Abstract
The Rhizobiaceae are a bacterial family of enormous agricultural importance due to the ability of its members to fix atmospheric nitrogen in an intimate relationship with plants. Their survival as naturally occurring soil bacteria in agricultural soils as well as popular seed inocula is affected directly by drought and salinity. Survival after desiccation in the presence of NaCl is enabled by underlying genetic mechanisms in the model organism Sinorhizobium meliloti 1021. Since salt stress parallels a loss in water activity, the identification of NaCl-responsive loci may identify loci involved in survival during desiccation. This approach enabled identification of the loci asnO and ngg by their reduced ability to grow on increased NaCl concentrations, likely due to their inability to produce the osmoprotectant N-acetylglutaminylglutamine (NAGGN). In addition, the mutant harboring ngg::Tn5luxAB was affected in its ability to survive desiccation and responded to osmotic stress. The desiccation sensitivity may have been due to secondary functions of Ngg (N-acetylglutaminylglutamine synthetase)-like cell wall metabolism as suggested by the presence of a d-alanine-d-alanine ligase (dAla-dAla) domain and by sensitivity of the mutant to β-lactam antibiotics. asnO::Tn5luxAB is expressed during the stationary phase under normal growth conditions. Amino acid sequence similarity to enzymes producing β-lactam inhibitors and increased resistance to β-lactam antibiotics may indicate that asnO is involved in the production of a β-lactam inhibitor.
INTRODUCTION
The Rhizobiaceae are a bacterial family of enormous agricultural importance due to their ability to fix atmospheric nitrogen in an intimate relationship with plants (1). They occur naturally in most agricultural soils, and their survival is affected directly by both drought and salinity (2). Unfortunately, changes in climate patterns are occurring and, as a direct consequence, salinification and desertification are some of the major threats to agricultural land use. It is estimated that over 40% of arable land will be affected by desiccation and salinity by 2025 (2). Furthermore, production of seed inocula often includes a drying phase negatively affecting CFU of added rhizobia (3, 4), potentially resulting in desiccation-induced viable but nonculturable cells (5).
A multitude of conditions affecting survival during desiccation of agriculturally important Rhizobiaceae have been studied (6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20). These conditions include the intrageneric differences to cope with desiccation stress which affect survival (5, 6, 9, 10, 13, 18, 21, 22). These data suggest that no single gene affects the ability of rhizobia to survive desiccation but that several mechanisms are likely responsible.
One of the conditions affecting the ability of Sinorhizobium meliloti to survive desiccation is NaCl availability (10, 13); hence, we studied the response of S. meliloti 1021 to drought in conjunction with different salt stresses (13). We showed that survival of the strain in the presence of desiccation when exponentially growing cells were resuspended in water was worse than under conditions of resuspension in water containing 400 mM NaCl. In contrast, increasing amounts (200 and 400 mM) of sodium chloride added to standard media such as yeast mannitol broth (YMB) and phosphate mannitol medium (PMM) were found to enhance survival. This effect was mainly caused by the presence of anions rather than cations in the growth medium (10, 13). These observations indicate that the response to desiccation in conjunction with the presence of NaCl is physiological in origin (4, 13) and that the underlying genetic mechanisms are important for the survival of the cell during desiccation. The in situ relevance of this physiological effect is further demonstrated by the fact that NaCl and the availability of osmoprotectants affect survival of rhizobia in soil (23, 24) as well as in seed inocula (25).
Examples of genetic mechanisms involved in survival during desiccation have been identified previously and include the operon agl for trehalose-maltose and sucrose uptake (smb03060 to smb03065) (26) and the trehalose-maltose-sucrose operon (thu; smb20324 to smb20330) identified by Jensen et al. (27). The thu operon is expressed to a higher level during an osmotic upshift (28), and McIntyre et al. (29) showed that loci involved in trehalose metabolism affect survival during desiccation. The above-described uptake systems involve osmoprotectants, which accumulate during salt stress, thus potentially affecting NaCl-mediated survival during desiccation.
In addition to trehalose uptake, a target of desiccation responses is the cell wall, which is also affected by the presence of NaCl (15, 30, 31). These responses include exopolysaccharide production (32, 33, 34), which is assumed to affect desiccation survival (4). Furthermore, Wei et al. (35) and Miller-Williams et al. (36) have shown that responses to NaCl and osmotic stress affect genes encoding proteins potentially involved in central metabolism such as elongation factors, DNA ligases, chaperones, and cell division proteins. It is well known that DNA is a target for desiccation stress, and recently Humann (37) confirmed this for rhizobia.
In this study, the hypothesis that certain NaCl-responsive loci are involved in survival during desiccation was further tested. We identified Tn5luxAB-tagged loci that are responsive to increased concentrations of NaCl. Some of these mutants were also tested for their ability to survive desiccation, and NaCl-responsive loci involved in survival during desiccation were characterized for their response to water stress. Finally, we also tested their response to β-lactam antibiotics to test for potential involvement in cell wall function.
MATERIALS AND METHODS
Materials.
S. meliloti 1021 was obtained from our strain collection (38). An Escherichia coli strain containing plasmid pRK2013 (39) was obtained from T. Lessie (University of Massachusetts) and used for triparental matings. Phage ΦM12, employed to reconstruct Tn5luxAB transcriptional fusions, was supplied by Graham Walker (40). A Tn5luxAB transcriptional fusion mutant bank was created for S. meliloti strain 1021 as described previously (41). S. meliloti strains Sce1 and Sce12 were identified and characterized in this study. S. meliloti strain CV2, which serves as the positive control for NaCl-dependent luciferase expression, was previously described by Milcamps et al. (41). A negative control for NaCl-dependent luciferase expression (strain 1D1) was randomly chosen from the mutant bank. E. coli DH5α- and JM109-competent cells were obtained from Invitrogen or prepared following standard protocols (42). All strains were maintained on tryptone-yeast extract (TY) plates with the appropriate antibiotics. Media used were TY (43), GTS (41), LB (42), and YMB and PMM (5, 13, 44). Alfalfa seeds were obtained from the seed company Outsidepride (BS-ALFALFA-5; lot no. A2N-1769-3; Outsidepride, Salem, OR). According to the manufacturer, these seeds had not been treated with any chemicals.
Induction studies using Tn5luxAB transcriptional fusions.
The screening of the Tn5luxAB transcriptional fusion mutant bank was carried out using a photonic camera (Hamamatsu C1966-20 [45]) as described previously by Milcamps et al. (41) and adapted to screen for luciferase fusions induced during exposure to NaCl as follows. The induction screenings were performed on PMM plates (PMM with 15 g/liter agar) containing 400 mM NaCl, and luciferase expression was measured after 4 and 8 h of incubation. Strains were selected based on an increase in luciferase expression compared to the same strain not exposed to NaCl. Those strains with increased luciferase expression in four replicate screenings were considered further. The luciferase activity assays using a luminometer were performed as described by Phillips et al. (46), with the following modification: PMM was employed. One culture (optical density [OD] = ∼0.2) was split into six culture tubes (23 mm in diameter, 5 ml of culture per tube) and diluted with the same amount (1:1) of PMM or PMM with 800 mM NaCl, resulting in PMM media containing 0 and 400 mM NaCl in triplicate. Light emission was measured using a TD 20/20 luminometer by adding a 100-μl subsample to a 10-μl bovine serum albumin (BSA) (Sigma Co.) (2%)–aldehyde (n-decanal) (Sigma Co.) (0.2%) solution. Strains with a positive response during 8 h in three replicate experiments were explored further.
