Abstract
Hypoxia has critical effects on the physiology of organisms. In the yeast Saccharomyces cerevisiae, glycolytic enzymes, including enolase (Eno2p), formed cellular foci under hypoxia. Here, we investigated the regulation and biological functions of these foci. Focus formation by Eno2p was inhibited temperature independently by the addition of cycloheximide or rapamycin or by the single substitution of alanine for the Val22 residue. Using mitochondrial inhibitors and an antioxidant, mitochondrial reactive oxygen species (ROS) production was shown to participate in focus formation. Focus formation was also inhibited temperature dependently by an SNF1 knockout mutation. Interestingly, the foci were observed in the cell even after reoxygenation. The metabolic turnover analysis revealed that [U-13C]glucose conversion to pyruvate and oxaloacetate was accelerated in focus-forming cells. These results suggest that under hypoxia, S. cerevisiae cells sense mitochondrial ROS and, by the involvement of SNF1/AMPK, spatially reorganize metabolic enzymes in the cytosol via de novo protein synthesis, which subsequently increases carbon metabolism. The mechanism may be important for yeast cells under hypoxia, to quickly provide both energy and substrates for the biosynthesis of lipids and proteins independently of the tricarboxylic acid (TCA) cycle and also to fit changing environments.
INTRODUCTION
Hypoxia is a condition of inadequate oxygen supply. Many studies define hypoxia at ≤2 mg/liter dissolved oxygen (DO) in a water environment (1, 2). In mammalian culture cells, 1% and 21% atmospheric oxygen are considered to be hypoxia and normoxia, respectively (3, 4). Hypoxia in mammalian cells in vivo often occurs when the oxygen supply is limited (5) and has been reported to correlate with many diseases, including heart attack, cancer, and stroke (6). In response to hypoxia, some tumor cells were shown to gain increased metastatic activity (7), radiation resistance (8), and drug resistance (9). Responses of yeast cells to hypoxia have also attracted attention, because they are important in infections by pathogenic fungi such as Candida albicans (10) and Aspergillus fumigatus (11). Under hypoxia, mammalian and yeast cells share common sensing mechanisms and physiological responses to some extent (12). Hypoxia induces the release of reactive oxygen species (ROS) from mitochondria via the participation of complex III (13). The mechanisms of ROS generation under hypoxia, although not completely understood, are known to depend on a proton gradient across the inner mitochondrial membrane (14). In addition to activating hypoxia-responsive element-regulated genes in the nucleus (14), ROS also triggers the AMP-activated protein kinase (AMPK) signaling pathway (15–18) independently of the cellular AMP/ATP ratio (19).
Under hypoxia, mammalian and yeast cells produce energy and substrates for glycolysis-dependent biosynthesis (4, 20). Specifically, in response to hypoxia, mammalian culture cells produce increased amounts of amino acids, fatty acids, and phospholipids, in addition to lactate (4, 21), whereas Saccharomyces cerevisiae cells produce increased amounts of ethanol, glycerol, succinate, and alanine (22, 23) as the end products of glycolysis. The production of pyruvate, which is associated with the synthetic pathways of fatty acids, nucleotides, and other amino acids (24), is necessary for cell growth under hypoxia.
For the rapid production of glycolytic end products, the assembly of glycolytic enzymes into a complex has been considered to be effective (25, 26), in addition to the transcriptional regulation. Indeed, the specific intracellular localization of glycolytic enzymes has been reported in some organisms and cells. In a few protozoan species, including Trypanosoma brucei, glycolytic enzymes are contained in the membrane-enclosed organelle called the glycosome (27, 28). Some glycolytic enzymes, such as GAPDH (glyceraldehyde-3-phosphate dehydrogenase), aldolase, and lactate dehydrogenase, have been found to be associated with the cytoskeleton (29, 30), erythrocyte membrane (31), muscle (32), or each other (33–35). The association of glycolytic enzymes has been considered to contribute to metabolic efficiency by increasing pyruvate production (25, 26); however, this notion is still under debate (36, 37). To date, the regulation of metabolic pathways via the association of metabolic enzymes has been found only in plants, in which 10 glycolytic enzymes were identified to be associated with mitochondria for supporting respiration (38, 39).
An intracellular assembly of a glycolytic enzyme under hypoxia was recently reported in mammalian cells; that is, the glycolytic enzyme GAPDH conjugated with green fluorescent protein (GFP) was found to form fluorescent foci under hypoxia (40). Given that changes in carbon metabolism under hypoxia have been reported (4, 41, 42), the regulation of metabolic pathways by the spatial reorganization of glycolytic enzymes under hypoxia is plausible. In S. cerevisiae, 203 proteins, including heat shock proteins, have been shown to change their cellular localization under hypoxia (43). In those experiments, a hypoxic condition was achieved with 95% N2 and 5% H2. Although localization changes of glycolytic enzymes in hypoxic yeast cells have never been reported, it is nonetheless possible that glycolytic enzymes change their localization under hypoxia as a result of excess CO2 during fermentation (44, 45).
Spatial reorganization of proteins and organelles is often a sign of unexpected phenomena in the cell. Some researchers have found novel phenomena by tracking the fluorescence of protein-conjugated GFP (46). For example, in several organisms, the synthesis of purine and CTP is promoted by the formation of protein complexes (47–50). We previously found that recombinant enhanced GFP (EGFP) conjugated with N-terminal amino acid residues (aa 1 to 28) of yeast enolase (Eno2p) can form fluorescent foci in the cell (51). We speculated that the N-terminal (1 to 28) amino acid sequence might be the region regulating the intracellular localization of Eno2p. Full-length Eno2p conjugated with EGFP was localized uniformly in cells grown in an aerobic culture (51). If the N-terminal region of Eno2p participates in its localization, full-length Eno2p conjugated with fluorescent proteins would be expected to form foci under unknown environmental stimuli or during a specific phase of the cell life cycle. Moreover, amino acid substitutions that inhibit the focus formation by the Eno2p N-terminal region would also inhibit the focus formation by full-length Eno2p. The comparison of focus-forming and -nonforming cells, in conjunction with the use of reagents that inhibit focus formation by interfering with specific cellular processes, may reveal the mechanisms for the regulation and biological functions of focus formation. Here, we demonstrated hypoxia-triggered focus formation of Eno2p and investigated the mechanisms for the regulation and physiological effects of spatial reorganization of Eno2p.
MATERIALS AND METHODS
Strains and media.
The Escherichia coli DH5α strain [F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1] was used as host cells in the cloning experiments. The yeast strain BY4741 (MATa his3Δ1 leu2Δ met15Δ, ura3Δ) and the derived deletion strains of HOG1 (hog1Δ), SCH9 (sch9Δ), SNF1 (snf1Δ), and UPC2 (upc2Δ) were purchased from EUROSCARF (Frankfurt, Germany). The yeast GFP clones (Invitrogen, Carlsbad, CA) with GFP-tagged endogenous proteins (Eno2p, Glk1p, Pgi1p, Pfk1p, Fba1p, Tpi1p, Tal1p, Tkl1p, Tdh3p, Pgk1p, Gpm1p, Cdc19p [Pyk1p], Pyc1p, Pdc1p, Ald6p, Fas1p, Fas2p, and Eno1p) and the HIS3 marker in the parent BY4741 strain were used to determine changes in protein localizations. E. coli was grown in lysogeny broth containing 1% (wt/vol) tryptone, 0.5% (wt/vol) yeast extract, 0.5% (wt/vol) sodium chloride, and 100 μg/ml ampicillin. Yeast cells were grown in yeast extract peptone dextrose (YPD) medium (1% [wt/vol] yeast extract, 2% [wt/vol] polypeptone, and 2% [wt/vol] glucose), YPD+G418 medium (YPD medium supplemented with 0.2 mg/ml G418 disulfate; Nacalai Tesque, Kyoto, Japan), SDC+HM agar medium (0.67% [wt/vol] yeast nitrogen base without amino acids, 2% [wt/vol] glucose, 0.002% l-histidine-HCl, 0.003% l-methionine, 2% Casamino Acids [BD, Franklin Lakes, NJ], and 2% [wt/vol] agar), SDC+HM medium (0.67% [wt/vol] yeast nitrogen base without amino acids, 2% [wt/vol] glucose, 0.002% l-histidine-HCl, 0.003% l-methionine, 2% Casamino Acids [BD], 50 mM MES [2-(morpholino)ethanesulfonic acid], pH 6.0), or SC+ML medium (0.67% [wt/vol] yeast nitrogen base without amino acids, 2% [wt/vol] glucose, 0.003% l-methionine, 0.003% l-leucine, 0.13% SD multiple drop out [-Ade, -His, -Leu, -Lys, -Trp, -Ura; Funakoshi Co., Ltd., Tokyo, Japan], and 2% [wt/vol] agar). Glucose solutions were added to media after autoclaving to avoid the Maillard reaction.
Construction of plasmids.
Plasmids pULI1 and pUL-ATG-EGFP were used not only to adjust growth conditions for different cell types but also as controls. To determine amino acid residues of Eno2p important for focus formation, plasmids encoding Eno2p fragments as well as fragments carrying alanine substitutions (see Table S1 in the supplemental material) were constructed. iProof DNA Polymerase (Bio-Rad, Richmond, CA), Ligation High (Toyobo, Tokyo, Japan), and synthetic oligonucleotides (Japan Bio Services, Asaka, Japan) were used for the plasmid construction. DNA sequencing was performed using the BigDye Terminator v3.1 Cycle Sequencing kit and the ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). All other chemicals were of analytical grade. Primers and restriction enzymes used are listed in Table S1 in the supplemental material. In brief, nucleotide sequences were amplified or mixed (for pUL-ATG-EGFP construction) and ligated with restriction fragments of plasmids (pULSG1 [52] and pRS423 [ATCC]).
Plasmid transformation.
Yeast cells were transformed with plasmids using the Frozen-EZ Yeast Transformation II kit (Zymo Research, Orange, CA) and grown on SDC+HLM agar plates. Transformants were selected as single colonies and inoculated into 10 ml of SDC+HM medium with 50 mM MES (pH 6.0) for preculture at 25°C with shaking. At the late log phase, precultures were subcultured in 100 ml of SD+HM medium at an optical density at 600 nm (OD600) of 0.01 and incubated at 25°C with shaking for 24 h. Cultures were then subcultured in 100 ml of SDC+HM medium with 50 mM MES (pH 6.0) at an OD600 of 0.1 for aerobic or semianaerobic (CO2 bubbled) cultures at indicated temperatures.
Preparation of yeast genome.
The Gentoru-kun High Recovery kit (TaKaRa, Otsu, Japan) was used to extract genomic DNA of yeast GFP clones and knockout mutants. Resulting genome samples were used as the templates for preparing nucleotide fragments to be transformed into cells.
