Abstract
STK_08120 is a member of the thermoacidophile-specific DUF3211 protein family from Sulfolobus tokodaii strain 7. Its molecular function remains obscure, and sequence similarities for obtaining functional remarks are not available. In this study, the crystal structure of STK_08120 was determined at 1.79-Å resolution to predict its probable function using structure similarity searches. The structure adopts an α/β structure of a helix-grip fold, which is found in the START domain proteins with cavities for hydrophobic substrates or ligands. The detailed structural features implied that fatty acids are the primary ligand candidates for STK_08120, and binding assays revealed that the protein bound long-chain saturated fatty acids (>C14) and their trans-unsaturated types with an affinity equal to that for major fatty acid binding proteins in mammals and plants. Moreover, the structure of an STK_08120-myristic acid complex revealed a unique binding mode among fatty acid binding proteins. These results suggest that the thermoacidophile-specific protein family DUF3211 functions as a fatty acid carrier with a novel binding mode.
INTRODUCTION
Thermoacidophiles are defined as organisms that can survive at extremely low pH (optimal pH for growth: typically 0.7 to 3.0) and at high temperatures (above 50°C). These organisms are found exclusively in archaeal species, and complete genome sequences have been determined for several species in the past 2 decades (1–3). Comparative genome analyses among thermoacidophilic archaea have been applied to better understand their acidophilic survival strategies, which are based on the known or annotated functions of the genes of interest. This approach has shed light on the possible mechanisms underlying acid adaptation, such as a maintenance system of genome structure and integrity (4), energy metabolism with unique respiratory chain compositions (1, 5), and membrane transports for tolerating a huge pH gradient across the membrane (6). Comparative genome analyses have also identified various genes specific to thermoacidophilic archaea and functionally unknown, and their functional aspects could help us understand the mechanisms by which these archaea adapt to acidic environments.
STK_08120 is a member of the thermoacidophile-specific domain unknown function 3211 (DUF3211) family of proteins encoded by the STK_08120 gene of Sulfolobus tokodaii strain 7 and consists of 136 amino acid residues. According to a BLAST (7) search, homologous sequences are specifically found in thermoacidophilic archaea such as Sulfolobus, Acidianus, and Metallosphaera. There are no structural and functional findings on the DUF3211 family proteins and no sequence similarities that could provide insight into their molecular function.
Structure-based function prediction for proteins is often superior to sequence-based approaches because the folding pattern is retained even if the sequence similarity is not found (8). Proteins with similar functions often have similar folds, and hence, the first step toward structure-based function prediction is to find structural neighbors. There are now over 90,000 entries in the Protein Data Bank (PDB) and several methods for structural alignment, such as the DALI server (9). In fact, the functions of some targeted proteins have been elucidated either by global structure similarity to well-characterized proteins (10, 11) or by the combination of structural comparisons and biochemical experiments (12, 13).
In this report, we characterized the molecular function of STK_08120 by a structural alignment approach coupled with biochemical experiments. We found that STK_08120 bound to long-chain saturated fatty acids (>C14) and their trans-unsaturated form with an affinity equal to that of major fatty acid binding proteins (FABPs) in mammals and plants. A unique binding mode is here proposed on the basis of the structure of the STK_08120 complex with myristic acid.
MATERIALS AND METHODS
Plasmid construction and protein expression.
STK_08120 was overexpressed in Escherichia coli Rosetta (DE3) by using the pET system (Novagen). The DNA sequence encoding full-length STK_08120 (STK_08120) was amplified from the genome of Sulfolobus tokodaii strain 7 by using PCR and was then inserted between the NdeI and BamHI sites of the pET-28a vector (Novagen). The cells were transformed with the plasmid and then cultured in lysogeny broth (LB) medium containing 20 mg liter−1 kanamycin and 34 mg liter−1 chloramphenicol at 37°C. The protein expression was induced by the addition of 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at the mid-log phase. The selenomethionine (SeMet) derivative of STK_08120 was overexpressed in M9 medium containing 0.4% (wt/vol) glucose; 0.1 mg liter−1 thiamine; 1 mM MgSO4; 4.2 mg liter−1 FeSO4; 20 mg liter−1 kanamycin; 34 mg liter−1 chloramphenicol; 50 mg liter−1 of isoleucine, leucine, valine, and l-selenomethionine; and 100 mg liter−1 of lysine and phenylalanine by using the methionine biosynthesis inhibition method (14).
Protein purification.
The harvested cells were disrupted by sonication in a buffer containing 20 mM Tris-HCl (pH 7.2), 0.5 M NaCl, 30 mM imidazole, and 1 mM dithiothreitol (DTT). The supernatant after centrifugation was incubated at 80°C for 15 min and was then centrifuged once more. The supernatant was applied onto a Ni-Sepharose column (GE Healthcare). His6-tagged STK_08120 was trapped and purified on the column. The fusion protein was then cleaved between the sequences of His6 tag and STK_08120 by thrombin protease to release only the untagged STK_0812 from the column. The untagged protein was dialyzed against 20 mM morpholineethanesulfonic acid (MES)-NaOH (pH 5.6) and then applied onto a Mono-S cation-exchange column (GE Healthcare). The protein was eluted with a linear gradient of 0 to 200 mM NaCl in the MES buffer. The protein solution was dialyzed against 10 mM Tris-HCl (pH 7.0) and was concentrated to 32 mg ml−1 by using VivaSpin (Sartorius AG). The protein concentration was estimated by the absorbance at 280 nm with an absorption coefficient of 7,450 M−1 cm−1 (15). The SeMet derivative was purified and concentrated using the method mentioned above.
Crystallization.
