Abstract
Many nematophagous fungi use morphological structures called traps to capture nematodes by adhesion or mechanically. To better understand the cellular functions of adhesive traps, the trap cell proteome of the fungus Monacrosporium haptotylum was characterized. The trap of M. haptotylum consists of a unicellular structure called a knob that develops at the apex of a hypha. Proteins extracted from knobs and mycelia were analyzed using SDS-PAGE and liquid chromatography-tandem mass spectrometry (LC–MS-MS). The peptide sequences were matched against predicted gene models from the recently sequenced M. haptotylum genome. In total, 336 proteins were identified, with 54 expressed at significantly higher levels in the knobs than in the mycelia. The upregulated knob proteins included peptidases, small secreted proteins with unknown functions, and putative cell surface adhesins containing carbohydrate-binding domains, including the WSC domain. Phylogenetic analysis showed that all upregulated WSC domain proteins belonged to a large, expanded cluster of paralogs in M. haptotylum. Several peptidases and homologs of experimentally verified proteins in other pathogenic fungi were also upregulated in the knob proteome. Complementary profiling of gene expression at the transcriptome level showed poor correlation between the upregulation of knob proteins and their corresponding transcripts. We propose that the traps of M. haptotylum contain many of the proteins needed in the early stages of infection and that the trap cells can tightly control the translation and degradation of these proteins to minimize the cost of protein synthesis.
INTRODUCTION
Nematode-trapping fungi have the unique ability to capture and infect free-living nematodes (1). Given their potential use as biological control agents for plant- and animal-parasitic nematodes (2), there is much interest in studying their infection biology. To enter the parasitic stage, nematode-trapping fungi develop a unique morphological structure called traps. These traps develop from hyphal branches, either spontaneously or in response to signals from the environment, such as peptides or other compounds released by the host nematode (3). Molecular phylogeny studies have shown that the majority of nematode-trapping fungi belong to a monophyletic group in the order Orbiliales (Ascomycota). Within this clade, the trapping mechanisms have evolved along two major lineages, one leading to species with constricting rings and the other to species with adhesive traps, including three-dimensional networks, knobs, and branches (4–6).
Despite large variation in their morphology, adhesive traps share a unique ultrastructure that clearly separates them from vegetative hyphae (3). One feature that is common to all traps is the presence of numerous cytosolic organelles called dense bodies. These organelles have catalase and d-amino acid oxidase activities, which indicates that they are peroxisome-like organelles (3). However, the function of these organelles is not yet fully understood. Another feature of the trap cells is the presence of a fibrillar layer of extracellular polymers, which are believed to be important for attachment of the trap cell to the nematode surface (7). Following adhesion, the infection proceeds by formation of a penetration tube that pierces the nematode cuticle. At this stage, the nematode becomes paralyzed (killed), and the internal tissues are rapidly colonized by fungal hyphae (3).
A unique opportunity to examine the molecular features of adhesive traps is provided by the fungus Monacrosporium haptotylum. This fungus infects nematodes by using unicellular structures called knobs, which develop on the apices of hyphal branches. During growth in liquid culture with heavy aeration, knobs detach from the mycelium and can be separated by filtration. The isolated knobs retain their function as infection structures, i.e., they can trap and kill nematodes (8). A microarray analysis based on a limited set of expressed sequence tag (EST) probes compared the gene expression in knobs with that in vegetative mycelia (9). The result showed that 23% of the genes were differentially expressed; many were homologous to genes involved in the regulation of morphogenesis and cell polarity, stress response, protein synthesis and protein degradation, transcription, and carbon metabolism.
We have recently sequenced the genome of M. haptotylum (T. Meerupati, K.-M. Andersson, E. Friman, D. Kumar, A. Tunlid, and D. Ahrén, submitted for publication) and detected many similarities to the genome of the net-forming nematode-trapping fungus Arthrobotrys oligospora (10). The two genomes are similar in size and consist of ∼62% core genes that are shared with other fungi, ∼20% genes that are specific to the two species, and ∼16% genes that are unique to each genome. Transcriptional analysis showed that M. haptotylum expresses a unique set of genes during the early stages of infection of the nematode Caenorhabditis briggsae. A large proportion of these genes belong to gene families that are significantly expanded in the genomes of M. haptotylum and A. oligospora, including genes that encode subtilisins, tyrosinases, and extracellular proteins with WSC and mucin domains. In addition, transcripts encoding secreted proteins, in particular small secreted proteins (SSPs), were highly expressed during infection (Meerupati et al., submitted).
