Abstract
VanRS two-component regulatory systems are key elements required for the transcriptional activation of inducible vancomycin resistance genes in bacteria, but the precise nature of the ligand signal that activates these systems has remained undefined. Using the resistance system in Streptomyces coelicolor as a model, we have undertaken a series of in vivo studies which indicate that the VanS sensor kinase in VanB-type resistance systems is activated by vancomycin in complex with the d-alanyl-d-alanine (d-Ala-d-Ala) termini of cell wall peptidoglycan (PG) precursors. Complementation of an essential d-Ala-d-Ala ligase activity by constitutive expression of vanA encoding a bifunctional d-Ala-d-Ala and d-alanyl-d-lactate (d-Ala-d-Lac) ligase activity allowed construction of strains that synthesized variable amounts of PG precursors containing d-Ala-d-Ala. Assays quantifying the expression of genes under VanRS control showed that the response to vancomycin in these strains correlated with the abundance of d-Ala-d-Ala-containing PG precursors; strains producing a lower proportion of PG precursors terminating in d-Ala-d-Ala consistently exhibited a lower response to vancomycin. Pretreatment of wild-type cells with vancomycin or teicoplanin to saturate and mask the d-Ala-d-Ala binding sites in nascent PG also blocked the transcriptional response to subsequent vancomycin exposure, and desleucyl vancomycin, a vancomycin analogue incapable of interacting with d-Ala-d-Ala residues, failed to induce van gene expression. Activation of resistance by a vancomycin–d-Ala-d-Ala PG complex predicts a limit to the proportion of PG that can be derived from precursors terminating in d-Ala-d-Lac, a restriction also enforced by the bifunctional activity of the VanA ligase.
INTRODUCTION
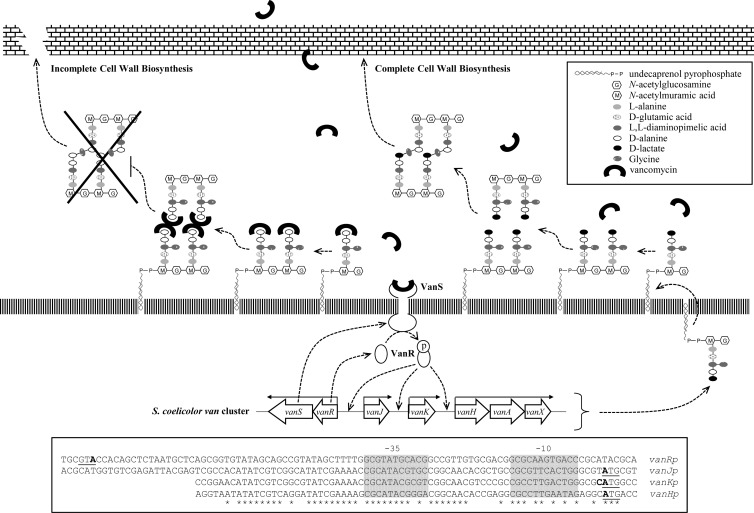
The glycopeptide family antibiotic vancomycin is a front-line therapy for the treatment of problematic bacterial infections, particularly those caused by methicillin-resistant Staphylococcus aureus (MRSA). Vancomycin inhibits bacterial cell wall biosynthesis by binding to the d-alanyl-d-alanine (d-Ala-d-Ala) terminus of lipid-attached peptidoglycan (PG) precursors, blocking formation of the mature PG that gives the cell wall its rigidity (1) (Fig. 1). The spread of resistance to glycopeptide antibiotics through bacterial populations is an acute public health issue, as highlighted by the emergence of vancomycin-resistant MRSA (VRSA) in hospitals (2). The first clinical isolates of vancomycin-resistant infections emerged in enterococcal strains in the mid-1980s and were shown to result from the replacement of the d-Ala-d-Ala dipeptide terminus of cell wall PG precursors by d-alanyl-d-lactate (d-Ala-d-Lac), which reduces the affinity of vancomycin binding by ∼1,000-fold. This alteration in cell wall biosynthesis requires the expression of at least three genes, vanHAX, which are transcribed as a single polycistronic message that is usually dependent on a VanRS two-component regulatory system. The genes encoding VanR, a response regulator, and VanS, a sensor kinase, are themselves transcribed as a single transcription unit and are typically located adjacent to vanHAX on the chromosome. The total numbers and types of genes present in such vancomycin resistance clusters can vary, but the majority contain vanSRHAX as their core components (3–5).
Fig 1.
Model illustrating the mode of action of vancomycin and the vancomycin resistance mechanism in S. coelicolor. Vancomycin binds to the d-Ala-d-Ala dipeptide termini of non-cross-linked peptidoglycan pentapeptide side chains, preventing the formation of mature peptidoglycan and lowering its strength. Resistance is initiated when vancomycin is directly perceived by the sensor kinase, VanS, which then phosphorylates its cognate partner response regulator, VanR. Phospho-VanR activates the expression of the van gene cluster and results in the replacement of the d-Ala-d-Ala dipeptide terminus of cell wall PG precursors by d-Ala-d-Lac, which significantly reduces the affinity of vancomycin binding by ∼1,000-fold, consequently rendering the modified bacteria resistance to vancomycin. The boxed area presents the promoter sequences upstream of each transcription unit with the ATG translational start sites underlined and the −10 and −35 consensus sequences indicated. Transcription start sites are written in bold, and highly conserved bases are marked with asterisks.
Vancomycin resistance gene clusters were first found either associated with glycopeptide biosynthetic gene clusters in antibiotic-producing bacteria or as part of transferrable multiple-drug-resistance islands in pathogenic bacteria. However, the explosion in microbial genome sequencing facilitated by recent advances in sequencing technologies has revealed the presence of these genes in many more bacterial sources, interestingly also including many nonpathogenic or non-glycopeptide-producing species (6). This type of organism offers a safe and convenient system for the in vivo study of important aspects of vancomycin resistance, as exemplified by recent work using Streptomyces coelicolor (7–10). S. coelicolor serves as the model species for Streptomyces, a genus of Gram-positive, mycelial soil bacteria responsible for the production of two-thirds of the commercially important antibiotics. It does not synthesize any glycopeptide antibiotic, but it does possess a cluster of seven genes, vanRSJKHAX, that confer inducible resistance to vancomycin (Fig. 1) (7, 8). On exposure to vancomycin in S. coelicolor, VanS activity switches from being a phosphatase to a kinase, and the resulting accumulation of phospho-VanR activates transcription from van promoters and induces vancomycin resistance. In the absence of drug, however, the constitutive phosphorylation of the response regulator VanR mediated by the small-molecule phosphodonor acetyl phosphate is suppressed by the phosphatase activity of VanS (9). Deletion of VanS therefore results in constitutive expression of the van genes in S. coelicolor, consistent with VanS functioning to negatively regulate VanR activity in the absence of antibiotic.