Crystals of STK_08120 and the SeMet derivative were obtained under the reservoir solution containing 15% (wt/vol) polyethylene glycol (PEG) 4000, 20% (vol/vol) isopropanol, and 0.1 M MES-NaOH (pH 6.6) at 20°C by the sitting-drop vapor diffusion method. In the case of an STK_08120-myristic acid complex, the crystallization solution was prepared by adding myristic acid at a final concentration of 2 mM to protein solution (diluted to 20 mg ml−1). The STK_08120-myristic acid crystals were grown at 20°C in sitting drops containing 1 μl of the protein solution and an equal volume of the reservoir solution, which contained 22 to 25% (vol/vol) isopropanol, 0.2 M ammonium acetate, and 0.1 M buffer (Tris-HCl or HEPES-NaOH, pH 7.5 to 8.5).
X-ray data collection and processing.
Reservoir solution containing 40% (vol/vol) ethylene glycol was piled up on the drops with protein crystals as a cryoprotectant. The crystals were picked up in a nylon loop (Hampton Research) and mounted for flash-cooling with a liquid nitrogen stream. X-ray diffraction data were collected with synchrotron radiation by using an ADSC Quantum-210 charge-coupled device (CCD) detector system on the AR-NW12 beamline at the Photon Factory (Tsukuba, Japan). For single-wavelength anomalous diffraction (SAD) data for the SeMet derivative crystal, the peak wavelength (0.97924 Å) was determined from the selenium absorption spectrum. For the STK_08120-myristic acid complex, the data were collected with synchrotron radiation by using an ADSC Quantum-210 CCD detector system on the BL-17A beamline at the Photon Factory. All diffraction data were indexed, integrated, and scaled with the HKL2000 program suite (16).
Structure determination and refinement.
The structure of STK_08120 was determined by the single-wavelength anomalous dispersion (SAD) method and the data set of the SeMet derivative crystal. Experimental SAD phasing and density modification were performed using the SHELXC/D/E program (17, 18). The initial model for the SeMet derivative was built and refined with the programs ARP/wARP (19) and Refmac5 (20) in the CCP4 program suite (21). The refined model was used to determine the tertiary structure of STK_08120 by the molecular replacement method of the CCP4 program MOLREP (22) and the X-ray diffraction data of the native crystal. The final model was rebuilt and refined using the CCP4 programs XtalView (23) and Refmac5 (20). The tertiary structure of the STK_08120-myristic acid complex was determined by the molecular replacement method of MOLREP (22) and the coordinates of STK_08120. A model was manually rebuilt using the COOT program (24) and was refined using the Refmac5 program (20).
Bioinformatics analyses.
Homologous proteins with STK_08120 were searched using the BLAST (7) server at the NCBI. Distribution analysis of START domain proteins was performed using the SUPERFAMILY database (25). The signal peptide sequence was searched using the PRED-SIGNAL server (26). The DALI (9) and Pocket-Finder (27) servers were used for structure similarity and ligand-binding pocket searches, respectively. Protein-ligand interactions were analyzed using the program LIGPLOT (28). Computer graphic representations were prepared using the software PyMOL (http://www.pymol.org/).
Gel filtration analysis.
Analytical gel filtration chromatography was carried out using a Superdex 75 HR 10/30 (GE Healthcare) column in a buffer containing 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 1 mM DTT. The standard protein mixture was supplied from the gel filtration calibration kit (GE Healthcare).
Fatty acid binding assay.
The fatty acid binding assay of STK_08120 was carried out using 2-p-toluidinylnaphthalene-6-sulfonate (TNS) as a fluorescence probe. All fluorescence experiments were performed on a Shimadzu RF-5300PC spectrofluorophotometer at 25°C as described previously (29) with some modifications. The excitation and emission wavelengths were set at 355 and 430 nm, respectively. Before the initial fluorescence was recorded, 0.5 μM STK_08120 and 15 μM TNS were incubated for 1 min in a measurement buffer containing 175 mM mannitol, 0.5 mM K2SO4, 0.5 mM CaCl2, and 5 mM MES (pH 7.0). Each fatty acid was prepared at a concentration of 0.05 to 25 mM in dimethyl sulfoxide (DMSO) and was then titrated by 12 successive 5-μl injections into the reaction solution. The equilibrated fluorescence per injection was recorded for 30 s. The binding affinity of STK_08120 may be somewhat affected by the unfavorable interaction of TNS with the tested fatty acids. To at least avoid the incorporation of TNS into the micelles of the fatty acids, each fatty acid was used in the range of concentration lower than the critical micelle concentration (CMC). Control injections of DMSO into the reaction solution containing protein were performed to determine background corrections. The data set of decreased fluorescence intensities was analyzed as a single-site binding function by using GraphPad Prism 4 (GraphPad Software).
Protein structure accession numbers.
Coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with accession codes 2EJX and 3W9K for STK_08120 and its complex with myristic acid, respectively.
RESULTS
Structure determination and overall structure of STK_08120.
The crystal structure of STK_08120 was determined at 1.79-Å resolution by the single-wavelength anomalous dispersion (SAD) phasing method by using the SeMet-substituted protein. The final model with 0.22/0.26 of R/Rfree contains a protein molecule (Lys3 to Ile136) and 102 water molecules in an asymmetric unit. SFCHECK (30) analysis revealed that 97.7% of all the residues were in the most favored region and that the remaining residues were in the additionally allowed region of a Ramachandran plot (31). The data collection and refinement statistics are summarized in Table 1.
Table 1.