When the response to water activity and osmotic stress was tested, media with double the amount of the final concentration of NaCl were mixed 1:1 with the growing cell culture. Except in the screening in which luciferase expression is reported as relative light units (RLU)/ml/min, all of the study results are reported as RLU/ml/min/OD at 595 nm (OD595). Concentrations mimicking a reduction in water activity were calculated from empirically obtained data presented by Leistner and Rodel (47) and by Brown (48). The addition to PMM of 400 mM NaCl, 520 mM glycerol, 222 mM sucrose, or 780 mM polyethylene glycol 200 (PEG 200) results in a final water activity value (Aw) of 0.986.
Growth experiments.
Initial growth experiments were performed as follows. Five milliliters of PMM and PMM amended with 400 mM NaCl in culture tubes was inoculated with 50 μl of a 3-day-old TY culture and incubated at 28°C with shaking at 220 rpm. Over the course of 5 days, growth was checked twice daily and compared to that of S. meliloti 1021. Growth curves were generated using 30 ml media in 250-ml flasks, inoculated with 1/100 (vol/vol) 3-day-old TY culture, and incubated at 28°C and agitated at 220 rpm. Antibiotics were used at the following concentrations: kanamycin at 25 μg/ml, streptomycin at 25 μg/ml, spectinomycin at 25 μg/ml, and chloramphenicol at 10 μg/ml. For the amino acid complementation studies, 5 ml of PMM–400 mM NaCl in culture tubes was inoculated with 50 μl of full-density, 3-day-old TY cultures. Amino acids were added at a final concentration of 50 μg/ml. Cultures were incubated at 28°C and agitated at 220 rpm.
Molecular methods.
All molecular procedures were based on protocols of Sambrook and Russell (42) or performed as previously described by Wolk et al. (45) and Milcamps et al. (41). The copy number of Tn5luxAB was determined using Southern hybridization, and the rescue, sequencing, and insertion site determinations were performed as described by Milcamps et al. (41) and Wolk et al. (45). Fragments resulting from BglII and EcoRI restriction digests were separated on 0.7% agarose gels and transferred to nitrocellulose filters using Southern blotting. EcoRI-digested, digoxigenin (DIG)-labeled pRL1063a served as the probe (Boehringer Mannheim, Richfield, CT). The insertion site was determined by sequencing outward from the insertion sequence using primers as previously described by Milcamps et al. (41) and comparing the sequences to the S. meliloti 1021 database (http://iant.toulouse.inra.fr/bacteria/annotation/cgi/rhime.cgi/). The Tn5luxAB insertions were transduced using ΦM12 to reconstruct the same mutation in a new genetic background to reduce the possibility of secondary mutations (40). Transductants were selected on GTS with kanamycin and streptomycin, and the copy number of Tn5luxAB was determined as described above.
Molecular sequence analysis.
The amino acid sequences from the loci tagged in strain Sce10 and Sce11 were searched using BLAST and the cDart and the conserved domain database at NCBI (http://www.ncbi.nlm.nih.gov). The promoter prediction programs employed were (i) Neural Networks (http://www.fruitfly.org/seq_tools/promoter.html; 49), (ii) Sequence Alignment Kernel (50), and (iii) Virtual Footprint (http://prodoric.tu-bs.de/vfp/vfp_promoter.php; 51).
Antibiotic susceptibility.
A full-density TY culture was diluted in sterile water to OD595 = 0.05. A volume of 100 μl of diluted cells was spread on PMM plates, and a filter paper with a defined amount of antibiotics was placed on the plate surface. Antibiotic discs were generated from filter paper or supplied by Benton-Dickinson (Sparks, MD). Plates were incubated for 3 days at 28°C, and growth was checked twice daily. The antibiotics tested were streptomycin, kanamycin, bacitracin, cycloserine, penicillin, and vancomycin. Inhibition with lysozyme was also tested by dropping 5 μl of a 50 mg/ml solution onto a lawn of cells. Plates were checked daily for inhibition of the lawn.
Survival during desiccation.
This method was previously described by Vriezen et al. (13) and used with the following modifications. Three culture tubes containing 5 ml liquid TY medium with the appropriate antibiotics were inoculated with one isolated colony from fresh TY plates and grown to full OD at 28°C and agitated at 220 rpm. A 50-μl volume of these cultures were transferred to three culture tubes containing 5 ml PMM–400 mM NaCl and antibiotics and incubated until OD595 values of 0.2 to 0.4 were reached. Equal amounts of cells, estimated using the OD595 values, were concentrated in a microcentifuge (13,000 × g for 2 min), the supernatant was removed, and the pellets were washed in 1 ml of PMM–400 mM NaCl. Suspensions (100 μl) were pipetted onto a membrane filter in a microcentrifuge tube. Six tubes were stored in a 450-ml glass jar containing 100 ml oversaturated KCl solution, which resulted in a relative humidity in the air phase of 22%. After a storage time of at least 3 days at room temperature in the dark, samples were removed and exposed to 100% relative humidity (RH) for 1 h. YMB (1 ml) was added to resuspend the cells, and surviving CFU were established. The relative survival rates were calculated using the CFU at T = 0 as 100%.
Symbiotic phenotypes.
The mutants selected were tested for their symbiotic phenotypes as described by Milcamps et al. (41). Inoculated seedlings were incubated for 4 weeks at 20°C with a 12-h light and dark cycle. Alfalfa roots were checked for the occurrence of nodules.
RESULTS
Identification of a NaCl-responsive putative operon.
The goal of this work was to identify genetic loci in S. meliloti 1021 involved in survival during desiccation. Previous studies have shown that survival during desiccation of S. meliloti increases in the presence of NaCl (10, 13). Therefore, a S. meliloti Tn5luxAB transcriptional fusion mutant bank (41) was screened for an increase in luciferase expression during exposure to 400 mM NaCl followed by a screen for loci affecting the desiccation phenotype of mutant strains. After 8 h of exposure to NaCl, 12 mutants whose expression was significantly (P < 0.001) induced in the presence of NaCl were identified (Fig. 1) that had a single copy of Tn5luxAB integrated (data not shown). These strains were annotated sodium chloride expressed-strains, abbreviated as Sce1 to Sce12. Except for Sce5 and Sce12, all strains showed a significant increase in luciferase expression after 3 h of incubation in the presence of 400 mM NaCl (fold increase ≥ 2) (P < 0.001; n ≥ 6). The genomic sequence of regions immediately adjacent to Tn5luxAB was determined for 10 strains and was used to identify the tagged loci in S. meliloti 1021 (Table 1). Expression of Sce10 (asnO) in response to NaCl was confirmed using reverse transcription-PCR (RT-PCR) (data not shown).
Fig 1.