In the present study, we focused on the proteome of the knob cells in M. haptotylum and used liquid chromatography-tandem mass spectrometry (LC–MS-MS) to identify and quantify the proteins expressed differentially in knobs and mycelia. The upregulated knob proteome was characterized by an overrepresentation of secreted proteins (including SSPs) and extracellular proteins containing the carbohydrate-binding domain WSC or GLEYA and peptidases and proteins involved in stress response. The knob proteome was significantly different from the transcriptome expressed in the mycelium, knob, and penetrating hyphae.
MATERIALS AND METHODS
Culture.
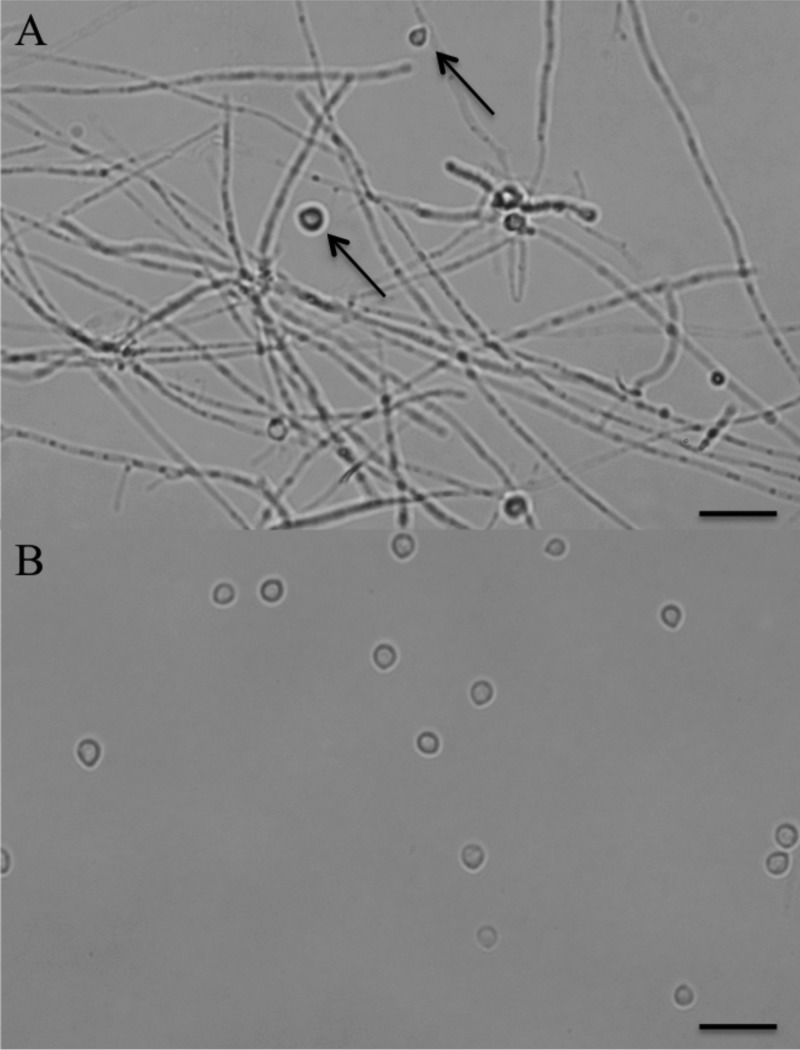
M. haptotylum (CBS 200.50) was grown in aerated 5-liter cultures containing 0.01% soya peptone, 0.005% phenylalanine, and 0.005% valine. Knobs and mycelia were separated by filtering, as described previously (8). A micrograph of the mycelia and isolated knobs is shown in Fig. 1.
Fig 1.
Micrograph of mycelia and isolated knobs of M. haptotylum. M. haptotylum was grown in aerated liquid cultures. After 10 days, when sufficient numbers of traps (knobs) had formed and detached from the mycelia, the knobs were separated as previously described (8). (A) Mycelia after knobs had been filtered; a small number of knobs (arrow) are still attached to the mycelia. Bar, 20 μm. (B) Knobs separated from mycelia. Bar, 20 μm.
Proteome analysis. (i) Protein extraction.
Knobs and mycelia were resuspended in phosphate-buffered saline (PBS), and phenylmethanesulfonylfluoride (PMSF) (final concentration, 1 mM) was added. The cells were sonicated on ice 5 times for 10 s each time at 50% amplitude on a Vibra Cell VC 50 (Sonics), allowing 1 min of cooling between sonications. The homogenates were centrifuged at 16,000 × g for 10 min at 4°C, and the supernatant was collected. The proteins were precipitated by adding 4 volumes of ice-cold acetone, incubating overnight at −20°C, and centrifuging at 16,000 × g for 10 min at 4°C. The pellet was air dried and resuspended in PBS containing 6 M urea and 1 mM PMSF. The protein concentration was determined by the Bradford method (11). In total, four biological replicates of both knobs and mycelia were analyzed.
(ii) Sample preparation and LC–MS-MS.