Crystal parameters, data collection statistics, and refinement statistics
| Parameter or statistic | Value for proteina: |
||
|---|---|---|---|
| STK_08120 | SeMet derivative | STK_08120-myristic acid complex | |
| Data collection | |||
| Space group | P21212 | P21212 | P21212 |
| Lattice constants (Å) | a = 54.9, b = 70.4, c = 35.2 | a = 53.3, b = 70.0, c = 34.8 | a = 53.9, b = 70.6, c = 35.3 |
| Wavelength (Å) | 1.00000 | 0.97924 | 1.00000 |
| Resolution range (Å) | 50.0–1.79 (1.85–1.79) | 50.0–2.00 (2.07–2.00) | 50.0–1.80 (1.86–1.80) |
| No. of observed reflections | 73,265 | 44,721 | 89,716 |
| Data completeness (%) | 97.8 (96.6) | 96.8 (99.1) | 99.9 (99.6) |
| Redundancy | 5.5 (6.0) | 5.2 (6.0) | 6.9 (6.1) |
| Rsym | 0.072 (0.255) | 0.062 (0.158) | 0.073 (0.214) |
| <I>/<σ(I)> | 43.9 (10.8) | 48.1 (23.8) | 45.4 (8.5) |
| Wilson B factor (Å2) | 24.5 | 45.0 | 18.5 |
| Refinement | |||
| Resolution range (Å) | 20.0–1.79 | 20.0–1.80 | |
| No. of reflections | 12,526 | 12,356 | |
| R/Rfree (%) | 22.2/26.2 | 18.3/23.2 | |
| No. of atoms | |||
| Protein | 1,090 | 1,098 | |
| Ligand | 16 | ||
| Water | 102 | 103 | |
| B factor | |||
| Protein (Å2) | 29.9 | 17.5 | |
| Ligand (Å2) | 34.9 | ||
| Water (Å2) | 41.1 | 27.6 | |
| RMSDs | |||
| Bond lengths (Å) | 0.017 | 0.023 | |
| Bond angles (°) | 1.61 | 1.98 | |
| Ramachandran plot | |||
| Favored (%) | 92.5 | 99.2 | |
| Allowed (%) | 7.5 | 0.8 | |
| Outliers (%) | 0 | 0 | |
Values in parentheses are for the highest-resolution shell.
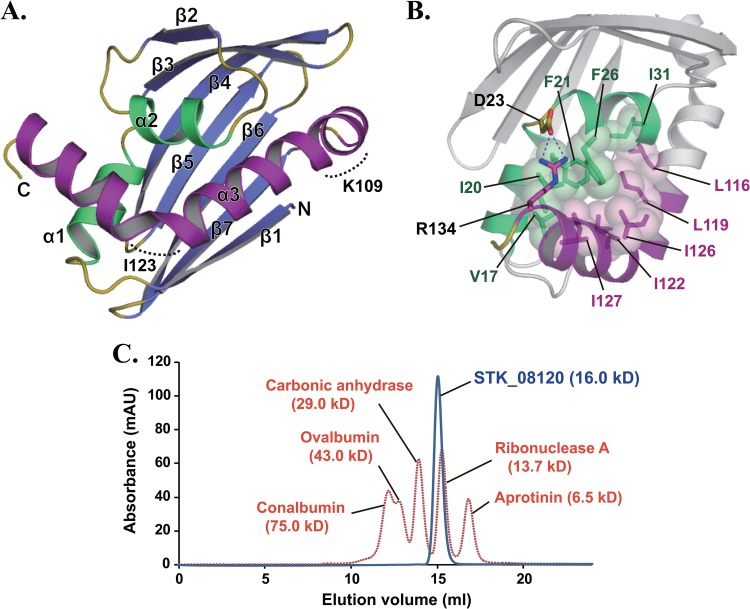
The tertiary structure of STK_08120 was comprised of three α-helices and a seven-stranded β-sheet with the following topology: β1-α1-α2-β2-β3-β4-β5-β6-β7-α3 (Fig. 1A). The order of the strands was 1-7-6-5-4-3-2, and all strands were in antiparallel orientations. The β-sheet bends into an unclosed β-barrel and surrounds α1- and α2-helices. These helices also form tight hydrophobic contacts with the C-terminal half (residues Leu116 to Arg134) of the α3-helix (Fig. 1B). In addition, Arg134 on the α3-helix forms a salt bridge with Asp23 on a loop connecting the α1- and α2-helices (L2). The contact region among the three helices seems to be more rigid, with a lower average B factor of Cα atoms (23.9 Å2) compared with the other regions (average B factor of Cα atoms, 28.8 Å2). The C-terminal long α3-helix shows a unique twisted-S-shaped feature caused by two kinks of approximately 40 and 75 degrees at the positions around Lys109 and Ile123, respectively (Fig. 1A). Gel filtration experiments showed that STK_08120 existed as a monomer in solution (Fig. 1C).
Fig 1.
Crystal structure of STK_08120. (A) The structure is colored blue and yellow for the β-sheet and loops, respectively. Among the three α-helices, the α3-helix and the other helices (α1 and α2) are shown in purple and green, respectively. Dashed curves indicate kinked regions around Lys109 and Ile123. (B) Stick-and-sphere models and dashed lines represent hydrophobic contacts among the residues on the three α-helices and a salt bridge between Asp23 and Arg134, respectively. The purple region corresponds to the C-terminal half (residues Leu116 to Arg134) of the α3-helix. (C) Elution patterns of STK_08120 (blue) and standard proteins (orange) on a Superdex 75 HR 10/30 gel filtration column. The absorption wavelength was 280 nm for detecting protein peaks. Values in parentheses are molecular masses estimated from the amino acid sequence for STK_08120.
Structural similarity to START domain proteins.
Based on the BLAST search using the STK_08120 sequence, all the sequences with alignment scores of >40 are annotated as hypothetical proteins and do not provide any structural information. Among the sequences with lower alignment scores, levansucrase (alignment score, 38.9; E value, 0.24) is the only protein whose function is known. The enzyme exhibits a five-bladed β-propeller fold (32), which is quite different from the structure of STK_08120.
A structural homology search by the DALI server showed that there were 22 structures homologous to STK_08120 with Z values of >10.0, where the Z value represents the degree of structural similarity. Most of the searched proteins with high Z values were uncharacterized proteins, whereas the functions of some proteins are understood: polyketide aromatase/cyclase (PAC) ZhuI (Z value, 10.6; root mean square deviation [RMSD], 3.1 Å; number of Cα atoms, 123; sequence identity, 9%; PDB code, 3TFZ [33]), abscisic acid (ABA) receptor pyrabactin resistance 1 (PYR1)-like 2 (PYL2) (Z value, 10.3; RMSD, 3.1 Å; number of Cα atoms, 123; sequence identity, 7%; PDB code, 3NS2 [34]), and phenolic oxidative coupling protein Hyp-1 (Z value, 10.1; RMSD, 3.4 Å; number of Cα atoms, 122; sequence identity, 7%; PDB code, 3IE5 [35]).