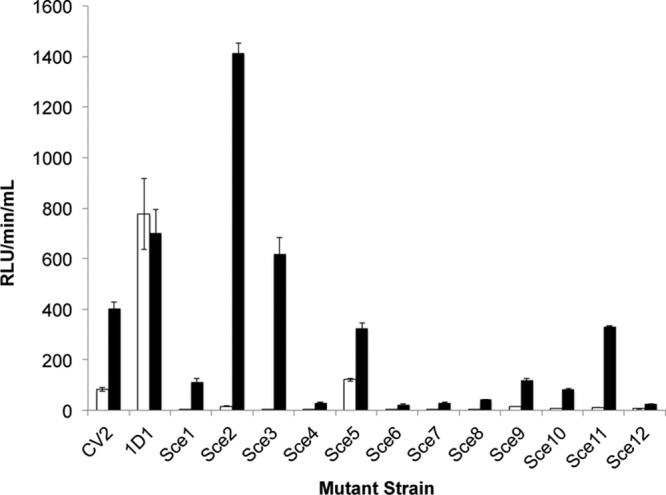
Identification of S. meliloti 1021 mutants carrying NaCl-responsive Tn5luxAB transcriptional fusions after 8 h of exposure to 400 mM NaCl. RLU, relative light units; white bar, PMM; black bar, PMM–400 mM NaCl; Sce, sodium chloride expressed. Error bars represent standard errors of the means (SEM); n = 9 except for CV2 (n = 6) and 1D1 (n = 12). Using the one-sided t test with equal variances, all luciferase fusions were significantly expressed on the P < 0.001 level.
Table 1.
Selected NaCl responsive Tn5luxAB transcriptional fusions in S. meliloti 1021 after 8 h of incubation
| S. meliloti strain | Sequencing and BLAST result(s) |
Reference(s) or source | |||
|---|---|---|---|---|---|
| Descriptiona | Locus | Gene/locus name | Tn5luxAB locationc | ||
| 1021 | 38 | ||||
| Sce1 | Hypothetical protein with SH3 region | smc01590 | 256–265 | This study | |
| Sce2 | Putative dipeptide transporter permease and ATP binding protein | smb20478 | 572–580 | This study | |
| Sce3 | Putative dipeptide ABC transporter ATP binding protein | smb20479 | 124–133 | This study | |
| Sce4 | Putative nutrient deprivation induced | smb20227 | ndiA1 | 299–307 | 67, 68 |
| Sce5 | Hypothetical protein HAD superfamily | smc04299 | loe3b | −1–7 | 56 |
| Sce6 | Conserved hypothetical protein | smc01445 | 4–12 | This study | |
| Sce7 | NS | This study | |||
| Sce8 | Putative methyl transferase | smb20238 | loe2b/C1 | 17–26 | 41, 56 |
| Sce9 | NS | This study | |||
| Sce10 | Asparagine synthase | smb20481 | loe4b/asnO | 979–987 | 44, 53, 55, 56, 69, 70 |
| Sce11 | GCN5-related N-acetyltransferase | smb20482 | loe6b/ngg | 1058–1066 | 44, 53, 56, 69, 70 |
| Sce12 | Short-chain alcohol dehydrogenase | smb20073 | 273–281 | This study | |
HAD, haloacid dehalogenase; NS, not sequenced.
Expressed under conditions of low oxygen.
Location of the Tn5luxAB insertion relative to upstream ATG.
Four of the 10 Tn5luxAB-tagged loci identified in strains Sce2, Sce3, Sce10, and Sce11 mapped to the same part of the genome and had inserted into what appears to be one operon (Fig. 2A). The first four genes are involved in dipeptide uptake, the fifth is a potential regulator of this operon, and the last two are hypothetically involved in the synthesis of NAGGN, a compatible solute known to result in growth at increased osmolarity upon osmotic shock (52, 53).
Fig 2.
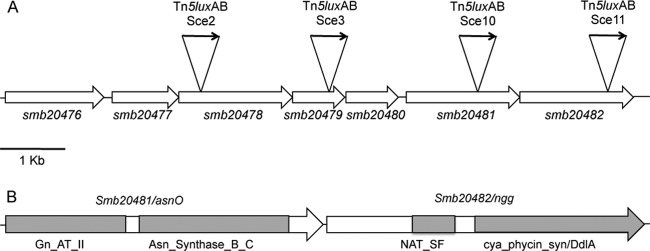
Physical map of 4 of the 12 identified NaCl-responsive Tn5luxAB-tagged loci, which may form one continuous operon. (A) Large arrows indicate an identified open reading frame (ORF); the smb numbers designate the Sinorhizobium meliloti SymB ORF reference numbers. The insertion sites of the Tn5luxAB in the respective strains are indicated. Locations are approximate, but sizes are to scale. Smb20476 to Smb20479 are involved in ABC transport of dipeptides. Smb20476 is a putative periplasmic dipeptide binding protein, Smb20477 a putative permease, Smb20478 the putative ATP binding protein of the permease, Smb20479 a putative ATP binding protein, Smb20480 a putative transcriptional regulator, presumable cis-acting, Smb20481 asparagin synthase, and Smb20482 a hypothetical protein. (B) Domain structure of AsnO and Ngg. Only domains with the highest BLAST scores are given. Abbreviations for the different domains are as follows: Gn_At_II for glutamine aminotransferase, Asn_Synthase_B_C for asparagine synthase, NAT_SF for A-acetyltransferase, cya_phycin_syn for cyanophycin synthase, and DdlA for dAla-dAla ligase.
Ngg (N-acetylglutaminylglutamine synthetase)::Tn5luxAB affects the ability to respond to water stress.
To test if the tagged mutants are indeed affected in their ability to grow on increased concentrations of NaCl, eight mutants (Sce1, Sce2, Sce3, Sce4, Sce6, Sce9, Sce10, and Sce11) were grown in PMM and in PMM amended with 400 mM NaCl. No mutant showed differences in growth compared to the reference strain in PMM, indicating the absence of auxotrophs under this condition. However, in the presence of 400 mM NaCl, the ability of strains Sce10 and Sce11 to grow was reduced (Fig. 3A). The Tn5 mutations in these two strains were reconstructed in the reference strain and their geno- and phenotypes confirmed. Furthermore, complementation by a plasmid borne asnO in Sce10 was verified (data not shown).
Fig 3.