Equal amounts (8.0 μg) of the samples were loaded onto a one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (1D SDS-PAGE) gel (see Fig. S1 in the supplemental material). The proteins were separated by an 8% stacking, 12.5% separation gel in Tris-glycine buffer. Upon electrophoresis, the gel was stained with Coomassie brilliant blue G-250 (CBB). Each lane was cut into five slices, which were destained with 50% acetonitrile and 25 mM ammonium bicarbonate. After reduction with 10 mM dithiothreitol (DTT) in 100 mM ammonium bicarbonate at 55°C for 1 h, the proteins were alkylated with 55 mM iodoacetamide in 100 mM ammonium bicarbonate in the dark for 45 min. The slices were washed twice, first with 100 mM ammonium bicarbonate and then with acetonitrile, and thereafter completely dried using a SpeedVac (Savant). The proteins were cleaved by adding trypsin (12.5 ng/μl) in 50 mM ammonium bicarbonate in a volume until the gel slices were fully rehydrated. The samples were then incubated overnight at 37°C. The peptides were extracted by adding 1 volume of 75% acetonitrile, 5% trifluoroacetic acid, followed by incubation for 30 min. The peptides were separated by nano-high-performance liquid chromatography (nano-HPLC) performed on a CapLC (Waters) with online connection to a Micromass QTOF Ultima mass spectrometer (Waters).
For each gel slice extract, a liquid chromatography-mass spectrometry (LC-MS) run for protein quantification was followed by an LC–MS-MS run for protein identification. MS was performed in positive-ion mode, with a scan range of 400 to 1,600 in MS mode and 50 to 1,800 in MS-MS mode. The scan time was set to 1.9 s for MS spectra and to 1.0 s for MS-MS spectra, with an interscan delay of 0.1 s. MS-MS spectra were acquired for a maximum of 7 s for peaks of charge 2, 3, and 4, using data-dependent acquisition for a maximum of three peaks at a time, with an intensity threshold of 20. Real-time exclusion with a 120-s inclusion time was used, with an exclusion window of ±1,500 mDa. LC separation was performed on an Atlantis 75-μm by 150-mm column (Waters) at an approximate flow rate of 300 nl/min using a 65-min gradient from 4% to 40% acetonitrile in 0.1% formic acid.
(iii) Data analysis and protein identification.
The raw data files were converted to mzData format using Mascot Distiller 2.3.2 (Matrix Science, London, United Kingdom) with default settings for MassLynx QTOF and to mzML format using msconvert from the ProteoWizard package (12). The resulting files were uploaded to Proteios SE version 2.16 (13; http://www.proteios.org). The mzData and mzML files are available at the Swestore repository (http://webdav.swegrid.se/snic/bils/lu_proteomics/pub/). MS-MS-based protein identification was performed in Proteios with the mzData files using Mascot Server 2.3.01 (Matrix Science) and X!Tandem Tornado 2008.12.01 (14) in the in-house-generated M. haptotylum database. The database contains 10,959 gene models and open reading frames (ORFs) from all six potential reading frames predicted in the genome of M. haptotylum (NCBI accession no. PRJNA186729). These sequences were concatenated with the reverse sequences, yielding a total of 3,959,574 protein sequence entries. The search settings were as follows: 70-ppm precursor tolerance and 0.1-Da fragment tolerance, and up to one missed cleavage. Amino acid modifications that were considered in the analysis comprised fixed carbamidomethylation of cysteine and variable oxidation of methionine. The search results were combined in Proteios at the peptide level and filtered at a false-discovery rate of 1%.
(iv) Peptide quantification.
The peptides were quantified in Proteios using MS1 peak intensities, with a workflow similar to that described by Sandin et al. (15) but with fixed settings. First, potential peptide features were extracted from the MS spectra in the mzML files using msInspect (16) with default settings, and the resulting features were imported into Proteios. For each sample, features from the LC-MS run and MS-MS identifications from the LC–MS-MS run were matched using a procedure implemented in Proteios build 4196. Initially, all features were retained that unambiguously matched an MS-MS identification within 4 min from the start or end of the MS feature with an m/z tolerance of 0.04. Then, the feature apex retention times and the MS-MS identification retention times were fitted using a Loess interpolation to yield a polynomial spline function. The function was used to recalibrate the retention times when matching the LC-MS and LC–MS-MS files from the same sample. After this initial recalibration, features and MS-MS identifications were considered matched if the MS-MS was within the feature time boundary of 0.5 min, with an m/z tolerance of 0.04, and the MS-MS-derived peptide identities were assigned to the matching features.
