Abstract
A carbapenem-resistant Acinetobacter baumannii strain was isolated from the peritoneal fluid of a patient with complicated intra-abdominal infection and evaluated at the Multidrug-resistant Organism Repository and Surveillance Network by whole-genome sequencing and real-time PCR. The isolate was sequence type 25 and susceptible to colistin and minocycline, with low MICs of tigecycline. blaNDM-1 was located on a plasmid with >99% homology to pNDM-BJ02. The isolate carried numerous other antibiotic resistance genes, including the 16S methylase gene, armA.
TEXT
Carbapenem-resistant Gram-negative organisms are emerging pathogens that threaten global public health and seriously challenge infection control and therapy efforts (1–3). Though still relatively uncommon, the carbapenemase encoded by blaNDM has quickly established itself as one of the most important (4). First described in 2009 (5), the gene has since been identified in nearly every continent and has been detected in species as diverse as Acinetobacter baumannii and Vibrio cholerae (6–8). Furthermore, the gene has been associated with a diverse range of plasmids that often harbor multiple antibiotic resistance genes (9).
In 2012, the Multidrug-resistant Organism Repository and Surveillance Network (MRSN) (10) received a request from a Ministry of Health hospital in Tegucigalpa, Honduras, for assistance in identifying the genetic mechanism associated with an extensively drug-resistant (XDR) isolate of A. baumannii. The isolate was recovered from a 76-year-old male patient admitted for ongoing dialysis in the setting of dysfunctional hemodialysis access. On the second day of admission, a peritoneal catheter was placed and the patient was prescribed empirical ciprofloxacin (250 mg) and a single dose of cephalothin (2 g). Two days later, the patient developed a fever (38.2°C) and experienced right lower abdominal pain. Peritoneal fluid cultures revealed moderate growth of Gram-negative bacilli. The patient was prescribed ceftriaxone (2 g/day) followed by intravenous ciprofloxacin (200 mg every 12 h) and intraperitoneal ceftazidime (250 mg). The organism was identified as A. baumannii with resistance to the fluoroquinolones and β-lactams, including carbapenems. Prompt clinical improvement followed removal of the peritoneal dialysis catheter and the initiation of tigecycline (50 mg every 12 h). He was discharged 17 days later and completed a 10-day course of therapy with tigecycline.
The isolate was forwarded to the MRSN and tested for carbapenemase genes by real-time PCR (11). Identification and antibiotic susceptibility tests were performed on three automated systems: the Vitek 2 (bioMérieux, Inc., NC), the BD Phoenix (BD Diagnostics Systems, MD), and the Microscan Walk-Away (Siemens Healthcare Diagnostics Inc., IL). MICs of colistin, minocycline, polymyxin B, and tigecycline were determined by Etest; colistin susceptibility was also assayed using broth microdilution. Whole genome sequencing was performed using an Ion Torrent Personal Genome Machine (PGM) with 200-bp chemistry.
The isolate (MRSN 12227) was identified as A. baumannii. It was resistant to amikacin, ampicillin-sulbactam, aztreonam, cefepime, cefotaxime, ceftazidime, ceftriaxone, ciprofloxacin, gentamicin, imipenem, levofloxacin, meropenem, tetracycline, tobramycin, and trimethoprim-sulfamethoxazole using CLSI breakpoints (12). The isolate was susceptible to minocycline (MIC = 3 μg/ml) and to colistin by broth microdilution (MIC = 0.25 μg/ml) and Etest (MIC = 0.125 μg/ml). It had low MICs of tigecycline (1.5 μg/ml) and polymyxin B (0.25 μg/ml), which correlate with the successful treatment of the patient with tigecycline.
MRSN 12227 was assigned to sequence type 25 (ST25) (Pasteur scheme), an ST that has been sporadically identified in a number of countries, including The Netherlands, Turkey, Greece, Italy, Sweden, and Singapore, and is not associated with any clonal complex (13, 14). Though sporadic, ST25 appears to be a stable clone and has been implicated in clinical infections since 1985 (14).