Construction of GFP-encoding yeast cells.
To construct a GFP clone of ENO2 containing the V22A substitution, an ENO2 knockout strain was constructed (see Table S1 in the supplemental material). After transformation of gene fragments, cells were grown at 25°C. Successful knockout of ENO2 was confirmed by sequencing the genomic sequence. Oligonucleotide fragments containing ENO1-GFP-HIS3 and ENO2V22A-GFP-HIS3 were prepared (see Table S1 in the supplemental material) and then inserted into the genome of the ΔENO2 strain at the position of ENO2. Yeast cells were transformed with nucleotide fragments and grown on SC+MLU agar plates. Single colonies were picked and again cultured on SC+ML or SC+MLU agar plates. Resulting cells were inoculated into SDC+HM media with 50 mM MES (pH 6.0) and cultured. Construction of GFP-encoding yeast cells was confirmed by microscopic observation of fluorescence.
Preparation of knockout mutants of GFP clones.
The primers used are listed in Table S1 in the supplemental material. The following 2 methods were adopted. For the first method, KanMX4-containing gene fragments were amplified from genomic DNA of yeast knockout mutant strains and transformed into yeast GFP clones. Transformants were cultivated on YPD+G418 agar plates, and resulting single colonies were again plated on SC+MLU+G418 agar media. For the second method, target gene fragments conjugated with GFP-HIS3 were amplified and transformed into knockout strains. Transformants were cultured on SC+MLU agar plates, and resulting single colonies were again plated on the same media. Constructed yeast strains were cultured in YPD+G418 liquid media and transformed with the plasmid pULI1. Transformants were cultured on SDC+HM agar plates, and the resulting colonies were used.
Culture conditions.
For aerobic cultivation, a 500-ml Erlenmeyer flask with 100 ml of media was used. For semianaerobic cultivation, a modified method of Katahira et al. (53) was used. In brief, culture vials with 100 ml of media and stir bar were used. For the introduction of CO2 before cultivation, CO2 was bubbled for 2 min into culture media to remove DO. To provide air in the culture vial, a small air pump (Ei-bukubuku set; Kotobuki-kogei, Matsubara, Japan) equipped with a needle-connected tube was used. Yeast cells were cultivated at indicated temperatures with stirring at 130 rpm.
pH and DO measurement.
pH measurement of culture media was performed using an F-52 pH meter (Horiba, Kyoto, Japan). Time course measurement of DO (mg/liter) was performed using a luminescent DO (LDO) meter (HQ30d; Hach Co., Loveland, CO). Measurements were recorded automatically every 15 min for 8 h.
Treatment of cells with reagents.
Stock solutions of 100 mM farnesol (Sigma) and 1 mg/ml rapamycin (Santa Cruz Biotechnology, Santa Cruz, CA) were prepared in ethanol. To determine whether mitochondria participate in focus formation, stock solutions of 5 mM carbonyl cyanide m-chlorophenylhydrazone (CCCP; Sigma) and 10 mM antimycin (Sigma) were prepared in ethanol and added to media. Antioxidant N-acetyl-l-cysteine (NAC; Nacalai Tesque) was directly added to media.
Fluorescence microscopy.
For confocal microscopy, cells were fixed with 4% (wt/vol) paraformaldehyde-containing phosphate-buffered saline (PBS) buffer for 1 h. For the observation of foci, fixed or unfixed cells were immediately mounted on a glass slide and observed. For confocal microscopy, a Carl Zeiss LSM 700 laser scanning microscope (Carl Zeiss, Oberkochen, Germany) with a 100× objective (oil immersion numerical aperture [NA], 1.40), ZEN Software (Carl Zeiss), and Imaris Software (Bitplane AG, Zurich, Switzerland) were used. Otherwise, an inverted fluorescence microscope IX71 (Olympus, Tokyo, Japan) with BP 470 to 490 excitation filter and BA 510 to 550 emission filter (Olympus), as well as a 100× objective (oil immersion NA, 1.40) and Aquacosmos software (Hamamatsu Photonics, Hamamatsu, Japan), were used.
Measuring focus-forming cells.
The proportion of focus-forming cells was calculated as follows. More than 100 cells in total were counted each time in two-dimensional pictures taken by an epifluorescence microscope, using Katikati Counter (GTSOFT; http://www.geocities.jp/gen_0715/). The proportion of focus-forming cells was presented as means ± standard errors of the means (SEM) (n ≥ 3). The significance of the difference in the ratio of focus-forming cells was tested using the one-shoulder t test.
FACS analysis.
For fluorescence activated cell sorter (FACS) analysis, cells were suspended in PBS (pH 7.4) and assayed immediately using a cell sorter (JSAN; Bay Bioscience, Kobe, Japan) with the detection channel FLT1 (535DF45). In each case, fluorescence data for 10,000 cells were acquired.
Preparation of yeast proteins for proteomic analysis.
Saccharomyces cerevisiae strain BY4741 transformed with pYEX-ENO2G or pYEX-ENO2V22AG was cultivated aerobically or semianaerobically at 30°C. Cells were lysed, and proteins were extracted as follows. Briefly, 250 μl of 25 mM Tris-HCl buffer (pH 7.8) was added to frozen cells. After homogenization (3 times) at 4,000 rpm for 60 s using glass beads (GB-05; diameter, 0.5 mm; TOMY, Tokyo, Japan) and Bead Smash 12 (Wakenyaku, Kyoto, Japan), sample solutions were centrifuged at 9,700 × g at 4°C for 5 min. Supernatant aliquots (500 μl) were filtrated using 0.45-μm spin column filter membranes (Durapore polyvinylidene difluoride [PVDF] membrane; Millipore, Eschborn, Germany) and then set still on ice. Purification of proteins was carried out immediately after protein extraction using Anti-FLAG M2 affinity gel (Sigma) and column (Poly-Prep Chromatography Columns; Bio-Rad) according to the manufacturer's protocol. After purification, samples were washed with 20 mM triethylammonium bicarbonate using the Microcon YM-3 concentrator (Millipore). Collected proteins were reduced with 10 mM Tris(2-carboxyethyl)phosphine (Thermo Scientific, Bremen, Germany) for 30 min and alkylated with 20 mM iodoacetamide (Thermo Scientific) for 60 min in the dark at room temperature. After acetone precipitation, proteins were solubilized in 200 mM triethylammonium bicarbonate (Sigma). Protein digestion (trypsin/protein, 1:50) was performed overnight at 37°C, and tryptic digests were applied to a proteome analysis system.
LC/MS-MS analysis and MS data analysis.
Protein identification was performed with a liquid chromatography-tandem mass spectrometry system (LC/MS-MS) as described elsewhere (54). Proteolytic digests were separated by reversed-phase chromatography using the UltiMate3000 nano LC system (Dionex). A monolithic silica capillary column (200 cm long, 0.1-mm inside diameter [i.d.]) prepared with a mixture of tetramethoxysilane and methyltrimethoxysilane was used at a flow rate of 500 nl/min. The gradient was provided by changing the mixing ratio of the two following eluents: A, 0.1% (vol/vol) formic acid; and B, 80% acetonitrile containing 0.1% (vol/vol) formic acid. The gradient was started with 5% B and increased to 50% B for 600 min. Separated analytes were detected on an LTQ Velos linear ion trap mass spectrometer (Thermo Scientific). An electrospray ionization (ESI) voltage of 2.4 kV was applied directly to the LC buffer distal to the chromatography column using a MicroTee (Upchurch Scientific, Oak Harbor, WA). The ion transfer tube temperature on the LTQ Velos ion trap was set to 300°C. For data-dependent acquisition, the method was set to automatically analyze the 5 most intense ions observed in the MS scan. Mass spectrometry data were used for protein identification by the Mascot search engine on Protein Discoverer software (Thermo Scientific) against the information in the Saccharomyces Genome Database (SGD; http://www.yeastgenome.org). Search parameters for peptide identification included a precursor mass tolerance of 1.2 Da, a fragment mass tolerance of 0.8 Da, a minimum of 1 tryptic terminus, and a maximum of 1 internal trypsin cleavage site. Cysteine carbamidomethylation (+57.021 Da) and methionine oxidation (+15.995 Da) were set as differential amino acid modifications. Data were then filtered at a q value of ≤0.01, corresponding to a 1% false-discovery rate (FDR) at the spectral level.
Extraction of cellular metabolites.
Cells incubated at 30°C in 500 μl media (OD600, 8.0) containing 2% [U-13C]glucose (U-13C6, 99%; Cambridge Isotope Laboratories Inc., Andover, MA) for 0, 2, 5, and 10 min were immediately injected into 5 ml of 60% methanol at −40°C. After centrifugation at 5,000 × g at −9°C for 5 min, supernatants were discarded, and 3 ml of 75% ethanol was added. After heating at 100°C for 30 min and cooling on ice and then at −40°C, cells were lyophilized and stored at −80°C. For sample preparation, 1 ml of MilliQ water and 60 μl of 0.2-mg/ml ribitol were added to lyophilized cells and heated at 37°C for 30 min in a 1.5-ml test tube. After samples were centrifuged at 16,000 × g for 5 min at 4°C, 900 μl of supernatants was transferred to new tubes. Next, 400 μl of MilliQ water was added to each sample, followed by centrifugation at the same rate. Finally, 400 μl of supernatants was transferred to new tubes, lyophilized, and used for the metabolite analysis.
For oximation, 100 μl of 20 mg/ml methoxyamine hydrochloride (Sigma) in pyridine (Wako, Osaka, Japan) was added and incubated at 30°C for 90 min. For trimethylsilylation, 50 μl of N-methyl-N-(trimethylsilyl) trifluoroacetamide (GL Science Inc., Tokyo, Japan) was added, followed by incubation at 37°C for 30 min. Insoluble residues were removed by centrifugation at 10,000 × g for 10 min at 4°C, and supernatants were transferred to clean vials.
GC/MS analysis.
Cells were incubated at 30°C in 500 μl of media (OD600, 8.0) containing 2% [U-13C]glucose (Cambridge Isotope Laboratories Inc., Andover, MA) for 0, 2, 5, and 10 min. Cellular metabolites extracted using modified methods of Mashego et al. (55) were derivatized as previously described (56). Derivatized metabolites were analyzed using a GCMS-QP2010 Ultra (Shimadzu, Kyoto, Japan) equipped with a 30-m by 0.25-μm (i.d.) fused silica capillary column coated with 0.25-μm CP-SIL 8 CB low bleed (Agilent Technologies, Palo Alto, CA). Aliquots (1 μl) were injected in a split mode (25/1) at 230°C using helium as the carrier gas at a flow rate of 1.12 ml/min. The column temperature was maintained isothermally at 80°C for 2 min, raised to 130°C at a rate of 4°C/min and then to 330°C at a rate of 25°C/min, and maintained for 6 min isothermally. Temperatures of the interface and MS were 250°C and 200°C, respectively, and ion voltage was 1 kV. Data were collected by GC-MS solution software (Shimadzu), and identified metabolites are shown in Table S2 in the supplemental material. Mass isotopomer distributions were corrected for natural isotope abundance as previously described (57).
