Abstract
Colanic acid (CA) is a common exopolysaccharide produced by many genera in the Enterobacteriaceae. It is critical for biofilm formation on HEp-2 cells and on chicken intestinal tissue by Salmonella. In this study, we generated different CA synthesis gene mutants and evaluated the immune responses induced by these mutants. One of these mutations, Δ(wza-wcaM)8, which deleted the whole operon for CA synthesis, was introduced into two Salmonella vaccine strains attenuated by auxotrophic traits or by the regulated delayed attenuation strategy (RDAS). The mice immunized with the auxotrophic Salmonella vaccine strain with the deletion mutation Δ(wza-wcaM)8 developed higher vaginal IgA titers against the heterologous protective antigen and higher levels of antigen-specific IgA secretion cells in lungs. In Salmonella vaccine strains with RDAS, the strain with the Δ(wza-wcaM)8 mutation resulted in higher levels of protective antigen production during in vitro growth. Mice immunized with this strain developed higher serum IgG and mucosal IgA antibody responses at 2 weeks. This strain also resulted in better gamma interferon (IFN-γ) responses than the strain without this deletion at doses of 108 and 109 CFU. Thus, the mutation Δ(wza-wcaM)8 will be included in various recombinant attenuated Salmonella vaccine (RASV) strains with RDAS derived from Salmonella enterica serovar Paratyphi A and Salmonella enterica serovar Typhi to induce protective immunity against bacterial pathogens.
INTRODUCTION
Colanic acid (CA) is a common exopolysaccharide (EPS) structure loosely associated with the surfaces of a wide variety of bacteria (1), especially in the Enterobacteriaceae, which are normally found as inhabitants of the intestine. The CA structure contains repeating subunits of d-glucose, l-fucose, d-galactose, and d-glucuronic acid sugars that are decorated with O-acetyl and pyruvate side chains and are assembled by essentially identical processes as lipopolysaccharide (LPS) O antigen (1–5). The CA biosynthetic gene cluster is composed of 20 genes in Escherichia coli and Salmonella (6, 7). The complex transcriptional regulation of CA production is controlled through the Rcs (regulation of capsule synthesis) proteins, Lon protease and RpoS (8–12). Overexpression of CA caused by either rcsB or lon mutation causes a mucoid colony phenotype on agar surfaces and attenuation (13).
CA is normally produced in small amounts constitutively, whereas large amounts can be synthesized in response to specific mutations or environmental factors (14). It is generally not produced at temperatures above 30°C in typical laboratory media; however, E. coli K92 is able to produce CA even at 42°C (15), suggesting a role of CA when E. coli is outside mammalian hosts. A substantial amount of the CA produced by a culture is secreted into the growth medium (5, 16). The biological role of CA lies primarily outside the host (14), especially in regard to the bacterial survival under adverse physicochemical and environmental conditions (17–19). CA biosynthesis genes are coordinately regulated in response to a variety of environmental factors that modulate or damage cell envelope structure (5), such as temperature (15), desiccation (20), β-lactam antibiotics (21–23), osmotic shock (18, 24, 25), oxidative stress, acid and heat stress (15, 18, 26, 27), metal ion exposure (28), growth on solid surfaces (29), carbon and nitrogen sources (15), and a combination of different factors. In E. coli K-12 and uropathogenic E. coli, CA synthesis is not important for attachment to abiotic surfaces but is critical to bacteria growing in biofilms on abiotic surfaces at 30°C (17, 29–33). CA can attach through a covalent linkage to LPS to form a novel LPS glycoform containing CA repeats, which respond to certain environmental stimuli (34).
CA has no known role in virulence in E. coli (14, 35). The inability to produce CA in an extraintestinal pathogenic E. coli (ExPEC) strain showed no effect on resistance to the bactericidal effects of serum and bactericidal/permeability-increasing protein in vitro or on virulence in an abscess model or in a systemic infection model in vivo (35). The CA-deficient E. coli strain had less viability in the human gastrointestinal tract environment and upon exposure to acid but not to bile (36). Most of this research on CA was carried out with E. coli strains (5), such as different laboratory K-12 strains, including MC4100, MG1655, and W3310 (6, 8, 17, 30, 31, 37–40), K1 (41), K5 (41), K92 (42), extraintestinal isolate O4/K54/H5 (35, 43), different O8 and O9 pathovars (44), enterohemorrhagic E. coli O157:H7 (18, 26, 27, 36, 37, 45), and uropathogenic E. coli (33). Although it was reported that CA in Salmonella is not important for biofilm formation in vitro (46), some reports indicated that CA in Salmonella may play a role in vivo (4, 47, 48). The divergence of CA gene clusters between Salmonella and E. coli is slightly higher than the average for other genes of the two species (4). The six genes involved in the synthesis of GDP-fucose in Salmonella were replaced recently by genes from a close relative of the original donor species (4). The mucoid phenotype, caused by CA overexpression, is highly variable in different Salmonella subspecies and strains, even with induction of synthesis by p-fluorophenylalanine (1). Mucoid Salmonella strains were not lysed by P22, indicating the inaccessibility to the P22 receptor LPS O antigen (49). A mucoid variant of Salmonella has been isolated from a clinical specimen, which indicated that CA overexpression in vivo may help Salmonella escape antibiotic treatment (48). Although the physiological stimuli in vivo that induce CA synthesis are not fully understood (50), mutation of wcaM, one of the CA synthesis genes in Salmonella, resulted in reduced biofilm formation on HEp-2 cells and on chicken intestinal tissue at 37°C in vitro (47), which indicated that the CA can be produced at 37°C when bacteria are exposed to eukaryotic cells.
Recently, it was reported that the production and secretion of recombinant proteins in an E. coli strain with a whole CA operon deletion mutation and other mutations increases by about ∼16% and ∼ 25%, respectively, by an unknown mechanism (51). Thus, we speculated whether the attenuated Salmonella vaccine vectors with a CA operon deletion mutation could have the same phenotype and, hopefully, lead to enhanced immune responses to recombinant protective protein antigens.
We developed an in vivo regulated delayed attenuation strategy (RDAS) to construct safe and efficacious attenuated Salmonella vaccine vectors (52), with Salmonella enterica serovar Typhimurium strain χ9558 and its isogenotype Salmonella enterica serovar Typhi strains χ9633, χ9639, and χ9640 as representatives. These vaccine vectors are safe and effective in adult, newborn, and infant mice (53–56). The pmi mutation is one of the main ways to achieve the in vivo regulated delayed attenuation phenotype by reversible synthesis of LPS O-antigen side chains. The pmi gene encodes phosphomannose isomerase that interconverts mannose 6-phosphate (mannose 6-P) and fructose 6-phosphate (57). Mannose 6-P can be converted to GDP-mannose and used for synthesis of LPS O-antigen side chains (58). The combination of the Δpmi-2426 (Δ = deletion) mutation, which deletes the whole pmi gene, with exogenous mannose during in vitro growth results in a smooth phenotype with the synthesis of wild-type LPS O antigen to facilitate successful colonization of lymphoid tissues by the mutant strain (59, 60). In vivo, there is an absence of nonphosphorylated mannose so that the synthesis of LPS O-antigen side chain ceases to result in a rough phenotype and avirulence (60). This mutation has been shown to significantly but not completely attenuate S. Typhimurium (60). S. Typhimurium pmi mutants are highly immunogenic (61), with enhanced abilities to induce antibody titers to cross-protective outer membrane proteins (OMPs) (61), to produce outer membrane vesicles (OMVs) that can also deliver recombinant protective antigens to enhance induction of protective immunity (62, 63), and to induce antibody responses against the LPS core (64, 65) that is common to all S. enterica serotypes (66, 67).
In S. Typhimurium strain χ9558 and its isogenotypic S. Typhi vaccine strains, we described the deletion mutation Δ(gmd-fcl)26, which eliminates two enzymes needed to synthesize GDP-fucose, which is required for colanic acid synthesis. The mutation can preclude GDP-mannose from being converted into GDP-fucose so that all added mannose would be incorporated into LPS O antigen; thus, it can be used in combination with the Δpmi-2426 mutation to tightly regulate O-antigen synthesis by exogenous mannose (61). This deletion does not alter attenuation, tissue colonization, or immunogenicity of a strain with the Δpmi-2426 mutation alone (61). It was also used in regulated delayed lysis strains to block the potential synthesis of CA, which could help cells undergoing diaminopimelic acid-less and muramic acid-less death to survive (68, 69), as well as in our balanced-lethal vector system. Our lab also proved that the lrp gene is an antivirulence gene (70). The deletion of the lrp gene results in an increase in epithelial cell invasion and enhanced transcription of the hilA and ssrA genes, the master regulators of the Salmonella pathogenicity island 1 (SPI-1) type III secretion system and the SPI-2 type III secretion system, respectively (70). In this paper, we report the construction of different CA operon deletion mutations in place of the Δ(gmd-fcl)26 mutation in Salmonella vaccine strains. Our goals were to evaluate the effects on protein synthesis and immune responses by including the CA operon deletion mutation and lrp mutation individually or combined.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. S. Typhimurium vaccine strains were derived from the highly virulent strain UK-1 (71). S. Typhimurium cultures were grown statically overnight at 37°C in LB broth or on LB agar (72). When necessary, arabinose (0.05%) and/or mannose (0.1%) was added to the broth as indicated. Bacterial growth curves were obtained using optical density measurements with a Genesys 10 UV spectrophotometer (Thermo Scientific) and by plating serial dilutions of bacterial cultures on LB agar (72). LPS profiles were examined using silver staining as previously described (73). The genotypes of the strains were verified using corresponding primer sets (Table 2). The phenotype characterizations associated with mutations in the strains were described elsewhere (52, 74). LB agar without NaCl and containing 5% sucrose was used for sacB gene-based counterselection in allelic exchange to generate mutations. Streptococcus pneumoniae strain WU2 was cultured on brain heart infusion agar containing 5% sheep blood or in Todd-Hewitt broth plus 0.5% yeast extract (75).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| χ6212 | F− λ− ϕ80 Δ(lacZYA-argF) endA1 recA1 hsdR17 deoR thi-1 glnV44 gyrA96 relA1 ΔasdA4 | 104 |
| χ7213 | thi-1 thr-1 leuB6 fhuA21 lacY1 glnV44 ΔasdA4 recA1 RP4 2-Tc::Mu (λ pir) Kmr | 105 |
| S. enterica serovar Typhimurium | ||
| χ3761 | UK-1 | 71 |
| χ8650 | χ3761 Δpmi-2426 | 61 |
| χ8831 | χ3761 Δ(gmd-fcl)26 | 52 |
| χ8868 | Δpmi-2426 Δ(gmd-fcl)26 | 81 |
| χ9241 | ΔpabA1516 ΔpabB232 ΔasdA16 ΔrelA198::araC PBAD lacI TT ΔaraBAD23 | 74 |
| χ9535 | χ3761 Δ(wza-wcaL)6 | This study |
| χ9536 | χ3761 Δ(wza-wcaL)7 | This study |
| χ9537 | χ3761 Δ(wza-wcaM)8 | This study |
| χ9540 | Δpmi-2426 Δ(wza-wcaM)8 | This study |
| χ9558 | Δpmi-2426 ΔPfur81::TT araC PBAD fur ΔPcrp527::TT araC PBAD crp ΔasdA27::TT araC PBAD c2 ΔaraE25 ΔaraBAD23 ΔrelA198::araC PBAD lacI TT ΔsopB1925 ΔagfBAC811 Δ(gmd-fcl)26 | 56 |
| χ9837 | χ9558 Δlrp-23 | This study |
| χ9902 | χ9558 Δ(wza-wcaM)8 | This study |
| χ9903 | χ9837 Δ(wza-wcaM)8 | This study |
| χ11370 | χ9241 Δ(wza-wcaM)8 | This study |
| S. pneumoniae WU2 | Wild-type virulent, encapsulated type 3 strain | 75 |
| Plasmids | ||
| pRE112 | Suicide plasmid, λ pir dependent, oriT, oriV, sacB, Cmr | 106 |
| pYA3493 | Periplasmic secretion plasmid based on β-lactamase N-terminal signal sequence, pBR ori, Ptrc, AsdA+ | 84 |
| pYA4088 | 849-bp DNA encoding the α-helical region of PspA Rx1 from amino acid 3 to 285 in pYA3493 | 76 |
| pYA4366 | pRE112 derivative for constructing Δ(wza-wcaL)6 mutation | This study |
| pYA4367 | pRE112 derivative for constructing Δ(wza-wcaL)7 mutation | This study |
| pYA4368 | pRE112 derivative for constructing Δ(wza-wcaM)8 mutation | This study |
TT, transcription terminator.