One of the most important questions yet to be answered in the study of vancomycin resistance is the nature of the specific ligand recognized by the VanS sensor protein. Two distinct models have been proposed: (i) direct induction, in which the sensor kinase is activated by direct binding of antibiotic to the sensor domain; and (ii) indirect induction, in which the sensor kinase is activated by binding a cell wall metabolite that is either intermediate in cell wall biosynthesis or accumulates as a result of antibiotic action (11). These models are, however, not mutually exclusive, since an intermediate possibility exists whereby VanS is induced by the glycopeptide antibiotic when it is bound to a d-Ala-d-Ala-containing cell wall metabolite. A significant amount of effort has been put into studying the different VanR-VanS two-component systems in clinical isolates of enterococcal strains, but the exact nature of the direct molecular ligand that activates VanS has remained elusive. VanA-type enterococci are resistant to both vancomycin and teicoplanin, and VanS in these strains has been shown to be responsive not only to these two drugs but also to some nonglycopeptide cell wall-specific antibiotics such as moenomycin (4, 12–20). This is supportive of the indirect induction model since it is considered unlikely that the VanS sensor domain could recognize such structurally unrelated antibiotics. In contrast, however, experiments analyzing the activity of VanS in VanB-type strains which are resistant to vancomycin but not teicoplanin have been supportive of the direct induction model. Only glycopeptide antibiotics structurally related to vancomycin were capable of inducing VanB-type VanS, while all the nonglycopeptide antibiotics tested failed to act as inducers (21, 22). S. coelicolor M600 is resistant to vancomycin but susceptible to teicoplanin, similar to VanB-type enterococci. There have been several attempts to elucidate the nature of the direct ligand sensed by the S. coelicolor VanS sensor protein. In previous work, a range of antibiotics were tested for their ability to induce the VanR-VanS system in S. coelicolor using bioassay systems (8, 9). The inducers identified are all structurally closely related glycopeptide antibiotics (including vancomycin, chloroeremomycin, ristocetin, A47934, and balhimycin), suggesting that S. coelicolor VanS interacts directly with glycopeptide antibiotics. In addition, Koteva et al. (10) recently showed that a vancomycin photoaffinity probe directly binds to S. coelicolor VanS and that this binding is required for the expression of the van genes and for resistance to vancomycin. These observations strongly favor the direct induction mechanism over the indirect one, but since the membrane protein preparations used may still contain cell wall precursors, such as lipid II, they do not exclude a requirement for binding as a vancomycin-cell wall metabolite complex. In this study, we use the van cluster in S. coelicolor as a model for the VanB resistance system and present the first in vivo evidence that vancomycin requires binding to the d-Ala-d-Ala termini of cell wall PG precursors to be perceived by the VanS sensor protein.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and chemicals.
All E. coli and S. coelicolor strains, plasmids, and oligonucleotides used in this study are described in Table S1 in the supplemental material (23, 24). To bypass the methyl-specific restriction system of S. coelicolor, cosmid and plasmid DNAs were introduced by transformation into the dam dcm hsdS Escherichia coli strain ET12567 containing the driver plasmid pUZ8002. Conjugal transfer of unmethylated cosmid or plasmid DNA between E. coli ET12567 and S. coelicolor was carried out as described by Kieser et al. (25). Desleucyl vancomycin was a kind gift from Daniel Kahne. Other antibiotics and chemicals used for this study were purchased from Sigma-Aldrich.
RNA preparation.
RNA was prepared from NMMP liquid cultures according to the method described previously (9, 25). Spores of S. coelicolor strains were germinated by heat shock treatment in TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] buffer and incubated in double-strength germination medium at 37°C for 3 h. Germinated spores were then inoculated and grown to mid-log phase in NMMP at 30°C. Immediately after the first sample was taken, inducer antibiotic was added to the culture broth, and samples were taken in appropriate subsequent time intervals. For RNA preparation, the cell pellet was resuspended in 1 ml of ice-cold Kirby mix (25), sonicated twice for 4 to 5 s each, and then extracted with 0.8 ml of phenol-chloroform (pH 8.0) by vortexing. Samples were reextracted with phenol-chloroform (pH 8.0), and nucleic acids were precipitated at −20°C using 0.3 M sodium acetate (pH 6.0) and an equal volume of isopropanol. After centrifugation, the nucleic acid pellet was washed with 70% ethanol, dissolved in DNase I buffer (27), and then treated with 5 U of DNase I at 37°C for 30 min. Samples were extracted with phenol-chloroform (pH 8.0) and precipitated again as described above. After centrifugation, RNA pellets were dissolved in RNase-free distilled water and stored at −80°C.
S1 nuclease protection assay.
The vanH and sigE probes for S1 nuclease protection analysis were generated by PCR from S. coelicolor M600 chromosomal DNA using a [γ-32P]ATP-labeled (3,000 Ci/mmol) 5′-end oligonucleotide internal to each gene and an unlabeled upstream primer as described previously (7, 9, 25, 28). Primer pair vanH S1 FOR and vanH S1 REV produced a 270-bp vanH probe, while a 400-bp sigE probe was generated using primers G1680 and G1681 (28) (see Table S1 in the supplemental material) (23, 24). For all assays, 30 μg of RNA and 25 pmol of labeled probe were dissolved in 20 μl of Na trichloroacetic acid (TCA) buffer and hybridized together at 45°C overnight after initial denaturation at 65°C for 15 min. Digestion with S1 nuclease at 37°C for 45 min was followed by PAGE analysis of the protected fragment as described previously (29).
qRT-PCR.
The modified procedure for quantitative real-time PCR (qRT-PCR) analysis was performed according to Hesketh et al. (30, 31). Primers for qRT-PCR analysis of vanH and vanK transcription (assigned names beginning with q in Table S1) (23, 24) were designed using Primer3 (http://frodo.wi.mit.edu/). Aliquots of total RNA (2.5 μg) were subjected to RNase-free DNase I treatment (amplification grade; Invitrogen) in a 20-μl reaction volume, and 8 μl of the digest was used directly in a 20-μl cDNA synthesis reaction using Superscript III first strand synthesis supermix (Invitrogen). PCR cycling was carried out as described by Hesketh et al. (31). After RNase H treatment, the samples were diluted 100 times in DNase-free distilled water, and 2.5-μl aliquots subjected to qRT-PCR analysis in 25-μl reaction volumes comprising 12.5 μl of SYBR GreenER quantitative PCR (qPCR) supermix (Invitrogen), 200 nM forward and reverse primers, 4.8% dimethyl sulfoxide (DMSO), and 250 nM ROX. qRT-PCR cycling was performed in a 7300 real-time PCR system (Applied Biosystems) at 50°C for 2 min and 95°C for 10 min, followed by 30 cycles of 95°C for 15 s and 57°C for 1 min. Parallel reactions were performed in the same 96-well plate using different dilutions of genomic DNA to generate a standard curve for each gene. For vanH, this yielded a curve with formula CT = −3.754 (log copy no.) + 44.071 (R2 = 0.991), and for vanK, CT = −3.636 (log copy no.) + 44.538 (R2 = 0.997). (CT is threshold cycle.) All determinations were performed in triplicate, and the results were analyzed using 7300 System SDS software (version 1.4; Applied Biosystems). Results were normalized to an endogenous control gene, SCO4742, selected from previous microarray data as being constantly and constitutively expressed under the conditions used (31, 32).
Preparation and HPLC-MS/MS analysis of cell wall peptidoglycan precursors.
Cytoplasmic peptidoglycan precursors were prepared based on the method described by Ruzin et al. (33). Spores were germinated and inoculated into NMMP liquid medium (with vancomycin for the dependent strain H2027). The cultures were grown to mid-log phase and where necessary were exposed to d-lactate (10 mM) for 3 h before harvesting. Cell pellets were washed twice with 0.9% NaCl and extracted with 5% ice-cold trichloroacetic acid at 4°C for 30 min. The extract was desalted using a Sephadex G-25 column (PD-10 columns; Amersham Biosciences) according to the manufacturer's recommendations and then concentrated by rotary evaporation. The samples were resuspended in high-performance liquid chromatography (HPLC)-grade deionized water and analyzed by ion-trap HPLC-tandem mass spectrometry (MS/MS) as described previously (8).
Construction of the S. coelicolor ΔddlA mutant.
For PCR-directed replacement of the ddlA (SCO5560) gene with an apramycin-resistance cassette (apr), cosmid St7A1 in E. coli was targeted with a disruption cassette created by PCR using primers ddlA KO F and ddlA KO R and with pIJ773 as the template. The resulting cosmid was verified by PCR and restriction digestion and transferred into S. coelicolor M600 by conjugation. Double-crossover integrants were sought as apramycin-resistant, kanamycin-sensitive colonies from plates also containing 20 μg/ml of vancomycin. No apramycin-resistant, kanamycin-sensitive colonies could be obtained from conjugation plates lacking vancomycin, indicating that deletion of ddlA is lethal when van gene expression is not induced, analogous to the ΔfemX mutant (8). Correct deletion of ddlA was confirmed by PCR using primers ddlA KO Test F and ddlA KO Test R.
Complementation of the ΔddlA mutant by constitutive expression of ddlA or vanA under the control of ermEp.