Protein structure accession number.
The data determined in this study have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (58) with the data set identifier PXD000173.
RESULTS
V22 residue-dependent focus formation by full-length Eno2p under hypoxia.
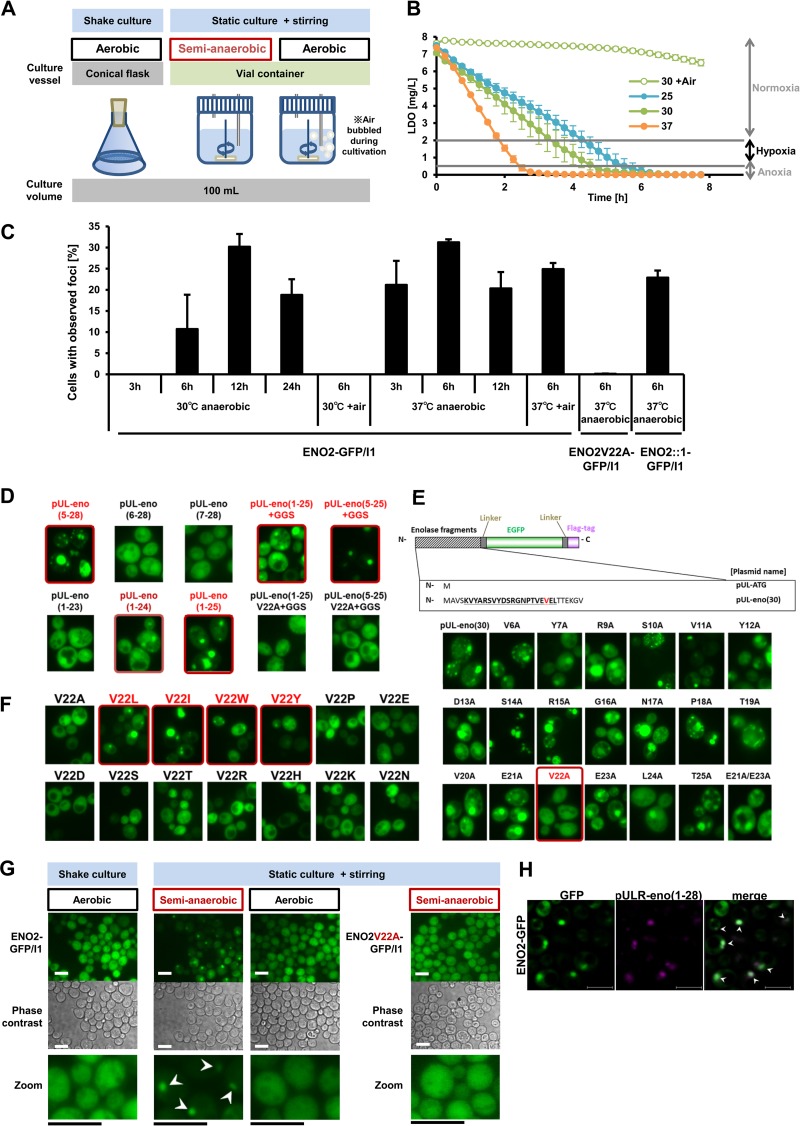
Fermentation vials were used to culture cells under semianaerobic conditions (hypoxia) (Fig. 1A). During cultivation, pH did not fall below 5 (see Fig. S1A in the supplemental material), whereas the amount of DO decreased with time (Fig. 1B). At 30°C, DO in the culture media decreased to a hypoxic level within 6 h (Fig. 1B). We found that a GFP clone, in which ENO2 was fused with GFP (strain ENO2-GFP), formed foci under the semianaerobic condition after 6 h of culture in vials at 30°C. At 37°C, foci were formed after 3 h of culture in vials, but at 25°C, foci were not observed after 24 h of culture (Fig. 1C; see Fig. S1B and C in the supplemental material). When air was introduced into the vial, foci were not observed at 30°C but were observed at 37°C (Fig. 1C; see Fig. S1D in the supplemental material). When cells were observed two-dimensionally, the maximum ratio of focus-forming cells was around 35% (Fig. 1C), whereas almost all the cells were found to form GFP foci when being observed three-dimensionally, as the number of foci formed by strain ENO2-GFP and DAPI (4′,6-diamidino-2-phenylindole) were 143 and 147, respectively (see Movie S1 in the supplemental material).
Fig 1.
Focus formation by the glycolytic enzyme enolase (Eno2p) under hypoxia (see also Fig. S1 in the supplemental material). (A) Illustration of fermentation vials for semianaerobic and anaerobic fermentative cultures. (B) Amount of luminescent dissolved oxygen (LDO) in culture medium at indicated temperatures (25, 30, and 37°C). 30 + air, culture media at 30°C with aeration during culture. Data show the DO level (mean ± SEM; n = 3) in culture media. Normoxia, LDO ≥ 2 mg/liter; hypoxia, 2 > LDO ≥ 0.5 mg/liter; anoxia, LDO < 0.5 mg/liter. (C) Temperature-dependent focus formation by Eno2p-GFP under hypoxia. Values represent mean values of the proportion of cells with foci (% total cells) ± SEM (n = 3). 30°C anaerobic, cells grown at 30°C semianaerobically in culture vials; 30°C + air, cells grown at 30°C semianaerobically in culture vials with aeration; 37°C anaerobic, cells grown at 37°C semianaerobically in culture vials; 37°C + air, cells grown at 37°C semianaerobically in culture vials with aeration. ENO2-GFP/I1, the ENO2-GFP strain transformed with plasmid pULI1; ENO2::1-GFP/I1, a pULI1-transformed strain carrying genome-integrated ENO1 conjugated with GFP at the position of ENO2 (ENO2::1-GFP strain). Cells were cultivated under indicated conditions for 3, 6, 12, or 24 h and observed. (D) Determination of the focus-forming region in the Eno2p N terminus conjugated with EGFP and a FLAG tag. pUL-eno(X-Y), cells transformed with various pUL-eno(X-Y) plasmids; pUL-eno(X–25)+GGS, cells transformed with various pUL-eno(X–25)+GGS plasmids. Cells were aerobically cultivated and observed. Highlighted pictures show cells with foci: bright red, cells with clear foci; dark red, cells with unclear foci. (E) Single alanine substitution of N-terminal amino acids of Eno2p conjugated with EGFP and FLAG. pUL-eno(30), cells transformed with plasmid pUL-eno(30); XxA, cells transformed with plasmids pUL-eno(30)XxA, where X indicates a single letter of amino acid residues and x indicates the position of amino acid residues substituted with alanine. Cells were aerobically cultivated and observed. (F) Amino acid substitutions of the V22 residue. V22X, cells transformed with plasmids pUL-eno(30) V22X, in which the Eno2p N-terminal (aa 1 to 30) amino acid sequences carrying V22X substitution were conjugated with EGFP and a FLAG tag. Highlighted pictures show a cell's successful single amino acid substitution that blocked the focus formation. (G) Inhibition of the focus formation by substitution of alanine at residue V22. Cells were cultivated at 30°C semianaerobically for 6 h and observed. Bar, 10 μm. ENO2-GFP/pULI1, the ENO2-GFP strain transformed with pULI1; ENO2V22A-GFP/pULI1, the ENO2V22A-GFP strain transformed with pULI1. Representative data of at least 3 independent experiments are shown. White arrowheads indicate observed foci. (H) Colocalization of foci formed by N-terminal amino acids of Eno2p and full-length Eno2p under hypoxia. Cells were cultivated at 30°C semianaerobically for 12 h and observed. Bar, 5 μm. White arrowheads indicate colocalized foci.
To determine the key region and residue of Eno2p for focus formation, the N-terminal focus-forming Eno2p region (51) was investigated. We found that the shortest focus-forming region of N-terminal Eno2p fused with EGFP included amino acid residues 5 to 25 (Fig. 1D). The N-terminal region of Eno2p forms conserved β-hairpin-like structure, in which R9, E21, E23, and V22 residues seem to stack the β-hairpin (see Fig. S1E in the supplemental material). We speculated that the conserved structure might have a role in forming foci. When alanine replacements were introduced in amino acid residues 6 to 25, substitutions at V22 inhibited the focus formation (Fig. 1E). Replacement of V22 with P, E, D, S, T, R, H, K, or N also inhibited the focus formation by the N-terminal region; in contrast, substitution of L, I, W, or Y conserved foci (Fig. 1F). To determine whether the V22 residue is necessary for the focus formation, conservation of the focus-forming ability of the N-terminal region was further investigated. The N-terminal region of Eno2p is conserved across species (see Fig. S1F in the supplemental material). In Escherichia coli enolase, although V22 is not conserved, its N-terminal region still exhibited the focus-forming ability (see Fig. S1G in the supplemental material). In the case of mouse β- and γ-enolase, the focus-forming ability was lost, although V22 residues were conserved (see Fig. S1F and G in the supplemental material). Interestingly, mouse α-enolase retained the focus-forming ability (see Fig. S1G in the supplemental material). Taken together, these results suggest that the focus-forming ability of the N-terminal region of S. cerevisiae Eno2p is conserved across species and is not always dependent on V22.
To test whether the V22 residue contributes to the focus formation of full-length Eno2p, a single alanine substitution for the Eno2p V22 residue was introduced into the ENO2-GFP strain (see Table S1 in the supplemental material). After 6 h of culture in vials at 30°C or 37°C, the V22A mutant of ENO2-GFP (ENO2V22A-GFP strain) did not form foci (Fig. 1C and G). The fluorescent foci formed by DsRed-conjugated N-terminal Eno2p were colocalized with foci of the ENO2-GFP strain at 30°C after 12 h of fermentative culture (Fig. 1H), indicating the similar localization of foci formed by both full-length Eno2p and its N-terminal region.
These results suggest that Eno2p-dependent focus formation in yeast cells depended on not only the V22 residue but also culture temperatures and DO concentrations.
Investigations on the regulatory mechanisms of the focus formation.
To determine the regulatory mechanisms of the focus formation, possible regulatory mechanisms such as protein synthesis, cell signaling, mitochondrial activity, and protein association were investigated by chemicals, gene knockouts, or proteomic analysis.
(i) Temperature-independent inhibition of focus formation by cycloheximide and rapamycin.
