/biofilm fraction after 48 h (Fig. 5). The TNO-09.006 strain grows well in milk in the absence of TNO-09.020, reaching approximately 107 CFU/ml in the milk medium and 105 CFU/biofilm after 8 h. However, no CFU of this strain could be detected after 48 h in either biofilm or milk medium when TNO-09.020 was present (Fig. 5).
Fig 5.
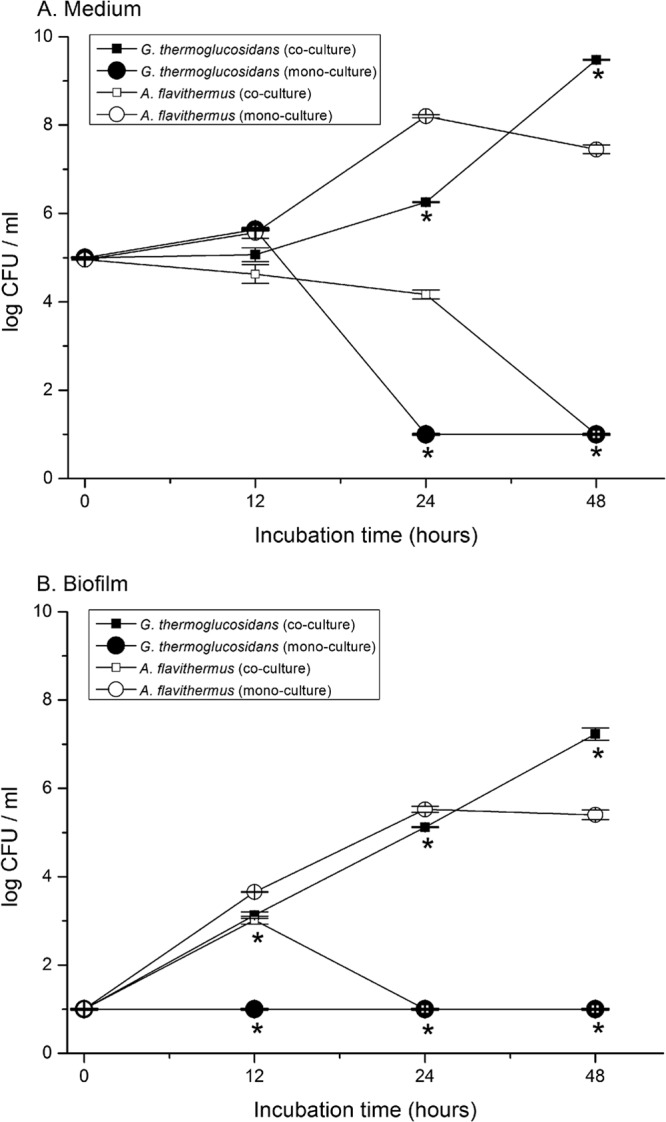
CFU of planktonic cells and biofilms in coculture of thermophiles. Panels A and B show the results of a total of three batch cultivation experiments in milk determined using the standing steel biofilm model system, including one coculture and two monocultures. (A) Bacterial cell counts in the 3-ml milk medium fraction of a coculture of planktonic cells of Geobacillus thermoglucosidans TNO-09.020 (■) and Anoxybacillus flavithermus TNO-09.006 (□). For reference, the results determined for monocultures of TNO-09.020 (●) and TNO-09.006 (○) in milk were plotted. (B) Bacterial cell counts of the biofilm attached to stainless steel in a coculture of TNO-09.020 cells (■) and TNO-09.006 cells (□). For reference, the results determined for biofilms obtained using monocultures of TNO-09.020 cells (●) and TNO-09.006 cells (○) were plotted. The bacteria were enumerated at 4 different time points; each bar represents the mean and the error bar the standard deviation from two experiments of triplicate measurements. *, significant difference for growth (log CFU) of G. thermoglucosidans TNO-09.020 in the presence or absence of A. flavithermus TNO-09.006 during coculture (t test; P < 0.01).
DISCUSSION
In this study, we developed and applied a number of novel approaches to study the growth and biofilm-forming capacity of sporeformers associated with the dairy industry. This work included a cultivation-independent approach to study contaminants in milk, factory fouling samples, and end products and enrichments thereof. We have grown biofilms with milk samples as an inoculum and screened factory isolates for their ability to form biofilms under laboratory conditions in multiwell plates. We have characterized three of these thermophilic biofilm-forming isolates and their spores in detail. Three major findings resulted from this work: (i) dairy processing environments harbor species-rich microbial communities, (ii) the thermophilic sporeformers studied preferentially form biofilms at air-liquid interfaces, and (iii) the thermophilic sporeformer Geobacillus thermoglucosidans depends on the presence of other thermophilic species for growth and biofilm formation in milk-based media.