To test whether constitutive expression of ddlA or vanA can complement the ddlA mutant, both the ddlA and vanA genes were amplified by PCR and cloned individually into pIJ10257 (8) downstream of the ermE* promoter (ermEp). Primers ndeI-ddlA F and pacI-ddlA R were used to amplify ddlA from cosmid St7A1, producing the gene flanked by NdeI and PacI restriction sites. vanA flanked by NdeI and BlnI restriction sites was similarly amplified from cosmid StH66 using primers ndeI-vanA F1 and bln-vanA R. Both PCR products were first cloned into Promega's pGEM-T Easy vector system (creating plasmids pHJH5 and pGN15) before being subcloned into pIJ10257 using the NdeI-PacI or NdeI-BlnI restriction sites. The resulting constructs, pGN8 (pIJ10257-ddlA) and pGN17 (pIJ10257-vanA), were verified by sequencing and introduced into the ΔddlA mutant background (H2004) by conjugal transformation, selecting exconjugants by hygromycin resistance.
Construction of pMK2: a vancomycin-inducible kanamycin-resistant reporter system.
The van gene cluster in S. coelicolor controls four highly conserved VanR-dependent promoter sequences (Fig. 1) known to respond identically following induction by vancomycin (7). To create pMK2, a conjugative and integrative vector carrying a promoter-gene fusion encoding vancomycin-inducible neomycin/kanamycin resistance, a 0.5-kb DNA fragment containing the tsr (thiostrepton resistance) gene was first excised from plasmid pIJ6902 (34) by digestion with NheI. This was ligated into XbaI-digested pSET152 (27), producing pMK1. A 1.2-kb DNA fragment containing a vancomycin-inducible promoter sequence (for this work, the vanJ promoter sequence was used) fused with the neo gene, vanJp-neo, was obtained by digestion of pHJH4 with BamHI and BglII, and this was then ligated into pMK1 digested with BamHI. The resulting plasmid, designated pMK2 (see Fig. S1 in the supplemental material), was verified by restriction digest mapping and sequencing and introduced into the appropriate S. coelicolor strains by conjugal transformation. A 50-μg/ml concentration of apramycin was used for the selection of pMK2 in E. coli, and 50 μg/ml thiostrepton was used for selection in apramycin-resistant S. coelicolor strains.
RESULTS
Induction of van gene expression by vancomycin in S. coelicolor is inhibited by preexposure to antibiotics which bind the d-Ala-d-Ala termini of PG precursors.
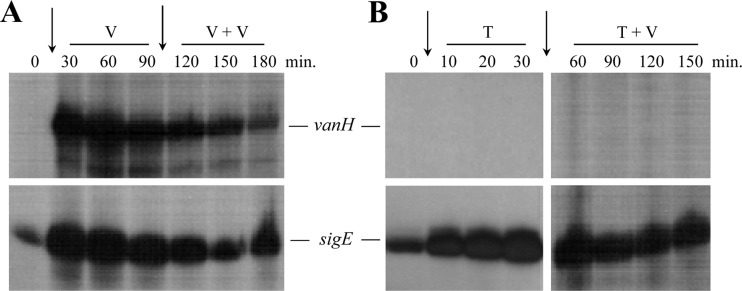
Transcriptional analysis of the vanRSJKHAX gene cluster in S. coelicolor has previously characterized their organization as four VanR-dependent transcription units, vanRS, vanJ, vanK, and vanHAX (Fig. 1) (7). Expression of all four transcripts is induced in liquid cultures of S. coelicolor M600 exposed to vancomycin, but the upregulation is known to be transient (7). Transcription rapidly reaches a maximum level 15 to 30 min after exposure but decreases in strength thereafter, concomitant with the decrease in abundance of PG precursors terminating in d-Ala-d-Ala and increase in d-Ala-d-Lac PG precursors brought about by expression of the van genes (8). To determine whether S. coelicolor VanS is capable of responding to a second dose of vancomycin administered during this period of decline in van transcription, liquid cultures of S. coelicolor M600 were exposed to two successive doses of vancomycin (10 μg/ml) spaced 90 min apart. Mycelial samples were taken from the cultures for RNA extraction at regular intervals commencing from the first dose, and vanH transcript abundance was monitored by S1 nuclease protection analysis (Fig. 2A). Transcription of the sigE gene, part of a signal transduction system that senses and responds to cell wall stress in S. coelicolor (35), was also analyzed as a positive control. As expected vanH transcription increased dramatically in the 30 to 60 min immediately following the first dose of vancomycin, but it then began a decline which continued unchecked even after addition of the second dose at 90 min. sigE transcription showed the same trend, indicating that the second dose of vancomycin is not detected by either the VanS or SigE signal transduction systems. To further characterize this lack of response, the experiment was repeated using teicoplanin (50 μg/ml) as the initial antibiotic treatment. Teicoplanin is a glycopeptide antibiotic with a very similar structure to vancomycin and also binds to the d-Ala-d-Ala termini of extracellular PG precursors (26). In contrast to vancomycin, however, it does not inhibit the binding of a vancomycin photoaffinity probe to VanS in vitro (10), nor does it act as a positive inducer of VanS activity in bioassays (7, 9). Vancomycin should still therefore be able to access VanS in the presence of teicoplanin, but interaction with d-Ala-d-Ala-containing PG precursor will be inhibited. Strikingly, not only did teicoplanin fail to activate any detectable vanH transcription, it also blocked induction of vanH in response to the subsequent addition of vancomycin (Fig. 2B). Transcription of sigE was strongly induced by teicoplanin and was also readily detectable at all times following the vancomycin addition, confirming the viability of the cells and the integrity of the RNA samples. Although alternative explanations exist, these experiments suggest that vancomycin may need to be bound to the d-Ala-d-Ala termini of extracellular PG precursors in order to interact productively with VanS since in each case unbound vancomycin present in the second dose was incapable of activating the sensor. To test this hypothesis further, we attempted to construct a strain that cannot synthesize any d-Ala-d-Ala-containing cell wall PG precursors.
Fig 2.
Saturation of the d-Ala-d-Ala termini of immature PG by pretreatment of cells with (A) vancomycin or (B) teicoplanin prevents the sensing of vancomycin by VanS. S. coelicolor M600 cells were grown to the mid-late exponential phase in NMMP liquid medium and pretreated at time zero by addition of 10 μg/ml vancomycin (A) or 50 μg/ml teicoplanin (B), as indicated by the vertical arrows. Samples were taken at the intervals shown before addition of a further 10 μg/ml vancomycin (arrowed) after 90 min (A) or 30 min (B). RNA was extracted from each sample, and vanH transcript abundance was determined by S1 nuclease protection. Transcription of sigE is shown as a positive control for cell viability and RNA integrity and as an indicator of cell wall stress. Where shown, “V” represents vancomycin and “T” indicates teicoplanin.
The S. coelicolor ΔddlA mutant is dependent on vancomycin for viability but still produces d-Ala-d-Ala-containing cell wall precursors.
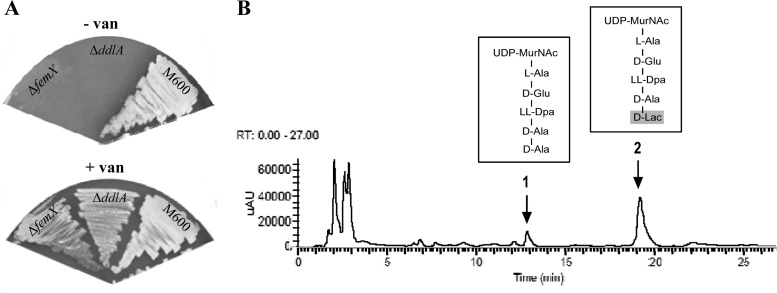
ddlA (SCO5560) in the S. coelicolor genome is predicted to encode the essential d-Ala-d-Ala ligase enzyme required for PG biosynthesis, a protein homologous to the VanA d-Ala-d-Lac ligase (37% amino acid identity). In a mutational analysis, deletion of ddlA was only possible in the presence of vancomycin, analogous to a previous study analyzing mutation of femX (8). The S. coelicolor ΔddlA strain, like the ΔfemX strain, is only viable when grown in the presence of vancomycin, suggesting that the ddlA gene is essential in the absence of vancomycin but dispensable in its presence (Fig. 3A). This is consistent with vancomycin-induced vanA expression compensating for the loss of ddlA function. To determine whether vancomycin-dependent ddlA mutant cells only produce cell wall precursors terminating in d-Ala-d-Lac, PG precursors were extracted from mutant cells grown in the presence of vancomycin and analyzed using HPLC-MS/MS. Interestingly, precursors terminating in both d-Ala-d-Ala (peak 1, UDP-MurNAc-pentapeptide) and d-Ala-d-Lac (peak 2, UDP-MurNAc-pentadepsipeptide) were readily detectable, in a ratio of ca. 1:9, respectively (Fig. 3B). These data indicated that the VanA d-Ala-d-Lac ligase also possesses significant d-Ala-d-Ala ligase activity.