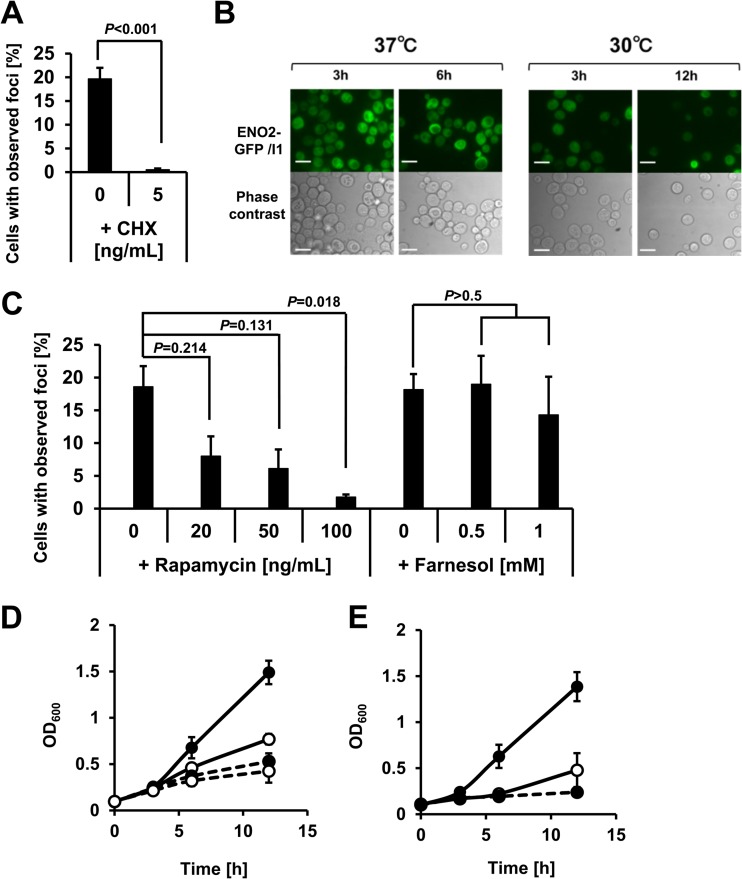
To determine the mechanisms for the regulation of focus formation, several reagents were used to inhibit the focus formation. We demonstrated that cycloheximide blocked the focus formation by the ENO2-GFP strain at 30°C and 37°C in semianaerobic cultures (Fig. 2A and B), whereas rapamycin at a growth-inhibiting dose hindered the focus formation at 37°C (Fig. 2C and D; see Fig. S2 in the supplemental material). In contrast, the focus formation was maintained with the application of a growth-inhibiting dose of farnesol, an inhibitor of the cyclic AMP (cAMP), protein kinase A (PKA), and mitogen-activated protein kinase (MAPK) signaling pathways in C. albicans (59–62) and a mitochondrial ROS generator (63) and a growth inhibitor (64) in S. cerevisiae (Fig. 2C and E; see Fig. S2 in the supplemental material). These results suggest the DO-independent participation of both the de novo protein synthesis and TORC1-dependent regulation at 37°C in the focus formation.
Fig 2.
Temperature-independent inhibition of the Eno2p focus formation by chemicals (see also Fig. S2 in the supplemental material). (A) Inhibition of focus formation by the addition of cycloheximide (CHX) at 37°C. A final concentration of 0.5 ng/ml CHX was added to culture vials before semianaerobic cultivation. Values represent mean values of the proportion of cells with foci (% total cells) ± SEM (n = 3) after 12 h of semianaerobic culture at 37°C. Data were analyzed using Student's t test. (B) Inhibition of focus formation by the addition of CHX at 30°C and 37°C. A final concentration of 5 ng/ml CHX was added to culture vials before semianaerobic cultivation at indicated temperatures. Bar, 10 μm. (C) Effects of rapamycin and farnesol on focus formation. Values represent mean values of the proportion of cells with foci (% total cells) ± SEM (n = 3) after 12 h of semianaerobic culture at 37°C. Data were analyzed using Student's t test with the Bonferroni correction for multiple comparisons. (D) Growth inhibition by the addition of rapamycin. Filled circle with solid line, 0 ng/ml rapamycin; open circle with solid line, 20 ng/ml rapamycin; filled circle with dashed line, 50 ng/ml rapamycin; open circle with dashed line, 100 ng/ml rapamycin. (E) Growth inhibition by the addition of farnesol. Filled circle with solid line, 0 mM farnesol; open circle with solid line, 0.5 mM farnesol; filled circle with dashed line, 1 mM farnesol.
(ii) Identification of SNF1 as a regulator of focus formation at 30°C.
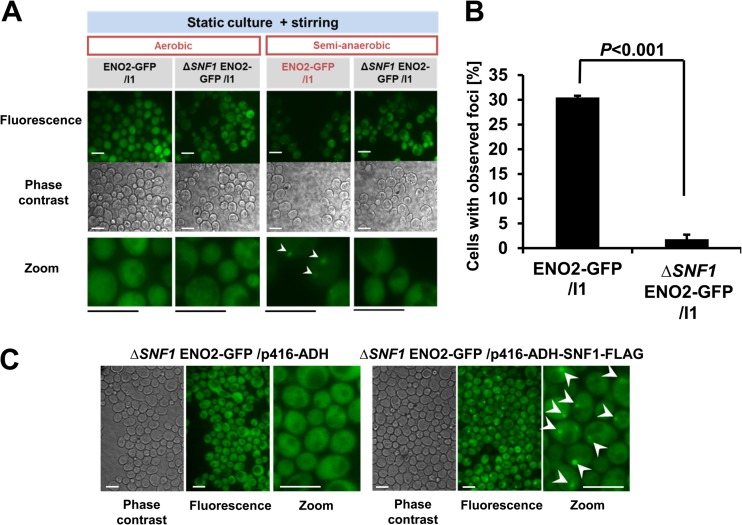
To determine the signaling pathway regulating the focus formation, knockout mutations of genes participating in various signaling pathways, namely, HOG1 (MAPK pathway), SCH9 (PI3K-AKT pathway), and SNF1 (SNF1/AMPK pathway), were introduced into the ENO2-GFP strain. In semianaerobic cultures, the focus formation by the ENO2-GFP strain without SNF1 (ΔSNF1 ENO2-GFP strain) was inhibited at 30°C, in contrast to all other strains tested (Fig. 3A and B; see Fig. S3A in the supplemental material). To assess the involvement of Upc2p, which is a known regulator of hypoxia-responding transcription in yeast C. albicans (65) and S. cerevisiae (66), a UPC2 knockout mutation was introduced in the same manner. As a result, the focus formation was not inhibited (see Fig. S3A in the supplemental material), suggesting no involvement of Upc2p in the focus formation. In semianaerobic cultures at 37°C, the ΔSNF1 ENO2-GFP strain formed foci (see Fig. S3B in the supplemental material), suggesting the temperature-dependent regulation of focus formation by SNF1. A strain carrying plasmid-reintegrated SNF1 regained the focus-forming ability in semianaerobic cultures at 30°C (Fig. 3C), confirming a role of SNF1 in the focus formation at 30°C. Taken together, these results suggest that foci were formed at 30°C in response to hypoxia via the participation of SNF1/AMPK. In general, the optimum temperature for cultivating S. cerevisiae strain BY4741 is 30°C. Accordingly, we focused on the focus formation induced at 30°C under hypoxia.
Fig 3.
Temperature-dependent inhibition of the Eno2p focus formation in SNF1 knockout mutants (see also Fig. S3 in the supplemental material). (A) Inhibition of focus formation in the ΔSNF1 ENO2-GFP strain at 30°C. Plasmid pULI1-transformed cells were cultivated at 30°C semianaerobically for 12 h and observed. Bar, 10 μm. White arrowheads indicate observed foci. (B) Values represent mean values of the proportion of cells with foci (% total cells) ± SEM (n = 3) after 24 h of semianaerobic culture at 30°C. Data were analyzed using Student's t test. (C) Compensation of the focus formation by reintroduction of SNF1. Cells were observed after 12 h of semianaerobic culture. Bar, 10 μm. Representative data of 3 independent experiments are shown. White arrowheads indicate observed foci.
(iii) Involvement of mitochondrial ROS production in focus formation.
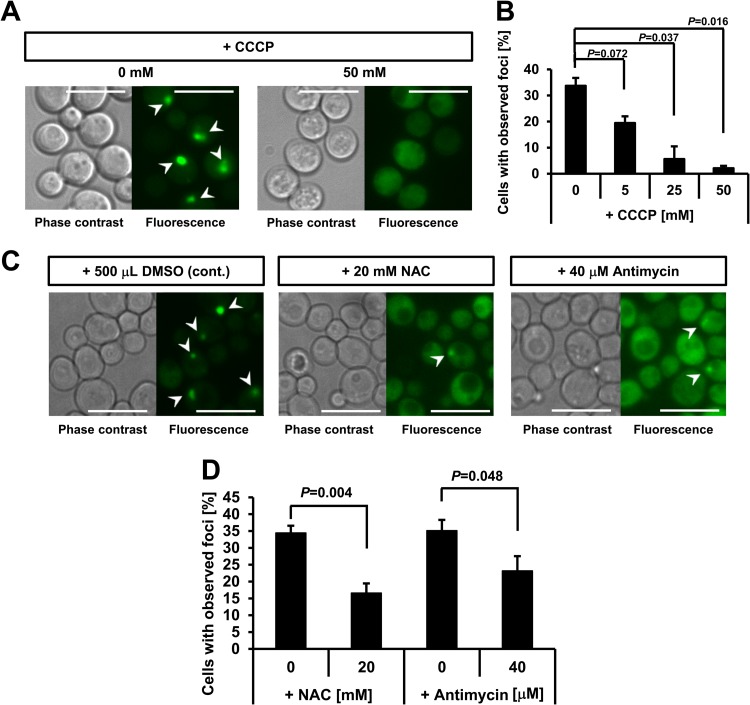
The involvement of mitochondrial ROS production, which is known to activate AMPK, was investigated using mitochondrial inhibitors and an antioxidant. The mitochondrial uncoupler CCCP was found to inhibit focus formation (Fig. 4A and B), indicating mitochondrial involvement. Antimycin, which is an inhibitor of mitochondrial complex III, also inhibited focus formation (Fig. 4C and D). Importantly, the antioxidant NAC inhibited focus formation, indicating the involvement of mitochondrial ROS released into the cytoplasm under hypoxia (Fig. 4C and D).
Fig 4.