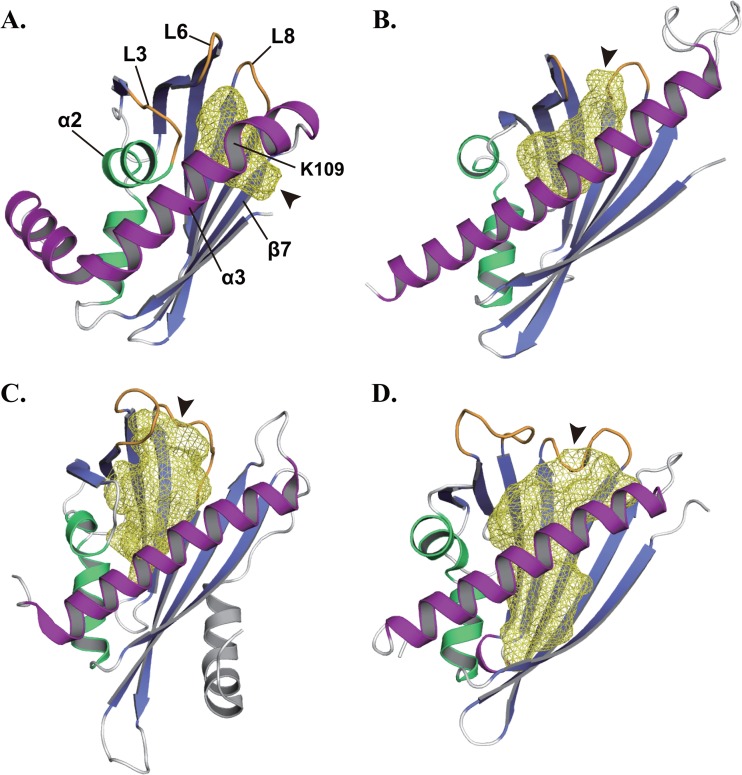
PAC catalyzes a cyclization reaction of polyketide with diverse cyclization patterns (33, 36, 37). ABA receptors receive ABA, a plant hormone, to inhibit a target phosphatase activity (38). Hyp-1 is involved in the dimerization reaction of emodin in hypericin biosynthesis with no enzymatic function (35, 39). These proteins are classified as steroidogenic acute regulatory protein (StAR)-related lipid transfer (START) domain proteins, which share the helix-grip fold of a β-α-α-β-β-β-β-β-β-α topology and a cavity with a preference for hydrophobic substrates or ligands (40). Like START domain proteins, STK_08120 had not only a helix-grip fold composed of 3 α-helices and 7 β-strands but also an internal cavity formed as the β-barrel wraps around the α3-helix (Fig. 2A). The database search using the SUPERFAMILY server (25) showed that 52 genes in 49 bacterial genomes encoded START domain proteins and there was no gene for START domain protein in archaeal genomes. However, a previous distribution analysis has reported that 2 archaeal genes encode START domain proteins (40). Compared to the 4,600 genes for START domain proteins in eukaryotes, the number of START domain proteins is extremely low in archaea and eubacteria, which may be caused by the considerable diversity in the sequence of bacterial START domain proteins, as in the instance of STK_08120.
Fig 2.
Cavities of STK_08120 and START domain proteins. (A) STK_08120. (B) ZhuI polyketide aromatase/cyclase (PDB identifier, 3TFZ [33]). (C) ABA receptor PYL2 (PDB identifier, 3NS2 [34]). (D) Phenolic oxidative coupling protein Hyp-1 (PDB identifier, 3IE5 [35]). Cavities are represented by the yellow mesh model, and arrowheads indicate the entrances to the cavities. Gate loops are shown by orange color in the ZhuI, PYL2, and Hyp-1 structures. Purple regions correspond to the α3-helix of STK_08120 and the C-terminal long helices of ZhuI, PYL2, and Hyp-1.
Compared with the cavities of ZhuI, PYL2, and Hyp-1, there are two major differences in the cavity of STK_08120. One is the difference in the size of cavity, and the other is the difference in the direction of entrance into the cavity. The α2-helix and the following loop (L3) of STK_08120 deeply penetrate the protein molecule to narrow the cavity (Fig. 2A). The cavity volume is 340 Å3 as calculated by the Pocket-Finder server (27), whereas the START domain proteins generally each have a cavity with a typical volume of >600 Å3 (Fig. 2B to D). In the cavity of STK_08120, an entrance is located between the C-terminal region of the β7-strand (Leu97-His-99) and the N-terminal region of the α3-helix (Lys102-Val110). The first kink in the α3-helix, around Lys109, appears to provide enough space for the entrance (Fig. 1A and 2A). In the START domain proteins, an entrance to the cavity is generally formed by the C-terminal long helix and two loops (Fig. 2B to D), corresponding to the α3-helix and loops L6 and L8 of STK_08120, respectively. The loops often act as a gate that regulates the access of ligands into the cavity, which has been well characterized in ABA receptors like PYL2 (34, 38). In contrast to this typical feature, the cavity of STK_08120 is closed at the positions around loops L6 and L8 in a ligand-free form by tight contacts among the residues on the α3-helix and loops L6 and L8.
Function prediction based on the detailed properties of the cavity.