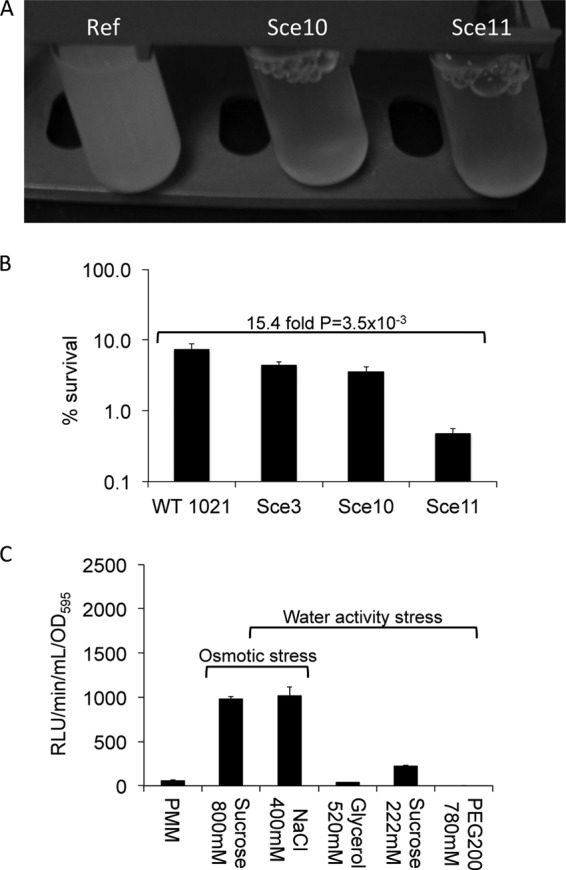
Responses of strains S. meliloti 1021, Sce3, Sce10, Sce11, and ngg::Tn5luxAB to water stresses. (A) Growth of S. meliloti 1021 (Ref [reference strain]), Sce10, and Sce11 in the presence of 400 mM NaCl. (B) Survival during desiccation of strains S. meliloti 1021, Sce3, Sce10, and Sce11. The error bars represent standard errors of the means (SEM) measured twice in three independent incubations (n = 6 for all cases except wild-type [WT] 1021, for which n = 12). (C) Responses of ngg::Tn5luxAB to a decrease in water activity (Aw = 0.986; 400 mM NaCl, 520 mM glycerol, 222 mM sucrose, and 780 mM PEG 200) and osmotic stress (0.800 osmol, 400 mM NaCl, and 800 mM sucrose). Luciferase activity was measured after 3 h of incubation. The error bars represent standard errors of the means (SEM) of the result determined for one independent incubation measured trice (n = 3).
To determine if loci asnO and ngg also affect the ability of S. meliloti 1021 to survive desiccation, strain Sce10 and Sce11 were dried at 22% RH for 3 days in PMM in the presence of 400 mM NaCl. As controls, the survival rates of S. meliloti 1021 and strain Sce3 were also determined. As the data indicate, strains Sce3 and Sce10 were able to survive desiccation to a degree similar to that seen with the reference strain (P > 0.05), while survival of strain Sce11 was 15.4-fold reduced relative to that of the reference strain (P = 3.5 × 10−3; Fig. 3B). The ability of strains Sce3 and Sce10 to survive desiccation shows that the presence of the transposon, the expression of luciferase in the presence of NaCl, the ability to grow in the presence of NaCl, the presence of NAGGN, or polar effects cannot account for the reduced ability of strain Sce11 to survive desiccation.
To determine if a reduction in water activity affects expression of ngg::Tn5luxAB, strain Sce11 was exposed to several compounds in concentrations that lead to the same reduction in water activity (Aw). The presence of 400 mM NaCl, 520 mM glycerol, 222 mM sucrose, or 780 mM PEG 200 in PMM results in an Aw of 0.986 (47, 48). The results depicted in Fig. 3C indicate that a reduction in the Aw does not affect expression of ngg::Tn5luxAB since in that case luciferase expression levels would be similar under all four conditions. Although ngg::Tn5luxAB does not respond to a reduction in water activity, the responses to the same osmotic stress caused by 400 mM NaCl or 800 mM sucrose, a nonaccumulating osmoprotectant in rhizobia (54), are similar under both conditions. This indicates that ngg::Tn5luxAB responds to osmotic stress and that the response to NaCl is more likely caused by osmotic stress than by ionic stress. Thus, a reduction in the Aw and an increase in ionic stress do not affect expression of ngg::Tn5luxAB.
Glutamine and glutamate complementation of growth in the presence of NaCl.
asnO and ngg are genetically linked in a diverse range of microorganisms and are involved in the production of NAGGN, a dipeptide that accumulates during osmotic stress (13, 44, 52, 53). The possibility that strains Sce10 and Sce11 are limited in their response to grow in the presence of glutamine under inducing and growth-limiting conditions was tested since biosynthesis of NAGGN uses glutamine as the substrate. The response of strain Sce11 resembles that of the reference strain, indicating that disruption of ngg does not lead to a malfunction in the response to NaCl in the presence of glutamine (Fig. 4). However, strain Sce10 had lost its ability to grow on 400 mM NaCl in the presence of glutamine, suggesting that disruption of asnO leads to the inability to use glutamine in a mechanism involved in the response to increased NaCl concentrations. Although the addition of glutamate did not affect growth in S. meliloti 1021 (P = 0.32), it stimulated growth slightly in Sce11 and reduced growth slightly in Sce10 (P = 0.02 and P = 0.04, respectively).
Fig 4.
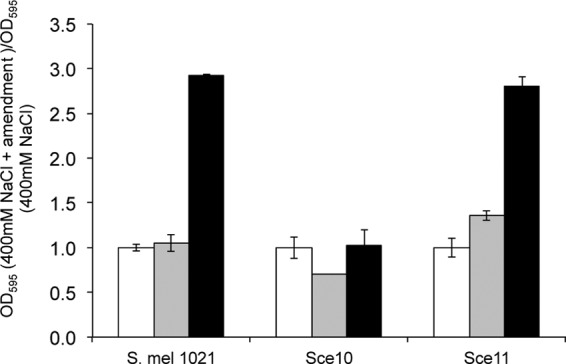
Growth of S. meliloti 1021, Sce10, and Sce11 in the presence of 400 mM NaCl with and without amino acid supplementation. Data are expressed as the ratio of the OD595 in PMM–400 mM NaCl. Error bars represent the SEM of three growing cultures measured once (n = 3). White bars, PMM–400 mM NaCl; gray bars, PMM–400 mM NaCl–glutamate; black bars, PMM–400 mM NaCl–glutamine.
asnO::Tn5luxAB and ngg::Tn5luxAB are differentially expressed during the stationary phase.
Previous work indicated that the survival of S. meliloti increases 2.9-fold in the stationary phase compared to the exponential phase (13). To test if asnO::Tn5luxAB and ngg::Tn5luxAB are expressed during the stationary phase, luciferase activity levels were measured over the course of a growth curve (Fig. 5). When strains Sce10 and Sce11 grew in PMM, the growth curves were similar to those of S. meliloti 1021, suggesting the absence of auxotrophs under permissive conditions, although exponential growth started earlier in Sce11. When luciferase induction levels were measured during the course of growth, differential responses of asnO::Tn5luxAB and ngg::Tn5luxAB were found during the lag, exponential, and stationary phases. Expression of asnO::Tn5luxAB drastically decreased during exponential growth of strain Sce10 and increased again in the stationary phase. If the organism was responsive at all, expression of ngg::Tn5luxAB slightly increased very early during exponential growth; however, expression of asnO::Tn5luxAB showed a decrease in luciferase activity in the early stages of growth, while activity increased again during the stationary phase.
Fig 5.