Inhibition of focus formation by antioxidant and inhibitors of mitochondrial ROS production at 30°C. (A) Inhibition of focus formation by addition of the mitochondrial uncoupler CCCP. A final concentration of 0 or 50 mM CCCP was added to culture vials before cultivation. Cells were cultured semianaerobically at 30°C for 12 h and observed. Representative data of 3 independent experiments are shown. Bar, 10 μm. White arrowheads indicate observed foci. (B) Calculation of the rate of inhibition by CCCP. Values represent mean values of the proportion of cells with foci (% total cells) ± SEM (n = 3) after 12 h of semianaerobic culture at 30°C. Data were analyzed using Student's t test with the Bonferroni correction for multiple comparisons. (C) Effects of the mitochondrial inhibitor NAC and the antioxidant antimycin on focus formation. After the addition of specific reagents, cells were semianaerobically cultured at 30°C for 12 h and observed. Representative data of at least 3 independent experiments are shown. Bar, 10 μm. White arrowheads indicate observed foci. DMSO, dimethyl sulfoxide. (D) Calculation of the inhibition rate by NAC and antimycin. Values represent mean values of the proportion of cells with foci (% total cells) ± SEM (n = 3) after 12 h of semianaerobic culture at 30°C. Data were analyzed using Student's t test.
(iv) Detection of proteins coimmunoprecipitated with focus-forming Eno2p.
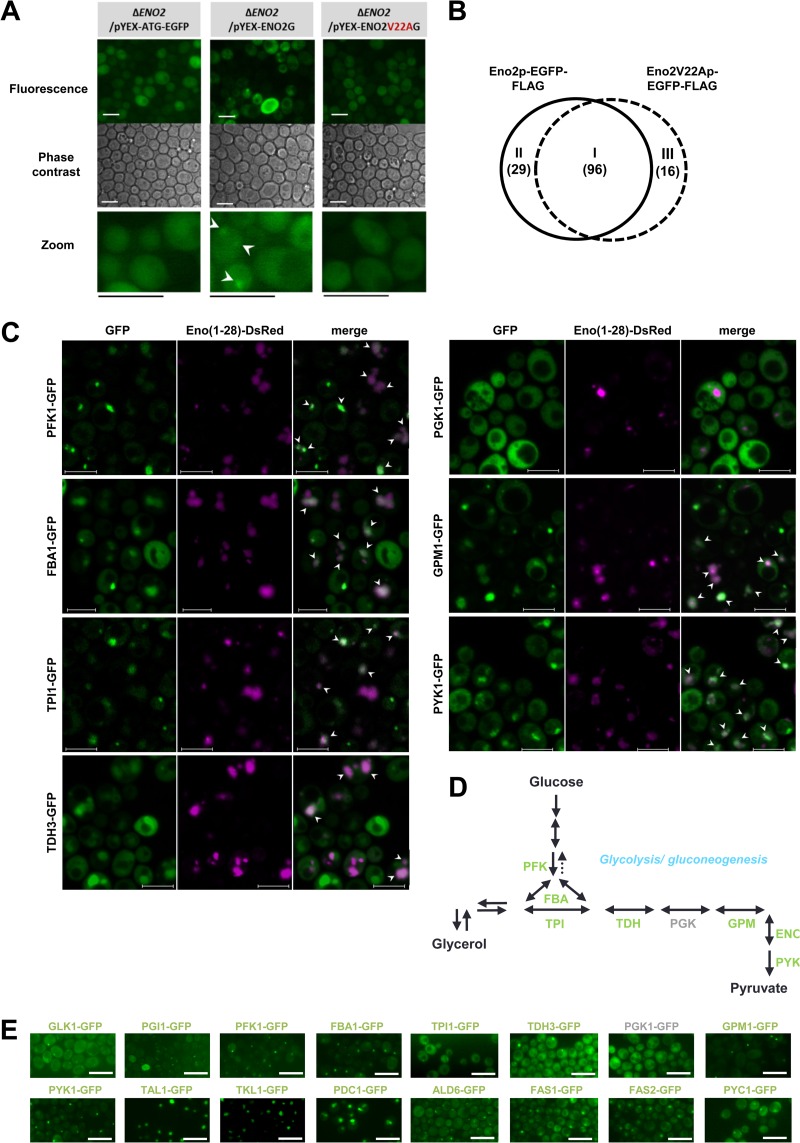
To detect proteins involved in foci, an ENO2 knockout (ΔENO2) strain and plasmids for producing recombinant Eno2p-EGFP-FLAG or its V22A mutant (Eno2V22Ap-EGFP-FLAG) were prepared (see Table S1 in the supplemental material). The fluorescence intensities of ΔENO2 strains producing recombinant proteins were comparable to those of strain ENO2-GFP in aerobic culture (see Fig. S4A in the supplemental material). After 12 h of semianaerobic culture, the Eno2p-EGFP-FLAG protein was able to form foci (Fig. 5A). To investigate proteins associated with focus-forming Eno2p, Eno2p-EGFP-FLAG and Eno2V22Ap-EGFP-FLAG proteins were immunoprecipitated, and their interacting proteins were identified by LC/MS-MS (see Fig. S4B in the supplemental material). As a result, 96 proteins, including 35 metabolic proteins, were detected in samples coimmunoprecipitated with Eno2p-EGFP-FLAG and Eno2V22Ap-EGFP-FLAG (Fig. 5B, area I; see Fig. S4C and Table S3 in the supplemental material). We also identified that 29 proteins, including 15 metabolic proteins, were coimmunoprecipitated only with Eno2p-EGFP-FLAG (Fig. 5B, area II; see Fig. S4D and Table S3 in the supplemental material). Area I included 17 glycolytic enzymes. To test whether glycolytic enzymes form foci under hypoxia, individual GFP clones were used. We observed the focus formation by glycolytic enzymes Pfk1p, Fba1p, Tpi1p, Tdh3p, Gpm1p, and Cdc19p (Pyk1p) under hypoxia (Fig. 5C). In addition, foci formed by these glycolytic enzymes were colocalized with foci formed by the N-terminal Eno2p (aa 1 to 28) fragment conjugated with the DsRed monomer (Fig. 5C). Pgk1p, though included in area I, did not form foci under hypoxia (Fig. 5C). Among detected metabolic enzymes in area I, Pdc1p-GFP formed foci under normoxia (see Fig. S4E in the supplemental material). Among metabolic enzymes that were detected by the proteomic analysis, GFP-conjugated Tal1p (area I) and Tkl1p (area II), enzymes of the pentose phosphate pathway, and Pdc1p (area I), Ald6p (area II), Fas1p (area I), and Fas2p (area I), enzymes involved in the fatty acid synthesis, were detected to form foci under hypoxia (Fig. 5C; see Fig. S4F in the supplemental material). Foci formed by Pdc1p, Ald6p, Fas1p, and Fas2p were colocalized with those formed by the N-terminal Eno2p (aa 1 to 28) fragment conjugated with the DsRed monomer (see Fig. S4F). Colocalization of foci formed by Tkl1p and Tal1p with those formed by the N-terminal Eno2p (aa 1 to 28) fragment conjugated with the DsRed monomer was also observed; however, not all the foci were colocalized (see Fig. S4F). An overview of focus-forming proteins in glycolytic pathway under hypoxia is shown in Fig. 5D.
Fig 5.
Detection of focus-forming metabolic proteins under hypoxia (see also Fig. S4 and Table S3 in the supplemental material). (A) Focus formation by plasmid-introduced recombinant Eno2p. The ΔENO2 strain transformed with various plasmids was semianaerobically cultured at 30°C for 12 h and observed. pYEX-ATG-EGFP, the plasmid used for producing EGFP-FLAG tag; pYEX-ENO2G, the plasmid for producing Eno2p conjugated with the EGFP-FLAG tag; pYEX-ENO2V22AG, the plasmid for producing the Eno2p V22A mutant conjugated with the EGFP-FLAG tag. Bar, 10 μm. Representative data of at least 3 independent experiments are shown. White arrowheads indicate observed foci. (B) Overview of identified proteins coimmunoprecipitated with recombinant Eno2p-EGFP-FLAG and Eno2V22Ap-EGFP-FLAG. Before protein extraction, cells were cultured for 12 h at 30°C semianaerobically. Data represent 2 biological replicates and 2 technical replicates (n = 4). I, 96 proteins coimmunoprecipitated with recombinant Eno2p-EGFP-FLAG and Eno2V22Ap-EGFP-FLAG; II, 29 proteins coimmunoprecipitated with recombinant Eno2p-EGFP-FLAG; III, 16 proteins coimmunoprecipitated with recombinant Eno2V22Ap-EGFP-FLAG. (C) Focus formation by metabolic enzymes and their colocalization with foci formed by N-terminal amino acid sequences conjugated with DsRed. Various GFP strains (of Pfk1p, Fba1p, Tpi1p, Tdh3p, Pgk1p, Gpm1p, and Pyk1p) or strain BY4741 transformed with the plasmid pULR-eno(1-28) were cultured semianaerobically at 30°C for 12 h and observed. Eno(1-28)-DsRed, red fluorescence from the N-terminal (aa 1 to 28) amino acid sequence of Eno2p conjugated with the DsRed monomer (shown in magenta); GFP, green fluorescence from the proteins conjugated with GFP. Bar, 5 μm. White arrowheads indicate colocalized foci (shown in white). (D) Overview of focus-forming proteins in glycolytic pathway under hypoxia. Green or gray characters indicate enzymes that formed or did not form, respectively, foci under hypoxia. (E) Focus formation by metabolic enzymes under hypoxia. Various GFP strains (of Glk1p, Pgi1p, Pfk1p, Fba1p, Tpi1p, Tdh3p, Pgk1p, Gpm1p, Pyk1p, Tal1p, Tkl1p, Pdc1p, Ald6p, Fas1p, Fas2p, and Pyc1p) transformed with the plasmid pULI1 were cultured semianaerobically at 30°C for 12 h (for Pfk1p, Fba1p, Tpi1p, Tdh3p, Pgk1p, Gpm1p, Pyk1p, Tal1p, Tkl1p, Pdc1p, Fas1p, Fas2p, and Pyc1p) or 24 h (for Pgi1p, Glk1p, and Ald6p) and observed. Bar, 10 μm. Green or gray characters indicate enzymes that formed or did not form, respectively, foci under hypoxia.
Investigation of the effects of focus-inhibiting mutations on the carbon metabolic pathway in hypoxia-treated cells.