Table 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| P1 | 5′ ATATAGGTACCGCGAACATCCAGCGTCACATTG 3′ |
| P2 | 5′ ACGCGAGATCTCAGCCTGCTACAAACGATATAAAC 3′ |
| P3 | 5′ CGCGAGATCTGATTATTTATCACTTTGGCAG 3′ |
| P4 | 5′ ACGAGGAGCTCCTTGCCTGTCATTAGGTTAG 3′ |
| P5 | 5′ ATATGAGATCTATGCCCGCGACTAAATTCTCCCG 3′ |
| P6 | 5′ GTGAAGGTACCAAGTTCATAAGAGGTGTCGAAGTG 3′ |
| P7 | 5′ CGCTGAGATCTGTACCGCTATTTTTACGAAAATTC 3′ |
Construction of plasmids and strains.
Plasmid pYA4366 for deletion of the CA gene cluster to generate the Δ(wza-wcaL)6 mutation was constructed as follows. A 423-bp fragment at the 3′ end of wcaL was generated by PCR by using primers P1 and P2 (Table 2), and a 405-bp fragment of the 5′ end of wza was generated by PCR by using primers P3 and P4 (Table 2). These two fragments were cloned into the KpnI and SacI sites of pRE112 to generate suicide vector pYA4366. The plasmid pYA4367 for deletion of the CA gene cluster to generate the Δ(wza-wcaL)7 mutation was constructed essentially as described above except that the 423-bp fragment was replaced with a 391-bp fragment at the 3′ end of wcaL, generated by PCR with primers P1 and P5 (Table 2). The 391-bp and 405-bp fragments were cloned into the KpnI and SacI sites of pRE112 to generate suicide vector pYA4367. Plasmid pYA4368 for deletion of the CA gene cluster to generate the Δ(wza-wcaM)8 mutation was constructed essentially as described above except that the 423-bp fragment at the 3′ end of wcaL was replaced with a 418-bp fragment at the 3′ end of wcaM, generated by PCR with primers P6 and P7 (Table 2). The 418-bp and 405-bp fragments were cloned into the KpnI and SacI sites of pRE112 to generate suicide vector pYA4368.
The Δ(wza-wcaM)8 mutation deletes 20 genes from the start codon of wza to the stop codon of wcaM. It was introduced into S. Typhimurium strains χ9241, χ9558, and χ9837 by allelic exchange using the suicide vector pYA4368 to generate strains χ11370, χ9902, and χ9903, respectively. The presence of the 22,624-bp deletion was confirmed by PCR with a primer set flanking the deletion region, primers P4 and P6 (Table 2).
SDS-PAGE, immunoblot analyses, and GFP detection.
Vaccine strains from static cultures grown overnight at 37°C in LB broth with 0.05% arabinose and/or 0.1% mannose were diluted 1:100 into the same medium at 37°C. The culture was grown with aeration at 37°C to an optical density at 600 nm (OD600) of 0.5 with continued growth for 4 h after addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Equal numbers of cells were collected for each culture. All samples were subjected to SDS-PAGE and Western blotting as previously described (76).
For detection of green fluorescent protein (GFP), cells were grown as described above. After addition of 1 mM IPTG to induce the GFP synthesis, the cells were cultured for an additional 5 h at 25°C with shaking. The fluorescence intensity and OD600 were measured using a SpectraMax M2e microplate reader (Molecular Devices). The relative expression levels were calculated by the ratio of fluorescence to OD600.
Animals.
Six-week-old female BALB/c mice were obtained from Charles River Laboratories (Wilmington, MA). All animal protocols were approved by the ASU IACUC and complied with the rules and regulations of the American Association for Accreditation of Laboratory Animal Care. The mice were acclimated for 7 days after arrival before the experiments were started.
Virulence, immunogenicity, and protection tests in mice.
Virulence tests for determination of the 50% lethal dose (LD50) were as described previously (76). LB was supplemented with 0.05% arabinose and/or 0.1% mannose when needed. Static overnight cultures of recombinant attenuated Salmonella vaccine (RASV) strains were diluted 1:100 into LB broth with 0.05% arabinose and/or 0.1% mannose at 37°C. Each culture was grown with aeration at 37°C to an OD600 of 0.85 to 0.9. Procedures for cell collection, immunization, blood and vaginal wash sample collection, and storage have been described previously (76). Groups of mice were orally inoculated with approximately 1 × 109 CFU of vaccine strains. The mice were boosted with approximately 1 × 109 CFU of the same strain at week 6. At week 10, mice were challenged by intraperitoneal (i.p.) injection with 2 × 104 CFU of S. pneumoniae WU2 in 100 μl of BSG (33), which is equivalent to 100 times the LD50. Challenged mice were monitored for death daily for 30 days.
Antigen preparation and ELISA.
Recombinant PspA (rPspA) protein and Salmonella outer membrane proteins (SOMPs) were purified as described previously (76). The rPspA clone was a kind gift from Susan Hollingshead at the University of Alabama at Birmingham. Enzyme-linked immunosorbent assay (ELISA) was used to assay antibodies to S. Typhimurium SOMPs and rPspA in serum and to rPspA in vaginal washes as described previously (76).
IL-4, IFN-γ, and IL-17 ELISPOT and ASC assays.
One week after boosting at 6 weeks, spleen and lung cells were harvested from three or four mice from each group. Enzyme-linked immunospot (ELISPOT) assays were performed to test interleukin-4 (IL-4)-, gamma interferon (IFN-γ)-, and IL-17-secreting cells in triplicate wells as previously described (77). IgG and IgA antibody-secreting cells (ASCs) in spleens and lungs were detected in triplicate as previously described (78). All antibody pairs used were from BD Biosciences.
Statistics.
Statistical analyses were performed by using the GraphPad Prism 5 software package (Graph Software, San Diego, CA). Antibody titers were expressed as means ± standard errors. The means were evaluated with two-way analysis of variance (ANOVA) and Bonferroni tests for multiple comparisons among groups. Differences were considered significant at a P value of <0.05.
RESULTS
Construction of strains with CA operon deletion.
We constructed three different mutations in the CA biosynthetic operon, Δ(wza-wcaL)6, Δ(wza-wcaL)7 and Δ(wza-wcaM)8 (Fig. 1). There were 19 genes deleted in the Δ(wza-wcaL)6 and Δ(wza-wcaL)7 mutations and 20 genes deleted in the Δ(wza-wcaM)8 mutation. The Δ(wza-wcaL)6 mutation still has the SD sequence of wcaM, while Δ(wza-wcaL)7 does not. We also had another mutation, Δ(gmd-fcl)26, with two genes, gmd and fcl, deleted in the CA operon (Fig. 1). The Δ(gmd-fcl)26 mutation deletes only genes related to the GDP-mannose conversion to GDP-fucose, one of the precursors of CA, whereas the Δ(wza-wcaL)6, Δ(wza-wcaL)7, and Δ(wza-wcaM)8 mutations also remove the genes important to colanic acid synthesis, modification, polymerization, export (wzc and wzxC), and translocation (wza) and the GDP-mannose pathway, which is related to O-antigen synthesis (cpsG and manC) (6, 79). The Wza, Wzb, and Wzc proteins are highly conserved in bacteria that produce capsular polysaccharides (CPS) and extracellular polysaccharide (EPS) (80). They are required for translocation and surface assembly of EPS or CPS, which are tightly associated with the surface of the bacterial cell (5, 80). There is no difference between the growth of strains with the Δ(wza-wcaL)6, Δ(wza-wcaL)7, or Δ(wza-wcaM)8 mutation (data not shown). Because wcaM was shown to be important for biofilm formation on tissue cultures (47), we decided to use the larger deletion mutation Δ(wza-wcaM)8, which deletes the wcaM gene, to continue our work.
Fig 1.
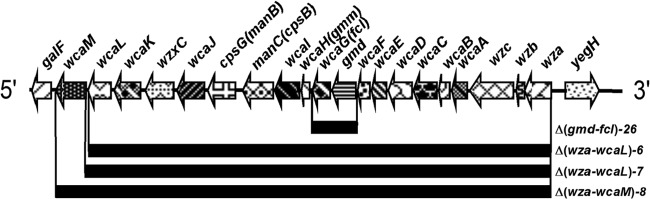
Schematic map showing chromosome deletion of different CA gene mutations. The mutation Δ(gmd-fcl)26 deletes 2 genes, gmd and fcl. Mutations Δ(wza-wcaL)6 and Δ(wza-wcaL)7 delete 19 genes of the CA gene cluster; the former keeps the SD sequence of wcaM, while the latter does not. Mutation Δ(wza-wcaM)8 deletes all 20 genes in the CA gene cluster.
Virulence of strains with CA operon deletion mutations in mice.
The presence or absence of CA did not affect the virulence of the human blood isolate E. coli O4/K54/H5 strain in the rat granuloma pouch, an in vivo model for localized infection, and after intraperitoneal inoculation into mice, a systemic infection model (35). To evaluate the effect of the CA operon deletion mutants on Salmonella virulence, we determined the LD50s of strains with or without the CA mutation in mice (Table 3). The wild-type strain χ3761 was highly virulent, with an LD50 of 1 × 104 CFU, which is consistent with previous reports (61). The strain χ8831 [Δ(gmd-fcl)26] was not effected in virulence (Table 3) (81). The LD50 of strain χ9537 [Δ(wza-wcaM)8] was similar to that of the wild-type strain (Table 3). Strain χ8650 (Δpmi-2426) has an LD50 of over 0.76 × 108 (Table 3) (61). Strain χ9540 [Δpmi-2426 Δ(wza-wcaM)8] has an LD50 of about 107, which is about a 10-times-higher virulence than that of its parent strain χ8650 (Δpmi-2426) (Table 3). The LD50 of χ8868 [Δpmi-2426 Δ(gmd-fcl)26] was between 107 and 108 CFU (Table 3). These results showed that CA operon deletion alone does not affect virulence; however, it slightly increased the virulence when combined with the Δpmi-2426 mutation.
Table 3.
Virulence of Δ(wza-wcaM)8 mutants in 7-week-old BALB/c mice infected by oral inoculationa
| Strain | LD50 (CFU) | Dose (CFU) | No. of survivors/total | Groupb |
|---|---|---|---|---|
| χ3761 (wild type) | < 1 × 104 | 0.9 × 106 | 0/5 | a |
| 0.9 × 105 | 1/5 | |||
| 0.9 × 104 | 0/5 | |||
| 0.9 × 103 | 1/5 | |||
| χ8650 (Δpmi-2426) | >0.76 × 108 | 0.76 × 109 | 0/5 | a |
| 0.76 × 108 | 4/5 | |||
| 0.76 × 107 | 3/5 | |||
| 1.0 × 108 | 3/5 | b | ||
| 1.0 × 107 | 4/5 | |||
| χ8831 [Δ(gmd-fcl)26] | < 8.6 × 104 | 5.9 × 105 | 1/4 | c |
| 5.9 × 104 | 4/4 | |||
| 5.9 × 103 | 4/4 | |||
| 5.9 × 102 | 4/4 | |||
| 8.6 × 106 | 0/4 | d | ||
| 8.6 × 105 | 0/4 | |||
| 8.6 × 104 | 0/4 | |||
| 8.6 × 103 | 1/4 | |||
| χ9537 [Δ(wza-wcaM)8] | <1 × 104 | 0.9 × 106 | 0/2 | e |
| 0.9 × 105 | 0/2 | |||
| 0.9 × 104 | 1/2 | |||
| 1.8 × 106 | 0/5 | a | ||
| 1.8 × 105 | 0/5 | |||
| 1.8 × 104 | 1/5 | |||
| χ8868 [Δ(gmd-fcl)26 Δpmi-2426] | ∼108 | 1.08 × 109 | 2/5 | f |
| 1.08 × 108 | 0/5 | |||
| 1.08 × 107 | 5/5 | |||
| χ9540 [Δpmi-2426 Δ(wza-wcaM)8] | <1.0 × 107 | 1.8 × 108 | 0/2 | e |
| 1.8 × 107 | 0/2 | |||
| 1.8 × 106 | 0/2 | |||
| 1.2 × 109 | 1/5 | a | ||
| 1.2 × 108 | 2/5 | |||
| 1.2 × 107 | 2/5 | |||
| 1.04 × 109 | 1/5 | f | ||
| 1.04 × 108 | 2/5 | |||
| 1.04 × 107 | 2/5 |
All strains were grown in LB broth except strains with the Δpmi-2426 mutation, which were grown in LB broth with 0.1% mannose. The data are from different experiments.