Fig 3.
S. coelicolor ΔddlA is dependent on vancomycin for viability (A) but still produces cell wall PG precursors terminating in d-Ala-d-Ala (B). (A) The ΔddlA mutant strain requires the presence of vancomycin for growth, analogous to the ΔfemX mutant previously reported (8). The M600 parent strain is shown as a control. (B) HPLC-MS/MS analysis of cytoplasmic PG cell wall precursors isolated from the ΔddlA null mutant grown in the presence of 20 μg/ml vancomycin. The analysis was performed on biological replicates, and a representative result is shown. Peaks eluting after 11.92 min (peak 1) and 17.75 min (peak 2) using detection by UV absorbance at 254 nm were identified as UDP-MurNAc-pentapeptide (the d-Ala-d-Ala-containing PG precursor [1,193 Da]) and UDP-MurNAc-pentadepsipeptide (the d-Ala-d-Lac-containing PG precursor [1,194 Da]), respectively. uAU, microabsorbance units.
Manipulating the amounts of d-Ala-d-Ala- and d-Ala-d-Lac-containing cell wall precursors in the cell.
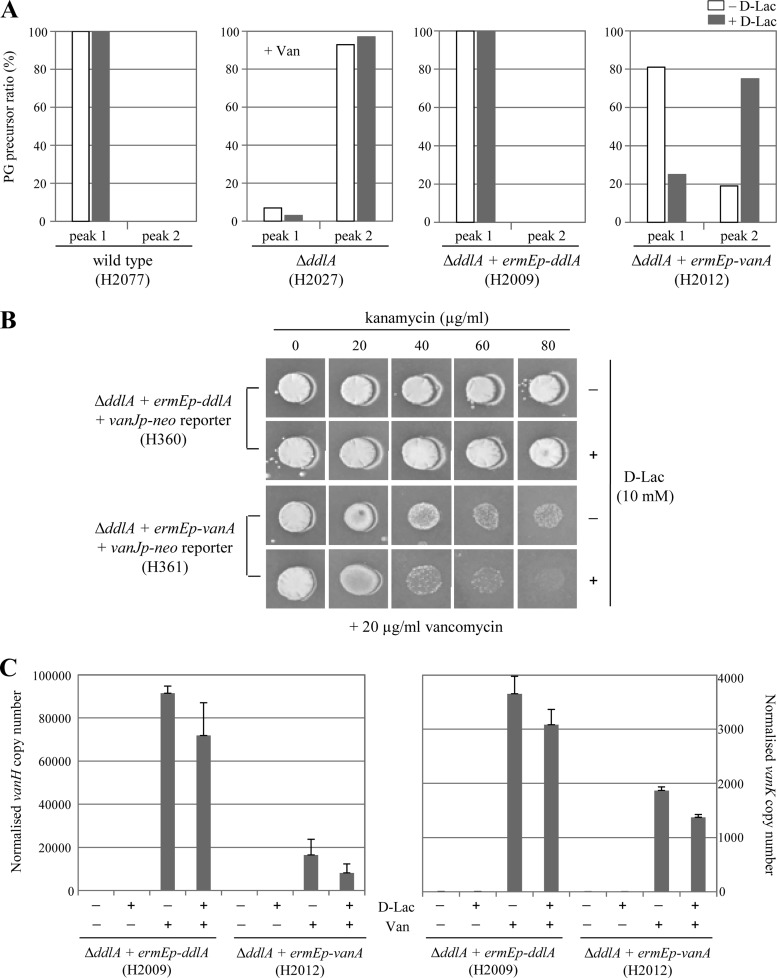
Because it was not possible to construct a strain completely devoid of d-Ala-d-Ala-containing cell wall PG precursors, we next attempted to manipulate the relative amounts of d-Ala-d-Ala- and d-Ala-d-Lac-containing cell wall PG precursors in the cell genetically. First, we complemented the ΔddlA mutant with S. coelicolor ddlA or vanA genes expressed from the constitutively active but non-vancomycin-dependent ermE* promoter (ermEp). Conjugative and integrative (ϕBT1 attP-int) plasmid DNAs carrying ermEp-ddlA or ermEp-vanA were constructed, and these were introduced into the vancomycin-dependent ΔddlA null mutant strain by conjugation (see Table S1 in the supplemental material) (23, 24). Constitutive expression of both the ddlA and the vanA genes allowed the ΔddlA mutant to grow vigorously even in the absence of vancomycin, indicating that both genes complement ddlA (see Fig. S2 in the supplemental material). Analysis of the relative abundance of d-Ala-d-Ala-containing PG precursors (equivalent to UDP-MurNAc-pentapeptide in Fig. 3B, peak 1) and d-Ala-d-Lac-containing PG precursors (equivalent to UDP-MurNAc-pentadepsipeptide in Fig. 3B, peak 2) in these strains indicated that while complementation with ermEp-ddlA (H2009) yielded 100% d-Ala-d-Ala-containing PG precursors, similar to the wild-type strain (H2077), the ermEp-vanA-complemented strain (H2012) produced both d-Ala-d-Ala-containing PG precursors and d-Ala-d-Lac-containing PG precursors in a ratio of ca. 4:1 (Fig. 4A, white bars). This is markedly different from the ca. 1:9 ratio observed in the vancomycin-dependent ΔddlA strain (H2027; necessarily grown in the presence of vancomycin), and this presumably reflects the activity of the other van genes whose expression is coinduced by vancomycin in the ΔddlA strain. vanX encodes a d-Ala-d-Ala dipeptidase, while vanH encodes an α-ketoacid dehydrogenase that synthesizes d-lactate (d-Lac) from pyruvate (36). Both activities would be expected to increase the proportion of d-Ala-d-Lac-containing PG precursors. The ermEp-vanA-complemented strain grown in the absence of vancomycin expresses only vanA (from the constitutive ermE promoter), not vanX or vanH (which would require vancomycin to activate their transcription), and the d-Ala-d-Ala-containing PG precursor is the major cell wall PG precursor present. The most likely explanation for this is that the intracellular concentration of d-alanine (d-Ala) is much higher than that of d-Lac, resulting in preferential use of d-Ala by the VanA ligase. Consistent with this hypothesis, when the growth medium for the strains was supplemented with 10 mM d-Lac, the ratio of d-Ala-d-Ala-containing PG precursors to d-Ala-d-Lac-containing PG precursors dramatically changed from 4:1 to 1:4 in the ermEp-vanA-complemented strain but was unaffected in the wild type or the strain complemented with ermEp-ddlA (Fig. 4A, gray bars). Even in the vancomycin-dependent ΔddlA strain grown in the presence of vancomycin, d-Lac supplementation altered the ratio in favor of the d-Ala-d-Lac-containing PG precursors, changing it from 1:9 to 1:20 (Fig. 4A, gray bars). Interestingly, this also coincided with an increase in the concentration of vancomycin required to produce viability in this strain (see Fig. S2 in the supplemental material), which is consistent with the hypothesis that VanS senses vancomycin when bound to the d-Ala-d-Ala termini of an extracellular PG precursor: d-Lac supplementation further decreases the abundance of d-Ala-d-Ala-containing PG precursors, resulting in reduced activation of VanS and a lower level of vanA transcription. Under these circumstances, a higher vancomycin concentration than expected would be required to produce sufficient VanA for viability in the ΔddlA mutant background.
Fig 4.