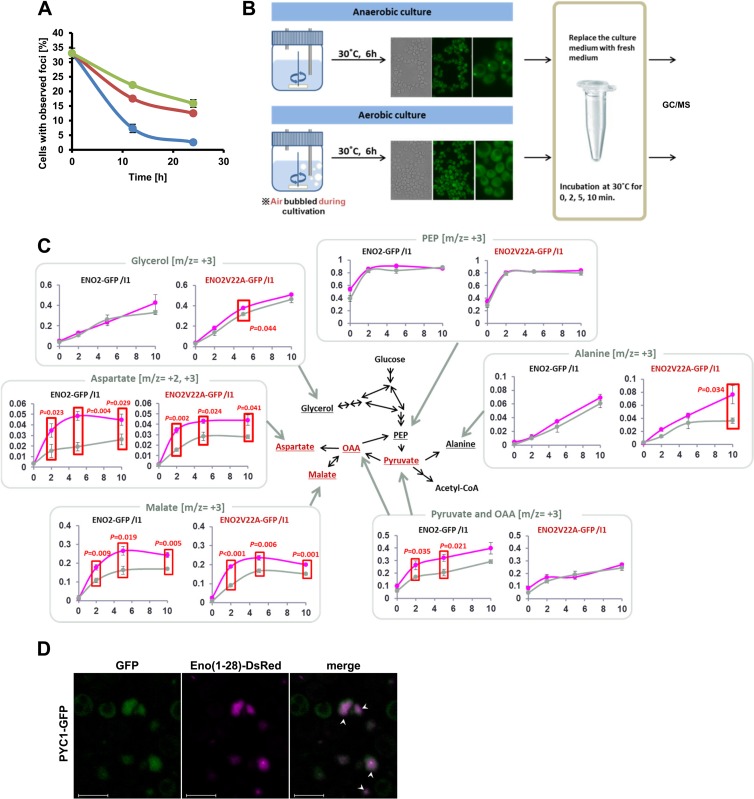
The ENO2-GFP V22A mutant strain (ENO2V22A-GFP) grew slower than the ENO2-GFP strain in semianaerobic cultures (see Fig. S5A in the supplemental material), suggesting changes in cell metabolism. After semianaerobic culture for 12 h, foci formed by the ENO2-GFP strain were retained following 24 h of aerobic culture in fresh media (Fig. 6A). To investigate the effects of focus formation on cellular carbon metabolism, a metabolic turnover analysis of strains ENO2-GFP and ENO2V22A-GFP was carried out using [U-13C]glucose after semianaerobic (focus-forming condition) or aerobic (focus-nonforming condition) culture (Fig. 6B). The ratios of 13C-labeled pyruvate and oxaloacetate in focus-forming cells were higher than those in focus-nonforming cells after 2 and 5 min of intake (Fig. 6C). For glycerol and alanine in strain ENO2V22A-GFP, the ratios of 13C-labeled metabolites were slightly higher in cells grown in semianaerobic cultures, whereas the ratios remained unchanged in strain ENO2-GFP. In both strains, the ratios of aspartate, malate, citrate, and succinate, all of which have 3 13C atoms, were higher in cells subjected to semianaerobic cultures, indicating the incorporation of oxaloacetate-derived 13C into the TCA cycle (Fig. 6C; see Fig. S5B in the supplemental material). These results suggest that cells forming foci accelerated the incorporation of glucose-derived 13C into pyruvate and oxaloacetate and preferentially produced aspartate and malate, rather than alanine, from pyruvate. To test the involvement of Pyc1p, which converts pyruvate to oxaloacetate (see Fig. S5C in the supplemental material), the focus-forming ability of Pyc1p under hypoxia was investigated. We observed that GFP fused with Pyc1p formed foci under hypoxia (Fig. 6D), suggesting the coordinated synthesis of pyruvate and oxaloacetate by focus-forming enzymes.
Fig 6.
Changes in the carbon metabolic pathway of focus-forming cells (see also Fig. S5 in the supplemental material). (A) Conservation of foci after semianaerobic culture. Cells were cultivated semianaerobically at 30°C for 12 h, collected, and suspended in fresh media to an OD600 of 8.0 (green line), 4.0 (red line), or 1.0 (blue line), followed by aerobic cultivation at 25°C for 12 and 24 h. Values represent mean values of the proportion of cells with foci (% total cells) ± SEM (n = 3). (B) Scheme for the measurement of 13C incorporated in metabolites. (C) Incorporation of glucose-derived 13C into metabolites of focus-forming and -nonforming cells. Values represent mean values of 3 (for glycerol) or 4 (for other metabolites) biological replicates ± SEM (n = 3 or 4). ENO2-GFP/I1, the ENO2-GFP strain transformed with pULI1; ENO2V22A-GFP/I1, the ENO2V22A-GFP strain transformed with pULI1. Magenta and gray lines, metabolites extracted from cells after semianaerobic culture and after aerobic culture, respectively. x axis, labeled fraction; y axis, time (min). (D) Focus formation by PYC1-GFP under hypoxia. The PYC1-GFP strain transformed with pULI1 was cultured semianaerobically at 30°C for 12 h and observed. Bar, 5 μm. White arrowheads indicate colocalized foci.
DISCUSSION
Focus formation by proteins, including Eno2p, under hypoxia.
Spatial reorganization of glycolytic enzymes, including Eno2p, was detected for the first time in S. cerevisiae under hypoxic fermentation conditions. As the foci formed by full-length Eno2p showed localization similar to those of the foci formed by the N-terminal region of Eno2p (Fig. 1H), which colocalized with proteins of cytoplasmic or nucleus membrane in addition to proteins of the Golgi body or endosome (51), the foci formed by Eno2p seem to localize in the cytoplasm near intracellular organelles. The assembly of glycolytic enzymes via spatial reorganization is considered to be effective for accelerating glycolysis. Therefore, spatial reorganization of metabolic enzymes may be a novel mechanism for efficient energy production, without the TCA cycle, under hypoxia. Moreover, the novel changes in the intracellular localization detected in this study may enable glycolytic proteins to perform unforeseen functions in response to specific environmental stimuli, in addition to their roles in regulating cell metabolism. It is also noteworthy that many metabolic enzymes were coimmunoprecipitated with Eno2p-EGFP-FLAG-formed foci under hypoxia (Fig. 1H, 5C, and 6D; see Fig. S4F and Table S3 in the supplemental material); these enzymes are involved in the pentose phosphate pathway, fatty acid synthesis, tRNA synthesis, and amino acid synthesis. It is possible that the flow rate of each metabolic pathway may be regulated via the control of spatial reorganization of proteins, in addition to the modulation of the amount of proteins. Of the detected proteins, some have roles in protein transport (see Table S3 in the supplemental material), indicating that the spatial reorganization of glycolytic enzymes is possibly regulated by these proteins. Given that S. cerevisiae has many biological processes in common with other eukaryotic cells, these mechanisms may be conserved across species.
Importance of single amino acids in Eno2p for focus formation.
The colocalization of foci formed by full-length Eno2p-GFP and an N-terminal fragment fused with DsRed (Fig. 1H) supports our speculation that an N-terminal focus-forming region regulates spatial reorganization of full-length Eno2p. Amino acids V, I, L, Y, and W have also been reported to be important for the stacking of β-hairpin structures of tau proteins (67). Accordingly, the three-dimensional structure of the Eno2p N-terminal region might be important for focus formation. We conclude that spatial reorganization by the specific amino acid sequence of Eno2p may promote spatial reorganization of the whole protein. Screening peptide sequences would be a useful approach for discovering such amino acid sequences in other focus-forming proteins under hypoxia.
Focus formation by Eno2p in response to ROS produced by hypoxic mitochondria, SNF1/AMPK, TORC1, and de novo protein synthesis.
The temperature-independent inhibition of focus formation by cycloheximide suggests that de novo protein synthesis is an important factor, in addition to the regulation by the signaling pathway. While the inhibition of focus formation by rapamycin was also temperature independent, the rapamycin doses that blocked the focus formation were found to inhibit cell growth. However, considering that the focus formation was not hindered when farnesol was added at a growth-inhibiting dose, TORC1 incorporation is still important. The finding that HOG1 and SCH9 knockout failed to inhibit the focus formation is reasonable, because farnesol was reported to be an inhibitor of the MAPK and PKC/Akt pathways in the pathogenic fungus C. albicans (65).
The inhibition of focus formation by knocking out SNF1 was unexpected, because the SNF1/AMPK pathway is known to be activated by a glucose-limiting state, in which glycolytic enzymes are downregulated. However, the inhibition of focus formation by mitochondrial inhibitors and an antioxidant supports the involvement of SNF1/AMPK in hypoxia-responsive focus formation. The focus formation was strongly dependent on temperatures and decreased DO in culture media. Together, these results suggest that the focus formation by Eno2p was dependent on more than 1 pathway (Fig. 7). Although AMPK is known to inhibit the TOR pathway, there are some instances in which both the AMPK and TOR pathways regulate cell physiology (68, 69). For example, in S. cerevisiae, both Snf1p and TORC1 have been suggested to have roles in regulating fatty acids via unknown mechanisms (70). Such unknown regulatory mechanisms for these 2 pathways may be revealed by future studies.
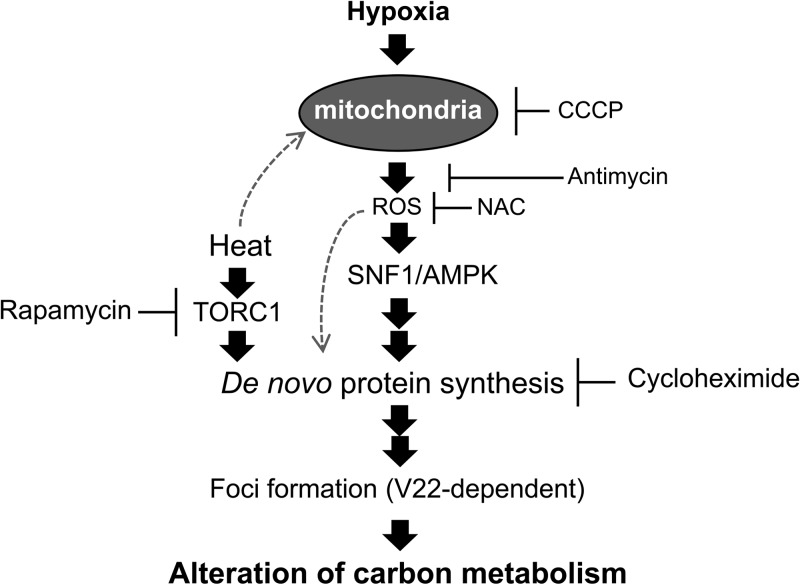
Fig 7.
Schematic illustration of the proposed regulation and biological role of focus formation. Dotted arrows show possible mechanisms of focus formation at higher temperatures by the involvement of increased mitochondrial electron transport activity.
The detailed mechanisms that enable rapid focus formation at higher temperature (37°C) should also be revealed by future studies. One possible explanation may be that increased amount of mitochondrial ROS was produced at higher temperature via increased mitochondrial transport chain activity, as previously reported (71–73).
Shifting the carbon metabolic pathway by spatial reorganization.
Given that the glycolytic pathway has many branches connected to various metabolic pathways, including the synthesis of nucleotides, amino acids, and lipids, and to energy production, the effective use of carbon sources according to cellular needs in the struggle for survival is expected to be extremely important. The regulation of the carbon metabolic pathway has been reported to be accomplished by the transcriptional modulation of various regulators (74–77). However, it has never been reported that the central carbon metabolic pathway may be regulated by spatial reorganization or association of glycolytic enzymes in vivo.