The same letter indicates that experiments were carried on the same batch of mice.
Synthesis of rPspA in attenuated Salmonella vaccine strains with different attenuation characteristics.
To evaluate the effect of the CA operon deletion in recombinant attenuated Salmonella vaccine (RASV) strains, we introduced the Δ(wza-wcaM)8 mutation into two sets of strains derived from either χ9241 or χ9558. RASV strains χ9241 and χ9558 have been successfully used to deliver the pneumococcal surface protein PspA and to induce protective immunity against S. pneumoniae challenge in mice (53, 76). They represented two kinds of attenuation strategies. Strain χ9241 adopts the auxotrophic character of ΔpabA ΔpabB mutations (82). Genes pabA and pabB are two unlinked genes that encode the two subunits of 4-amino-4-deoxy-chorismate synthase, which is required for the production of folic acid in Salmonella. Mutations in either of these genes cause attenuation due to the fact that Salmonella cannot assimilate folic acid from the environment (83). Strain χ9558 is based on using the regulated delayed attenuation strategy (RDAS) (52). In strains with this strategy, bacteria are expected to display features of wild-type virulence of Salmonella at the time of oral vaccination to enable strains to first effectively colonize lymphoid tissues and then exhibit attenuation gradually in vivo. Thus, χ9241 and χ9558 provided two kinds of attenuation backgrounds for evaluating the efficacy of the CA operon deletion in vaccine strains. Both χ9241 and χ9558 carry a ΔasdA deletion to facilitate use of the Asd+ balanced-lethal antigen-encoding plasmid. The Δ(wza-wcaM)8 mutation was introduced into χ9241 and χ9558 to generate the corresponding strains χ11370 and χ9902, respectively. This mutation was also introduced into χ9837, a derivative of χ9558 with the Δlrp-23 mutation, which enhances the invasion of epithelial cells (70), to yield the corresponding strain χ9903. In strains χ9902 and χ9903, the larger CA operon deletion Δ(wza-wcaM)8 replaced the small deletion Δ(gmd-fcl)26 in χ9558 and χ9837. All these strains were then transformed with the recombinant Asd+ plasmid pYA4088, carrying a recombinant pspA gene fused to DNA encoding the β-lactamase signal sequence (76), or the control empty Asd+ plasmid pYA3493 (84).
We then evaluated PspA synthesis by these strains carrying plasmid pYA4088 or pYA3493. There was no PspA synthesis in strains carrying the control plasmid pYA3493. All strains with plasmid pYA4088 can synthesize a 37-kDa protein that specifically reacts with rabbit anti-rPspA antibody, as expected (Fig. 2A). Although in our initial test, we found that PspA synthesis in strain χ11370 [Δ(wza-wcaM)8] decreased 26% compared with that in parent strain χ9241 [(wza-wcaM)+] (Fig. 2A), repeated experiments showed that they produced similar levels of PspA synthesis (Fig. 2B). The PspA level synthesized in strain χ9558 [Δ(gmd-fcl)26] was 18% less than that in strain χ9902 [Δ(wza-wcaM)8] (Fig. 2A). We repeated this experiment 8 times with duplicates. The results confirmed that strain χ9558 produced 18% less PspA than strain χ9902 [Δ(wza-wcaM)8] (P < 0.05) (Fig. 2B). However, strain χ9837 [Δlrp-23 Δ(gmd-fcl)26] produced similar amounts of PspA as did χ9903 [Δlrp-23 Δ(wza-wcaM)8] (P = 0.48) (Fig. 2B). Next, we used GFP to evaluate whether the Δ(wza-wcaM)8 mutation can increase the level of protein synthesis by measuring the ratio of relative fluorescence to OD600 (Table 4). Strain χ9902 [Δ(wza-wcaM)8] generally gave about 3 to 6% more GFP fluorescence than strain χ9558 [Δ(gmd-fcl)26] (P < 0.05). In another comparison, strain χ9903 [Δlrp-23 Δ(wza-wcaM)8] produced 3 to 7% more GFP than its parent strain χ9837 [Δlrp-23 Δ(gmd-fcl)26] (P < 0.05). However, strains χ9241 and χ11370 [Δ(wza-wcaM)8] showed similar levels of GFP syntheses. These results showed that the mutation Δ(wza-wcaM)8 can increase recombinant protein production in RDAS strains, but the amount of increase varied according to the antigen and strain background. Considering the methods for detection of these two antigens, i.e., Western blotting for PspA and fluorescence for GFP, the latter is more sensitive than the former. Thus, a precise method and enough repeated tests are needed to measure the marginal improvement of antigen synthesis in strains with the CA deletion.
Fig 2.
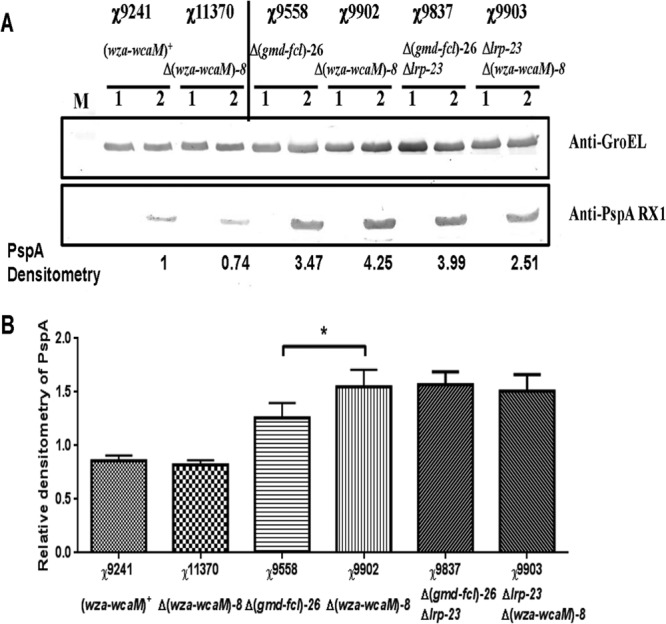
PspA synthesis in S. Typhimurium vaccine strains. (A) The Western blots show PspA synthesis in χ9241, χ11370 [χ9241 Δ(wza-wcaM)8], χ9558, χ9837 (χ9558 Δlrp-23), χ9902 [χ9558 Δ(wza-wcaM)8], and χ9903 [χ9558 Δlrp-23 Δ(wza-wcaM)8] carrying plasmid pYA3493 (vector control) or pYA4088 (specifying PspA amino acids 3 to 285). After induction of PspA synthesis with 1 mM IPTG, strains were continually grown for 4 h at 37°C. Equal numbers of cells from each culture were subjected to SDS-PAGE, transferred to nitrocellulose membranes, and probed with different polyclonal antibodies specific for either PspA or GroEL. GroEL was used as a standardization marker. Relevant portions of each blot are shown. The bands were normalized according to the densitometry of χ9241. Lanes 1, control vector pYA3493; lanes 2, pspA expression vector pYA4088. (B) Relative densitometry of PspA synthesis in different strains. The results are averages from 8 independent experiments with duplicates. The densitometry of χ9241 or χ9558 was used for normalizing bands from strains derived from χ9241 or χ9558, respectively. *, P < 0.05 by paired two-tailed t test.
Table 4.
GFP protein synthesis levelsa
| Strain (with gfp expression plasmid) | Relevant genotype | Relative synthesis level (fluorescence/OD600)b in expt: |
||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| Control | 48.2 ± 0.6 | 32.8 ± 2.4 | 56.8 ± 2.5 | |
| χ9241 | Wild type | 748.6 ± 22.0 | 860.7 ± 26.2 | 743.4 ± 20.3 |
| χ11370 | Δ(wza-wcaM)8 | 789.9 ± 17.9 | 853.0 ± 30.1 | 754.9 ± 24.7 |
| χ9558 | Δ(gmd-fcl)26 | 998.9 ± 57.4 | 1,221.1 ± 28.0 | 947.1 ± 40.0 |
| χ9902 | Δ(wza-wcaM)8 | 1,031.2 ± 53.9 | 1,287.6 ± 47.3** | 1,008.7 ± 46.0* |
| χ9837 | Δlrp Δ(gmd-fcl)26 | 987.0 ± 59.9 | 1,146.6 ± 21.5 | 938.1 ± 39.1 |
| χ9903 | Δlrp Δ(wza-wcaM)8 | 1,059.1 ± 41.9* | 1,224.9 ± 22.3** | 1,035.4 ± 51.2** |
The synthesis of the GFP proteins in strains with or without the Δ(wza-wcaM)8 mutation was compared. After induction of GFP synthesis with 1 mM IPTG, cultures were continually grown at 25°C for 5 h with agitation, and readings were taken every hour to monitor the fluorescence intensity and cell density (optical density at 600 nm [OD600]).
The values represent average fluorescence/OD600 values at 4 h calculated from sextuple or octuple samples from three independent experiments; errors are standard deviations. *, P< 0.05; **, P< 0.01 [for comparison between strains with or without mutation Δ(wza-wcaM)8].
Immune responses in strains attenuated by auxotrophic traits.
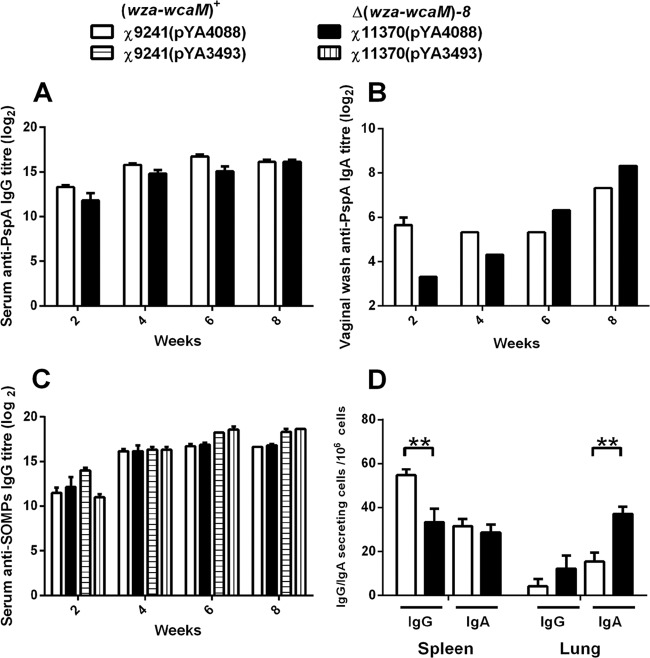
We first evaluated the effect of the CA operon deletion mutation on the immunogenicity of auxotrophic attenuated strain χ9241. Strain χ11370 is derived from χ9241 with the CA operon deletion mutation Δ(wza-wcaM)8. Both RASV strains carrying pYA4088 induced similar anti-PspA serum IgG responses at 8 weeks (Fig. 3A). Strain χ11370 [Δ(wza-wcaM)8] induced 4- or 2-times-lower vaginal IgA antibody responses at 2 and 4 weeks, respectively, but 2-times-higher responses at 6 and 8 weeks (Fig. 3B). The vaginal IgA responses in mice induced by χ11370 [Δ(wza-wcaM)8] slowly increased during 8 weeks, and the increase of IgA responses were faster in mice immunized with χ11370 than in mice immunized with its parent strain χ9241 (Fig. 3B). These results showed that strains with the CA operon deletion can slightly increase the mucosal immune responses against a heterologous antigen delivered by RASV strains. No anti-PspA antibodies were detected in sera or vaginal secretions of mice immunized with strains with the control plasmid pYA3493, which did not specify synthesis of rPspA (data not shown). Although the CA operon was deleted in χ11370, this did not change the anti-SOMP responses compared with those for strain χ9241 with the wild-type CA operon (Fig. 3C), which indicated that the CA operon deletion does not compromise the ability of the vaccine strain to induce antibody responses against Salmonella. At 6 and 8 weeks, the anti-SOMP IgG titer in mice immunized with the same strains containing empty vectors is slightly higher than that in mice immunized with the strains containing PspA expression vectors, which was observed in other papers published from our lab (85). This was mainly due to the growth advantage that the strain harboring the empty vector has over that harboring a plasmid specifying antigen synthesis.