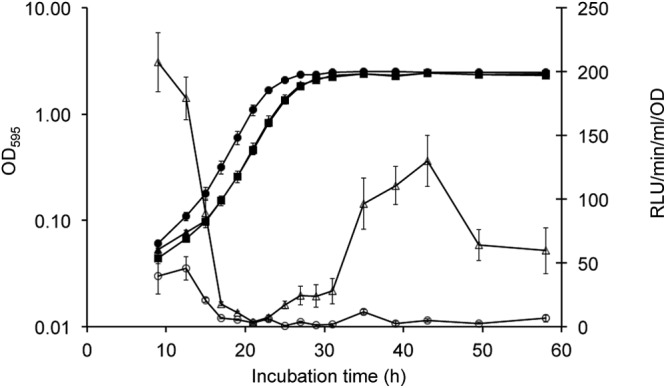
(A) Response of asnO::Tn5luxAB and ngg::Tn5luxAB during growth of strains S. meliloti 1021, Sce10, and Sce11 in liquid PMM. Error bars for OD measurements represent standard errors of the means (SEM) determined from three independent cultures (n = 3). RLU error bars represent the SEM of the results of three independent cultures performed in duplicate (n = 6). Black squares, growth of S. meliloti 1021 expressed in optical density (OD595) units; black circles, growth of Sce11 expressed in OD units; black triangles, growth of Sce10 expressed in OD units; open circles, luciferase activity of ngg::Tn5luxAB expressed in units of RLU/min/ml/OD; open triangles, luciferase activity of asnO::Tn5luxAB expressed in units of RLU/min/ml/OD.
The domain structures of AsnO and Ngg relate to antibiotic resistance.
A BLAST analysis of the amino acid sequence against the cDart and the conserved domain database at NCBI revealed that AsnOS. meliloti 1021 contains a GnAT domain between amino acids 2 and 224 (Evalue = 6e−32) and is associated with a conserved Asn synthase domain located between amino acids 246 and 524 (Evalue = 1.7e−55) (Fig. 2B). AsnOS. meliloti 1021 is in a class of sequences most closely related to AsnBE. coli (Evalue = 5.6e−118), a glutamine-hydrolyzing asparagine synthase, as reported earlier (53, 55, 56). More diverse members of this group of proteins include AsnOBacillus subtilis and LtsACorynebacterium glutamicum (55). Although previously not appreciated, AsnOS. meliloti also has similarity to β-LSStreptomyces clavuligerus, a protein with the ability to synthesize clavulanic acid, an inhibitor of β-lactamases (57).
An amino acid sequence comparison of Ngg to entries in cDart and the conserved domain databases revealed two conserved domains. The first is an acetyltransferase domain between amino acids 170 and 240 (Evalue = 9e−6). The second domain has highest sequence similarity to RimK RelE ligase (Evalue = 5.2e−7), cyanophycin synthase (CphA; Evalue = 1.6e−56), and d-alanine-d-alanine (dAla-dAla) ligase (Ddl domain; peptidoglycan synthesis) between amino acids 287 and 595 (Evalue = 4.2e−47) (Fig. 2B). Potential involvement in dAla-dAla ligase activity and peptidoglycan synthesis suggests that strain Sce11 is cycloserine sensitive (58, 59) and/or penicillin sensitive (60).
Because of these similarities, we hypothesized that strains Sce10 and Sce11 are affected in their ability to deal with β-lactam antibiotics since β-lactam antibiotics target the cell wall (60). To put these hypotheses to the test, Kirby-Bauer experiments were performed (Table 2). Both strains show the expected sensitivities to several antibiotics, including streptomycin (all insensitive) and kanamycin (Tn5luxAB carries kanamycin resistance in Sce10 and Sce11, making them less sensitive to kanamycin). Sce10 is less sensitive to the β-lactam antibiotic penicillin and, in contrast, Sce11 is more sensitive to penicillin than reference strain S. meliloti 1021.
Table 2.
Antibiotic sensitivity assaya
| Antibiotic | Amt/disc |
S. meliloti 1021 |
Sce10 |
Sce11 |
|||
|---|---|---|---|---|---|---|---|
| Diam (mm) | SEM | Diam (mm) | SEM | Diam (mm) | SEM | ||
| Streptomycin | 10 μg | 7.0 | 0.0 | 7.0 | 0.0 | 7.0 | 0.0 |
| Kanamycin | 30 μg | 11.3 | 0.3 | 7.0* | 0.0 | 7.0* | 0.0 |
| Bacitracin | 2 IU | 7.0 | 0.0 | 7.0 | 0.0 | 7.0 | 0.0 |
| Lysozyme | 250 μg | 7.0 | 0.0 | 7.0 | 0.0 | 7.0 | 0.0 |
| Cycloserine | 100 μg | 9.7 | 0.3 | 10 | 0.6 | 9.7 | 0.7 |
| Penicillin | 10 IU | 14.3 | 0.9 | 7.7* | 0.7 | 22.7* | 1.5 |
| Vancomycin | 5 μg | 21.3 | 1.2 | 19 | 0.0 | 22.7** | 0.3 |
The diameters of clearance around the disc (7 mm across) are presented together with the associated standard errors of the means (SEM). For each experiment, the average of the results determined for three plates is presented (n = 3). *, significant differences (P < 0.01; two-sided t test with equal variances) between the reference strain and strains Sce10 and Sce11; **, significant differences (P < 0.01; two-sided t test with equal variances) between strains Sce10 and Sce11.
Symbiotic characterization.
To further characterize the mutants, alfalfa seedlings were infected with the Tn5luxAB mutant strains and the ability to nodulate was recorded. All strains formed nodules; thus, no locus is essential for nodulation. When strains Sce10 and Sce11 were used to infect alfalfa roots, the strains formed pink nodules, indicating that they are nitrogen fixation proficient. Furthermore, alfalfa plants appeared the same when inoculated with S. meliloti 1021 or with Sce10 and Sce11 and did not appear nitrogen limited.
DISCUSSION
To address our hypothesis that a genetic mechanism is in S. meliloti 1021 that is inducible by NaCl and affecting this organism's ability to survive desiccation (13), a genetic screen for NaCl-inducible loci tagged by Tn5luxAB was performed. Twelve mutants harboring single transcriptional fusions that express luciferase at higher levels with NaCl than without NaCl were found (Fig. 1). Four of these Tn5luxAB-tagged loci in strains Sce2, Sce3, Sce10, and Sce11 form what appears to be one operon (Fig. 2A). Parts of this operon were previously identified and described (53, 55, 56, 70). Genetic loci that are part of this operon are known to be responsive to a decrease in oxygen availability (56) as well as to an increase in NaCl (61) and to PhoB-independent phosphate limitation (62).
Our hypothesis that certain NaCl-responsive loci are involved in survival during desiccation was supported by the results showing that the ability of two of the mutants (Sce10 and Sce11, with asnO and ngg tagged, respectively) to grow at increased NaCl concentrations was reduced (Fig. 3A) and that the ngg locus was also involved in NaCl-mediated survival during desiccation (Fig. 3B). Induction of this locus is mainly osmotic stress related and is not a consequence of a reduction in water activity or of ionic stress (Fig. 3C). Other loci in S. meliloti must exist that affect survival during desiccation, since ngg::Tn5luxAB is not predominantly responsive to NaCl. This conclusion is supported by the fact that survival of reference strain S. meliloti 1021 is mainly affected by the presence of the chloride anion rather that its cation (13). AsnO, however, is not involved in NaCl-mediated desiccation resistance.