The focus formation by GFP-conjugated Eno2p and other glycolytic enzymes (Glk1p, Pgi1p, Pfk1p, Fba1p, Tpi1p, Tdh3p, Gpm1p, and Pyk1p) under hypoxia (Fig. 1H and 5C, D, and E; see Fig. S4F in the supplemental material) and their colocalization with foci formed by N-terminal Eno2p (Fig. 5C) suggest the complex formation of glycolytic enzymes in the cytosol. As predicted by a simulation study of glycolytic flux, under focus-forming conditions, the incorporation of glucose-derived 13C into pyruvate and oxaloacetate was accelerated. The inhibition of focus formation by introducing the V22A mutation and its metabolic turnover analysis demonstrated the participation of Eno2p focus formation in controlling carbon metabolism. Moreover, the increased ratios of 13C-labeled pyruvate, oxaloacetate, aspartate, malate, citrate, and succinate in focus-forming cells suggest that foci participate in increasing the flux of pyruvate and oxaloacetate synthesis, at the same time increasing the synthesis of the other metabolites. Taken together, these results support the hypothesis that under hypoxia, certain glycolytic enzymes are spatially reorganized to alter the carbon metabolic pathway. Fluxes and concentrations of metabolites in glycolysis are rapid and small, respectively, especially in reactions catalyzed by Eno2p, even though Eno2p is one of the most abundant proteins in a cell. However, changing the amount of Eno2p seems to have no significant effect on cell metabolism, as indicated by previous results in E. coli (78). In S. cerevisiae, the amounts of Eno2p and other glycolytic enzymes under hypoxia were reported to be increased significantly (79). Based on these observations, forming a complex of metabolic enzymes in a particular cellular location would be a reasonable and effective way to switch the carbon metabolic pathway.
In addition to hypoxia, higher temperatures (37°C) also induced the Eno2p focus formation. The involvement of high temperatures and hypoxia in the induction of focus formation remains unclear. The finding that focus formation at 37°C was inhibited by the addition of cycloheximide or rapamycin but not by the SNF1 knockout mutation suggests 2 possible ways of regulation: via oxygen concentration and by temperature increase. Postmus et al. recently reported that glycolytic flux was increased in fermenting S. cerevisiae at 38°C (42). They showed that the increased activity of glycolytic enzymes did not correlate with protein abundance and suggested the contribution of posttranslational regulation to enzyme activities. The focus formation by glycolytic enzymes may be efficient in regulating glycolytic enzymes posttranslationally.
The amino acid residues or domains important for the focus formation by each enzyme could be determined in the same manner that Eno2p was investigated in this study. The control of the carbon metabolic pathway in proliferating cells is an important issue. In particular, enolase and other glycolytic enzymes were shown to be overproduced in tumor cells in which the glycolysis rate was increased (80). If spatial reorganization of glycolytic enzymes occurs in mammalian cells, the results and methods demonstrated in this study could contribute to clarifying the regulatory mechanisms of carbon metabolism in proliferating cells, including tumor cells.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge valuable discussions and helpful suggestions from Wataru Fujibuchi of RIKEN (currently at Kyoto University) (on the effects of colocalization of glycolytic enzymes), Hiro-o Hamaguchi of Tokyo University (currently at Waseda University) (on the spatial reorganization of proteins in a cell), Hiroshi Harada of Kyoto University and Tetsuya Kadonosono of Tokyo Institute of Technology (on cellular hypoxic responses and biological meanings of foci), and Eiichiro Fukusaki of Osaka University (on the metabolic turnover analysis using S. cerevisiae). We also acknowledge the PRIDE Team for technical support.
This work was supported by a Grant-in-Aid for JSPS Fellows (09J02920 to N.M.).
Footnotes
Published ahead of print 7 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00093-13.
REFERENCES
- 1. Eby LA, Crowder LB. 2002. Hypoxia-based habitat compression in the Neuse River Estuary: context-dependent shifts in behavioral avoidance thresholds. Can. J. Fish. Aquat. Sci. 59:952–965 [Google Scholar]
- 2. Buzzelli CP, Luettich RA, Powers SP, Peterson CH, McNinch JE, Pinckney JL, Paerl HW. 2002. Estimating the spatial extent of bottom-water hypoxia and habitat degradation in a shallow estuary. Mar. Ecol. Prog. Ser. 230:103–112 [Google Scholar]
- 3. Hagen T, Taylor CT, Lam F, Moncada S. 2003. Redistribution of intracellular oxygen in hypoxia by nitric oxide: effect on HIF1α. Science 302:1975–1978 [DOI] [PubMed] [Google Scholar]
- 4. Frezza C, Zheng L, Tennant DA, Papkovsky DB, Hedley BA, Kalna G, Watson DG, Gottlieb E. 2011. Metabolic profiling of hypoxic cells revealed a catabolic signature required for cell survival. PLoS One 6:e24411. 10.1371/journal.pone.0024411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Denko NC. 2008. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat. Rev. Cancer 8:705–713 [DOI] [PubMed] [Google Scholar]
- 6. Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, Semenza GL. 1998. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 12:149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. 1999. Overexpression of hypoxia-inducible factor 1α in common human cancers and their metastases. Cancer Res. 59:5830–5835 [PubMed] [Google Scholar]
- 8. Eyler CE, Rich JN. 2008. Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J. Clin. Oncol. 26:2839–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Teicher BA. 1994. Hypoxia and drug resistance. Cancer Metastasis Rev. 13:139–168 [DOI] [PubMed] [Google Scholar]
- 10. Grahl N, Cramer RA., Jr 2010. Regulation of hypoxia adaptation: an overlooked virulence attribute of pathogenic fungi? Med. Mycol. 48:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grahl N, Puttikamonkul S, Macdonald JM, Gamcsik MP, Ngo LY, Hohl TM, Cramer RA. 2011. In vivo hypoxia and a fungal alcohol dehydrogenase influence the pathogenesis of invasive pulmonary aspergillosis. PLoS Pathog. 7:e1002145. 10.1371/journal.ppat.1002145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Netzer NC, Breitenbach M. 2010. Metabolic changes through hypoxia in humans and in yeast as a comparable cell model. Sleep Breath. 14:221–225 [DOI] [PubMed] [Google Scholar]
- 13. Guzy RD, Mack MM, Schumacker PT. 2007. Mitochondrial complex III is required for hypoxia-induced ROS production and gene transcription in yeast. Antioxid. Redox Signal. 9:1317–1328 [DOI] [PubMed] [Google Scholar]
- 14. Sena LA, Chandel NS. 2012. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 48:158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hong SP, Carlson M. 2007. Regulation of snf1 protein kinase in response to environmental stress. J. Biol. Chem. 282:16838–16845 [DOI] [PubMed] [Google Scholar]
- 16. Jung SN, Yang WK, Kim J, Kim HS, Kim EJ, Yun H, Park H, Kim SS, Choe W, Kang I, Ha J. 2008. Reactive oxygen species stabilize hypoxia-inducible factor-1α protein and stimulate transcriptional activity via AMP-activated protein kinase in DU145 human prostate cancer cells. Carcinogenesis 29:713–721 [DOI] [PubMed] [Google Scholar]
- 17. Zmijewski JW, Banerjee S, Bae H, Friggeri A, Lazarowski ER, Abraham E. 2010. Exposure to hydrogen peroxide induces oxidation and activation of AMP-activated protein kinase. J. Biol. Chem. 285:33154–33164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mungai PT, Waypa GB, Jairaman A, Prakriya M, Dokic D, Ball MK, Schumacker PT. 2011. Hypoxia triggers AMPK activation through reactive oxygen species-mediated activation of calcium release-activated calcium channels. Mol. Cell. Biol. 31:3531–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emerling BM, Weinberg F, Snyder C, Burgess Z, Mutlu GM, Viollet B, Budinger GR, Chandel NS. 2009. Hypoxic activation of AMPK is dependent on mitochondrial ROS but independent of an increase in AMP/ATP ratio. Free Radic. Biol. Med. 46:1386–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lunt SY, Vander Heiden MG. 2011. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 27:441–464 [DOI] [PubMed] [Google Scholar]
- 21. Chateil J, Biran M, Thiaudiere E, Canioni P, Merle M. 2001. Metabolism of [1-13C]glucose and [2-13C]acetate in the hypoxic rat brain. Neurochem. Int. 38:399–407 [DOI] [PubMed] [Google Scholar]
- 22. Chico E, Olavarria JS, Nunez de Castro I. 1978. l-Alanine as an end product of glycolysis in Saccharomyces cerevisiae growing under different hypoxic conditions. Antonie Van Leeuwenhoek 44:193–201 [DOI] [PubMed] [Google Scholar]
- 23. Gleason JE, Corrigan DJ, Cox JE, Reddi AR, McGinnis LA, Culotta VC. 2011. Analysis of hypoxia and hypoxia-like states through metabolite profiling. PLoS One 6:e24741. 10.1371/journal.pone.0024741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. 2008. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7:11–20 [DOI] [PubMed] [Google Scholar]
- 25. Beeckmans S, Van Driessche E, Kanarek L. 1990. Clustering of sequential enzymes in the glycolytic pathway and the citric acid cycle. J. Cell. Biochem. 43:297–306 [DOI] [PubMed] [Google Scholar]
- 26. Amar P, Legent G, Thellier M, Ripoll C, Bernot G, Nystrom T, Saier MH, Jr, Norris V. 2008. A stochastic automaton shows how enzyme assemblies may contribute to metabolic efficiency. BMC Syst. Biol. 2:27. 10.1186/1752-0509-2-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hannaert V, Michels PA. 1994. Structure, function, and biogenesis of glycosomes in kinetoplastida. J. Bioenerg. Biomembr. 26:205–212 [DOI] [PubMed] [Google Scholar]
- 28. Bakker BM, Mensonides FI, Teusink B, van Hoek P, Michels PA, Westerhoff HV. 2000. Compartmentation protects trypanosomes from the dangerous design of glycolysis. Proc. Natl. Acad. Sci. U. S. A. 97:2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Masters C. 1984. Interactions between glycolytic enzymes and components of the cytomatrix. J. Cell Biol. 99:222s–225s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stephan P, Clarke F, Morton D. 1986. The indirect binding of triose-phosphate isomerase to myofibrils to form a glycolytic enzyme mini-complex. Biochim. Biophys. Acta 873:127–135 [DOI] [PubMed] [Google Scholar]
- 31. Campanella ME, Chu H, Low PS. 2005. Assembly and regulation of a glycolytic enzyme complex on the human erythrocyte membrane. Proc. Natl. Acad. Sci. U. S. A. 102:2402–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brooks SP, Storey KB. 1988. Reevaluation of the “glycolytic complex” in muscle: a multitechnique approach using trout white muscle. Arch. Biochem. Biophys. 267:13–22 [DOI] [PubMed] [Google Scholar]