Feature matching between samples within each slice fraction was subsequently performed pairwise for all LC-MS files. An initial time recalibration was performed using a polynomial spline function derived using features with equal peptide identities in the file pair being matched. After this initial recalibration, features were matched between the two files, using a retention time tolerance for the apex intensity of 1.0 min and an m/z tolerance of 0.03. Peptide abundance comparison reports, based on the feature intensities, were finally exported.
(v) Protein quantification and annotation.
Protein levels were calculated by summing the intensities of all peptides matching the protein sequence. Peptides that matched more than one protein were not considered. Proteins that were detected in more than two gel slices that were not adjacent or proteins in two neighboring slices with large differences in the ratio between knobs and mycelia were considered to be protein isoforms. The counts were log2 transformed, and P values were calculated from pairwise comparisons of protein levels in knob and mycelium samples, using Omics Explorer version 2.2 (Qlucore AB, Lund, Sweden) software.
Protein sequences were annotated based on searches in the UniProt database (17) using the BLASTP algorithm (18) with an E value threshold of 1e−10. In addition, the Pfam protein family database (19) (E value threshold, 0.05), the pathogen-host interaction (PHI) protein database version 3.2 (E value < 1e−10) (20), and the KOG database (21) were searched to further annotate protein sequences. Target signals were analyzed with SignalP 4.0 (22) and PTS1 predictor (23). The PTS1 predictor uses an algorithm to predict peroxisomal targeting signal 1 (PTS1). The probability, P, of observing the number of proteins within a given category by chance was estimated using hypergeometric distribution. The Pearson correlation coefficient (r) was used to calculate the correlation between the transcriptome and the proteome.
Phylogenetic analysis of the WSC domain.
Proteins containing the WSC domain were retrieved from the genomes of M. haptotylum (33 proteins) (Meerupati et al., submitted), A. oligospora (16 proteins) (10), Metarhizium anisopliae (16 proteins) (24), Aspergillus fumigatus (5 proteins) (25), Emericella nidulans (4 proteins) (26), and Magnaporthe grisea (13 proteins) (27). In total, 87 sequences were retrieved, and regions corresponding to 173 WSC domains were extracted using the extractseq program from the EMBOSS package (28). In order to produce a more reliable alignment and tree, the first WSC domain for each of the 87 sequences was selected and aligned using the MAFFT multiple-alignment program (29). An unrooted maximum-likelihood (ML) tree was reconstructed from the 87 WSC domains using PhyML version 3.0 (30) with 1,000 bootstrap replications. The phylogenetic tree was visualized in iTOL (31), and the domain architecture of the Pfam domains for each sequence was marked on the tree.
Transcriptome analysis.
RNA was extracted from two mycelium samples using the RNeasy Plant Mini Kit (Qiagen) with the supplied buffer RLC (containing guanidine hydrochloride). After reverse transcription into double-stranded cDNA for tag preparation (32), the material was sequenced in single-read mode (50 bp) using Illumina HiSeq2000 by GATC Biotech AG, Konstanz, Germany. In total, more than 51 million reads were obtained. The reads were mapped using Burrows-Wheeler Aligner (BWA) software (33) with default settings. The R package DESeq (34) was used for data normalization and statistical analysis. The profiles of the expressed mycelium transcripts were compared with the transcripts obtained from knobs and from hyphae infecting the nematode C. briggsae (Meerupati et al., submitted).
RESULTS
Knob proteome.
The SDS-PAGE and LC–MS-MS analyses yielded 6,131 spectra that could be assigned to peptides within the M. haptotylum sequence database (applying a false-discovery rate of 1%). In total 6,105 of these peptides were assigned to 336 gene models predicted in the genome of M. haptotylum (see Table S1 in the supplemental material) (Meerupati et al., submitted). The remaining 26 peptides matched 26 ORFs that were not found among these gene models. Further analysis of these ORFs showed that 16 of them were found in the same locations as predicted gene models either in different reading frames (9 ORFs) or in alternative splicing variants (7 ORFs). The remaining 10 ORFs were either wrongly mapped (false positives) or matched ORFs not located among the predicted gene models. BLASTP searches of the sequences of these ORFs against the NCBI nr database (E value cutoff, <1e−5) showed that only one of them displayed sequence similarities to proteins in the database (best hit, EGX50602.1 from A. oligospora).