Mean coverage depth for genome assembly was 129-fold. MRSN 12227 carried blaNDM-1 on a plasmid that shared >99% homology to pNDM-BJ02, a plasmid initially identified in a clinical isolate of Acinetobacter lwoffii in Beijing, China (15). Interestingly, we also identified this plasmid in a clinical isolate of A. schindleri recovered from a surveillance groin swab of a U.S. service member wounded in Afghanistan (16). This plasmid is unclassifiable by PCR-based replicon typing, has a novel plasmid backbone sequence, and carries genes that encode a type IV secretion system (T4SS) that facilitates horizontal transmission (15). The composition of the genetic region surrounding blaNDM shares common elements with other plasmids carrying blaNDM in Enterobacteriaceae (17), including the putative bleomycin resistance gene bleMDL, the N-(5′-phosphoribosyl)anthranilate isomerase gene trpF, and the insertion sequence ISAba125 (Fig. 1). pNDM-BJ02 also carries a nonfunctional copy of aphA6, which normally encodes an aminoglycoside-modifying enzyme (AME) that confers resistance to amikacin. The same genetic architecture is apparent in the plasmid from MRSN 12227, with an upstream transposition event seemingly disrupting the promoter region of aphA6. However, unlike the A. lwoffii (15) and A. schindleri (16) isolates that also carried this plasmid, MRSN 12227 is resistant to amikacin. This discrepancy was resolved when further analysis of the MRSN 12227 genome using ResFinder (18) revealed a chromosomal copy of the 16S methylase gene, armA, which confers resistance to all aminoglycosides (19) (Table 1). pNDM-BJ02 has been reported to have a very high frequency of transfer (9.1 × 10−3 to 1.3 × 10−2 per donor cell) to Escherichia coli J53 Azir (15). Our experience with blaNDM-carrying strains of Acinetobacter provides further evidence for the promiscuity of this plasmid. In both instances in which the MRSN has encountered blaNDM-carrying strains of Acinetobacter, pNDM-BJ02 has been its vehicle of transmission.
Fig 1.
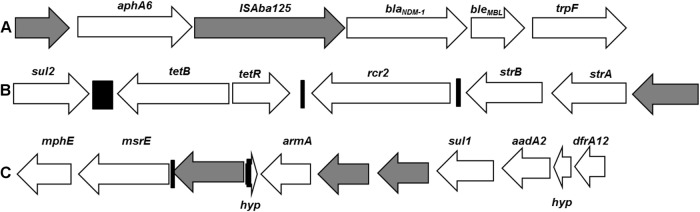
Composition of the genetic environment surrounding blaNDM-1 (A), the AbaR resistance island (B), and the 16S methylase gene armA (C). Arrows indicate gene orientation from 5′ to 3′. Relative gene sizes are proportional to the lengths of the arrows. White arrows indicate confirmed or putative functional genes with hypothetical genes, marked as hyp. Gray arrows indicate transposons, and black bars indicate repeat regions.
Table 1.
Antibiotic resistance genes carried by MRSN 12227a
| Geneb | Contigc | Locationd | Functione | Notes |
|---|---|---|---|---|
| blaOXA-64 | 1 | Chromosome | Class D β-lactamase | blaOXA-51-like |
| mphE | 1 | Chromosome | Macrolide phosphotransferase | Confers resistance to macrolides |
| msrE | 1 | Chromosome | Macrolide efflux protein | Confers resistance to macrolides |
| armA | 1 | Chromosome | 16S methylase | Confers resistance to all aminoglycosides |
| sul1 | 1 | Chromosome | Dihydropteroate synthase | Confers resistance to sulfonamides |
| ant(3″)-Ia | 1 | Chromosome | AME | aadA2; confers resistance to gentamicin and tobramycin |
| dfrA-12 | 1 | Chromosome | Dihydrofolate reductase | Confers resistance to trimethoprim |
| ceoA-like | 6 | Chromosome | RND efflux pump | |
| ceoB | 6 | Chromosome | RND efflux pump | 98% identity to CeoB (protein)f |
| opcM-like | 6 | Chromosome | RND efflux pump | |
| sul2 | 38 | Chromosome | Dihydropteroate synthase | Confers resistance to sulfonamides |
| tetB | 38 | Chromosome | Efflux pump | Confers resistance to tetracycline |
| aph(6)-Id | 38 | Chromosome | AME | strB; confers resistance to streptomycin |
| aph(3″)-Ib | 38 | Chromosome | AME | strA; confers resistance to streptomycin |
| blaCTX-M-15 | 44 | pCTXM360 | ESBL | Confers resistance to cephalosporins and monobactams |
| aph(3′)-VIa | 50 | pNDM-BJ02 | AME | Nonfunctional |
| blaNDM-1 | 50 | pNDM-BJ02 | Class B β-lactamase | Confers resistance to all β-lactams except aztreonam |
| aac(3)-IIa | 56 | Putative Plasmid | AME | Confers resistance to gentamicin and tobramycin |
| blaTEM-1 | 59 | Unknown | ESBL | 3 synonymous SNPsg; resistance to early cephalosporins |
Abbreviations: AME, aminoglycoside-modifying enzyme; ESBL, extended-spectrum β-lactamases; SNP, single-nucleotide polymorphism; RND, resistance/nodulation/division.