Fig 3.
Immune responses against PspA in mice immunized with RASV strains attenuated by mutations conferring auxotrophy. Serum and mucosal antibody titers in pooled samples from mice orally immunized with approximately 1 × 109 CFU of attenuated Salmonella vaccine strains harboring either control plasmid or pspA-encoding plasmid were determined by ELISA. (A) Serum IgG against rPspA; (B) mucosal IgA in vaginal wash against rPspA; (C) serum IgG against S. Typhimurium OMPs. (D) Numbers of IgG- and IgA-producing cells in spleens and lungs were determined by ELISPOT assay. Splenocytes and lung cells were harvested from 3 mice per group at 7 days after the boost immunization. The results from each well are expressed as spots per million splenocytes or lung cells minus background (typically ≈15 spots) from cells unstimulated with rPspA. Significant differences between groups are indicated and were determined using two-way ANOVA and Bonferroni tests. **, P < 0.01.
The serum immune responses to rPspA were further examined by measuring the levels of IgG isotype subclasses IgG1 and IgG2a, which are indicators of Th2 cells directing the humoral response or Th1 cells directing the cellular immunity (86, 87). Th1-type dominant immune responses are frequently observed after immunization with attenuated Salmonella strains (88–90). All strains induced high IgG1 and IgG2a responses, indicating a mixed Th1 and Th2 response (see Fig. S1 in the supplemental material). The IgG2a titers were slightly higher than IgG1 titers induced by both strains χ9241(pYA4088) and χ11370(pYA4088) at different weeks postimmunization, indicating that the reaction is slightly skewed to the Th1 response. These results showed that the CA operon deletion mutation did not affect the Th1/Th2 balance.
We further evaluated the presence of PspA-specific IgG and IgA ASCs in spleens and lungs at 7 weeks, i.e., 7 days after boosting at 6 weeks. Strain χ11370 [Δ(wza-wcaM)8] induced lower numbers of IgG ASCs and similar numbers of IgA ASCs as the numbers induced by the parent strain χ9241 in spleens. However, strain χ11370 induced similar numbers of IgG ASCs to and significantly higher numbers of IgA ASCs than strain χ9241 in lungs (P < 0.01) (Fig. 3D). The ASCs are responsible for recall immune responses that confer protection against infection. The generation of IgA ASCs in the lung is important to fight against S. pneumoniae infection since lungs are the primary invasion sites of S. pneumoniae, and mouse lung has IgA-bearing lymphocytes that help against pneumococcal infection through an IgA-driven mechanism (91). The higher IgA ASC responses in lungs at 7 weeks were correlated with the slightly enhanced IgA responses in vaginal washes at 6 and 8 weeks (Fig. 3B). Thus, these results suggested that the CA operon deletion mutation helps to generate better mucosal immune responses.
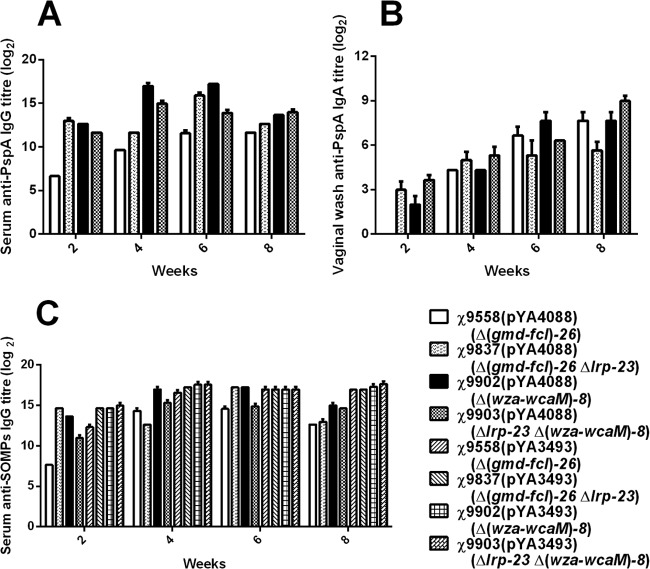
Antibody responses in strains with RDAS.
We further evaluated the effects of the CA operon deletion mutation on immunogenicity in strains with RDAS. At 8 weeks, all RASV strains carrying pYA4088 induced strong anti-PspA serum IgG responses (Fig. 4A). Strain χ9902(pYA4088) [Δ(wza-wcaM)8] induced 64-times-higher anti-PspA antibody titers than its parent strain χ9558(pYA4088) at all weeks except at 8 weeks, when it induced a 4-times-higher titer (Fig. 4A). Strain χ9837(pYA4088) with the Δlrp-23 mutation also induced 64-times-higher anti-PspA antibody responses than parent strain χ9558(pYA4088) at 2 and 6 weeks. Strain χ9903(pYA4088) [Δlrp-23 Δ(wza-wcaM)8] induced 32-times-higher antibody responses than χ9558(pYA4088) at 2 and 4 weeks but lower responses than strain χ9902(pYA4088) except at week 8 and lower responses than strain χ9937(pYA4088) at weeks 2 and 6 (Fig. 4A). Strain χ9558(pYA4088) induced undetectable IgA responses against PspA at week 2, whereas all other strains induced detectable responses. At week 8, strain χ9903(pYA4088) induced 8 times more IgA than χ9837(pYA4088) (Fig. 4B). Notably, strain χ9902 [Δ(wza-wcaM)8] with the CA operon deletion induced higher anti-PspA IgG and IgA antibody titers than χ9558 at 2 weeks (Fig. 4A and B). No anti-PspA antibodies were detected in sera or vaginal secretions of mice immunized with control strains that did not express pspA. All immunized mice developed high titers against S. Typhimurium SOMPs (Fig. 4C). The SOMP responses showed trends similar to those for the anti-PspA antibody responses, with strain χ9902(pYA4088) inducing higher antibody response than any other strain harboring plasmid pYA4088 at 8 weeks (Fig. 4C). Strains χ9902(pYA4088), χ9837(pYA4088), and χ9903(pYA4088) induced higher anti-SOMP responses than χ9558(pYA4088) at 2 weeks. The anti-SOMP antibody titers induced by strain χ9902(pYA4088) were similar to those induced by the same strain harboring the empty plasmid pYA3493 at 2, 4, and 6 weeks. The same trend was seen in strain χ9837 at 2 and 6 weeks.
Fig 4.
Immune responses against PspA in mice immunized with RASV strains attenuated by RDAS. Serum IgG responses to rPspA (A) and to S. Typhimurium OMPs (C) and vaginal wash IgA responses to rPspA (B) were measured by ELISA. The data represent reciprocal anti-IgG antibody levels in pooled sera from mice orally immunized with attenuated Salmonella carrying either plasmid pYA3493 (control) or pYA4088 (specifying rPspA) at the indicated weeks after immunization. The mice were inoculated with 2.135 × 109 CFU χ9558(pYA3493), 1.275 × 109 CFU χ9558(pYA4088), 1.94 × 109 CFU χ9837(pYA3493), 2.075 × 109 CFU χ9837(pYA4088), 2.16 × 109 CFU χ9902(pYA3493), 1.645 × 109 CFU χ9902(pYA4088), 2.12 × 109 CFU χ9903(pYA3493), or 2.075 × 109 CFU χ9837(pYA4088) and boosted at 6 weeks with 2.22 × 109 CFU χ9558(pYA3493), 2.14 × 109 CFU χ9558(pYA4088), 2.24 × 109 CFU χ9837(pYA3493), 2.47 × 109 CFU χ9837(pYA4088), 1.99 × 109 CFU χ9902(pYA3493), 2.19 × 109 CFU χ9902(pYA4088), 2.025 × 109 CFU χ9903(pYA3493), or 2. 5 × 109 CFU χ9837(pYA4088), respectively. Error bars represent variation between triplicate wells.
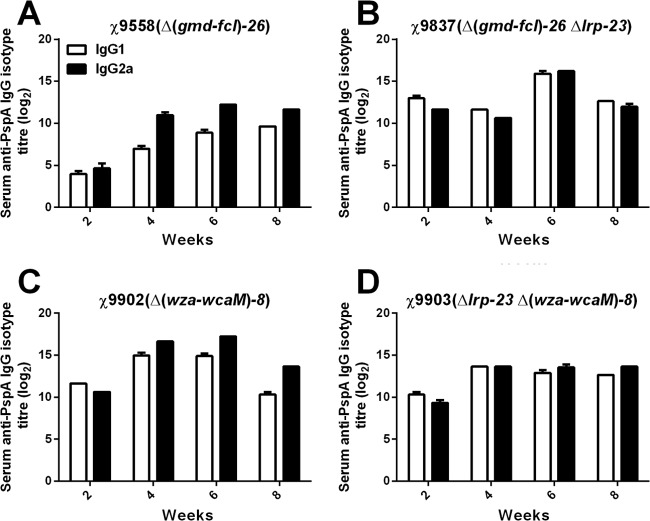
IgG1 and IgG2a in strains with RDAS.
We further measured the IgG1/IgG2a responses against PspA (86, 87) in strains with RDAS. Mice immunized with attenuated Salmonella strains usually generate a Th1-type dominant immune response (88–90). We observed that all strains induced high IgG1 and IgG2a responses, indicating a mixed Th1 and Th2 response against PspA (Fig. 5A to D). The IgG2a titers were slightly higher than IgG1 titers in strains χ9558(pYA4088) and χ9902(pYA4088) at weeks 4, 6, and 8 postimmunization, indicating that the reaction is slightly skewed to the Th1 response (Fig. 5A and C). Strain χ9837(pYA4088) induced higher IgG1 than IgG2a titers at 2 weeks, indicating a Th2 type response at early times, and then it changed to a balanced Th1/Th2 response (Fig. 5B); similar results were seen with χ9903(pYA4088) (Fig. 5D). These results showed that deletion of the CA operon did not affect the Th1/Th2 balance; however, inclusion of the Δlrp-23 mutation slightly skewed to a Th2 response at 2 weeks and resulted in a more balanced Th1/Th2 response at 8 weeks. Consistent with previous results, strains χ9902(pYA4088), χ9837(pYA4088), and χ9903(pYA4088) induced higher IgG1 and IgG2a responses than χ9558(pYA4088) at 2 weeks after vaccination (Fig. 5).
Fig 5.
Serum IgG1 and IgG2a responses to rPspA measured by ELISA. The data represent IgG1 and IgG2a subclass antibody levels to rPspA in pooled sera from orally immunized mice at various numbers of weeks after immunization. Error bars represent variation between triplicate wells. The inoculation doses were same as for Fig. 4.