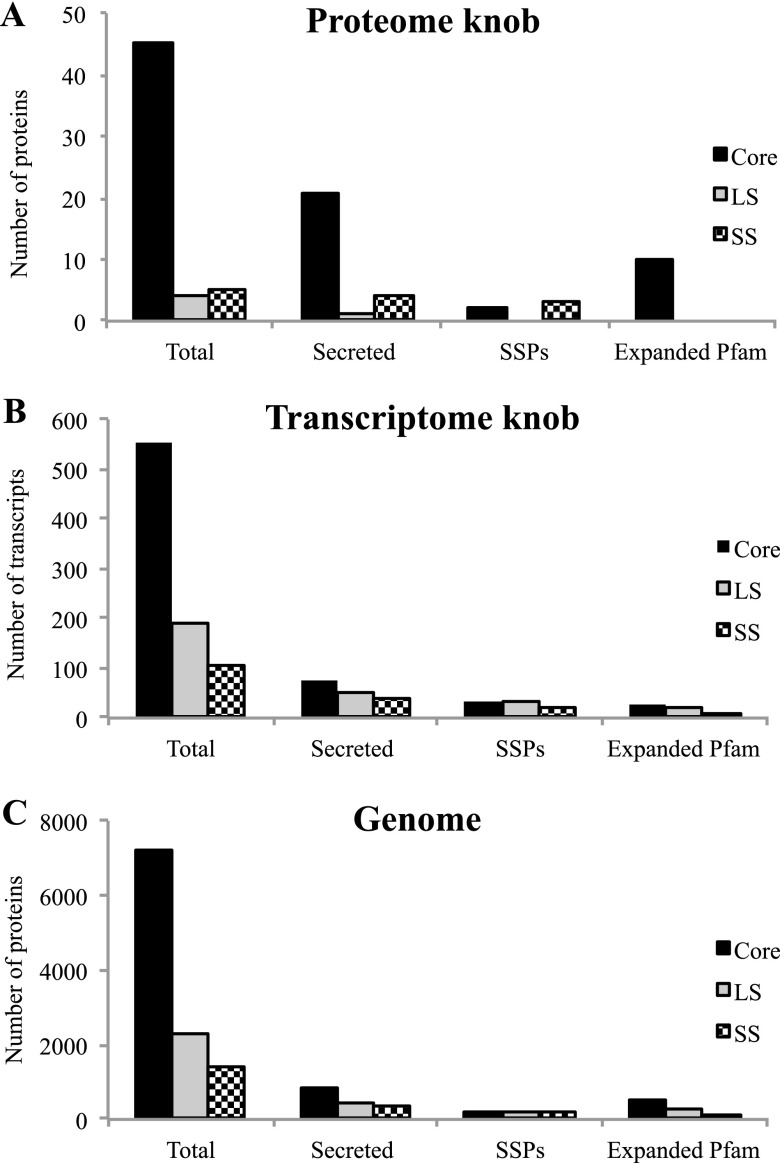
In total, 54 of the 336 predicted proteins among the M. haptotylum genome proteins were significantly more highly expressed in the knobs than in the mycelia (Table 1). Compared with the proteins in the genome overall, the overexpressed knob proteins showed several unique characteristics (Fig. 2A and C). First, the upregulated knob proteome was significantly enriched with core proteins (P = 0.003). Second, secreted proteins were overrepresented: secretion signals were predicted in 26 sequences, i.e., 48% of the proteins identified to be upregulated in the knobs, which is significantly (P = 8.4e−9) higher than the proportion of secreted proteins in the proteome overall (15%) (Meerupati et al., submitted). Five of the secreted proteins were short (<300 amino acids) and were therefore considered to be SSPs. Three of the SSPs—found among the species-specific proteins—were orphans, i.e., they shared no homology with proteins in the NCBI database and lacked Pfam domains (35).
Table 1.
Proteins identified by quantitative mass spectrometry as significantly upregulated in knobs versus mycelia from M. haptotyluma
| NCBI locus IDb | Featurec | Ratiod | Pe | Pfamf | Secg | Putative functionh |
|---|---|---|---|---|---|---|
| Cell surface proteins | ||||||
| H072_6833 | Core | 53.0 | 0.0025 | WSC | SiP | Cell surface protein, adhesion (I) |
| H072_973 | Core | 11.9 | 0.0183 | WSC | SiP | Cell surface protein, adhesion (I) |
| H072_10254 | Core | + | PA14_2/GLEYA | SiP | Cell surface protein, adhesion (K) | |
| H072_6835a | Core | + | WSC | SiP | Cell surface protein, adhesion (I) | |
| H072_6835c | Core | + | WSC | SiP | Cell surface protein, adhesion (I) | |
| H072_10205 | Core | + | WSC | SiP | Cell surface protein, adhesion (I) | |
| H072_8804 | Core | + | WSC | SiP | Cell surface protein, adhesion (I) | |
| H072_1293 | Core | 10.4 | 0.0370 | SiP | Extracellular serine-rich protein (K) | |
| H072_5601 | Core | 2.6 | 0.0487 | Cupin_1 | SiP | Spherulin (SSP) |
| Peptidases | ||||||
| H072_11420 | Core | 7.1 | 0.0112 | Peptidase_S8; Inhibitor_I9 | SiP | Serine endopeptidase. cuticle degrading subtilisin |
| H072_11104 | Core | 7.3 | 0.0114 | Peptidase_S10 | SiP | Carboxypeptidase O |
| H072_4030 | Core | 3.2 | 0.0286 | Peptidase_S10; Carbpep_Y_N | SiP | Carboxypeptidase Y |
| H072_4639 | Core | 9.6 | 0.0033 | Peptidase_M28;PA | SiP | Aminopeptidase |
| H072_6217 | Core | 5.5 | 0.0474 | Peptidase_M18 | Aminopeptidase I | |
| H072_8764 | SS | + | Peptidase_M36; FTP | - | Metalloendopeptidase, fungalysin/thermolysin | |
| H072_3605 | Core | 11.7 | 0.0063 | Asp | SiP | Aspartic endopeptidase |
| Stress response | ||||||
| H072_1780 | Core | 3.8 | 0.0038 | Thioredoxin | SiP | Protein disulfide isomerase (I) |
| H072_5104 | Core | 3.8 | 0.0026 | 2-Hacid_dh_C; Pyr_redox_2 | Thioredoxin reductase | |
| H072_7628 | Core | 2.7 | 0.0245 | GST_C | Glutathione S-transferase (K) | |
| H072_6664 | Core | 2.5 | 0.0136 | GST_N;GST_C | Glutathione S-transferase | |
| H072_6484 | Core | 5.5 | 0.0273 | HSP70 | SiP | Chaperone, HSP 70 family (I) |
| Extracellular enzymes (not peptidases) | ||||||