The results revealed a wide diversity of genera in the processing lines. Fouling samples taken from the processing line where high temperatures were applied were not dominated by thermophilic sporeformers, but significant numbers mesophilic bacteria were identified. The mass sequencing applied here detects DNA molecules that encode 16S rRNA molecules and thus not necessarily viable bacteria, so this may lead to overestimation of the viable microbiota present (see also reference 22). Dairy-associated microbiota shows remarkable diversity, as previously reported and reflected in the assignment of dairy farm isolates to seven spore-forming genera, i.e., Aneurinibacillus, Bacillus, Brevibacillus, Geobacillus, Paenibacillus, Ureibacillus, and Virgibacillus (3). In the current study, we confirmed the presence of the spore-forming genera Aneurinibacillus, Bacillus, Brevibacillus, Geobacillus, and Anoxybacillus.
Enrichment at 55°C of the fouling materials resulted in selection of thermophilic genera and sporeformers. After enrichment at 65°C, the thermophilic genera (sporeformers or nonsporeformers) were dominant in most samples. The reason that these thermophiles are not always detected by this method in the fouling samples is that their numbers were below the detection limit of the method used. The enrichment of fouling samples in biofilm model systems shows that the predominant spoilage genera associated with biofilm formation are Geobacillus and Anoxybacillus, species of which have been isolated from milk powders and dairy concentrate-processing factories (2, 4, 23). According to the bar-coded 16S amplicon sequencing data from this study, a number of other thermophilic genera, including Thermus, Brevibacillus, and Aneuribacillus, are present in the fouling samples, even after enrichments, but do not appear among the cultured isolates; possibly these species are easily outcompeted by Anoxybacillus and Geobacillus on TSB plates at 55°C.
Concerning the abiotic conditions which control the biofilm formation of the thermophiles studied here, we identified in this study no evident correlation between the composition of surface-attached microbiota and the nature of the surface, including steel and plastic. However, a clear difference in the preferred environment for biofilm formation of microbial genera was identified, as Anoxybacillus and Geobacillus preferentially resided at air-liquid interfaces, whereas Pseudomonas accumulated at the surface of submerged steel. We hypothesize that the oxygen concentration may play a crucial role in selective accumulation of bacteria and spores on the stainless steel surface at the air-liquid interface. This suggests that biofilms of thermophilic sporeformers and associated spores may particularly develop at elevated temperature and in industrial piping systems that are only partly filled and as a result are exposed to oxygen during operation.
Finally, we present data from this study suggesting that the G. thermoglucosidans strain, which produces the most thermostable spores, is dependent on proteolytic strains for outgrowth in the dairy environment. Several observations support this interpretation. The G. thermoglucosidans was not enriched from the industrial milk or fouling samples in milk medium, probably due to its long lag phase before outgrowth (bottom row, Fig. 2). In addition, G. thermoglucosidans strain TNO-09.020 and strain TNO-09.023 did not readily grow or form biofilms in undigested casein or milk medium. However, they grew well and formed biofilms in predigested casein or milk medium or, alternatively, when the proteolytic strain A. flavithermus TNO-09.006 was also present in undigested casein or milk medium. Although there is no evidence for a mutual relationship between TNO-09.020 and TNO-09.006, our observation may bear some resemblance to that of the yogurt consortium, where the proteolytic activity of L. bulgaricus results in the supply of amino acids for Streptococcus thermophilus (24). The ecology and interrelationship between the selected isolates will be elucidated using gene trait-matching approaches based on whole-genome sequence information (20, 21). Such information would be relevant because our results suggest that the presence of proteolytic microorganisms in the dairy concentrate production line may contribute to the diversity and spore load of specific thermophiles in end products.
Supplementary Material
ACKNOWLEDGMENTS
We thank Carlo Brouwer, RuiRui Liu, and Rob Leer for expert assistance.
This work was supported by the Top Institute Food and Nutrition (TIFN).
Footnotes
Published ahead of print 12 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00949-13.