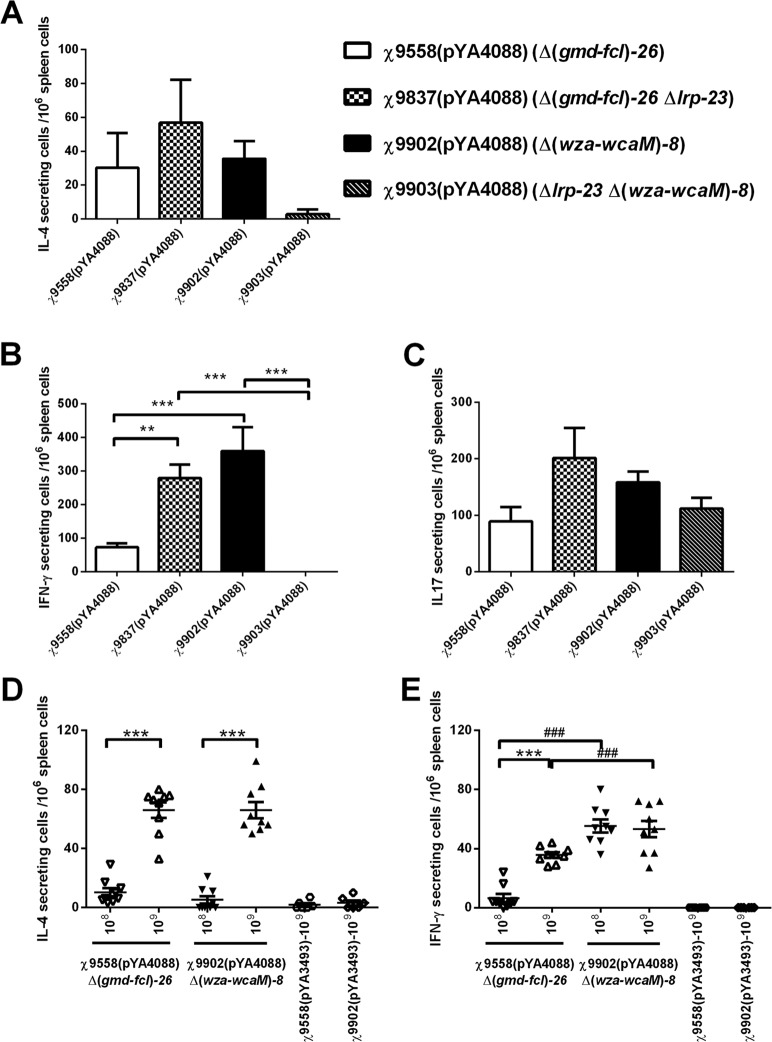
Antigen-specific stimulation of IL-4 or IFN-γ production in strains with RDAS.
We further examined PspA-specific responses in mice immunized with strains χ9558 and χ9837 and their derivatives χ9902 and χ9903 with the CA operon deletion mutation. ELISPOT assays were used to compare PspA stimulation of IFN-γ (Th1 associated), IL-4 (Th2 associated), and IL-17 (Th17 associated) production by spleen cells taken from immunized and control mice at week 7 (Fig. 6). The numbers of PspA-specific IL-4-secreting cells were similar in spleens from mice immunized with different strains, with χ9837 giving slightly higher numbers than others (Fig. 6A), while the number of PspA-specific IFN-γ-secreting cells in mice immunized with strain χ9902 was significantly higher than that in mice immunized with strains χ9558 and χ9903 (P < 0.05) (Fig. 6B). Strain χ9837 also generated more IFN-γ-secreting cells than strains χ9558 and χ9903 (P < 0.05) (Fig. 6B). The four strains induced similar numbers of IL-17-secreting cells, which were similar to the numbers of IL-4 secreting cells (Fig. 6C).
Fig 6.
PspA-specific stimulation of IL-4 (A and D)-, IFN-γ (B and E)-, and IL-17 (C)-producing cells in mice immunized with χ9558(pYA4088), χ9837(pYA4088), χ9902(pYA4088), or χ9903(pYA4088). Numbers of secreting cells were determined by ELISPOT assays. Splenectomies were performed on euthanized mice 7 days after the boosting immunization. Mice immunized with the same strains harboring the control plasmid pYA3493 were included as controls. Splenocytes were harvested from 3 mice per group, and cells from each spleen were assayed in triplicate. Each symbol represents the results from a single well. The results from each well are expressed as spots per million splenocytes or lung cells minus background (typically ≈15 spots) from cells unstimulated with rPspA. The inoculation doses for panels A, B, and C were same as for Fig. 4. The inoculation doses for panels D and E were 2.02 × 109 CFU χ9558(pYA3493), 2.18 × 108 or 2.18 × 109 CFU χ9558(pYA4088), 2.13 × 109 CFU χ9902(pYA3493), and 2.47 × 108 or 2.47 × 109 CFU χ9902(pYA4088), and boost doses were 1.59 × 109 CFU χ9558(pYA3493), 1.46 × 108 or 1.46 × 109 CFU χ9558(pYA4088), 1.43 × 109 CFU χ9558(pYA3493), and 1.18 × 108 or 1.18 × 109 CFU χ9902(pYA4088), respectively. **, P < 0.01; ***, P < 0.001. ###, compared with same dose, P < 0.001.
We further checked whether mice immunized with different doses (108 to 109 CFU) of strains χ9558 and χ9902 could generate different numbers of IL-4- and IFN-γ-secreting cells. The number of PspA-specific IL-4 secreting cells induced by both strains increased with escalating dose. In each strain, the dose of 109 CFU induced significantly higher numbers of IL-4-secreting cells than the dose of 108 CFU. Strain χ9902 induced similar numbers of IL-4-secreting cells as did χ9558 at the same dose (Fig. 6D) but induced significantly higher numbers of PspA-specific IFN-γ-secreting cells than χ9558 at the same dose (Fig. 6E). The number of IFN-γ-secreting cells induced by χ9558 increased as the dose increased from 108 to 109 CFU, while for χ9902, the dose of 108 CFU was enough to induce the highest IFN-γ response. The two strains with the control vector pYA3493 did not induce either PspA-stimulated IL-4- or IFN-γ-secreting cells. Thus, strain χ9902 is more potent to induce IFN-γ responses.
Protection.
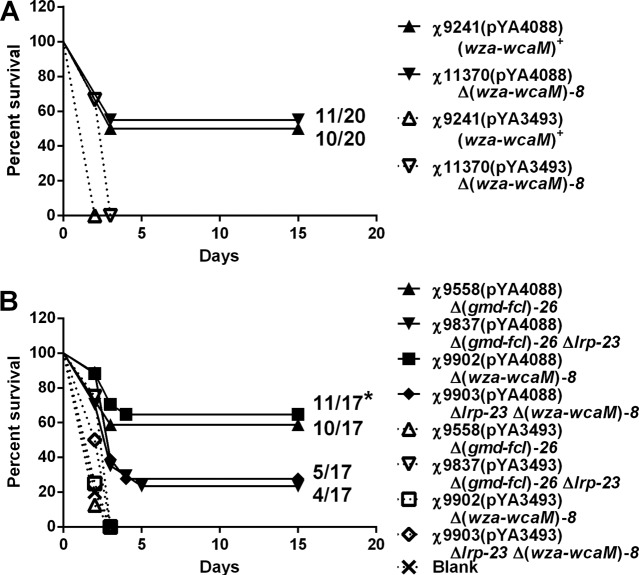
When vaccinated mice were challenged with virulent S. pneumoniae WU2, all groups that received strains carrying the pspA expression vector pYA4088 were significantly protected compared to those receiving the same strain carrying the empty vector pYA3493, except for χ9837(pYA4088) (P < 0.01) (Fig. 7). In the first group of experiments, two strains, χ9241 and χ11370 [Δ(wza-wcaM)8], induced similar protection in mice, with survival of 50% and 55%, respectively, at the endpoint (Fig. 7A). In the second experiment, mice were immunized with strain χ9558 and its derivatives. Protection levels in mice vaccinated with strains χ9558(pYA4088) and χ9902(pYA4088) were similar, with about 58.8% and 64.7%, respectively, surviving the challenge (Fig. 7B). However, only 23.5% and 29.4%, respectively, of the mice vaccinated with strains χ9837(pYA4088), carrying a Δlrp-23 mutation in χ9558, and χ9903(pYA4088), carrying a Δ(wza-wcaM)8 mutation in χ9837, survived the challenge (Fig. 7B). The percentage of survival in mice vaccinated with strain χ9902(pYA4088) was significantly greater than that in mice vaccinated with χ9903(pYA4088) (P < 0.05). These results showed that strains with the CA operon deletion mutation did not affect protection; however, strains with the Δlrp-23 mutation resulted in reduced protection.
Fig 7.
Survival curves for intraperitoneal challenge with virulent S. pneumoniae strain WU2. Female BALB/c mice were immunized with the indicated strains. Ten weeks after immunization, mice were challenged with 100 times the LD50 of virulent S. pneumoniae WU2. (A) Mice immunized with χ9241 or χ11370 [Δ(wza-wcaM)8] harboring different plasmids. The protection levels are similar in mice immunized with strains with or without CA operon deletion and are significantly different from those for the vector control (P < 0.001 for χ9241, P < 0.02 for χ11370). (B) Mice immunized with χ9558 [Δ(gmd-fcl)26], χ9837 (Δlrp-23), χ9902 [Δ(wza-wcaM)8], or χ9903 [Δ(wza-wcaM)8 Δlrp-23] harboring different plasmids. PspA-expressing strains were significantly protected compared with the same strain containing the control vector pYA3493 (P < 0.05), except for χ9837(pYA4088), which induced protection but at levels not statistically significantly different from those for the controls that gave no protection. Mice vaccinated with strain χ9902(pYA4088) showed significantly greater protection than mice vaccinated with χ9903(pYA4088) or χ9837(pYA4088) (P < 0.05). *, P < 0.05.
DISCUSSION
CA is a polysaccharide comprised of repeating subunits. It is believed to be required for biofilm formation on tissue culture cells in S. Typhimurium (47). It was reported that a CA-deficient mutant of an extraintestinal E. coli O4/K54/H5 pathovar was not affected in virulence in two in vivo models (35). We first compared the virulence of strains with a CA operon deletion mutation. Compared with the parent strain χ3761, the single CA operon deletion strain χ9537 with the Δ(wza-wcaM)8 mutation did not have a change in virulence. However, compared with the parent strain χ8650 (Δpmi-2426), which lack O-antigen synthesis in vivo, the double mutation [Δpmi-2426 Δ(wza-wcaM)8] increased the virulence about 10-fold. CA affects the surface of the bacteria. It was reported that the overproduction of CA in S. Typhimurium inhibits phagocytosis (92), which is one of the relevant factors that contributes to the attenuation of constitutively activated rcsC mutants (13). Considering that CA is encoded by a large operon containing 20 genes and confers a strong negative charge to the cell surface (35), the deletion of the CA operon may have two effects on virulence. It may reduce the metabolic burden of CA synthesis in vivo and may increase the interaction between bacteria and host cells, which facilitate the bacteria entering cells to result in increased virulence in vivo. However, we did not observe significant differences in attachment and invasion between wild-type and CA mutant strains in cell culture experiments (data not shown). Thus, further experiments are needed to address these hypotheses.