| H072_5927 | Core | 5.9 | 0.0109 | Pectate_lyase_3; mucin | SiP | Plant cell wall degradation, glycoside hydrolase |
| H072_448 | Core | + | Tannase | SiP | Plant cell wall degradation, feruloyl esterase | |
| H072_4515 | Core | 18.3 | 0.0436 | Tyrosinase | SiP | Oxidation of phenols |
| Metabolism | ||||||
| H072_9427 | Core | 11.3 | 0.0071 | Glyco_hydro_65N; glyco_hydro_65 m | SiP | Carbohydrate metabolism, trehalase |
| H072_7564 | Core | 3.7 | 0.0169 | Glyco_hydro_31 | SiP | Carbohydrate metabolism (PHI) |
| H072_10613 | Core | + | PRKCSH; DUF1403 | SiP | Carbohydrate metabolism (I) | |
| H072_6506b | Core | 5.5 | 0.0014 | Transaldolase; PTE | Pentose-phosphate shunt | |
| H072_6506a | Core | 4.0 | 0.0112 | Transaldolase; PTE | Pentose-phosphate shunt | |
| H072_6506d | Core | 2.2 | 0.0498 | Transaldolase; PTE | Pentose-phosphate shunt | |
| H072_4602 | Core | 3.4 | 0.0267 | F_bP_aldolase | Carbohydrate metabolism | |
| H072_3423 | Core | 3.5 | 0.0253 | Aminotran_3 | Metabolism | |
| H072_6416d | Core | 14.2 | 0.0086 | NmrA | SiP | Metabolism |
| H072_6416c | Core | 2.2 | 0.0291 | NmrA | SiP | Metabolism |
| H072_10483 | Core | 3.1 | 0.0146 | NUDIX | Isoprenoid synthesis | |
| H072_8576b | Core | 6.6 | 0.0012 | NDK | Nucleotide metabolism | |
| H072_8576a | Core | 4.6 | 0.0024 | NDK | Nucleotide metabolism | |
| H072_8576c | Core | 4.5 | 0.0026 | NDK | Nucleotide metabolism | |
| H072_8576d | Core | 3.9 | 0.0058 | NDK | Nucleotide metabolism | |
| H072_8285 | Core | 2.3 | 0.0362 | NUDIX | Phosphohydrolase | |
| H072_6884 | Core | 3.2 | 0.0407 | RRM_1; eIF2A | Protein synthesis | |
| H072_9479b | Core | 16.3 | 0.0002 | Mt_ATP-synt_D | Proton transporter | |
| H072_9479a | Core | 4.7 | 0.0025 | Mt_ATP-synt_D | Proton transporter | |
| H072_6761a | Core | 10.1 | 0.0302 | FMN_red | Oxidoreductase | |
| H072_6761b | Core | 8.3 | 0.0165 | FMN_red | Oxidoreductase | |
| H072_9515 | Core | 3.0 | 0.0285 | adh_short | Oxidoreductase (PHI) (K) | |
| H072_10514b | Core | 8.3 | 0.0220 | Ribonuc_L-PSP | PTS1 | Endoribonuclease |
| H072_10514c | Core | 8.1 | 0.0017 | Ribonuc_L-PSP | PTS1 | Endoribonuclease |
| H072_10514a | Core | 8.0 | 0.0015 | Ribonuc_L-PSP | PTS1 | Endoribonuclease |
| H072_10514d | Core | 7.4 | 0.0064 | Ribonuc_L-PSP | PTS1 | Endoribonuclease |
| Actin and DNA binding proteins | ||||||
| H072_6869c | Core | 6.7 | 0.0055 | Tropomyosin_1 | Actin binding protein (I) | |
| H072_6869d | Core | 3.5 | 0.0137 | Tropomyosin_1 | Actin binding protein (I) | |
| H072_6869a | Core | 3.3 | 0.0126 | Tropomyosin_1 | Actin binding protein (I) | |
| H072_4754 | Core | 5.3 | 0.0321 | HMG_box | DNA binding (PHI) | |
| Others | ||||||
| H072_9522 | Core | + | DUF297 | Endo alpha-1,4 polygalactosaminidase | ||
| H072_940 | Core | 4.0 | 0.0282 | OSCP | SiP | ATP synthesis coupled proton transport (SSP) |
| H072_10638 | Core | + | SiP | Outer membrane autotransporter | ||
| Hypothetical proteins | ||||||
| H072_1845 | SS | + | SiP/PTS1 | Hypothetical protein (SSP) (I) | ||
| H072_2504 | LS | + | SiP | Hypothetical protein (K) | ||
| H072_6857 | SS | + | SiP | Hypothetical protein (SSP) (K) | ||
| H072_8717 | SS | + | SiP | Hypothetical protein (SSP) (I) | ||
| H072_4225 | LS | 74.1 | 0.030 | DUF1921 | Hypothetical protein | |