Based on closest match to BLAST (http://blast.ncbi.nlm.nih.gov) search.
The MRSN 12227 whole-genome sequence was assembled into 95 contigs. The relative sizes of the reported contigs (in base pairs) are as follows: contig 1, 383,184; contig 6, 197,316; contig 38, 23,159; contig 44, 13,686; contig 50, 8,127; contig 56, 1,720; and contig 59, 1,375.
Putative or confirmed location of the respective antibiotic resistance gene based on whole-genome sequencing.
Confirmed or putative function of protein.
Confers resistances to chloramphenicol, trimethoprim, and ciprofloxacin.
blaTEM-1contains 3 synonymous mutations compared to the reference blaTEM-1 sequence (26). pCTXM-360 contains blaTEM in close proximity to blaCTX-M-15, but the exact location of blaTEM-1 is still uncertain.
MRSN 12227 also carries plasmid pCTXM360, which harbors the β-lactamase gene blaCTX-M-15 (20), and has two additional chromosomal insertions carrying multiple antibiotic resistance genes (Table 1 and Fig. 1). The first of these carries six antibiotic resistance loci, including the aforementioned 16S methylase gene armA and the macrolide resistance genes mphE and msrE (Table 1 and Fig. 1). This structure has been identified on plasmids associated with Klebsiella oxytoca and Citrobacter freundii (GenBank accession numbers CP003684.1 and JX182975.1, respectively) and on the chromosome of A. baumannii MDR-TJ and TYTH-1 (21, 22). The second region also includes multiple antibiotic resistance genes, including the tetracycline efflux gene tetB, the streptomycin resistance genes strA and strB, and the dihydropteroate synthase gene sul2. This region most closely resembles the Acinetobacter resistance island AbaR1 (23), which is widely distributed in Acinetobacter baumannii strains. MRSN 12227 carries the chromosomally encoded blaOXA-51-like gene blaOXA-64 but does not carry any acquired class D carbapenemase genes, indicating that carbapenemase resistance in this strain is probably due to blaNDM-1.
This is the first report of blaNDM from Honduras, and it follows the recent identification of this gene in a strain of Klebsiella pneumoniae in neighboring Guatemala (24). The current epidemiological evidence from the hospital supports nosocomial transmission of MRSN 12227, but further investigations are ongoing. The modified Hodge test and the Carba NP tests were both negative for MRSN 12227, but this is not unexpected for Acinetobacter (25). Finally, pNDM-BJ02 appears to be emerging as a common vehicle for the horizontal transmission of blaNDM, particularly in Acinetobacter species. In vitro evidence suggests that this plasmid is highly promiscuous, and further research is warranted to see if this plasmid is capable of being transmitted and stably maintained in other clinically important bacteria.
ACKNOWLEDGMENTS
Major funding for this study was provided by the U.S. Army Medical Command (MEDCOM) and the Department of Defense Global Emerging Infections Surveillance and Response System (GEIS). We gratefully acknowledge the support of Guy Lemire and Bart Diaz at the Joint Task Force-Bravo, Soto Cano Air Base, Honduras. We also gratefully acknowledge the support and assistance of Douglas Lougee, a surgeon with the U.S. Southern Command (USSOUTHCOM). We thank Blanca Hernández and Fanny Hidalgo, Facultad de Microbiología, Universidad Nacional Autónoma de Honduras, for isolate collection and transport.
Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official, or reflecting the views of the Department of the Army or the Department of Defense.