Further, we found that the whole CA operon deletion Δ(wza-wcaM)8 resulted in increased levels of PspA synthesis in the χ9558 background and GFP synthesis in the χ9558 and χ9837 backgrounds but not in the χ9241 background. There are several possible reasons. First, it was reported (93) that eliminating synthesis of CA can increase gene expression from plasmids, because CA inhibits RNA polymerase activity and thus RNA synthesis. Removing CA leads to about 2.2- to 4.4-fold-increased reporter gene expression in organs of mice (93). Second, compared with the Δ(gmd-fcl)26 deletion mutation, with only 2 genes deleted, mutation Δ(wza-wcaM)8, the whole operon deletion with 20 genes deleted, directly eliminates expression of more genes and results in a reduced overall metabolic burden. Third, the increased protein synthesis may be related to the metabolic flow directly caused by the whole-operon deletion. The Δ(gmd-fcl)26 mutation deleted only two genes related to GDP-mannose convertible to GDP-fucose. The strain with this mutation may accumulate GDP-mannose and other possible intermediate products that are redirected to other metabolic pathways to be assimilated. However, strains with the CA operon deletion will not have this problem of redirection or assimilation of intermediates and lead to a reduced metabolic burden. All these factors may contribute to the increased capacity of RASV cells to synthesize more heterologous protective antigen. We also notice, however, that this phenotype is not universal in all strains. Although we could not see the increase of PspA synthesis in the χ9241 and χ9837 backgrounds, we did see the increase of GFP synthesis in the χ9558 and χ9837 backgrounds. We inferred that this may be related to the characterization of the protective antigen, strain backgrounds, and other, unknown gene regulation circuits. Most functions of the gene products specified by the Salmonella CA operon were deduced from sequence comparison with the CA operon in E. coli and were not fully characterized. As mentioned above, at least six genes involved in the synthesis of GDP-fucose in Salmonella have different donor species compared with E. coli (4). These genes might evolve slightly different regulations or functions other than their E. coli compartment. They may also specify synthesis of other membrane components unique to Salmonella. This needs to be probed in the future. Another possible reason may be the effect of Lrp. The function of Lrp is related to the cellular metabolism and the nutritional state of the environment. Lrp represses rRNA synthesis (94). Heterologous protein synthesis is inversely related to the transcription of lrp (95). Thus, the Δlrp-23 mutation allows the maximum upregulation of substrate uptake genes in rich medium, leading to greater antigen synthesis. This mechanism might have some overlap with Δ(wza-wcaM)8, leading to the results we observed.
There have been few studies reported concerning the effect of a CA deletion mutation on the induction of immunity. The presence of CA capsules on E. coli conferred little resistance to the bactericidal activity of human serum or phagocytic uptake and did not protect against intracellular killing by polymorphonuclear leukocytes (96). We wanted to see whether the effect of the CA deletion on immune responses was universal or not in different strain backgrounds. Thus, we evaluated the CA deletion mutation in strains with two different attenuation mechanisms, one through mutations in the auxotrophic genes and another through regulated delayed in vivo attenuation. Our results showed that the IgA levels in mice immunized with strain χ9241, attenuated by mutations in the auxotrophic genes, were similar to those in our previous report, with the antibody being highest at 8 weeks (76). However, strain χ11370 [χ9241 Δ(wza-wcaM)8] induced lower levels of IgA at 2 and 4 weeks although higher levels at 6 and 8 weeks. Strains without or with the CA deletion mutation have similar abilities to stimulate the systemic humoral immune responses, since mice immunized with both strains generated similar titers of anti-SOMP antibodies. However, the vaginal IgA is from both systemic and local production (97). The IgA levels in the genital tract are subject to a strong hormonal control that regulates the transportation of immunoglobulins, the level of cytokines, the distribution of various cell populations, and antigen presentation during the reproductive cycle as well as a compartmentalization of the immune response within the genital tract (97, 98). All these factors may affect the level of IgA at a given time point. To induce effective IgA responses, two doses were needed, while one dose failed by vaginal immunization (7). Thus, the lower antibody levels at 2 and 4 weeks in mice immunized with strain χ11370 (χ9241 Δ(wza-wcaM)8] may reflect that there is not enough interaction between the antigen carried by this strain with the host vaginal immune system. With the time lapses, there was more contact, which led to increments of the IgA responses.
We found that introducing the Δ(wza-wcaM)8 deletion into χ9558 to replace the Δ(gmd-fcl)26 mutation can significantly increase the IgG and secretory IgA antibody responses at 2 weeks (Fig. 6), a prominent advantage in developing RASVs for use in newborns, who require induction of early antibody responses. It also relates to the higher antibody response against SOMPs at 2 weeks (Fig. 6). These may be related to the increased antigen synthesis and/or to the reduced metabolic burden conferred by the Δ(wza-wcaM)8 mutation. We also found increased levels of vaginal IgA at 8 weeks and antigen-specific IgA-secreting cells in the mouse lung at 7 weeks after immunization with strain χ11370 [χ9241 Δ(wza-wcaM)8] (Fig. 3) and increased IFN-γ-secreting cells at 7 weeks in mice immunized with strain χ9902 [χ9558 Δ(wza-wcaM)8] (Fig. 6). At doses of 108 and 109 CFU, strain χ9902 (χ9558 Δ(wza-wcaM)8] induced higher IFN-γ levels than χ9558. Strain χ9837 also induced earlier antibody responses than χ9558 at 2 weeks (Fig. 6). This may relate to the hyperinvasive ability of strains with the Δlrp-23 mutation. We did not observe the expected superimposed positive effects of the Δ(wza-wcaM)8 and Δlrp-23 mutations on the immune responses. One explanation is that there may be some overlapping of their functions. As we mentioned above, we tried to see if strains with the Δ(wza-wcaM)8 mutation have enhanced invasion ability. However, we observed only a marginal difference between strains with or without the Δ(wza-wcaM)8 mutation (data not shown). Another reason may relate to Lrp. It is a dual transcriptional regulator and antivirulence gene product responsible for altering expression of 10% of the genes relating to biosynthesis, nutrient transportation, and DNA packaging (70, 99); some of the downstream genes may lead to the poor T cell responses and protection (Fig. 6 and 7) and even eliminate the benefit of the Δ(wza-wcaM)8 mutation. Compared to the benefit of including a single mutation, either Δlrp-23 or the Δ(wza-wcaM)8, the additional benefit of including both mutations in a strain was not prominent, with the observation that strain χ9903 showed antibody responses higher than those of χ9558 but similar to those of χ9837 at 2 weeks (Fig. 5). We observed the increment of IgA and IgG responses in mice immunized with χ9902; whether this could lead to better protection if we challenged the mice earlier with an S. pneumoniae strain that colonizes the nasal pharynx is not clear. Such experiments will give more evidence of the benefit of CA deletion mutation. Generally, the mutation Δ(wza-wcaM)8 helped to generate better immune responses, especially mucosal responses, though the kinetics of antibody responses in vaccine strains with different attenuation mechanisms varied.
Although inclusion of Δlrp-23 increases invasiveness in some strain backgrounds (70), strain χ9837 (χ9558 Δlrp-23) induced a lower vaginal IgA response at 8 weeks and strain χ9903 [χ9558 Δlrp-23 Δ(wza-wcaM)8] induced lower PspA-specific IFN-γ and IL-4 responses. Strains χ9902 and χ9903, with the same genotypes except the Δlrp-23 mutation, produced similar amounts the PspA and similar levels of anti-PspA responses; strain χ9903 induced a significantly lower level of IFN-γ secreting cells. These weaknesses in strains χ9837 and χ9903 may lead to the poor protection observed with these strains. Compared with them, strain χ9902 induced decent anti-PspA IgG and IgA responses, as well as a better IFN-γ response. Given these considerations, strain χ9902 is superior to strains χ9837 and χ9903.
When testing the strains with the Δ(wza-wcaM)8 mutation, the differences in protective antigen production in strains attenuated with autotrophic characters is not as much as observed in strains with RDAS. Also, only at 8 weeks, we could see a higher IgA antibody response in χ11370 with the Δ(wza-wcaM)8 mutation than in its parent. This contrasts to the case for strains with RDAS, which induced increased titers of anti-PspA and anti-SOMP IgG and IgA at 2 weeks. A possible reason is that this is related to the strain backgrounds. There are two hypotheses. (i) The function of the Δ(wza-wcaM)8 mutation may be pronounced only when combined with mutations that affect the cell surface. Strain χ9241 has only 5 mutations, with no mutation related to the modification of cell surface molecules; however, strain χ9558 with RDAS has 10 mutations, with a mutation, Δpmi-2426, affecting the cell surface molecule O antigen. Although both strains are used as attenuated vaccine vectors, strain χ9558 is more attenuated than strain χ9241. As we observed (Table 3), a strain with both the Δ(wza-wcaM)8 and Δpmi-2426 mutations showed higher virulence. This might be one of the reasons why the Δ(wza-wcaM)8 mutation performs differently in different backgrounds. (ii) The function of the Δ(wza-wcaM)8 mutation may be related to the number of mutations a strain has. The generation of mutations in the chromosome disrupts the normal physiological state of bacteria. The more mutations are added, the more stress is exerted on the bacteria. In this case, one mutation may negatively affect the performance of another mutation. Thus, the balance among multiple mutations should be carefully evaluated. Optimization of different combinations of mutations is necessary to achieve optimal results (100). More experiments are required to clarify which is the correct reason or whether both reasons play a role in the performance of the Δ(wza-wcaM)8 mutation.
It was reported that CA is a main contaminant present in plasmid compositions used as DNA vaccines or for gene therapy (93). The CA component in nucleic acid preparations used for gene therapy can induce toxic effects in humans and other mammals (93). It is difficult to separate CA from nucleic acids using current standard purification procedures, even from a clinical-grade current good manufacturing practice (cGMP) preparation. A CA-degrading enzyme from phage NST1 was therefore used to remove CA in nucleic acid preparations for gene therapy. There are several phages that encode an enzyme that can degrade CA (93, 101, 102). Since CA overproduction prevents phage infection (103), an additional benefit of deletion of the CA operon in Salmonella vaccine strains is to increase its lysis by bacteriophages, which help to reduce the persistence of bacteria in vitro to increase the biocontainment character. Since CA expression is induced by antibiotic treatments (21), deletion of the CA operon may facilitate elimination of the vaccine strain by antibiotics, if necessary.
We are focusing on developing vaccine strains with the RDAS background. RDAS is a creative and effective way to increase vaccine efficacy and safety (52, 56). Results from a phase I clinical trial with three RDAS S. Typhi vaccine strains showed that they were safe (data not shown). The three RDAS S. Typhi vaccine strains have same genotypes as in S. Typhimurium vaccine strain χ9558 (54). In continuing efforts to increase the vaccine efficacy and safety of S. Typhimurium vaccine strain χ9558 and its isogenotype S. Typhi vaccine strains, we found several candidate mutations. One is the Δ(wza-wcaM)8 mutation, which was used to replace the Δ(gmd-fcl)26 mutation. Another candidate mutation, Δlrp, also showed positive effects in our preliminary screenings (70). We evaluated them in S. Typhimurium vaccine strain χ9558, due to its complex genetic background, with the new mechanism to achieve attenuation as well as in strain χ9241, with a simpler genetic background, to get a systematic comparison. In this report, we showed that the Δ(wza-wcaM)8 mutant derived from χ9558 resulted in a higher recombinant protein synthesis level, higher IgG and IgA responses at 2 weeks after immunization, and better IFN-γ secretion (Fig. 3 to 6). In terms of protection, the immunized mice were challenged well after antibody titers induced by the two vaccine strains with and without the Δ(wza-wcaM)8 mutation were very similar (Fig. 7). However, based on the induction of higher levels of immune responses soon after immunization, we would expect to see a higher level of protection induced by strains with the Δ(wza-wcaM)8 mutation when challenge was 2 to 3 weeks after immunization. These results provide evidence of the benefits of including the Δ(wza-wcaM)8 mutation in S. Typhimurium χ9558 and support incorporation of the Δ(wza-wcaM)8 mutation into our final S. Typhi vaccine constructions with RDAS to further increase the vaccine efficiency.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (R01 AI24533, R01 AI057885, and R01 AI056289) and the Bill and Melinda Gates Foundation (no. 37863).
We thank Susan Hollingshead (University of Alabama at Birmingham) for providing the WU2 strain and the plasmid for purification of PspA protein. We thank Jacquelyn Kilbourne for her help with animal experiments and Tina Hartig for laboratory support.
Footnotes
Published ahead of print 17 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00097-13.