Although the loci tagged in Sce2, Sce3, Sce10, and Sce11 may form one large operon, experimental data supporting the idea of one large transcriptional unit are still lacking. Our expression data do indicate different dynamics between asnO and ngg during the stationary phase. If the dipeptide uptake system and asnO and ngg do not form one operon, we would expect promoter sequences just upstream of asnO, which were not found. Differential expression of asnO::Tn5luxAB and ngg::Tn5luxAB also indicated regulatory sites between these two open reading frames (ORFs), which were not found either; neither were terminator sites found as indicated by Kingsford et al. (63). Therefore, we expect these genes to form one large operon unless unknown genetic elements are present in this locus.
AsnO and Ngg are both involved in a pathway for the production of NAGGN, a powerful osmoprotectant. Its biochemical production involves two steps as reported by Sagot et al. (53). In this pathway, Ngg (N-acetylglutaminylglutamine synthetase) produces the dipeptide N-acetylglutaminylglutamine (NAGG) from glutamine. AsnO is a glutamine-dependent amidotransferase transferring the amide nitrogen from glutamine to N-acetylglutaminylglutamine to produce NAGGN. The disruption of one or both steps leads to slower growth in the presence of NaCl. However, the fact that mutant Sce11 shows the same response to glutamine addition in the presence of NaCl as the reference strain and mutant Sce10 does not suggests that even in the absence of functional Ngg, a substrate for AsnO is still present. Thus, (i) another pathway for NAGG production must be present in S. meliloti 1021 or (ii) enough NAGG is present to ensure production of NAGGN for at least a couple of generations. Although unlikely due to the reduced growth on NaCl and the postulated lack of NAGGN accumulation, a possible alternative is that (iii) the Ngg::Tn5luxAB fusion protein in Sce11 is only partially disrupted; e.g., the acetyltransferase domain may still function. The argument in interpretation ii has some merit since growth complementation takes place at OD < 0.6 but is reduced with higher ODs (data not shown). Regardless, potential accumulation of NAGG in the asnO mutant does not lead to increased growth and from the data it is clear that the postulated reduced levels of NAGGN do not explain the reduced ability to survive desiccation of strain Sce11. It is most likely that as-yet-unknown functions, e.g., involvement in cell wall metabolism for the possible reasons explained next, are associated with AsnO and Ngg.
The amino acid sequence of Ngg is most similar to that of cyanophycin synthase and contains two domains, a NAT (or GCN-5) domain and a Ddl domain. Ddl domains are involved in the synthesis of peptidoglycan, and cell wall integrity is one of the main factors affecting the ability to survive desiccation in rhizobia (15, 30, 31). Furthermore, Ddl domains are found in dAla-dAla ligases, targets for cycloserine and peptidoglycan cross-linking enzymes and for β-lactam antibiotics such as penicillin. Our observations are that strain Sce11 is indeed more sensitive to penicillin than the reference strain; however, it is not affected by cycloserine (Table 2). The reduced ability to survive desiccation of strain Sce11is explained by a weaker cell wall, which is less able to withstand the extreme hypo-osmotic stress upon rehydration. That this may occur was shown previously by Salema et al. (30) and by Bushby and Marshall (31), who found the cell wall and envelope to be a major target upon rehydration.
The amino acid sequence of AsnO is most similar to that of AsnB, or asparagine synthase, and contains a GnAT domain and an Asn domain (Fig. 2B). Previously not appreciated, the similarity of AsnO to β-LS allows the hypothesis that AsnO may be involved in the production of a β-lactamase inhibitor. In this case, dysfunction of the production of inhibitor leads to an increased resistance to antibiotics such as penicillin. The data in Table 2 show exactly that: strain Sce10 is less sensitive to the β-lactam antibiotic.
Even though strains Sce10 and Sce11 form effective nodules on alfalfa, it is unclear if both strains are effective during competition under conditions more closely matching natural environments. Soil is a harsh environment, and cells are continuously exposed to challenging environmental conditions (64). Soils frequently undergo drying and rewetting cycles. During drying, salts accumulate, which further reduces the ability of microbes to grow. Also, many soil organisms produce antimicrobials, which may have negative effects on strains lacking the ngg locus. The plants may reduce the competitiveness of strains lacking locus ngg by excreting toxic secondary metabolites. On the other hand, a slight growth advantage of strain Sce11 may allow faster invasion of hair roots and have a positive effect of nodule occupancy since growth may be a primary factor during infection.
In conclusion, the function of AsnO and Ngg is not limited to the production and accumulation of NAGGN alone. First, NAGGN accumulation is not involved in survival during desiccation. Second, AsnO and Ngg may have functions such as the production of a β-lactamase inhibitor and involvement in cell wall metabolism. Third, they are involved in responses to β-lactam antibiotics, and fourth, a regulatory function has been attributed to AsnO during nodule development (55, 70), although the nature of the regulation remains obscure. Alternatively, it is interesting to speculate about physical location and other roles; e.g., the opposite effects of AsnO and Ngg on antibiotic resistance may indicate a physical interaction between the proteins with antagonistic effects on the cell wall such as changes in permeability.
Finally, identification of 4 of 10 loci that are responsive to NaCl and oxygen deprivation is at least curious and was mentioned before (65). A possible explanation may be the reduced solvability of oxygen in media high in salt. It may also be explained by being part of the environmental niche of this organism. In the rhizosphere, salts accumulate and available oxygen may be in short supply. Most interestingly, evidence that asnO and ngg have undergone lateral gene transfer has been presented (J. A. C. Vriezen, unpublished data, and reference 66). Nevertheless, we have identified a system that warrants further investigation on both the molecular and the ecological levels.
ACKNOWLEDGMENTS
This work was partially funded by LiphaTech, Inc. (grant 74576 to F.J.D.B.) and by the USDA (CSREES grant MAS 00087 to K.N.).
We thank Peter Wolk (MSU-PRL-DOE) for use of the photonic camera, Pamela Green (MSU-PRL-DOE) for use of the luminometer, Christine White-Zeigler (Smith College) for assistance with RT-PCR, and, last, two anonymous reviewers who gave excellent recommendations to increase the quality of the manuscript.