Induction of van gene expression by vancomycin is significantly reduced in cells where d-Ala-d-Lac-containing PG precursors are more abundant than d-Ala-d-Ala-containing PG precursors. (A) Relative abundance of the d-Ala-d-Ala-containing PG precursor (UDP-MurNAc-pentapeptide, peak 1, as shown in Fig. 3B) and the d-Ala-d-Lac-containing PG precursor (UDP-MurNAc-pentadepsipeptide, peak 2, as shown in Fig. 3B) in the wild type (H2077), ΔddlA mutant (H2027), and the ΔddlA mutant complemented with ermEp-ddlA (H2009) or ermEp-vanA (H2012) when grown in the presence (gray bars) or absence (white bars) of a 10 mM d-Lac supplement. Cells grown to the mid-late exponential phase in NMMP liquid cultures were extracted and analyzed by HPLC-MS/MS. Relative abundances were calculated by normalizing each sample so that the sum of the two UDP-MurNAC derivatives equaled 100. Strain H2027 (ΔddlA) is viable only in the presence of vancomycin and was therefore grown in NMMP containing 20 μg/ml vancomycin, as indicated by “+Van.” (B) Bioassay for kanamycin resistance induced from the vancomycin-inducible vanJ promoter in the reporter strains. Approximately 105 spores were spotted onto agar plates containing 20 μg/ml vancomycin, kanamycin at a range of concentrations between 0 and 80 μg/ml in 20 μg/ml increments, and in the presence (+) or absence (−) of 10 mM d-Lac. The result was scored after 4 days of incubation at 30°C. (C) Induction of vanH and vanK transcription in ΔddlA mutant complemented with ermEp-ddlA (H2009) and ermEp-vanA (H2012) in response to vancomycin, as determined by qRT-PCR. Each strain was grown to the mid-late exponential phase in two different NMMP liquid media, one with 10 mM d-Lac (+) and one without (−). Samples were then taken immediately before the addition of 10 μg/ml vancomycin (−) and 30 min after treatment (+). Total RNAs were extracted from each sample, and qRT-PCR was carried out as described in Materials and Methods.
Vancomycin induction of van gene expression correlates with the abundance of d-Ala-d-Ala-containing cell wall PG precursors.
The model we propose for VanS induction where vancomycin is sensed only as a complex with d-Ala-d-Ala-containing extracellular PG precursors predicts that the transcriptional response induced by vancomycin should be markedly diminished in strains and conditions where the proportion of PG precursors terminating in d-Ala-d-Ala is reduced relative to PG precursors terminating in d-Ala-d-Lac. We therefore analyzed the strength of van gene expression induced by vancomycin in the ermEp-ddlA- and ermEp-vanA-complemented strains grown with and without d-Lac supplementation using a reporter plasmid, pMK2 (Fig. 4B), and qRT-PCR (Fig. 4C). pMK2 (see Fig. S1 in the supplemental material) is a conjugative and integrative (ϕC31 attP-int) vector containing a vanJp-neo fusion, which confers resistance to kanamycin in response to induction of the vanJ promoter by vancomycin. van gene expression in the ermEp-ddlA-complemented strain background (H2009) was strongly induced by vancomycin, and the level was independent of the presence of d-Lac supplementation in the medium (Fig. 4C). This strain exclusively produces d-Ala-d-Ala-containing PG precursors, even in the presence of 10 mM d-Lac (Fig. 4A, white and gray bars on H2009). Induction by vancomycin in the ermEp-vanA-complemented strain background (H2012), however, was significantly reduced compared to that in the ermEp-ddlA-complemented strain (H2009) and even further decreased when d-Lac was present as a supplement (Fig. 4C). This correlates with observed changes in PG precursor composition where precursors containing d-Ala-d-Ala comprise just 80% of the total in cells grown in NMMP medium and only 20% in NMMP plus 10 mM d-Lac (see Fig. 4A, white and gray bars on H2012). In the bioassay using the pMK2 reporter strains, vigorous growth of the ermEp-vanA-complemented reporter strain (H361) occurred in the presence of 20 μg/ml kanamycin or less, while the ermEp-ddlA complemented reporter strain (H360) grew strongly even with 80 μg/ml kanamycin (Fig. 4B). This suggests that induction by vancomycin is at least ca. 4-fold weaker in the ermEp-vanA-complemented reporter strain (H361) than in the strain complemented with ermEp-ddlA (H361) and is in good agreement with the qRT-PCR results for vanH and vanK transcript abundance in these strains (Fig. 4C). These data therefore show a correlation between d-Ala-d-Ala terminating PG precursor abundance and the response to vancomycin that is entirely consistent with the proposed model for drug sensing by VanS.
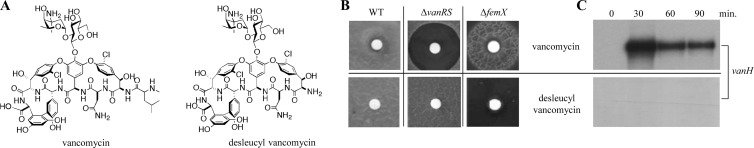
Desleucyl vancomycin does not induce van gene expression.
Desleucyl vancomycin (Fig. 5A) is a semisynthetic derivative of vancomycin in which the peptide-binding pocket has been modified so that it can no longer bind to the d-Ala-d-Ala termini of extracellular PG precursors (37, 38). Bioassays using a ΔfemX reporter strain dependent on van gene expression for its viability indicated that desleucyl vancomycin cannot induce the vancomycin resistance system and therefore is not sensed by VanS (Fig. 5B). The desleucyl derivative failed to stimulate the halo of growth of the ΔfemX strain that was clearly evident around the vancomycin disc. In addition, the large halo of growth inhibition produced by exposure of the ΔvanRS strain to a disc containing vancomycin was completely absent when a desleucyl vancomycin disc was used, indicating that the d-Ala-d-Ala binding pocket of vancomycin is important not only for the mode of action but also for inducing the vancomycin resistance system (Fig. 5B). The lack of any transcriptional response to desleucyl vancomycin was confirmed by monitoring the response to each drug of the vanH promoter in its native chromosomal context. The abundance of the vanH transcript was assessed in cells from liquid cultures of S. coelicolor wild-type M600 at 30-min intervals before and after exposure to 10 μg/ml of vancomycin or desleucyl vancomycin. The expected induction of vanH transcription in response to vancomycin was readily observable, but the vanH transcript was undetectable in the RNA samples extracted from cultures treated with desleucyl vancomycin (Fig. 5C). Basal transcription of the sigE control gene was readily detectable in the desleucyl vancomycin samples, verifying the integrity of the RNA.
Fig 5.
Desleucyl vancomycin, which does not bind to the d-Ala-d-Ala termini of extracellular PG precursors, fails to induce van gene expression. (A) Structures of vancomycin and desleucyl vancomycin. (B) Bioassay analyzing vancomycin and desleucyl vancomycin activities against the S. coelicolor wild-type (WT) M600 (vancomycin-resistant), ΔvanRS mutant (vancomycin-sensitive), and ΔfemX (vancomycin-dependent) strains. Paper discs containing 30 μg of each drug were placed on freshly spread lawns of each strain, and plates were incubated for 4 days at 30°C. (C) Response of the vanH promoter to vancomycin or desleucyl vancomycin in S. coelicolor M600. Cells were grown to the mid-late exponential phase in NMMP liquid medium and exposed to 10 μg/ml vancomycin or 10 μg/ml desleucyl vancomycin. Total RNAs were extracted from samples taken immediately before the addition of drug and at subsequent 30-min intervals and analyzed by S1 nuclease protection assays.
DISCUSSION
VanR and VanS comprise the response regulator and sensor kinase of a two-component signal transduction system which regulates transcription of the genes responsible for conferring resistance to vancomycin, a glycopeptide antibiotic with important clinical applications. The investigation of vancomycin resistance in bacteria has therefore been the subject of extensive research, particularly using vancomycin-resistant enterococci (VRE) as a model system, but the molecular details of the mechanism of VanS activation have so far remained undefined. Understanding the exact nature of the ligand which directly induces VanS activity is an important goal since it could enable the rational design of novel glycopeptide antibiotic structures that are still clinically active but which cannot be recognized by the VanS sensor. Such compounds would therefore be unable to turn on the existing bacterial resistance systems, consequently outsmarting glycopeptide resistance in clinical infection.