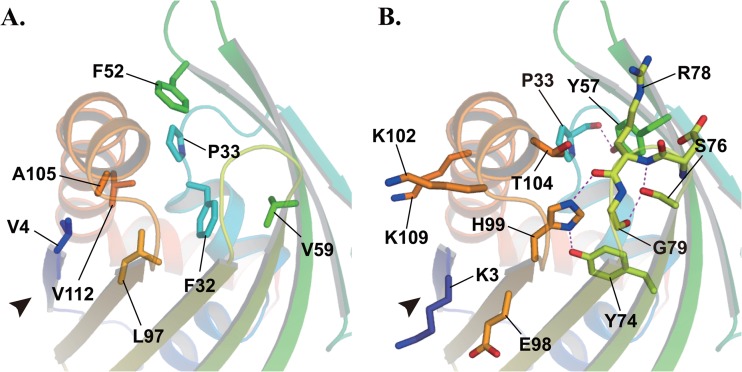
The cavity was comprised of eight nonpolar residues, Val4, Phe32, Pro33, Phe52, Val59, Leu97, Ala105, and Val112 (Fig. 3A), and seemed extremely hydrophobic. Several polar residues, Tyr57, Tyr74, Ser76, Arg78, His99, and Thr104, were also arranged in the cavity. All polar groups were either masked by forming hydrogen bonds with other polar groups (e.g., Pro33 [CO]—Tyr57 [ζ-OH], Tyr74 [ζ-OH]—His99 [δ-NH], Ser76 [γ-OH]—Arg78 [NH], Ser76 [γ-OH]—Gly79 [CO], and Arg78 [CO]—His99 [ε-NH]) or faced away from the cavity to the solvent (e.g., the guanidino group of Arg78 and Thr104 [γ-OH]) (Fig. 3B). These features help to maintain the hydrophobicity of the cavity. In addition, there were four charged residues, Lys3, Glu98, Lys102, and Lys109, at the entrance to the cavity (Fig. 3B), where the net charge was positive. These findings predict a ligand specificity of STK_08120 as follows. (i) The cavity may be able to bind a lipophilic molecule. (ii) A negatively charged group of ligands may be necessary for a lipophilic chain to access the cavity through a positively charged filter around the entrance.
Fig 3.
Residues composing a hydrophobic cavity of STK_08120. Nonpolar (A) and polar (B) residues are represented by stick models. Dashed lines and an arrowhead indicate a hydrogen bond and the cavity entrance, respectively.
Among the structurally homologous proteins to STK_08120, PAC functions as a single enzyme, and the activity requires three residues, Asp, Arg, and His, in the cavity (33, 36, 37), whereas STK_08120 has no aspartic acid residue in the cavity and the guanidino group of Arg78 flips out from the cavity as mentioned above. Therefore, STK_08120 might not be able to work as a PAC.
Fatty acid binding activity and selectivity.
To evaluate the predicted ligand specificities of STK_08120, ligand-binding assays were carried out using various natural fatty acids as candidate compounds. Table 2 summarizes the dissociation constants (Kd) of the fatty acids tested in this study. STK_08120 was able to bind to a saturated medium-chain or long-chain fatty acid with more than eight carbons (>C8). The affinities elevated in every elongation of two hydrocarbons and myristic acid (C14) showed the strongest affinity, with a Kd value of 0.48 ± 0.05 μM. The affinities are most effectively increased by adding two hydrocarbons at the C13 and C14 positions (∼30-fold increase). Palmitic acid (C16) and stearic acid (C18) each have a longer acyl chain than does myristic acid (C14), although the affinities are almost equal among these three fatty acids. These results imply that the cavity of STK_08120 is suitably sized for binding to an acyl chain with C14.
Table 2.
Dissociation constants (Kd) of STK_08120 toward several fatty acidsa
| Fatty acid | Construction | Kd (μM) |
|---|---|---|
| Hexanoic acid | C6:0 | NDb |
| Octanoic acid | C8:0 | 277 ± 23 |
| Decanoic acid | C10:0 | 29.8 ± 0.9 |
| Dodecanoic acid | C12:0 | 14.0 ± 1.8 |
| Myristic acid | C14:0 | 0.48 ± 0.05 |
| Palmitic acid | C16:0 | 0.67 ± 0.14 |
| Stearic acid | C18:0 | 0.59 ± 0.07 |
| Palmitoleic acid | C16:1, cis-9 | 4.32 ± 0.12 |
| Oleic acid | C18:1, cis-9 | 4.23 ± 0.32 |
| Elaidic acid | C18:1, trans-9 | 0.70 ± 0.10 |
| Linoleic acid | C18:2, cis-9,12 | 3.45 ± 0.26 |
| Linolenic acid | C18:3, cis-9,12,15 | 4.47 ± 0.19 |
All determinations were performed in triplicate, and results are expressed as mean values ± standard deviations.
ND, not detected.
Unsaturated fatty acids were also tested for comparison with saturated fatty acids (Table 2). Palmitoleic acid (C16:1, cis-9) and oleic acid (C18:1, cis-9) are unsaturated fatty acids with a cis double bond. Both showed lower affinities than their respective saturated forms, palmitic acid (C16:0) and stearic acid (C18:0). In contrast, the affinity of elaidic acid (C18:1, trans-9) with a trans double bond was almost equal to that of its saturated form (C18:0). These results indicate that saturated and trans-unsaturated fatty acids are preferred as ligands of STK_08120 over cis-unsaturated fatty acids among typical natural fatty acids. Furthermore, the comparison among oleic acid (C18:1, cis-9), linoleic acid (C18:2, cis-9,12), and linolenic acid (C18:1, cis-9,12,15) showed that the further addition of the cis double bonds at positions 12 and/or 15 had no apparent effect on the affinity to STK_08120. Fatty acids would partially lose their conformational flexibility by forming a cis double bond at the C9 position, which may restrict the arrangement of fatty acids in the cavity by a steric hindrance effect.
Binding mode for myristic acid.
To identify the precise binding mode between STK_08120 and fatty acids, we determined the structure of STK_08120 in complex with myristic acid at 1.80-Å resolution. The final model, with 0.18/0.23 of R/Rfree, contains a protein molecule (residues Met2 to Ile136), a myristic acid, and 103 water molecules in an asymmetric unit. SFCHECK (30) analysis revealed that 99.2% of all the residues were in the favored region and that the remaining residues were in the allowed region of the Ramachandran plot (31). The data collection and refinement statistics are summarized in Table 1.