- 33. Mowbray J, Moses V. 1976. The tentative identification in Escherichia coli of a multienzyme complex with glycolytic activity. Eur. J. Biochem. 66:25–36 [DOI] [PubMed] [Google Scholar]
- 34. Anderson LE, Wang X, Gibbons JT. 1995. Three enzymes of carbon metabolism or their antigenic analogs in pea leaf nuclei. Plant Physiol. 108:659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mazzola JL, Sirover MA. 2003. Subcellular localization of human glyceraldehyde-3-phosphate dehydrogenase is independent of its glycolytic function. Biochim. Biophys. Acta 1622:50–56 [DOI] [PubMed] [Google Scholar]
- 36. Fothergill-Gilmore LA, Michels PA. 1993. Evolution of glycolysis. Prog. Biophys. Mol. Biol. 59:105–235 [DOI] [PubMed] [Google Scholar]
- 37. Ginger ML, McFadden GI, Michels PA. 2010. Rewiring and regulation of cross-compartmentalized metabolism in protists. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:831–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Giege P, Heazlewood JL, Roessner-Tunali U, Millar AH, Fernie AR, Leaver CJ, Sweetlove LJ. 2003. Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15:2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Graham JW, Williams TC, Morgan M, Fernie AR, Ratcliffe RG, Sweetlove LJ. 2007. Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling. Plant Cell 19:3723–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Agbor TA, Cheong A, Comerford KM, Scholz CC, Bruning U, Clarke A, Cummins EP, Cagney G, Taylor CT. 2011. Small ubiquitin-related modifier (SUMO)-1 promotes glycolysis in hypoxia. J. Biol. Chem. 286:4718–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Feala JD, Coquin L, Zhou D, Haddad GG, Paternostro G, McCulloch AD. 2009. Metabolism as means for hypoxia adaptation: metabolic profiling and flux balance analysis. BMC Syst. Biol. 3:91. 10.1186/1752-0509-3-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Postmus J, Aardema R, de Koning LJ, de Koster CG, Brul S, Smits GJ. 2012. Isoenzyme expression changes in response to high temperature determine the metabolic regulation of increased glycolytic flux in yeast. FEMS Yeast Res. 12:571–581 [DOI] [PubMed] [Google Scholar]
- 43. Henke RM, Dastidar RG, Shah A, Cadinu D, Yao X, Hooda J, Zhang L. 2011. Hypoxia elicits broad and systematic changes in protein subcellular localization. Am. J. Physiol. Cell Physiol. 301:C913–C928. 10.1152/ajpcell.00481.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rosenfeld E, Beauvoit B, Blondin B, Salmon JM. 2003. Oxygen consumption by anaerobic Saccharomyces cerevisiae under enological conditions: effect on fermentation kinetics. Appl. Environ. Microbiol. 69:113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Simeonidis E, Murabito E, Smallbone K, Westerhoff HV. 2010. Why does yeast ferment? A flux balance analysis study. Biochem. Soc. Trans. 38:1225–1229 [DOI] [PubMed] [Google Scholar]
- 46. Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691 [DOI] [PubMed] [Google Scholar]
- 47. An S, Kumar R, Sheets ED, Benkovic SJ. 2008. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320:103–106 [DOI] [PubMed] [Google Scholar]
- 48. Noree C, Sato BK, Broyer RM, Wilhelm JE. 2010. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J. Cell Biol. 190:541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. An S, Deng Y, Tomsho JW, Kyoung M, Benkovic SJ. 2010. Microtubule-assisted mechanism for functional metabolic macromolecular complex formation. Proc. Natl. Acad. Sci. U. S. A. 107:12872–12876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. 2010. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat. Cell Biol. 12:739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Miura N, Kirino A, Endo S, Morisaka H, Kuroda K, Takagi M, Ueda M. 2012. Tracing putative trafficking of the glycolytic enzyme enolase via SNARE-driven unconventional secretion. Eukaryot. Cell 11:1075–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Matsui K, Kuroda K, Ueda M. 2009. Creation of a novel peptide endowing yeasts with acid tolerance using yeast cell-surface engineering. Appl. Microbiol. Biotechnol. 82:105–113 [DOI] [PubMed] [Google Scholar]
- 53. Katahira S, Mizuike A, Fukuda H, Kondo A. 2006. Ethanol fermentation from lignocellulosic hydrolysate by a recombinant xylose- and cellooligosaccharide-assimilating yeast strain. Appl. Microbiol. Biotechnol. 72:1136–1143 [DOI] [PubMed] [Google Scholar]
- 54. Aoki W, Ueda T, Tatsukami Y, Kitahara N, Morisaka H, Kuroda K, Ueda M. 2013. Time-course proteomic profile of Candida albicans during adaptation to a fetal serum. Pathog. Dis. 67:67–75 [DOI] [PubMed] [Google Scholar]
- 55. Mashego MR, van Gulik WM, Vinke JL, Heijnen JJ. 2003. Critical evaluation of sampling techniques for residual glucose determination in carbon-limited chemostat culture of Saccharomyces cerevisiae. Biotechnol. Bioeng. 83:395–399 [DOI] [PubMed] [Google Scholar]
- 56. Tsugawa H, Bamba T, Shinohara M, Nishiumi S, Yoshida M, Fukusaki E. 2011. Practical non-targeted gas chromatography/mass spectrometry-based metabolomics platform for metabolic phenotype analysis. J. Biosci. Bioeng. 112:292–298 [DOI] [PubMed] [Google Scholar]
- 57. Nanchen A, Fuhrer T, Sauer U. 2007. Determination of metabolic flux ratios from 13C-experiments and gas chromatography-mass spectrometry data: protocol and principles. Methods Mol. Biol. 358:177–197 [DOI] [PubMed] [Google Scholar]
- 58. Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O'Kelly G, Schoenegger A, Ovelleiro D, Pérez-Riverol Y, Reisinger F, Ríos D, Wang R, Hermjakob H. 2013. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41:D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sato T, Watanabe T, Mikami T, Matsumoto T. 2004. Farnesol, a morphogenetic autoregulatory substance in the dimorphic fungus Candida albicans, inhibits hyphae growth through suppression of a mitogen-activated protein kinase cascade. Biol. Pharm. Bull. 27:751–752 [DOI] [PubMed] [Google Scholar]
- 60. Rhome R, Del Poeta M. 2009. Lipid signaling in pathogenic fungi. Annu. Rev. Microbiol. 63:119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cho T, Nagao J, Imayoshi R, Kaminishi H, Aoyama T, Nakayama H. 2010. Quorum sensing and morphological regulation in the pathogenic fungus Candida albicans. J. Oral Biosci. 52:233–239 [Google Scholar]
- 62. Deveau A, Piispanen AE, Jackson AA, Hogan DA. 2010. Farnesol induces hydrogen peroxide resistance in Candida albicans yeast by inhibiting the Ras-cyclic AMP signaling pathway. Eukaryot. Cell 9:569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Machida K, Tanaka T, Yano Y, Otani S, Taniguchi M. 1999. Farnesol-induced growth inhibition in Saccharomyces cerevisiae by a cell cycle mechanism. Microbiology 145:293–299 [DOI] [PubMed] [Google Scholar]
- 64. Machida K, Tanaka T, Fujita K, Taniguchi M. 1998. Farnesol-induced generation of reactive oxygen species via indirect inhibition of the mitochondrial electron transport chain in the yeast Saccharomyces cerevisiae. J. Bacteriol. 180:4460–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Synnott JM, Guida A, Mulhern-Haughey S, Higgins DG, Butler G. 2010. Regulation of the hypoxic response in Candida albicans. Eukaryot. Cell 9:1734–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. González Siso MI, Becerra M, Maceiras ML, Vazquez AV, Cerdan ME. 2012. The yeast hypoxic responses, resources for new biotechnological opportunities. Biotechnol. Lett. 34:2161–2173 [DOI] [PubMed] [Google Scholar]
- 67. Margittai M, Langen R. 2006. Side chain-dependent stacking modulates tau filament structure. J. Biol. Chem. 281:37820–37827 [DOI] [PubMed] [Google Scholar]
- 68. Hardie DG. 2011. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 25:1895–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hardie DG. 2011. AMPK and autophagy get connected. EMBO J. 30:634–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhang J, Vaga S, Chumnanpuen P, Kumar R, Vemuri GN, Aebersold R, Nielsen J. 2011. Mapping the interaction of Snf1 with TORC1 in Saccharomyces cerevisiae. Mol. Syst. Biol. 7:545. 10.1038/msb.2011.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Li N, Ragheb K, Lawler G, Sturgis J, Rajwa B, Melendez JA, Robinson JP. 2003. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J. Biol. Chem. 278:8516–8525 [DOI] [PubMed] [Google Scholar]
- 72. Taylor ER, Hurrell F, Shannon RJ, Lin TK, Hirst J, Murphy MP. 2003. Reversible glutathionylation of complex I increases mitochondrial superoxide formation. J. Biol. Chem. 278:19603–19610 [DOI] [PubMed] [Google Scholar]
- 73. Elliott NA, Volkert MR. 2004. Stress induction and mitochondrial localization of Oxr1 proteins in yeast and humans. Mol. Cell. Biol. 24:3180–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Daran-Lapujade P, Jansen ML, Daran JM, van Gulik W, de Winde JH, Pronk JT. 2004. Role of transcriptional regulation in controlling fluxes in central carbon metabolism of Saccharomyces cerevisiae. A chemostat culture study. J. Biol. Chem. 279:9125–9138 [DOI] [PubMed] [Google Scholar]
- 75. Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. 2006. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126:107–120 [DOI] [PubMed] [Google Scholar]
- 76. Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. 2006. p53 regulates mitochondrial respiration. Science 312:1650–1653 [DOI] [PubMed] [Google Scholar]
- 77. Jones RG, Thompson CB. 2009. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 23:537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Usui Y, Hirasawa T, Furusawa C, Shirai T, Yamamoto N, Mori H, Shimizu H. 2012. Investigating the effects of perturbations to pgi and eno gene expression on central carbon metabolism in Escherichia coli using 13C metabolic flux analysis. Microb. Cell Fact. 11:87. 10.1186/1475-2859-11-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. de Groot MJ, Daran-Lapujade P, van Breukelen B, Knijnenburg TA, de Hulster EA, Reinders MJ, Pronk JT, Heck AJ, Slijper M. 2007. Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveals post-transcriptional regulation of key cellular processes. Microbiology 153:3864–3878 [DOI] [PubMed] [Google Scholar]
- 80. Pavlides S, Whitaker-Menezes D, Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, Casimiro MC, Wang C, Fortina P, Addya S, Pestell RG, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. 2009. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle 8:3984–4001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.