Footnotes
Published ahead of print 12 July 2013
REFERENCES
- 1.Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. 2007. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5:619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zahran HH. 1999. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol. Mol. Biol. Rev. 63:968–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deaker R, Roughley RJ, Kennedy IR. 2004. Legume seed inoculation technology—a review. Soil Biol. Biochem. 36:1275–1288 [Google Scholar]
- 4.Vriezen JAC, de Bruijn FJ, Nüsslein K. 2007. Responses of rhizobia to desiccation in relation to osmotic stress, oxygen, and temperature. Appl. Environ. Microbiol. 73:3451–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vriezen JAC, de Bruijn FJ, Nüsslein KR. 2012. Desiccation induces viable but non-culturable cells in Sinorhizobium meliloti 1021. AMB Express 2:6. 10.1186/2191-0855-2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Rensburg H, Strijdom B. 1980. Survival of fast- and slow-growing rhizobium spp under conditions of relatively mild desiccation. Soil Biol. Biochem. 12:353–356 [Google Scholar]
- 7.Dye M. 1982. A note on some factors affecting the survival of Rhizobium cultures during freeze drying and subsequent storage. J. Appl. Microbiol. 52:461–464 [Google Scholar]
- 8.Estrella M, Pieckenstain F, Marina M, Dìaz L, Ruiz O. 2004. Cheese whey: an alternative growth and protective medium for Rhizobium loti cells. J. Ind. Microbiol. Biotechnol. 31:122–126 [DOI] [PubMed] [Google Scholar]
- 9.Mary P, Ochin D, Tailliez R. 1985. Rates of drying and survival of Rhizobium meliloti strains during storage at different relative humidities. Appl. Environ. Microbiol. 50:207–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mary P, Ochin D, Tailliez R. 1986. Growth status of rhizobia in relation to their tolerance to low water activities and desiccation stresses. Soil Biol. Biochem. 18:179–184 [Google Scholar]
- 11.Bushby H, Marshall K. 1977. Some factors affecting the survival of root nodule bacteria on desiccation. Soil Biol. Biochem. 9:143–147 [Google Scholar]
- 12.Kosanke JW, Osburn RM, Shuppe GI, Smith RS. 1992. Slow rehydration improves the recovery of dried bacterial populations. Can. J. Microbiol. 38:520–525 [DOI] [PubMed] [Google Scholar]
- 13.Vriezen JAC, de Bruijn FJ, Nüsslein K. 2006. Desiccation responses and survival of Sinorhizobium meliloti USDA 1021 in relation to growth phase, temperature, chloride and sulfate availability. Lett. Appl. Microbiol. 42:172–178 [DOI] [PubMed] [Google Scholar]
- 14.Kremer RJ, Peterson HL. 1983. Effects of carrier and temperature on survival of Rhizobium spp. in legume inocula: development of an improved type of inoculant. Appl. Environ. Microbiol. 45:1790–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincent JT, Thompson J, Donovan K. 1962. Death of root-nodule bacteria on drying. Aust. J. Agr. Res. 13:258–270 [Google Scholar]
- 16.Salema M, Parker C, Kidby D, Chatel D. 1982. Death of rhizobia on inoculated seed. Soil Biol. Biochem. 14:13–14 [Google Scholar]
- 17.Smith R. 1992. Legume inoculant formulation and application. Can. J. Microbiol. 38:485–492 [Google Scholar]
- 18.Boumahdi M, Mary P, Hornez JP. 1999. Influence of growth phases and desiccation on the degrees of unsaturation of fatty acids and the survival rates of rhizobia. J. Appl. Microbiol. 87:611–619 [DOI] [PubMed] [Google Scholar]
- 19.Chenu C. 1993. Clay- or sand-polysaccharide associations as models for the interface between micro-organisms and soil: water related properties and microstructure. Geoderma 56:143–156 [Google Scholar]
- 20.Kaci Y, Heyraud A, Barakat M, Heulin T. 2005. Isolation and identification of an EPS-producing Rhizobium strain from arid soil (Algeria): characterization of its EPS and the effect of inoculation on wheat rhizosphere soil structure. Res. Microbiol. 156:522–531 [DOI] [PubMed] [Google Scholar]
- 21.Sadowski M, Graham P. 1998. Soil biology of the Rhizobiaceae, p 155–172 In Spaink HP, Kondorosi A, Hooykaas PJJ. (ed), The Rhizobiaceae. Kluwer Academic Publishers, Dordrecht, the Netherlands [Google Scholar]
- 22.Trotman AP, Weaver RW. 1995. Tolerance of clover rhizobia to heat and desiccation stresses in soil. Soil Sci. Soc. Am. J. 59:466–470 [Google Scholar]
- 23.Steinborn J, Roughley RJ. 1974. Sodium chloride as a cause of low numbers of Rhizobium in legume inoculants. J. Appl. Bacteriol. 37:93–99 [DOI] [PubMed] [Google Scholar]
- 24.Steinborn J, Roughley RJ. 1975. Toxicity of sodium and chloride ions to Rhizobium spp. in broth and peat culture. J. Appl. Bacteriol. 39:133–138 [DOI] [PubMed] [Google Scholar]
- 25.Kosanke JW, Osburn R, Smith R, LiphaTech Inc April 1999. Procedure for preparation of bacterial agricultural products. Canadian patent 2073507
- 26.Willis LB, Walker GC. 1999. A novel Sinorhizobium meliloti operon encodes an alpha-glucosidase and a periplasmic-binding-protein-dependent transport system for alpha-glucosides. J. Bacteriol. 181:4176–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen JB, Peters NK, Bhuvaneswari TV. 2002. Redundancy in periplasmic binding protein-dependent transport systems for trehalose, sucrose, and maltose in Sinorhizobium meliloti. J. Bacteriol. 184:2978–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Domínguez-Ferreras A, Pérez-Arnedo R, Becker A, Olivares J, Soto MJ, Sanjuán J. 2006. Transcriptome profiling reveals the importance of plasmid pSymB for osmoadaptation of Sinorhizobium meliloti. J. Bacteriol. 188:7617–7625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McIntyre HJ, Davies H, Hore TA, Miller SH, Dufour J-P, Ronson CW. 2007. Trehalose biosynthesis in Rhizobium leguminosarum bv. trifolii and its role in desiccation tolerance. Appl. Environ. Microbiol. 73:3984–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salema M, Parker C, Kidby D, Chatel D, Armitage T. 1982. Rupture of nodule bacteria on drying and rehydration. Soil Biol. Biochem. 14:15–22 [Google Scholar]
- 31.Bushby H, Marshall K. 1977. Desiccation induced damage to the cell envelope of root nodule bacteria. Soil Biol. Biochem. 9:149–152 [Google Scholar]
- 32.Zevenhuizen LP, Faleschini P. 1991. Effect of the concentration of sodium chloride in the medium on the relative proportions of poly- and oligo-saccharides excreted by Rhizobium meliloti strain YE-2SL. Carbohydr. Res. 209:203–209 [DOI] [PubMed] [Google Scholar]
- 33.Lloret J, Bolanos L, Lucas MM, Peart JM, Brewin NJ, Bonilla I, Rivilla R. 1995. Ionic stress and osmotic pressure induce different alterations in the lipopolysaccharide of a Rhizobium meliloti strain. Appl. Environ. Microbiol. 61:3701–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lloret J, Wulff BB, Rubio JM, Downie JA, Bonilla I, Rivilla R. 1998. Exopolysaccharide II production is regulated by salt in the halotolerant strain Rhizobium meliloti EFB1. Appl. Environ. Microbiol. 64:1024–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei W, Jiang J, Li X, Wang L, Yang SS. 2004. Isolation of salt-sensitive mutants from Sinorhizobium meliloti and characterization of genes involved in salt tolerance. Lett. Appl. Microbiol. 39:278–283 [DOI] [PubMed] [Google Scholar]