| H072_8026 | SS | 30.1 | 0.0001 | SiP | Hypothetical protein | |
| H072_11591 | Core | 8.0 | 0.0294 | SiP | Hypothetical protein | |
| H072_8078 | Core | 7.0 | 0.0075 | Hypothetical protein | ||
| H072_7877 | LS | 3.9 | 0.0289 | Hypothetical protein | ||
| H072_3089 | Core | 3.6 | 0.0232 | BAR_2 | Hypothetical protein | |
| H072_11515 | LS | 2.5 | 0.0113 | SiA | Hypothetical protein | |
Shown are proteins that were upregulated at least 2-fold in knobs versus mycelia or proteins that were detected in at least two of the four replicates of the knob. Peptides that matched more than one protein were excluded from the analysis. Information about protein identifications is provided in Table S1 in the supplemental material.
Gene models matching the peptide sequences. Before peptide sequencing, each lane in the SDS-PAGE gel was cut into five slices. Peptides generated from slices that were not adjacent, or with large differences in ratio were considered different (iso)forms of the proteins. An italic letter after the ID indicates such proteins.
Core, the protein has homologs in other fungi; LS (lineage specific), the protein is shared between M. haptotylum and A. oligospora; SS (species specific), the protein is unique to M. haptotylum (Meerupati et al., submitted). Proteins marked SS in boldface are orphans (i.e., they lack both Pfam domains and homologs in the NCBI database).
Ratio of protein levels in knobs and mycelia; +, the protein was present only in the knob samples.
P values estimated from pairwise comparisons of protein levels in knob and mycelium samples.
Pfam IDs in boldface designate gene families that were significantly expanded in the genome of M. haptotylum (Meerupati et al., submitted).
SiP, the protein has a predicted secretion signal; PTS1, the protein has a predicted peroxisomal target signal; SiA, the protein has a signal anchor sequence.
Assigment of the biological or enzymatic function based on manual annotations. PHI, the protein displayed sequence similarity to proteins found in the pathogen-host interaction protein database (20); SSP, small (<300 amino acids) secreted proteins; K and I, the gene models were at least 2-fold upregulated in the RNA-Seq analysis in the pairwise comparisons of knob versus mycelium and infecting hyphae versus knob.
Fig 2.
Features of proteins and genes that were significantly upregulated in knobs of M. haptotylum. Proteins and genes were classified as core (shared with other fungi), lineage specific (LS) (shared among the nematode-trapping fungi M. haptotylum and A. oligospora), and species specific (SS) (unique to M. haptotylum) (Meerupati et al., submitted). “Secreted” designates proteins with a predicted secretion signal; SSPs are small secreted proteins with a length of <300 amino acids; “Expanded Pfam” represents proteins found among 25 expanded gene families in M. haptotylum (Meerupati et al., submitted). (A) Proteins (54 total) that were at least 2-fold upregulated in knobs versus mycelia (Table 1). (B) Transcripts (851 total) that were at least 2-fold upregulated in knobs versus mycelia. (C) Distribution of predicted proteins (10,959 total) in the genome of M. haptotylum (Meerupati et al., submitted).