A new model for sensing vancomycin.
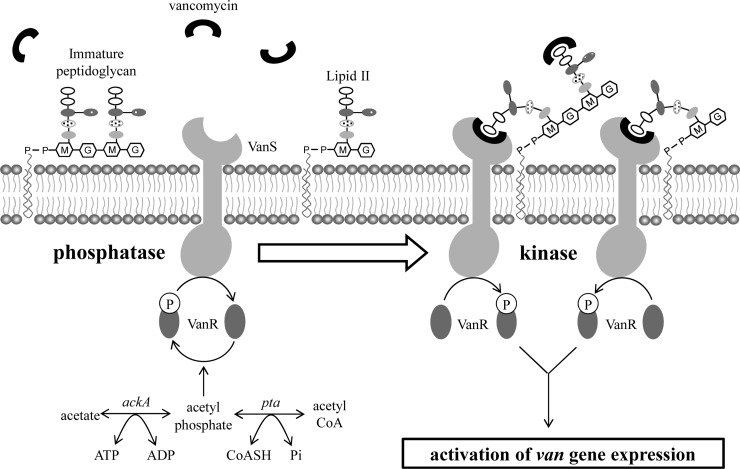
Using the VanB-type vancomycin resistance of S. coelicolor as a model system, recent studies have all favored a model in which the VanS sensor kinase is activated by directly binding the antibiotic over the alternative model in which the sensor kinase is activated by binding a cell wall metabolite accumulated as a result of antibiotic action (7–10). In this study, we have used in vivo approaches to extensively test the validity of the direct induction theory, and we now propose a revised model in which activation of resistance requires sensing of a complex between vancomycin and the d-Ala-d-Ala termini of extracellular cell wall PG precursors (Fig. 6). According to this new model, binding of vancomycin to lipid II or immature PG anchored in the lipid bilayer brings it into the vicinity of the membrane-localized VanS sensor and facilitates perception of the signal required to induce kinase activity in VanS. In contrast, unbound vancomycin is not perceived by the VanS sensor and VanS retains the phosphatase activity of its noninduced state. Importantly, the revised model is also fully consistent with the results of the earlier studies, activation of the resistance system only by antibiotics possessing closely related structures and labeling of VanS by a fluorescent photoaffinity vancomycin probe, which had been interpreted as indicating a simple direct induction (10).
Fig 6.
A model proposing that induction of the vancomycin resistance system in S. coelicolor via VanS kinase activity requires binding of the drug to d-Ala-d-Ala termini of the extracellular cell wall PG precursors. Vancomycin (black cup shapes) bound to d-Ala-d-Ala termini of extracellular PG precursor is perceived by VanS, which switches its activity from phosphatase to kinase, causing the accumulation of phospho-VanR, which in turn induces the expression of the van gene cluster and renders the cell resistant to vancomycin. Unbound vancomycin is not perceived by the VanS sensor, and VanS retains the phosphatase activity of its noninduced state.
Technical difficulties associated with working with a membrane-localized sensor protein and lipidated PG ligands have meant that obtaining definitive in vitro data has not yet been possible, but taken together, we believe that the in vivo studies presented here provide strong evidence for a mechanism by which VanS induction occurs via sensing of vancomycin in complex with a PG precursor terminating in d-Ala-d-Ala. Central to this are observations which indicate that the strength of induction of the resistance system by vancomycin correlates with the abundance of PG precursors terminating in d-Ala-d-Ala (as exemplified by UDP-MurNAc-pentapeptide) relative to those modified to end with d-Ala-d-Lac (as exemplified by UDP-MurNAc-pentadepsipeptide). Genetic engineering to replace the essential d-Ala-d-Ala ligase activity encoded by ddlA with the d-Ala-d-Lac ligase activity of vanA allowed construction of strains which synthesized intracellular PG precursors terminating in d-Ala-d-Lac (and consequently extracellular PG precursors terminating in d-Ala-d-Lac), even in the absence of vancomycin. In all cases, strains producing a higher proportion of d-Ala-d-Lac-containing PG precursors exhibited a lower response to vancomycin treatment in assays quantifying the induction of van gene expression. Interestingly, the response was not linear but was more sensitive to changes in the ratio of d-Ala-d-Ala-containing PG precursors to d-Ala-d-Lac-containing PG precursors in the range 100:0 to 80:20 (compare the ermEp-ddlA-complemented strain [H2009] with the ermEp-vanA-complemented strain [H2012] grown without d-Lac supplementation in Fig. 4A) than from 80:20 to 20:80 (compare the ermEp-vanA-complemented strain [H2012] grown without and with d-Lac supplementation in Fig. 4A). This may reflect a tuning of the sensory system that has evolved to keep the synthesis of d-Ala-d-Lac-containing PG precursors down to only the minimum required for survival in the prevailing vancomycin concentration detected in the environment (discussed further below). Consistent with these results, preexposure of cells to vancomycin desensitized them to a subsequent dose of the antibiotic such that little or no additional expression of the vanH reporter transcript was produced (Fig. 2A). While this could also be due to a saturation of the VanS sensor domain by the initial dose of vancomycin, pretreatment with teicoplanin, an antibiotic that binds to d-Ala-d-Ala residues in PG precursors but not to the VanS sensor (10), produced the same result (Fig. 2B), even in the absence of any reprogramming of cell wall biosynthesis toward synthesis of PG precursors containing the d-Ala-d-Lac termini. (Teicoplanin itself does not activate the resistance system.) Such reprogramming is, however, believed to be integral to the natural behavior of the sensory system, whereby initial exposure to vancomycin triggers a maximal upregulation of van gene expression in the first 15 to 30 min, which results in the synthesis of PG precursors terminating in d-Ala-d-Lac. As the proportion of these begins to increase in the nascent cell wall, the number of d-Ala-d-Ala sites available for vancomycin complex formation starts to decrease, leading to a gradual reduction in the output of the signal transduction pathway (7, 8). This diminution in the signaling response does not reflect any decrease in the concentration of vancomycin, which persists at high levels in the culture medium for at least 24 h. The proposal that it is a vancomycin-d-Ala-d-Ala PG precursor complex that is sensed rather than the drug itself is supported by the lack of any signaling response when cells are exposed to the desleucyl derivative of vancomycin (Fig. 5). The two compounds share virtually identical stereochemical structures and would be predicted to present themselves similarly for perception by the sensory domain of VanS. The absence of a leucine residue in the derivative, however, makes complex formation with d-Ala-d-Ala PG precursors impossible (37, 39), and according to the model, desleucyl vancomycin is as a consequence not sensed at all by VanS. We cannot, however, exclude the formal possibility that VanS senses vancomycin via interaction with its d-Ala-d-Ala binding pocket, but this seems unlikely given the inability of teicoplanin, which possesses an intact binding pocket, to activate the sensor.
The dual d-Ala-d-Lac and d-Ala-d-Ala ligase activities of VanA act in concert with VanH and VanX to set a limit to the proportion of d-Ala-d-Lac used in cell wall synthesis and establish a feedback loop for the perception of vancomycin.