- 36.Miller-Williams M, Loewen PC, Oresnik IJ. 2006. Isolation of salt-sensitive mutants of Sinorhizobium meliloti strain Rm1021. Microbiology 152:2049–2059 [DOI] [PubMed] [Google Scholar]
- 37.Humann JL, Ziemkiewicz HT, Yurgel SN, Kahn ML. 2009. Regulatory and DNA repair genes contribute to the desiccation resistance of Sinorhizobium meliloti 1021. Appl. Environ. Microbiol. 75:446–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meade HM, Long SR, Ruvkun GB, Brown SE, Ausubel FM. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ditta G, Stanfield S, Corbin D, Helinski DR. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. U. S. A. 77:7347–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finan TM, Hartweig E, LeMieux K, Bergman K, Walker GC, Signer ER. 1984. General transduction in Rhizobium meliloti. J. Bacteriol. 159:120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milcamps A, Ragatz DM, Lim P, Berger KA, de Bruijn FJ. 1998. Isolation of carbon- and nitrogen-deprivation-induced loci of Sinorhizobium meliloti 1021 by Tn5-luxAB mutagenesis. Microbiology 144:3205–3218 [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 43.Beringer JE. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188–198 [DOI] [PubMed] [Google Scholar]
- 44.Vriezen JAC. 2005. Responses of Sinorhizobium meliloti 1021 to water stress. Ph.D thesis University of Massachusetts, Amherst, MA [Google Scholar]
- 45.Wolk CP, Cai Y, Panoff JM. 1991. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc. Natl. Acad. Sci. U. S. A. 88:5355–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phillips D, Sande E, Vriezen JAC, de Bruijn FJ, LeRudulier D, Joseph CM. 1998. A new genetic locus in Sinorhizobium meliloti is involved in stachydrine utilization. Appl. Environ. Microbiol. 64:3954–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leistner L, Rodel L. 1976. Inhibition of micro-organisms in food by water activity, p 219–237 In Skinner FA, Hugo WB. (ed), Inhibition and inactivation of vegetative microbes. Academic Press, London, England [Google Scholar]
- 48.Brown AD. 1990. Microbial water stress physiology principles and perspectives. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 49.Reese MG. 2001. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 26:51–56 [DOI] [PubMed] [Google Scholar]
- 50.Gordon L, Chervonenkis AY, Gammerman AJ, Shahmuradov IA, Solovyev VV. 2003. Sequence alignment kernel for recognition of promoter regions. Bioinformatics 19:1964–1971 [DOI] [PubMed] [Google Scholar]
- 51.Münch R, Hiller K, Barg H, Heldt D, Linz S, Wingender E, Jahn D. 2003. Prodoric: prokaryotic database of gene regulation. Nucleic Acid Res. 31:266–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith LT, Smith GM. 1989. An osmoregulated dipeptide in stressed Rhizobium meliloti. J. Bacteriol. 171:4714–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sagot B, Gaysinski M, Mehiri M, Guigonis J-M, Le Rudulier D, Alloing G. 2010. Osmotically induced synthesis of the dipeptide n-acetylglutaminylglutamine amide is mediated by a new pathway conserved among bacteria. Proc. Natl. Acad. Sci. U. S. A. 107:12652–12657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gouffi K, Pichereau V, Rolland JP, Thomas D, Bernard T, Blanco C. 1998. Sucrose is a non-accumulated osmoprotectant in Sinorhizobium meliloti. J. Bacteriol. 180:5044–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bergès H, Checroun C, Guiral S, Garnerone AM, Boistard P, Batut J. 2001. A glutamine-amidotransferase-like protein modulates FixT anti-kinase activity in Sinorhizobium meliloti. BMC Microbiol. 1:6. 10.1186/1471-2180-1-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trzebiatowski JR, Ragatz DM, de Bruijn FJ. 2001. Isolation and regulation of Sinorhizobium meliloti 1021 loci induced by oxygen limitation. Appl. Environ. Microbiol. 67:3728–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller MT, Bachmann BO, Townsend CA, Rosenzweig AC. 2001. Structure of beta-lactam synthetase reveals how to synthesize antibiotics instead of asparagine. Nat. Struct. Biol. 8:684–689 [DOI] [PubMed] [Google Scholar]
- 58.Feng Z, Barletta RG. 2003. Roles of Mycobacterium smegmatis d-alanine:d-alanine ligase and d-alanine racemase in the mechanisms of action of and resistance to the peptidoglycan inhibitor d-cycloserine. Antimicrob. Agents Chemother. 47:283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bruning JB, Murillo AC, Chacon O, Barletta RG, Sacchettini JC. 2011. Structure of the Mycobacterium tuberculosis d-alanine:d-alanine ligase, a target of the antituberculosis drug d-cycloserine. Antimicrob. Agents Chemother. 55:291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bugg TDH, Braddick D, Dowson CG, Roper DI. 2011. Bacterial cell wall assembly: still an attractive antibacterial target. Trends Biotechnol. 29:167–173 [DOI] [PubMed] [Google Scholar]
- 61.Rüberg S, Tian Z-X, Krol E, Linke B, Meyer F, Wang Y, Pühler A, Weidner S, Becker A. 2003. Construction and validation of a Sinorhizobium meliloti whole genome DNA microarray: genome-wide profiling of osmoadaptive gene expression. J. Biotechnol. 106:255–268 [DOI] [PubMed] [Google Scholar]
- 62.Krol E, Becker A. 2004. Global transcriptional analysis of the phosphate starvation response in Sinorhizobium meliloti strains 1021 and 2011. Mol. Genet. Genomics 272:1–17 [DOI] [PubMed] [Google Scholar]
- 63.Kingsford CL, Ayanbule K, Salzberg SL. 2007. Rapid, accurate, computational discovery of rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 8:R22. 10.1186/gb-2007-8-2-r22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hirsch AM. 2010. How rhizobia survive in the absence of a legume host, a stressful world indeed, p 375–391 In Seckbach J, Grube M. (ed), Symbioses and stress. Springer, Houten, The Netherlands [Google Scholar]
- 65.Ni Bhriain N, Dorman CJ, Higgins CF. 1989. An overlap between osmotic and anaerobic stress responses: a potential role for DNA supercoiling in the coordinate regulation of gene expression. Mol. Microbiol. 3:933–942 [DOI] [PubMed] [Google Scholar]
- 66.Vriezen JAC. 2006. A genetic basis for the response to drought and antibiotic resistance in Sinorhizobium meliloti 1021: evidence for lateral gene transfer, p 7 Boston Bacterial Meeting, Harvard University, Boston, MA, 16 June 2006 [Google Scholar]
- 67.Davey ME, de Bruijn FJ. 2000. A homologue of the tryptophan-rich sensory protein TspO and FixL regulate a novel nutrient deprivation-induced Sinorhizobium meliloti locus. Appl. Environ. Microbiol. 66:5353–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sauviac L, Philippe H, Phok K, Bruand C. 2007. An extracytoplasmic function sigma factor acts as a general stress response regulator in Sinorhizobium meliloti. J. Bacteriol. 189:4204–4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bobik C, Meilhoc E, Batut J. 2006. FixJ: a major regulator of the oxygen limitation response and late symbiotic functions of Sinorhizobium meliloti. J. Bacteriol. 188:4890–4902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Bruijn FJ, Rossbach S, Bruand C, Parrish JR. 2006. A highly conserved Sinorhizobium meliloti operon is induced microaerobically via the FixLJ system and by nitric oxide (NO) via NnrR. Environ. Microbiol. 8:1371–1381 [DOI] [PubMed] [Google Scholar]