Footnotes
Published ahead of print 1 July 2013
REFERENCES
- 1.Livermore DM. 2012. Current epidemiology and growing resistance of gram-negative pathogens. Korean J. Intern. Med. 27:128–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 18:263–272 [DOI] [PubMed] [Google Scholar]
- 3.Patel G, Bonomo RA. 2011. Status report on carbapenemases: challenges and prospects. Expert Rev. Anti Infect. Ther. 9:555–570 [DOI] [PubMed] [Google Scholar]
- 4.Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol. 19:588–595 [DOI] [PubMed] [Google Scholar]
- 5.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushnell G, Mitrani-Gold F, Mundy LM. 15 January 2013, posting date Emergence of New Delhi metallo-beta-lactamase type 1-producing Enterobacteriaceae and non-Enterobacteriaceae: global case detection and bacterial surveillance. Int. J. Infect. Dis. [Epub ahead of print.] 10.1016/j.ijid.2012.11.025 [DOI] [PubMed] [Google Scholar]
- 7.Darley E, Weeks J, Jones L, Daniels V, Wootton M, MacGowan A, Walsh T. 2012. NDM-1 polymicrobial infections including Vibrio cholerae. Lancet 380:1358. [DOI] [PubMed] [Google Scholar]
- 8.Karthikeyan K, Thirunarayan MA, Krishnan P. 2010. Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J. Antimicrob. Chemother. 65:2253–2254 [DOI] [PubMed] [Google Scholar]
- 9.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11:355–362 [DOI] [PubMed] [Google Scholar]
- 10.Waterman P, Kwak Y, Clifford R, Julius M, Onmus-Leone F, Tsurgeon C, Riley M, Black C, McGann P, Lesho E. 2012. A multidrug-resistance surveillance network: 1 year on. Lancet Infect. Dis. 12:587–588 [DOI] [PubMed] [Google Scholar]
- 11.Milillo M, Kwak YI, Snesrud E, Waterman PE, Lesho E, McGann P. 2013. Rapid and simultaneous detection of blaKPC and blaNDM by use of multiplex real-time PCR. J. Clin. Microbiol. 51:1247–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing: 19th informational supplement. CLSI document M100-S21 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 13.Di Popolo A, Giannouli M, Triassi M, Brisse S, Zarrilli R. 2011. Molecular epidemiological investigation of multidrug-resistant Acinetobacter baumannii strains in four Mediterranean countries with a multilocus sequence typing scheme. Clin. Microbiol. Infect. 17:197–201 [DOI] [PubMed] [Google Scholar]
- 14.Karah N, Sundsfjord A, Towner K, Samuelsen O. 2012. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist. Updat. 15:237–247 [DOI] [PubMed] [Google Scholar]
- 15.Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, Hao Q, Yang X, Yang X, Xiao X, Luan C, Yang Y, Cui Y, Yang R, Gao GF, Song Y, Zhu B. 2012. Novel plasmid and its variant harboring both a bla(NDM-1) gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob. Agents Chemother. 56:1698–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGann P, Milillo M, Clifford RJ, Snesrud E, Stevenson L, Backlund MG, Viscount HB, Quintero R, Kwak YI, Zapor MJ, Waterman PE, Lesho EP. 3 April 2013, posting date Detection of New Delhi metallo-beta-lactamase (blaNDM-1) in Acinetobacter schindleri during routine surveillance. J. Clin. Microbiol. [Epub ahead of print.] doi:10.1128/JCM.00281-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 55:5403–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agerso Y, Lund O, Larsen MV, Aarestrup FM. 2013. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J. Antimicrob. Chemother. 68:771–777 [DOI] [PubMed] [Google Scholar]
- 19.Galimand M, Courvalin P, Lambert T. 2003. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 47:2565–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu WH, Luo L, Wang JY, Zhuang XH, Zhong L, Liao K, Zeng Y, Lu YJ. 2009. Complete nucleotide sequence of pCTX-M360, an intermediate plasmid between pEL60 and pCTX-M3, from a multidrug-resistant Klebsiella pneumoniae strain isolated in China. Antimicrob. Agents Chemother. 53:5291–5293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang H, Yang ZL, Wu XM, Wang Y, Liu YJ, Luo H, Lv X, Gan YR, Song SD, Gao F. 2012. Complete genome sequence of Acinetobacter baumannii MDR-TJ and insights into its mechanism of antibiotic resistance. J. Antimicrob. Chemother. 67:2825–2832 [DOI] [PubMed] [Google Scholar]
- 22.Liou ML, Liu CC, Lu CW, Hsieh MF, Chang KC, Kuo HY, Lee CC, Chang CT, Yang CY, Tang CY. 2012. Genome sequence of Acinetobacter baumannii TYTH-1. J. Bacteriol. 194:6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, Snyder M. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21:601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasteran F, Albornoz E, Faccone D, Gomez S, Valenzuela C, Morales M, Estrada P, Valenzuela L, Matheu J, Guerriero L, Arbizu E, Calderon Y, Ramon-Pardo P, Corso A. 2012. Emergence of NDM-1-producing Klebsiella pneumoniae in Guatemala. J. Antimicrob. Chemother. 67:1795–1797 [DOI] [PubMed] [Google Scholar]
- 25.Bonnin RA, Naas T, Poirel L, Nordmann P. 2012. Phenotypic, biochemical, and molecular techniques for detection of metallo-beta-lactamase NDM in Acinetobacter baumannii. J. Clin. Microbiol. 50:1419–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sirot D, Sirot J, Labia R, Morand A, Courvalin P, Darfeuille-Michaud A, Perroux R, Cluzel R. 1987. Transferable resistance to third-generation cephalosporins in clinical isolates of Klebsiella pneumoniae: identification of CTX-1, a novel beta-lactamase. J. Antimicrob. Chemother. 20:323–334 [DOI] [PubMed] [Google Scholar]