Strain H2012 carries a copy of vanA constitutively expressed from the strong, constitutive promoter ermEp to complement the otherwise lethal consequences of deleting the d-Ala-d-Ala ligase activity encoded by ddlA. HPLC-MS/MS analysis of soluble cell wall PG precursors extracted from H2012 revealed that in addition to the expected precursors terminating in d-Ala-d-Lac, those terminating in d-Ala-d-Ala were also readily detectable (Fig. 4A). Since the ΔddlA mutant is only viable when vanA is expressed (either through genetic engineering or by addition of vancomycin), and therefore there is no evidence for functional complementation by an as yet unidentified gene, this implies that S. coelicolor VanA must possess both d-Ala-d-Ala and d-Ala-d-Lac ligase activities. The A47934 glycopeptide antibiotic biosynthesis and resistance gene cluster in Streptomyces toyocaensis encodes a ligase, DdlM, that is highly homologous to VanA (78% amino acid identity over their 346-amino-acid length). In in vitro studies, Marshall and Wright (40) showed that DdlM can synthesize both d-Ala-d-Lac and d-Ala-d-Ala, but preferentially synthesizes the depsipeptide d-Ala-d-Lac. Surprisingly, an S. coelicolor ΔddlA mutant expressing vanA from a constitutive promoter (H2012) preferentially expressed d-Ala-d-Ala-containing PG precursors over d-Ala-d-Lac-containing PG precursors (4:1 ratio). The d-Lac synthetase and d-Ala-d-Ala hydrolase activities encoded by vanH and vanX, respectively, are, however, not expressed in this strain under these conditions, and it is therefore likely that d-Lac is in limited supply compared to d-Ala and that d-Ala-d-Ala is readily available. Interestingly, supplementation of the medium used to grow this strain with 10 mM d-Lac dramatically altered the relative abundance of d-Ala-d-Ala-containing PG precursors and d-Ala-d-Lac-containing PG precursors from 4:1 to 1:4, respectively. In the absence of either vanH expression or an exogenous d-Lac supply, we speculate that only a low level of intracellular d-Lac is available, presumably synthesized by the VanH homologue Ddlh2 (SCO2118). The proportion of d-Ala-d-Ala-containing PG precursors relative to those containing d-Ala-d-Lac is typically much smaller than 1:4 in S. coelicolor cells possessing a functional van resistance cluster when treated with vancomycin (e.g., 1:9 in H2027 in Fig. 4A), presumably reflecting the additional influence of VanX activity on the d-Ala-d-Ala/d-Ala-d-Lac metabolite pools. In contrast to vanA, constitutive ermEp-driven expression of ddlA in the ΔddlA mutant background only produced PG precursors terminating in d-Ala-d-Ala; d-Ala-d-Lac-containing PG precursors were never detected, even during growth with supplementation by 10 mM d-Lac (see the ermEp-ddlA-complemented strain H2009 in Fig. 4A). Attempts to complement a ΔvanA mutant using ddlA expressed from the constitutive ermEp or vancomycin-inducible van promoters were also unsuccessful, and DdlA therefore appears to possess d-Ala-d-Ala ligase activity exclusively. This is consistent with previous in vitro studies of the ddlA homologue in S. toyocaensis (40).
Expression of S. coelicolor ddlA is significantly downregulated in response to vancomycin (41), and the dual ligase activity of the inducible VanA enzyme can therefore help ensure that a proportion of PG precursors terminating in d-Ala-d-Ala are synthesized following vancomycin treatment. As a consequence, PG synthesis never takes place entirely via d-Ala-d-Lac-containing precursors, a possibility that has evidently been selected against during the process of evolution. Indeed, even the predominant production of d-Ala-d-Lac-containing cell wall precursors through constitutive expression of vanHAX comes at a severe fitness cost in bacteria, and this is believed to be why the majority of vancomycin resistance systems are inducible systems (42, 43). d-Lac is synthesized directly from the key glycolytic intermediate pyruvate, and production of the d-Ala-d-Lac depsipeptide is therefore likely to be energetically significantly more costly than d-Ala-d-Ala synthesis that is derived via isomerization of l-Ala (44), which can be salvaged from proteolytic pathways. The ester bond present in the d-Ala-d-Lac depsipeptide is inherently more reactive than the d-Ala-d-Ala amide linkage (due to the higher electronegativity of the O-atom compared to the N-atom) making it more susceptible to degradation by both chemical and enzymatic hydrolysis. In addition, d-Ala-d-Lac-containing cell wall precursors are unlikely to be the optimal substrates for all of the housekeeping enzymes involved in peptidoglycan biosynthesis and cell wall homeostasis. Hesketh et al. (32) showed that vancomycin treatment significantly altered the expression of five genes encoding penicillin-binding proteins (PBPs) in S. coelicolor, suggesting that a different spectrum of enzymes may be required to handle the changes induced in cell wall precursor substrate biosynthesis. For all of these reasons, synthesizing the minimal amount of d-Ala-d-Lac-containing PG precursors required for survival under the prevailing vancomycin concentration would be advantageous. The dual ligase activity of VanA coupled with the proposed complexation model for activating expression of the van resistance genes would help to ensure this is the case. Importantly, the observation that the combined activities of VanHAX always permit the synthesis of some d-Ala-d-Ala-containing PG precursors establishes a feedback loop whereby cells grown in the presence of vancomycin always retain the potential to respond to the drug. As discussed earlier, the extent of this response will, however, be dependent on the size of the d-Ala-d-Ala-containing PG precursor pool present at the time of exposure. Further detailed characterization of the properties and responses of the VanS vancomycin sensor will be dependent on establishing the appropriate in vitro assay systems. Given the small size of the putative extracytoplasmic sensor domain of VanS in actinomycete proteins (ca. 24 to 30 amino acid residues) and the observed importance of the transmembrane helix in signal transmission (41, 45, 46), this will require overcoming the technical challenges associated with working with full-length membrane proteins.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mark Buttner for helpful discussions and comments on the manuscript. We also thank Daniel Kahne for the kind gift of desleucyl vancomycin.
The work was supported by the Royal Society (516002.K5877/ROG) and partially funded through MRC grant G0700141.
Footnotes
Published ahead of print 8 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00523-13.
REFERENCES
- 1.Barna JCJ, Williams DH. 1984. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu. Rev. Microbiol. 38:339–357 [DOI] [PubMed] [Google Scholar]
- 2.Pearson H. 2002. ‘Superbug' hurdles key drug barrier. Nature 418:469–470 [DOI] [PubMed] [Google Scholar]
- 3.Arthur M, Molinas C, Courvalin P. 1992. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 174:2582–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur M, Depardieu F, Courvalin P. 1999. Regulated interactions between partner and non-partner sensors and response regulators that control glycopeptide resistance gene expression in enterococci. Microbiology 145:1849–1858 [DOI] [PubMed] [Google Scholar]
- 5.Bugg TD, Wright GD, Dutka-Malen S, Arthur M, Courvalin P, Walsh CT. 1991. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 30:10408–10415 [DOI] [PubMed] [Google Scholar]
- 6.Novotna G, Hill C, Vicent K, Liu C, Hong H-J. 2012. A novel membrane protein, VanJ, conferring resistance to teicoplanin. Antimicrob. Agents Chemother. 56:1784–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong H-J, Hutchings MI, Neu JM, Wright GD, Paget MS, Buttner MJ. 2004. Characterisation of an inducible vancomycin resistance system in Streptomyces coelicolor reveals a novel gene (vanK) required for drug resistance. Mol. Microbiol. 52:1107–1121 [DOI] [PubMed] [Google Scholar]
- 8.Hong H-J, Hutchings MI, Hill L, Buttner MJ. 2005. The role of the novel Fem protein VanK in vancomycin resistance in Streptomyces coelicolor. J. Biol. Chem. 280:13055–13061 [DOI] [PubMed] [Google Scholar]
- 9.Hutchings MI, Hong H-J, Buttner MJ. 2006. The vancomycin resistance VanRS signal transduction system of Streptomyces coelicolor. Mol. Microbiol. 59:923–935 [DOI] [PubMed] [Google Scholar]