REFERENCES
- 1.Grant WD, Sutherland IW, Wilkinson JF. 1969. Exopolysaccharide colanic acid and its occurrence in the Enterobacteriaceae. J. Bacteriol. 100:1187–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts IS. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50:285–315 [DOI] [PubMed] [Google Scholar]
- 3.Garegg PJ, Lindberg B, Onn T, Sutherland IW. 1971. Comparative structural studies on the M-antigen from Salmonella typhimurium, Escherichia coli and Aerobacter cloacae. Acta Chem. Scand. 25:2103–2108 [DOI] [PubMed] [Google Scholar]
- 4.Stevenson G, Lan R, Reeves PR. 2000. The colanic acid gene cluster of Salmonella enterica has a complex history. FEMS Microbiol. Lett. 191:11–16 [DOI] [PubMed] [Google Scholar]
- 5.Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75:39–68 [DOI] [PubMed] [Google Scholar]
- 6.Stevenson G, Andrianopoulos K, Hobbs M, Reeves PR. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 8.Ionescu M, Belkin S. 2009. Overproduction of exopolysaccharides by an Escherichia coli K-12 rpoS mutant in response to osmotic stress. Appl. Environ. Microbiol. 75:483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen MH, Takeda S, Yamada H, Ishii Y, Yamashino T, Mizuno T. 2001. Characterization of the RcsC→YojN→RcsB phosphorelay signaling pathway involved in capsular synthesis in Escherichia coli. Biosci. Biotechnol. Biochem. 65:2364–2367 [DOI] [PubMed] [Google Scholar]
- 10.Gottesman S, Stout V. 1991. Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol. Microbiol. 5:1599–1606 [DOI] [PubMed] [Google Scholar]
- 11.Stout V. 1994. Regulation of capsule synthesis includes interactions of the RcsC/RcsB regulatory pair. Res. Microbiol. 145:389–392 [DOI] [PubMed] [Google Scholar]
- 12.Takeda S, Fujisawa Y, Matsubara M, Aiba H, Mizuno T. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC → YojN → RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol. 40:440–450 [DOI] [PubMed] [Google Scholar]
- 13.García-Calderón CB, García-Quintanilla M, Casadesús J, Ramos-Morales F. 2005. Virulence attenuation in Salmonella enterica rcsC mutants with constitutive activation of the Rcs system. Microbiology 151:579–588 [DOI] [PubMed] [Google Scholar]
- 14.Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59:379–405 [DOI] [PubMed] [Google Scholar]
- 15.Navasa N, Rodríguez-Aparicio L, Martínez-Blanco H, Arcos M, Ferrero MA. 2009. Temperature has reciprocal effects on colanic acid and polysialic acid biosynthesis in E. coli K92. Appl. Microbiol. Biotechnol. 82:721–729 [DOI] [PubMed] [Google Scholar]
- 16.Reid AN, Whitfield C. 2005. Functional analysis of conserved gene products involved in assembly of Escherichia coli capsules and exopolysaccharides: evidence for molecular recognition between Wza and Wzc for colanic acid biosynthesis. J. Bacteriol. 187:5470–5481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danese PN, Pratt LA, Kolter R. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Lee SM, Mao Y. 2004. Protective effect of exopolysaccharide colanic acid of Escherichia coli O157:H7 to osmotic and oxidative stress. Int. J. Food Microbiol. 93:281–286 [DOI] [PubMed] [Google Scholar]
- 19.Prigent-Combaret C, Lejeune P. 1999. Monitoring gene expression in biofilms. Methods Enzymol. 310:56–79 [DOI] [PubMed] [Google Scholar]
- 20.Ophir T, Gutnick DL. 1994. A role for exopolysaccharides in the protection of microorganisms from desiccation. Appl. Environ. Microbiol. 60:740–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sailer FC, Meberg BM, Young KD. 2003. β-Lactam induction of colanic acid gene expression in Escherichia coli. FEMS Microbiol. Lett. 226:245–249 [DOI] [PubMed] [Google Scholar]
- 22.Laubacher ME, Ades SE. 2008. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J. Bacteriol. 190:2065–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stout V. 1996. Identification of the promoter region for the colanic acid polysaccharide biosynthetic genes in Escherichia coli K-12. J. Bacteriol. 178:4273–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sledjeski DD, Gottesman S. 1996. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 178:1204–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebel W, Vaughn GJ, Peters HK, Trempy JE., III 1997. Inactivation of mdoH leads to increased expression of colanic acid capsular polysaccharide in Escherichia coli. J. Bacteriol. 179:6858–6861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao Y, Doyle MP, Chen J. 2001. Insertion mutagenesis of wca reduces acid and heat tolerance of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:3811–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SM, Chen J. 2004. Survival of Escherichia coli O157:H7 in set yogurt as influenced by the production of an exopolysaccharide, colanic acid. J. Food Prot. 67:252–255 [DOI] [PubMed] [Google Scholar]
- 28.Pontel LB, Pezza A, Soncini FC. 2010. Copper stress targets the Rcs system to induce multiaggregative behavior in a copper-sensitive Salmonella strain. J. Bacteriol. 192:6287–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferrières L, Clarke DJ. 2003. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol. Microbiol. 50:1665–1682 [DOI] [PubMed] [Google Scholar]
- 30.Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993–6002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2:450–464 [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Zhao Y, McClelland M, Harshey RM. 2007. The RcsCDB signaling system and swarming motility in Salmonella enterica serovar Typhimurium: dual regulation of flagellar and SPI-2 virulence genes. J. Bacteriol. 189:8447–8457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanna A, Berg M, Stout V, Razatos A. 2003. Role of capsular colanic acid in adhesion of uropathogenic Escherichia coli. Appl. Environ. Microbiol. 69:4474–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meredith TC, Mamat U, Kaczynski Z, Lindner B, Holst O, Woodard RW. 2007. Modification of lipopolysaccharide with colanic acid (M-antigen) repeats in Escherichia coli. J. Biol. Chem. 282:7790–7798 [DOI] [PubMed] [Google Scholar]
- 35.Russo TA, Sharma G, Weiss J, Brown C. 1995. The construction and characterization of colanic acid-deficient mutants in an extraintestinal isolate of Escherichia coli (O4/K54/H5). Microb. Pathog. 18:269–278 [DOI] [PubMed] [Google Scholar]
- 36.Mao Y, Doyle MP, Chen J. 2006. Role of colanic acid exopolysaccharide in the survival of enterohaemorrhagic Escherichia coli O157:H7 in simulated gastrointestinal fluids. Lett. Appl. Microbiol. 42:642–647 [DOI] [PubMed] [Google Scholar]
- 37.Landau E, Shapira R. 2012. Effects of subinhibitory concentrations of menthol on adaptation, morphological, and gene expression changes in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 78:5361–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chao Y, Zhang T. 2011. Probing roles of lipopolysaccharide, type 1 fimbria, and colanic acid in the attachment of Escherichia coli strains on inert surfaces. Langmuir 27:11545–11553 [DOI] [PubMed] [Google Scholar]
- 39.May T, Okabe S. 2008. Escherichia coli harboring a natural IncF conjugative F plasmid develops complex mature biofilms by stimulating synthesis of colanic acid and curli. J. Bacteriol. 190:7479–7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez-Torres AJ, Stout V. 1996. Role of colanic acid polysaccharide in serum resistance in vivo and in adherence. Curr. Microbiol. 33:383–389 [DOI] [PubMed] [Google Scholar]
- 41.Keenleyside WJ, Bronner D, Jann K, Jann B, Whitfield C. 1993. Coexpression of colanic acid and serotype-specific capsular polysaccharides in Escherichia coli strains with group II K antigens. J. Bacteriol. 175:6725–6730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navasa N, Rodriguez-Aparicio LB, Ferrero MA, Moteagudo-Mera A, Martinez-Blanco H. 2011. Growth temperature regulation of some genes that define the superficial capsular carbohydrate composition of Escherichia coli K92. FEMS Microbiol. Lett. 320:135–141 [DOI] [PubMed] [Google Scholar]
- 43.Russo TA, Singh G. 1993. An extraintestinal, pathogenic isolate of Escherichia coli (O4/K54/H5) can produce a group 1 capsule which is divergently regulated from its constitutively produced group 2, K54 capsular polysaccharide. J. Bacteriol. 175:7617–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahn A, Whitfield C. 2003. Transcriptional organization and regulation of the Escherichia coli K30 group 1 capsule biosynthesis (cps) gene cluster. Mol. Microbiol. 47:1045–1060 [DOI] [PubMed] [Google Scholar]
- 45.Matthysse AG, Deora R, Mishra M, Torres AG. 2008. Polysaccharides cellulose, poly-β-1,6-N-acetyl-d-glucosamine, and colanic acid are required for optimal binding of Escherichia coli O157:H7 strains to alfalfa sprouts and K-12 strains to plastic but not for binding to epithelial cells. Appl. Environ. Microbiol. 74:2384–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prouty AM, Gunn JS. 2003. Comparative analysis of Salmonella enterica serovar Typhimurium biofilm formation on gallstones and on glass. Infect. Immun. 71:7154–7158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ledeboer NA, Jones BD. 2005. Exopolysaccharide sugars contribute to biofilm formation by Salmonella enterica serovar Typhimurium on HEp-2 cells and chicken intestinal epithelium. J. Bacteriol. 187:3214–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang TK, Yam WC, Yuen KY, Wong SS. 2006. Misidentification of a mucoid strain of Salmonella enterica serotype Choleraesuis as Hafnia alvei by the Vitek GNI+ card system. J. Clin. Microbiol. 44:4605–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guard-Petter J. 1999. Phage type and other outer-membrane characteristics associate with virulence. In Saeed A, Gast RK, Potter ME, Wall PG. (ed), Salmonella enterica serovar Enteritidis in humans and animals: epidemiology, pathogenesis, and control. Iowa State University Press, Ames, IA [Google Scholar]
- 50.Hagiwara D, Sugiura M, Oshima T, Mori H, Aiba H, Yamashino T, Mizuno T. 2003. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol. 185:5735–5746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sung BH, Lee CH, Yu BJ, Lee JH, Lee JY, Kim MS, Blattner FR, Kim SC. 2006. Development of a biofilm production-deficient Escherichia coli strain as a host for biotechnological applications. Appl. Environ. Microbiol. 72:3336–3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Curtiss R, III, Wanda SY, Gunn BM, Zhang X, Tinge SA, Ananthnarayan V, Mo H, Wang S, Kong W. 2009. Salmonella enterica serovar Typhimurium strains with regulated delayed attenuation in vivo. Infect. Immun. 77:1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Wang S, Scarpellini G, Gunn B, Xin W, Wanda SY, Roland KL, Curtiss R., III 2009. Evaluation of new generation Salmonella enterica serovar Typhimurium vaccines with regulated delayed attenuation to induce immune responses against PspA. Proc. Natl. Acad. Sci. U. S. A. 106:593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi H, Santander J, Brenneman KE, Wanda SY, Wang S, Senechal P, Sun W, Roland KL, Curtiss R., III 2010. Live recombinant Salmonella Typhi vaccines constructed to investigate the role of rpoS in eliciting immunity to a heterologous antigen. PLoS One 5:e11142. 10.1371/journal.pone.0011142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi H, Wang S, Roland KL, Gunn BM, Curtiss R., III 2010. Immunogenicity of a live recombinant Salmonella enterica serovar Typhimurium vaccine expressing pspA in neonates and infant mice born from naive and immunized mothers. Clin. Vaccine Immunol. 17:363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bollen WS, Gunn BM, Mo H, Lay MK, Curtiss R., III 2008. Presence of wild-type and attenuated Salmonella enterica strains in brain tissues following inoculation of mice by different routes. Infect. Immun. 76:3268–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Markovitz A, Sydiskis RJ, Lieberman MM. 1967. Genetic and biochemical studies on mannose-negative mutants that are deficient in phosphomannose isomerase in Escherichia coli K-12. J. Bacteriol. 94:1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosen SM, Zeleznick LD, Fraenkel D, Wiener IM, Osborn MJ, Horecker BL. 1965. Characterization of the cell wall lipopolysaccharide of a mutant of Salmonella typhimurium lacking phosphomannose isomerase. Biochem. Z. 342:375–386 [PubMed] [Google Scholar]