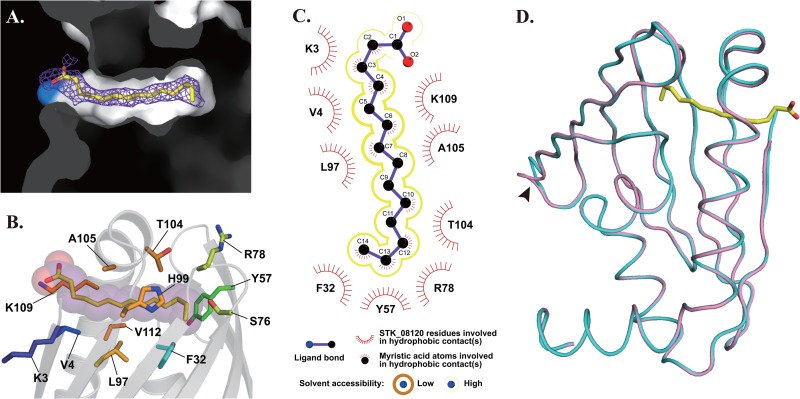
The electron density of myristic acid was observed in the cavity of STK_08120 (Fig. 4A), and the carboxyl group was located near the amino group of Lys109, probably through an electrostatic interaction. Thus, the linear structure of myristic acid could fit the cavity well. The length of myristic acid also appears to be almost equal to its depth from Lys109 to the bottom of the cavity, suggesting that no more interaction would be reinforced by the further elongation of the hydrocarbon chain in myristic acid (C14), as shown by the equal affinities of palmitic acid (C16) and stearic acid (C18) to myristic acid (Table 2). Myristic acid in the crystal was surrounded by six nonpolar residues (Val4, Phe32, Leu97, Ala105, Gly108, and Val112) and seven polar residues (Lys3, Tyr57, Ser76, Arg78, His99, Thr104, and Lys109) within the distance of 4 Å (Fig. 4B). Based on LIGPLOT (28) analysis, 9 of these 13 residues formed hydrophobic contacts with myristic acid (Fig. 4C). These interactions are suitable for trapping the hydrocarbon chain of myristic acid into the cavity, although solvent accessibility appears to be slightly high in the LIGPLOT analysis.
Fig 4.
Myristic acid-bound structure of STK_08120. (A) Myristic acid in the hydrophobic cavity is represented by a stick model along with the Fo-Fc electron density colored in violet (1.5σ). The light blue surface shows the amino group of Lys109. (B) Myristic acid is represented by a stick-and-sphere model. The other stick models indicate the residues surrounding the myristic acid within 4 Å. (C) Hydrophobic contacts are illustrated using a LIGPLOT diagram. Yellow lines surrounding the model of myristic acid represent solvent accessibility: dark thick line, low (tight hydrophobic contact between protein and ligand); light thin line, high (loose hydrophobic contact). (D) Superposed structures of STK_08120 are colored pink and cyan for apo and myristic acid-bound states, respectively. The arrowhead indicates a flexible loop (I41-E42) with comparatively high B factors (>50 Å2 for apo state and >30 Å2 for myristic acid-bound state). Myristic acid is represented by a yellow stick model.
DISCUSSION
STK_08120 has no signal sequence for secretion, according to the results of PRED-SIGNAL (26) analysis, indicating that it acts as an archaeal cytosolic protein. Several fatty acid binding proteins have been found in different compartments of cells and organisms (41). Among them, fatty acid binding protein (FABP) and lipid transfer protein (LTP) are known as major carrier proteins for fatty acids in the cytosol of mammalian and plant cells, respectively. They preferably bind to a molecule of saturated and unsaturated long-chain fatty acids, such as palmitic acid (C16:0) and oleic acid (C18:1, cis-9), with Kd values of approximately 0.5 μM (42, 43). Compared with these carrier proteins, STK_08120 has almost the same affinity for palmitic acid (Kd = 0.67 ± 0.14 μM) but binds oleic acid with lower affinity (Kd = 4.23 ± 0.32 μM). Thus, STK_08120 was revealed to have a reasonable binding affinity as a fatty acid carrier protein and unfavorable selectivity for cis-unsaturated fatty acids compared with FABP and LTP.
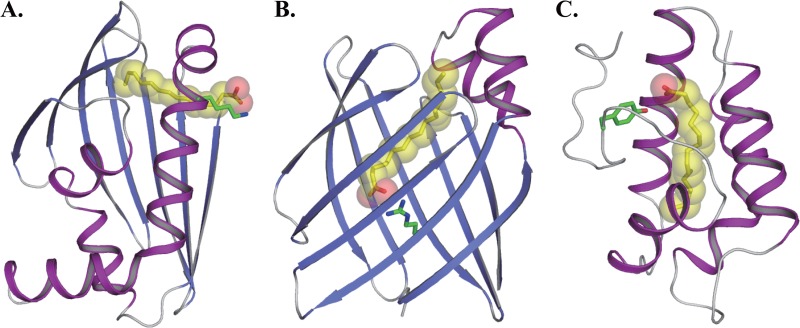
STK_08120 possesses an α/β structure of a helix-grip fold that is quite different from the structures of FABP and LTP (Fig. 5). The structure of FABP contains a β-barrel with 10 antiparallel β-strands and two short α-helices, which form the fatty acid binding cavity and a cap over one end of the cavity, respectively (Fig. 5B) (44). Some hydrophilic residues that contain arginine are at the bottom of the cavity, and they interact with the carboxyl group of fatty acids. On the other hand, LTP is formed with four α-helices and a long C-terminal tail region and has an internal hydrophobic cavity (Fig. 5C) (45). The cavity interacts with the hydrocarbon chain of fatty acids, and the carboxyl group is exposed to solvent in contact with a hydrophilic residue such as tyrosine. Based on the orientation of fatty acids, the binding mode of STK_08120 is similar to that of LTP rather than that of FABP, whereas the plasticity of the cavity appears to differ between STK_08120 and LTP. The cavity volume of LTP is estimated to increase from ∼400 Å3 in the free state to 547 to 620 Å3 in the lipidated state (41). In contrast, the structural studies on STK_08120 revealed that there is no significant change in the cavity upon binding of a myristic acid (RMSD of the Cα atoms of 3 to 136 residues, 0.25 Å) (Fig. 4D). The cavity of STK_08120 is relatively inflexible, which may cause an ∼7-fold decrease in binding affinity for cis-unsaturated fatty acids.