- 10.Koteva K, Hong H-J, Wang XD, Nazi I, Hughes D, Naldrett MJ, Buttner MJ, Wright GD. 2010. A vancomycin photoprobe identifies the histidine kinase VanSsc as a vancomycin receptor. Nat. Chem. Biol. 6:327–329 [DOI] [PubMed] [Google Scholar]
- 11.Hong H-J, Hutcings MI, Buttner MJ. 2008. Vancomycin resistance VanS/VanR two-component systems. Adv. Exp. Med. Biol. 631:200–213 [DOI] [PubMed] [Google Scholar]
- 12.Ulijasz AT, Grenader A, Weisblum BA. 1996. A vancomycin-inducible lacZ reporter system in Bacillus subtilis: induction by antibiotics that inhibit cell wall synthesis and by lysozyme. J. Bacteriol. 178:6305–6309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai MH, Kirsch DR. 1996. Induction signals for vancomycin resistance encoded by the vanA gene cluster in Enterococcus faecium. Antimicrob. Agents Chemother. 40:1645–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mani N, Sancheti P, Jiang ZD, McNaney C, DeCenzo M, Knight B, Stankis M, Kuranda M, Rothstein DM. 1998. Screening systems for detecting inhibitors of cell wall transglycosylation in Enterococcus. Cell wall transglycosylation inhibitors in Enterococcus. J. Antibiot. 51:471–479 [DOI] [PubMed] [Google Scholar]
- 15.Grissom-Arnold J, Alborn WE, Nicas TI, Jaskunas SR. 1997. Induction of VanA vancomycin resistance genes in Enterococcus faecalis: use of a promoter fusion to evaluate glycopeptide and nonglycopeptide induction signals. Microb. Drug Resist. 3:53–64 [DOI] [PubMed] [Google Scholar]
- 16.Baptista M, Depardieu F, Courvalin P, Arthur M. 1996. Specificity of induction of glycopeptide resistance genes in Enterococcus faecalis. Antimicrob. Agents Chemother. 40:2291–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen NE, Hobbs JN. 1995. Induction of vancomycin resistance in Enterococcus faecium by non-glycopeptide antibiotics. FEMS Microbiol. Lett. 132:107–114 [DOI] [PubMed] [Google Scholar]
- 18.Handwerger S, Kolokathis A. 1990. Induction of vancomycin resistance in Enterococcus faecium by inhibition of transglycosylation. FEMS Microbiol. Lett. 58:167–170 [DOI] [PubMed] [Google Scholar]
- 19.van Heijenoort J. 2001. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology 11:25R–36R [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Walker D, Sun B, Hu Y, Walker S, Kahne D. 2003. Vancomycin analogues active against vanA-resistant strains inhibit bacterial transglycosylase without binding substrate. Proc. Natl. Acad. Sci. U. S. A. 100:5658–5663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baptista M, Depardieu F, Reynolds P, Courvalin P, Arthur M. 1997. Mutations leading to increased levels of resistance to glycopeptide antibiotics in VanB-type enterococci. Mol. Microbiol. 25:93–105 [DOI] [PubMed] [Google Scholar]
- 22.Baptista M, Rodrigues P, Depardieu F, Courvalin P, Arthur M. 1999. Single-cell analysis of glycopeptide resistance gene expression in teicoplanin-resistant mutants of a VanB-type Enterococcus faecalis. Mol. Microbiol. 32:17–28 [DOI] [PubMed] [Google Scholar]
- 23.Paget MSB, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. 1999. Evidence that the extracytoplasmic function sigma factor, σE, is required for normal cell wall structure in Streptomcyes coelicolor A3(2). J. Bacteriol. 181:204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 18:1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics, The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 26.Billot-Klein D, Blanot D, Gutmann L, van Heijenoort J. 1994. Association constants for the binding of vancomycin and teicoplanin to N-acetyl-D-alanyl-D-alanine and N-acetyl-D-alanyl-D-serine. Biochem. J. 302:1021–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bierman M, Logan R, O'Brien K, Seno ET, Nagaraja Rao R, Schoner BE. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49 [DOI] [PubMed] [Google Scholar]
- 28.Paget MSB, Emmanuelle L, Buttner MJ. 1999. A putative two-component signal transduction system regulates σE, a sigma factor required for normal cell wall integrity in Streptomcyes coelicolor A3(2). Mol. Microbiol. 33:97–107 [DOI] [PubMed] [Google Scholar]
- 29.Jones GH, Paget MSB, Chamberlin L, Buttner MJ. 1997. σE is required for the production of the antibiotic actinomycin in Streptomyces antibioticus. Mol. Microbiol. 23:169–178 [DOI] [PubMed] [Google Scholar]
- 30.Hesketh A, Bucca G, Laing E, Flett F, Hotchkiss G, Smith CP, Chater KF. 2007. New pleiotropic effects of eliminating a rare tRNA from Streptomyces coelicolor, revealed by combined proteomic and transcriptomic analysis of liquid cultures. BMC Genomics 8:261. 10.1186/1471-2164-8-261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hesketh A, Kock H, Mootien S, Bibb M. 2009. The role of absC, a novel regulatory gene for secondary metabolism, in zinc-dependent antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 74:1427–1444 [DOI] [PubMed] [Google Scholar]
- 32.Hesketh A, Hill C, Mokhtar J, Novotna G, Tran N, Bibb M, Hong H-J. 2011. Genome-wide dynamics of a bacterial response to antibiotics that target the cell envelope. BMC Genomics 12:226. 10.1186/1471-2164-12-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruzin A, Severin A, Ritacco F, Tabei K, Singh G, Bradford PA, Siegel MM, Projan SJ, Shlaes DM. 2002. Further evidence that a cell wall precursor [C55-MurNAc-(peptide)-GlcNAc] serves as an acceptor in a sorting reaction. J. Bacteriol. 184:2141–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang J, Shi J, Molle V, Sohlberg B, Weaver D, Bibb MJ, Karoonuthaisiri N, Lih CJ, Kao CM, Buttner MJ, Cohen SN. 2005. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 58:1276–1287 [DOI] [PubMed] [Google Scholar]
- 35.Hong H-J, Paget MSB, Buttner MJ. 2002. A signal transduction system in Streptomyces coelicolor that activates the expression of a putative cell wall glycan operon in response to vancomycin and other cell wall-specific antibiotics. Mol. Microbiol. 44:1199–1211 [DOI] [PubMed] [Google Scholar]
- 36.Marshall CG, Zolli N, Wright GD. 1999. Molecular mechanism of VanHst, an α-ketoacid dehydrogenase required for glycopeptide antibiotic resistance from a glycopeptide producing organism. Biochemistry 38:8485–8491 [DOI] [PubMed] [Google Scholar]
- 37.Cristofaro MF, Beauregard DA, Yan H, Osborn NJ, Williams DH. 1995. Cooperativity between non-polar and ionic forces in the binding of bacterial cell wall analogues by vancomcyin in aqueous solution. J. Antibiot. 48:805–810 [DOI] [PubMed] [Google Scholar]
- 38.Jain RK, Trias J, Ellman JA. 2003. D-Ala-D-Lac binding is not required for the high activity of vancomycin dimers against vancomycin resistant enterococci. J. Am. Chem. Soc. 125:8740–8741 [DOI] [PubMed] [Google Scholar]
- 39.Kim SJ, Matsuoka S, Patti GJ, Schaefer J. 2008. Vancomycin derivative with damaged D-Ala-D-Ala binding cleft binds to cross linked peptidoglycan in the cell wall of Staphylococcus aureus. Biochemistry 47:3822–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marshall GG, Wright GD. 1997. The glycopeptide antibiotic producer Streptomyces toyocaensis NRRL15009 has both D-alanyl-D-alanine and D-alanyl-D-lactate ligases. FEMS Microbiol. Lett. 157:295–299 [DOI] [PubMed] [Google Scholar]
- 41.Hughson AG, Hazelbauer GL. 1996. Detecting the conformational change of transmembrane signaling in a bacterial chemoreceptor by measuring effects on disulphide cross-linking in vivo. Proc. Natl. Acad. Sci. U. S. A. 93:11546–11551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Foucault ML, Depardieu F, Courvalin P, Grillot-Courvalin C. 2010. Inducible expression eliminates the fitness cost of vancomycin resistance in enterococci. Proc. Natl. Acad. Sci. U. S. A. 107:16964–16969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foucault ML, Courvalin P, Grillot-Courvalin C. 2009. Fitness cost of VanA-type vancomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:2354–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walsh CT. 1989. Enzymes in the D-alanine branch of bacterial cell wall peptidoglycan assembly. J. Biol. Chem. 264:2393–2396 [PubMed] [Google Scholar]
- 45.Chervitz SA, Falke JJ. 1995. Lock on/off disulfides identify the transmembrane signaling helix of the aspartate receptor. J. Biol. Chem. 270:24043–24053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hall BA, Armitage JP, Sansom MSP. 2011. Transmembrane helix dynamics of bacterial chemoreceptors supports a piston model of signalling. PLoS Comp. Biol. 7:e1002204. 10.1371/journal.pcbi.1002204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.