- 59.Curtiss R, III, Xin W, Li Y, Kong W, Wanda SY, Gunn B, Wang S. 2010. New technologies in using recombinant attenuated Salmonella vaccine vectors. Crit. Rev. Immunol. 30:255–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Collins LV, Attridge S, Hackett J. 1991. Mutations at rfc or pmi attenuate Salmonella typhimurium virulence for mice. Infect. Immun. 59:1079–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Curtiss R, III, Zhang X, Wanda SY, Kang HY, Konjufca V, Li Y, Gunn B, Wang S, Scarpellini G, Lee IS. 2007. Induction of host immune responses using Salmonella-vectored vaccines, p 297–313 In Brogden KA, Minion FC, Cornick N, Stanton TB, Zhang Q, Nolan LK, Wannemuehler MJ. (ed), Virulence mechanisms of bacterial pathogens, 4th ed. ASM Press, Washington, DC [Google Scholar]
- 62.Kuehn MJ, Kesty NC. 2005. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 19:2645–2655 [DOI] [PubMed] [Google Scholar]
- 63.Alaniz RC, Deatherage BL, Lara JC, Cookson BT. 2007. Membrane vesicles are immunogenic facsimiles of Salmonella typhimurium that potently activate dendritic cells, prime B and T cell responses, and stimulate protective immunity in vivo. J. Immunol. 179:7692–7701 [DOI] [PubMed] [Google Scholar]
- 64.Nnalue NA. 1999. All accessible epitopes in the Salmonella lipopolysaccharide core are associated with branch residues. Infect. Immun. 67:998–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stanislavsky ES, Makarenko TA, Kholodkova EV, Lugowski C. 1997. R-form lipopolysaccharides (LPS) of Gram-negative bacteria as possible vaccine antigens. FEMS Immunol. Med. Microbiol. 18:139–145 [DOI] [PubMed] [Google Scholar]
- 66.Jansson PE, Lindberg AA, Lindberg B, Wollin R. 1981. Structural studies on the hexose region of the core in lipopolysaccharides from Enterobacteriaceae. Eur. J. Biochem. 115:571–577 [DOI] [PubMed] [Google Scholar]
- 67.Lüderitz O, Westphal O, Staub AM, Nikaido H. 1971. Isolation and chemical and immunological characterization of bacterial lipopolysaccharides, p 145–223 In Weinbaum G, Kadis S, Ajl S. (ed), Microbial toxins, vol 4 Bacterial endotoxins. Academic Press, Inc., New York, NY [Google Scholar]
- 68.Curtiss R, III, Pereira DA, Hsu JC, Hull SC, Clark JE, Maturin LJ, Sr Goldschmidt R, Moody R, Inoue M, Alexander L. 1977. Biological containment: the subordination of Escherichia coli K-12, p 45–56 In Roland Beers F, Bassett EG. (ed), Recombinant molecules: impact on science and society. Raven Press, New York, NY [Google Scholar]
- 69.Kong W, Wanda SY, Zhang X, Bollen W, Tinge SA, Roland KL, Curtiss R., III 2008. Regulated programmed lysis of recombinant Salmonella in host tissues to release protective antigens and confer biological containment. Proc. Natl. Acad. Sci. U. S. A. 105:9361–9366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baek CH, Wang S, Roland KL, Curtiss R., III 2009. Leucine-responsive regulatory protein (Lrp) acts as a virulence repressor in Salmonella enterica serovar Typhimurium. J. Bacteriol. 191:1278–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Curtiss R, III, Porter SB, Munson M, Tinge SA, Hassan JO, Gentry-Weeks C, Kelly SM. 1991. Nonrecombinant and recombinant avirulent Salmonella live vaccines for poultry, p 169–198 In Blankenship LC, Bailey JH.S, Cox N.A, Stern NJ, Meinersmann RJ. (ed), Colonization control of human bacterial enteropathogens in poultry. Academic Press, New York, NY [Google Scholar]
- 72.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hitchcock PJ, Brown TM. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang S, Li Y, Shi H, Scarpellini G, Torres-Escobar A, Roland KL, Curtiss R., III 2010. Immune responses to recombinant pneumococcal PsaA antigen delivered by a live attenuated Salmonella vaccine. Infect. Immun. 78:3258–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Briles DE, King JD, Gray MA, McDaniel LS, Swiatlo E, Benton KA. 1996. PspA, a protection-eliciting pneumococcal protein: immunogenicity of isolated native PspA in mice. Vaccine 14:858–867 [DOI] [PubMed] [Google Scholar]
- 76.Wang S, Li Y, Scarpellini G, Kong W, Shi H, Baek CH, Gunn B, Wanda SY, Roland KL, Zhang X, Senechal-Willis P, Curtiss R., III 2010. Salmonella vaccine vectors displaying delayed antigen synthesis in vivo to enhance immunogenicity. Infect. Immun. 78:3969–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang S, Li Y, Shi H, Sun W, Roland KL, Curtiss R., III 2011. Comparison of a regulated delayed antigen synthesis system with in vivo-inducible promoters for antigen delivery by live attenuated Salmonella vaccines. Infect. Immun. 79:937–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sedgwick JD, Holt PG. 1983. A solid-phase immunoenzymatic technique for the enumeration of specific antibody-secreting cells. J. Immunol. Methods 57:301–309 [DOI] [PubMed] [Google Scholar]
- 79.Whitfield C, Paiment A. 2003. Biosynthesis and assembly of group 1 capsular polysaccharides in Escherichia coli and related extracellular polysaccharides in other bacteria. Carbohydr. Res. 338:2491–2502 [DOI] [PubMed] [Google Scholar]
- 80.Rahn A, Drummelsmith J, Whitfield C. 1999. Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens: relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J. Bacteriol. 181:2307–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gunn BM, Wanda SY, Burshell D, Wang C, Curtiss R., III 2010. Construction of recombinant attenuated Salmonella enterica serovar Typhimurium vaccine vector strains for safety in newborn and infant mice. Clin. Vaccine Immunol. 17:354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stocker BA. 1988. Auxotrophic Salmonella typhi as live vaccine. Vaccine 6:141–145 [DOI] [PubMed] [Google Scholar]
- 83.Stocker BA. 2000. Aromatic-dependent salmonella as anti-bacterial vaccines and as presenters of heterologous antigens or of DNA encoding them. J. Biotechnol. 83:45–50 [DOI] [PubMed] [Google Scholar]
- 84.Kang HY, Srinivasan J, Curtiss R., III 2002. Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica serovar Typhimurium vaccine. Infect. Immun. 70:1739–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Juarez-Rodriguez MD, Yang J, Kader R, Alamuri P, Curtiss R, III, Clark-Curtiss JE. 2012. Live attenuated Salmonella vaccines displaying regulated delayed lysis and delayed antigen synthesis to confer protection against Mycobacterium tuberculosis. Infect. Immun. 80:815–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.DeKruyff RH, Rizzo LV, Umetsu DT. 1993. Induction of immunoglobulin synthesis by CD4+ T cell clones. Semin. Immunol. 5:421–430 [DOI] [PubMed] [Google Scholar]
- 87.Gor DO, Rose NR, Greenspan NS. 2003. TH1-TH2: a procrustean paradigm. Nat. Immunol. 4:503–505 [DOI] [PubMed] [Google Scholar]
- 88.Pascual DW, Hone DM, Hall S, van Ginkel FW, Yamamoto M, Walters N, Fujihashi K, Powell RJ, Wu S, Vancott JL, Kiyono H, McGhee JR. 1999. Expression of recombinant enterotoxigenic Escherichia coli colonization factor antigen I by Salmonella typhimurium elicits a biphasic T helper cell response. Infect. Immun. 67:6249–6256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pashine A, John B, Rath S, George A, Bal V. 1999. Th1 dominance in the immune response to live Salmonella typhimurium requires bacterial invasiveness but not persistence. Int. Immunol. 11:481–489 [DOI] [PubMed] [Google Scholar]
- 90.Ramarathinam L, Niesel DW, Klimpel GR. 1993. Salmonella typhimurium induces IFN-γ production in murine splenocytes. Role of natural killer cells and macrophages. J. Immunol. 150:3973–3981 [PubMed] [Google Scholar]
- 91.Sestini P, Nencioni L, Villa L, Boraschi D, Tagliabue A. 1988. IgA-driven antibacterial activity against Streptococcus pneumoniae by mouse lung lymphocytes. Am. Rev. Respir. Dis. 137:138–143 [DOI] [PubMed] [Google Scholar]
- 92.Mouslim C, Delgado M, Groisman EA. 2004. Activation of the RcsC/YojN/RcsB phosphorelay system attenuates Salmonella virulence. Mol. Microbiol. 54:386–395 [DOI] [PubMed] [Google Scholar]
- 93.Firozi P, Zhang W, Chen L, Quiocho FA, Worley KC, Templeton NS. 2010. Identification and removal of colanic acid from plasmid DNA preparations: implications for gene therapy. Gene Ther. 17:1484–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pul U, Wurm R, Lux B, Meltzer M, Menzel A, Wagner R. 2005. LRP and H-NS—cooperative partners for transcription regulation at Escherichia coli rRNA promoters. Mol. Microbiol. 58:864–876 [DOI] [PubMed] [Google Scholar]
- 95.Singh AB, Sharma AK, Mukherjee KJ. 2012. Analyzing the metabolic stress response of recombinant Escherichia coli cultures expressing human interferon-beta in high cell density fed batch cultures using time course transcriptomic data. Mol. Biosyst. 8:615–628 [DOI] [PubMed] [Google Scholar]
- 96.Allen PM, Fisher D, Saunders JR, Hart CA. 1987. The role of capsular polysaccharide K21b of Klebsiella and of the structurally related colanic-acid polysaccharide of Escherichia coli in resistance to phagocytosis and serum killing. J. Med. Microbiol. 24:363–370 [DOI] [PubMed] [Google Scholar]
- 97.Mestecky J, Lamm ME, Strober W, Bienenstock J, McGhee JR, Mayer L. 2005. Mucosal immunology, 3rd ed. Elsevier Academic Press, Amsterdam, The Netherlands [Google Scholar]
- 98.Cha HR, Ko HJ, Kim ED, Chang SY, Seo SU, Cuburu N, Ryu S, Kim S, Kweon MN. 2011. Mucosa-associated epithelial chemokine/CCL28 expression in the uterus attracts CCR10+ IgA plasma cells following mucosal vaccination via estrogen control. J. Immunol. 187:3044–3052 [DOI] [PubMed] [Google Scholar]
- 99.Brinkman AB, Ettema TJ, de Vos WM, van der Oost J. 2003. The Lrp family of transcriptional regulators. Mol. Microbiol. 48:287–294 [DOI] [PubMed] [Google Scholar]
- 100.Wang S, Kong Q, Curtiss R., III 2013. New technologies in developing recombinant attenuated Salmonella vaccine vectors. Microb. Pathog. 58:17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Santos SB, Kropinski AM, Ceyssens PJ, Ackermann HW, Villegas A, Lavigne R, Krylov VN, Carvalho CM, Ferreira EC, Azeredo J. 2011. Genomic and proteomic characterization of the broad-host-range Salmonella phage PVP-SE1: creation of a new phage genus. J. Virol. 85:11265–11273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sutherland IW, Wilkinson JF. 1965. Depolymerases for bacterial exopolysaccharides obtained from phage-infected bacteria. J. Gen. Microbiol. 39:373–383 [DOI] [PubMed] [Google Scholar]
- 103.Qimron U, Marintcheva B, Tabor S, Richardson CC. 2006. Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. Proc. Natl. Acad. Sci. U. S. A. 103:19039–19044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakayama K, Kelly SM, Curtiss R., III 1988. Construction of an Asd+ expression-cloning vector: stable maintenance and high level expression of cloned genes in a Salmonella vaccine strain. Nat. Biotechnol. 6:693–697 [Google Scholar]
- 105.Roland K, Curtiss R, III, Sizemore D. 1999. Construction and evaluation of a Δcya Δcrp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis. 43:429–441 [PubMed] [Google Scholar]
- 106.Edwards RA, Keller LH, Schifferli DM. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149–157 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.