Fig 5.
Binding modes for long-chain fatty acids. (A) STK_08120-myristic acid complex. (B) FABP-palmitic acid complex (PDB identifier, 2IFB [44]). (C) LTP-palmitic acid complex (PDB identifier, 1FK3 [45]). Green sticks are the interaction residues with the carboxyl group of myristic acid or palmitic acid.
The fatty acid binding affinity of STK_08120 corresponds to the affinities of major fatty acid carrier proteins in mammals and plants. These findings raise the possibility that STK_08120 functions as a carrier of fatty acids in their biosynthesis, metabolism, and/or intracellular transport processes. Based on this functional insight into STK_08120, we propose to designate it TaFACP (thermoacidophile-specific fatty acid carrier protein). The binding activity of TaFACP was not affected by the addition of cis double bonds at the C12 and C15 positions of oleic acid (Table 2). These results suggest that cis-unsaturated fatty acids bind relatively nonspecifically to the hydrophobic cavity with their low affinity. On the other hand, the hydrocarbon chain of myristic acid was not completely varied in the cavity by the solvent accessibility in the LIGPLOT analysis (Fig. 4C), and Lys109 would form electrostatic interactions with not only the carboxy group but also other positively charged groups. Therefore, our study could not exclude the possibility that TaFACP is capable of binding the analogs of fatty acids with other linear lipophilic chain and/or negatively charged groups. Although further studies are needed to develop lipophilic metabolites and their functions, fatty acids and their analogs may function through TaFACP in biological processes specific to thermoacidophilic archaea.
ACKNOWLEDGMENTS
We thank the scientists and staff at the Photon Factory. The synchrotron radiation experiments were performed at AR-NW12A and BL-17A at the Photon Factory, Tsukuba, Japan (proposal numbers 2006S2-006 and 2008S2-001).
This research was supported by the National Project on Protein Structural and Functional Analyses (Protein 3000) of the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) and by the Targeted Proteins Research Program (TPRP) of MEXT.
Footnotes
Published ahead of print 8 July 2013
REFERENCES
- 1.Kawarabayasi Y, Hino Y, Horikawa H, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, Kosugi H, Hosoyama A, Fukui S, Nagai Y, Nishijima K, Otsuka R, Nakazawa H, Takamiya M, Kato Y, Yoshizawa T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Masuda S, Yanagii M, Nishimura M, Yamagishi A, Oshima T, Kikuchi H. 2001. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain7. DNA Res. 8:123–140 [DOI] [PubMed] [Google Scholar]
- 2.She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, Erauso G, Fletcher C, Gordon PM, Heikamp-de Jong I, Jeffries AC, Kozera CJ, Medina N, Peng X, Thi-Ngoc HP, Redder P, Schenk ME, Theriault C, Tolstrup N, Charlebois RL, Doolittle WF, Duguet M, Gaasterland T, Garrett RA, Ragan MA, Sensen CW, Van der Oost J. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U. S. A. 98:7835–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fütterer O, Angelov A, Liesegang H, Gottschalk G, Schleper C, Schepers B, Dock C, Antranikian G, Liebl W. 2004. Genome sequence of Picrophilus torridus and its implications for life around pH 0. Proc. Natl. Acad. Sci. U. S. A. 101:9091–9096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu LJ, You XY, Zheng H, Wang S, Jiang CY, Liu SJ. 2011. Complete genome sequence of Metallosphaera cuprina, a metal sulfide-oxidizing archaeon from a hot spring. J. Bacteriol. 193:3387–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruepp A, Graml W, Santos-Martinez ML, Koretke KK, Volker C, Mewes HW, Frishman D, Stocker S, Lupas AN, Baumeister W. 2000. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407:508–513 [DOI] [PubMed] [Google Scholar]
- 6.Angelov A, Liebl W. 2006. Insights into extreme thermoacidophily based on genome analysis of Picrophilus torridus and other thermoacidophilic archaea. J. Biotechnol. 126:3–10 [DOI] [PubMed] [Google Scholar]
- 7.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 8.Whisstock JC, Lesk AM. 2003. Prediction of protein function from protein sequence and structure. Q. Rev. Biophys. 36:307–340 [DOI] [PubMed] [Google Scholar]
- 9.Holm L, Rosenström P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38:W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graille M, Quevillon-Cheruel S, Leulliot N, Zhou CZ, Gallay ILD, Jacquamet L, Ferrer JL, Liger D, Poupon A, Janin J, van Tilbeurgh H. 2004. Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family. Structure 12:839–847 [DOI] [PubMed] [Google Scholar]
- 11.Teplyakov A, Obmolova G, Sarikaya E, Pullalarevu S, Krajewski W, Galkin A, Howard AJ, Herzberg O, Gilliland GL. 2004. Crystal structure of the YgfZ protein from Escherichia coli suggests a folate-dependent regulatory role in one-carbon metabolism. J. Bacteriol. 186:7134–7140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanishvili R, Yakunin AF, Laskowski RA, Skarina T, Evdokimova E, Doherty-Kirby A, Lajoie GA, Thornton JM, Arrowsmith CH, Savchenko A, Joachimiak A, Edwards AM. 2003. Integrating structure, bioinformatics, and enzymology to discover function: BioH, a new carboxylesterase from Escherichia coli. J. Biol. Chem. 278:26039–26045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horita S, Ishibashi J, Nagata K, Miyakawa T, Yamakawa M, Tanokura M. 2010. Isolation, cDNA cloning, and structure-based functional characterization of oryctin, a hemolymph protein from the coconut rhinoceros beetle, Oryctes rhinoceros, as a novel serine protease inhibitor. J. Biol. Chem. 285:30150–30158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyakawa T, Sawano Y, Miyazono K, Hatano K, Tanokura M. 2007. Crystallization and preliminary X-ray analysis of ginkbilobin-2 from Ginkgo biloba seeds: a novel antifungal protein with homology to the extracellular domain of plant cysteine-rich receptor-like kinases. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 63:737–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 17.Schneider TR, Sheldrick GM. 2002. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58:1772–1779 [DOI] [PubMed] [Google Scholar]