REFERENCES
- 1.Burgess SA, Lindsay D, Flint SH. 2010. Thermophilic bacilli and their importance in dairy processing. Int. J. Food Microbiol. 144:215–225 [DOI] [PubMed] [Google Scholar]
- 2.Scott SA, Brooks JD, Rakonjac J, Walker KMR, Flint SH. 2007. The formation of thermophilic spores during the manufacture of whole milk powder. Int. J. Dairy Technol. 60:109–117 [Google Scholar]
- 3.Scheldeman P, Pil A, Herman L, De Vos P, Heyndrickx M. 2005. Incidence and diversity of potentially highly heat-resistant spores isolated at dairy farms. Appl. Environ. Microbiol. 71:1480–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flint SH, Bremer PJ, Brooks JD. 1997. Biofilms in dairy manufacturing plant—description, current concerns and methods of control. Biofouling 11:81–97 [Google Scholar]
- 5.Yuan D, Liu G, Ren D, Zhang D, Zhao L, Kan C, Yang Y, Ma W, Li Y, Zhang L. 2012. A survey on occurrence of thermophilic bacilli in commercial milk powders in China. Food Control 25:752–757. 10.1016/j.foodcont.2011.12.020 [DOI] [Google Scholar]
- 6.Nocker A, Richter-Heitmann T, Montijn R, Schuren F, Kort R. 2010. Discrimination between live and dead cells in bacterial communities from environmental water samples analyzed by 454 pyrosequencing. Int. Microbiol. 13:59–65 [DOI] [PubMed] [Google Scholar]
- 7.Bartholomew JW, Lechtman MD, Finkelstein H. 1965. Differential spore and lipid staining at room temperature by use of fluorescent dye. J. Bacteriol. 90:1146–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12:1889–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, The Human Microbiome Consortium. Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21:494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37(Database issue):D41–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gevers D, Huys G, Swings J. 2001. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 205:31–36 [DOI] [PubMed] [Google Scholar]
- 13.Ezaki T, Adnan S, Miyake M. 1990. Quantitative microdilution plate hybridization to determine genetic relatedness among bacterial strains. Nihon Saikingaku Zasshi 45:851–857. (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 14.Kort R, O'Brien AC, Van Stokkum IHM, Oomes SJCM, Crielaard W, Hellingwerf KJ, Brul S. 2005. Assessment of heat resistance of bacterial spores from food product isolates by fluorescence monitoring of dipicolinic acid release. Appl. Environ. Microbiol. 71:3556–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delbès C, Ali-Mandjee L, Montel MC. 2007. Monitoring bacterial communities in raw milk and cheese by culture-dependent and -independent 16S rRNA gene-based analyses. Appl. Environ. Microbiol. 73:1882–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ercolini D, Russo F, Ferrocino I, Villani F. 2009. Molecular identification of mesophilic and psychrotrophic bacteria from raw cow's milk. Food Microbiol. 26:228–231 [DOI] [PubMed] [Google Scholar]
- 17.Lafarge V, Ogier JC, Girard V, Maladen V, Leveau JY, Gruss A, Delacroix-Buchet A. 2004. Raw cow milk bacterial population shifts attributable to refrigeration. Appl. Environ. Microbiol. 70:5644–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Jonghe V, Coorevits A, Vandroemme J, Heyrman J, Herman L, De Vos P, Heyndrickx M. 2008. Intraspecific genotypic diversity of Bacillus species from raw milk. Int. Dairy J. 18:496–505 [Google Scholar]
- 19.Manaia CM, Moore ER. 2002. Pseudomonas thermotolerans sp. nov., a thermotolerant species of the genus Pseudomonas sensu stricto. Int. J. Syst. Evol. Microbiol. 52:2203–2209 [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y, Caspers MP, Abee T, Siezen RJ, Kort R. 2012. Complete genome sequence of Geobacillus thermoglucosidans TNO-09.020, a thermophilic sporeformer associated with a dairy-processing environment. J. Bacteriol. 194:4118. 10.1128/JB.00318-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caspers M, Boekhorst J, Abee T, Siezen R, Kort R. 2013. Complete genome sequence of Anoxybacillus flavithermus TNO-09.006, a thermophilic sporeformer associated with a dairy-processing environment. Genome Announc. 1:e00010-13. 10.1128/genomeA.00010-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ronaghi M. 2001. Pyrosequencing sheds light on DNA sequencing. Genome Res. 11:3–11 [DOI] [PubMed] [Google Scholar]
- 23.Burgess SA, Brooks JD, Rakonjac J, Walker KM, Flint SH. 2009. The formation of spores in biofilms of Anoxybacillus flavithermus. J. Appl. Microbiol. 107:1012–1018 [DOI] [PubMed] [Google Scholar]
- 24.Sieuwerts S, de Bok FA, Hugenholtz J, van Hylckama Vlieg JE. 2008. Unraveling microbial interactions in food fermentations: from classical to genomics approaches. Appl. Environ. Microbiol. 74:4997–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.