- 18.Sheldrick GM. 2002. Macromolecular phasing with SHELXE. Z. Kristallogr. 217:644–650 [Google Scholar]
- 19.Morris RJ, Perrakis A, Lamzin VS. 2002. ARP/wARP's model-building algorithms. I. The main chain. Acta Crystallogr. D Biol. Crystallogr. 58:968–975 [DOI] [PubMed] [Google Scholar]
- 20.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 21.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vagin A, Teplyakov A. 2010. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66:22–25 [DOI] [PubMed] [Google Scholar]
- 23.McRee DE. 1999. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125:156–165 [DOI] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 25.Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M, Chothia C, Gough J. 2009. SUPERFAMILY—sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 37:D380–D386. 10.1093/nar/gkn762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bagos PG, Tsirigos KD, Plessas SK, Liakopoulos TD, Hamodrakas SJ. 2009. Prediction of signal peptides in archaea. Protein Eng. Des. Sel. 22:27–35 [DOI] [PubMed] [Google Scholar]
- 27.Tan KP, Varadarajan R, Madhusudhan MS. 2011. DEPTH: a web server to compute depth and predict small-molecule binding cavities in proteins. Nucleic Acids Res. 39:W242–W248. 10.1093/nar/gkr356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallace AC, Laskowski RA, Thornton JM. 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8:127–134 [DOI] [PubMed] [Google Scholar]
- 29.Sawano Y, Hatano K, Miyakawa T, Komagata H, Miyauchi Y, Yamazaki H, Tanokura M. 2008. Proteinase inhibitor from ginkgo seeds is a member of the plant nonspecific lipid transfer protein gene family. Plant Physiol. 146:1909–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaguine AA, Richelle J, Wodak SJ. 1999. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D Biol. Crystallogr. 55:191–205 [DOI] [PubMed] [Google Scholar]
- 31.Ramachandran GN, Sasisekharan V. 1968. Conformation of polypeptides and proteins. Adv. Protein Chem. 23:283–438 [DOI] [PubMed] [Google Scholar]
- 32.Martínez-Fleites C, Ortíz-Lombardía M, Pons T, Tarbouriech N, Taylor EJ, Arrieta JG, Hernández L, Davies GJ. 2005. Crystal structure of levansucrase from the Gram-negative bacterium Gluconacetobacter diazotrophicus. Biochem. J. 390:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ames BD, Lee MY, Moody C, Zhang W, Tang Y, Tsai SC. 2011. Structural and biochemical characterization of ZhuI aromatase/cyclase from the R1128 polyketide pathway. Biochemistry 50:8392–8406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan X, Yin P, Hao Q, Yan C, Wang J, Yan N. 2010. Single amino acid alteration between valine and isoleucine determines the distinct pyrabactin selectivity by PYL1 and PYL2. J. Biol. Chem. 285:28953–28958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michalska K, Fernandes H, Sikorski M, Jaskolski M. 2010. Crystal structure of Hyp-1, a St. John's wort protein implicated in the biosynthesis of hypericin. J. Struct. Biol. 169:161–171 [DOI] [PubMed] [Google Scholar]
- 36.Ames BD, Korman TP, Zhang W, Smith P, Vu T, Tang Y, Tsai SC. 2008. Crystal structure and functional analysis of tetracenomycin ARO/CYC: implications for cyclization specificity of aromatic polyketides. Proc. Natl. Acad. Sci. U. S. A. 105:5349–5354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee MY, Ames BD, Tsai SC. 2012. Insight into the molecular basis of aromatic polyketide cyclization: crystal structure and in vitro characterization of WhiE-ORFVI. Biochemistry 51:3079–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyakawa T, Fujita Y, Yamaguchi-Shinozaki K, Tanokura M. 2012. Structure and function of abscisic acid receptors. Trends Plant Sci. 236:S1360–1385 [DOI] [PubMed] [Google Scholar]
- 39.Bais HP, Vepachedu R, Lawrence CB, Stermitz FR, Vivanco JM. 2003. Molecular and biochemical characterization of an enzyme responsible for the formation of hypericin in St. John's wort (Hypericum perforatum L.). J. Biol. Chem. 278:32413–32422 [DOI] [PubMed] [Google Scholar]
- 40.Iyer LM, Koonin EV, Aravind L. 2001. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins 43:134–144 [DOI] [PubMed] [Google Scholar]
- 41.Hamilton JA. 2004. Fatty acid interactions with proteins: what X-ray crystal and NMR solution structures tell us. Prog. Lipid Res. 43:177–199 [DOI] [PubMed] [Google Scholar]
- 42.Douliez JP, Michon T, Marion D. 2000. Steady-state tyrosine fluorescence to study the lipid-binding properties of a wheat non-specific lipid-transfer protein (nsLTP1). Biochim. Biophys. Acta 1467:65–72 [PubMed] [Google Scholar]
- 43.Zimmerman AW, van Moerkerk HT, Veerkamp JH. 2001. Ligand specificity and conformational stability of human fatty acid-binding proteins. Int. J. Biochem. Cell Biol. 33:865–876 [DOI] [PubMed] [Google Scholar]
- 44.Sacchettini JC, Gordon JI, Banaszak LJ. 1989. Crystal structure of rat intestinal fatty-acid-binding protein. Refinement and analysis of the Escherichia coli-derived protein with bound palmitate. J. Mol. Biol. 208:327–339 [DOI] [PubMed] [Google Scholar]
- 45.Han GW, Lee JY, Song HK, Chang C, Min K, Moon J, Shin DH, Kopka ML, Sawaya MR, Yuan HS, Kim TD, Choe J, Lim D, Moon HJ, Suh SW. 2001. Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography. J. Mol. Biol. 308:263–278 [DOI] [PubMed] [Google Scholar]