The upregulated knob proteome was also enriched with members of protein families that are expanded in M. haptotylum (Meerupati et al., submitted). In total, 10 (19%) of the 54 overexpressed proteins belonged to the expanded gene families compared with a proportion of 8% in the proteome overall (P = 0.009). Members belonging to expanded Pfam families were exclusively encoded by core genes and comprised proteins containing the WSC domain (5 proteins), proteins with the PA14_2 (GLEYA) domain (1 protein), mucin (1 protein), peptidase_S8 (subtilisin) (1 protein), aspartyl protease (1 protein), and tyrosinase (1 protein) (Tables 1 and 2). Three of the proteins displayed sequence similarity to proteins in the pathogen-host interaction (PHI base) database of experimentally verified pathogenicity and virulence genes from fungi (20) (Table 1).
Table 2.
Expanded gene families upregulated in the proteome and transcriptome of knobs from M. haptotyluma
| Pfam family | Genomeb | No. of transcripts or proteins upregulated |
||
|---|---|---|---|---|
| Proteome (knobc) | Trancriptome |
|||
| Knobd | Infectione | |||
| WSC | 33 | 5 | 2 | 13 |
| Peptidase_S8 | 59 | 1 | 3 | 8 |
| Asp | 38 | 1 | 2 | 1 |
| Tyrosinase | 29 | 1 | 5 | 8 |
| PA14_2 | 28 | 1 | 5 | 0 |
| Mucin | 16 | 1 | 3 | 3 |
| F box | 190 | 0 | 7 | 12 |
| Ank | 139 | 0 | 5 | 11 |
| CBM_1 | 108 | 0 | 2 | 17 |
| NACHT | 76 | 0 | 0 | 5 |
| PNP_UDP_1 | 49 | 0 | 0 | 4 |
| BTB | 36 | 0 | 2 | 0 |
| DUF3129 | 33 | 0 | 2 | 20 |
| SKG6 | 32 | 0 | 0 | 3 |
| Glyco_hydro_61 | 28 | 0 | 0 | 3 |
| Herpes_gE | 27 | 0 | 1 | 2 |
| CFEM | 26 | 0 | 8 | 5 |
| Glyco_hydro_28 | 20 | 0 | 0 | 3 |
| NB-ARC | 17 | 0 | 0 | 0 |
| SPRY | 17 | 0 | 0 | 2 |
| Pec_lyase_C | 10 | 0 | 0 | 0 |
| Peptidase_M10 | 9 | 0 | 1 | 2 |
| Pectinesterase | 8 | 0 | 0 | 2 |
| Ricin_B_lectin | 8 | 0 | 1 | 1 |
| Stig1 | 6 | 0 | 0 | 1 |
The table shows the numbers of proteins and transcripts from 25 expanded Pfam gene families (Meerupati et al., submitted) that were upregulated in the proteome and transcriptome of M. haptotylum. The five largest families in each column are in boldface.
Number of genes in the genome.
More than 2-fold upregulated (knob versus mycelium) or uniquely present in knobs; total, 54 proteins (Table 1).
More than 2-fold upregulated (knob versus mycelium; P < 0.01); total, 851 transcripts.
More than 2-fold upregulated (infected hyphae versus knob; P < 0.01); total, 842 transcripts (Meerupati et al., submitted).
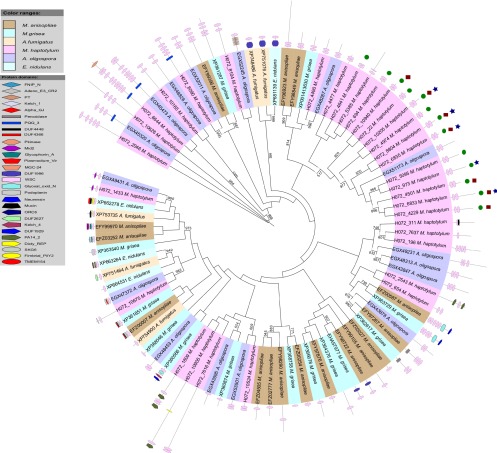
In the knob proteome, five proteins with the carbohydrate-binding domain WSC were upregulated compared with 33 genes with the domain in the M. haptotylum proteome overall (Meerupati et al., submitted). WSC is a cysteine-rich domain with eight conserved cysteine residues that are required for its function (36, 37). A sequence logo illustrating the prevalence of amino acids at specific positions showed that eight cysteine residues are highly conserved among the aligned WSC domains in this study (see Fig. S2 in the supplemental material). The WSC proteins varied extensively in length (223 to 3,063 amino acids). Phylogenetic analysis of the WSC proteins showed that 15 of the 33 WSC proteins in M. haptotylum form a large clade of expanded WSC paralogs (Fig. 3). All the proteins in this clade contain two WSC domains. Furthermore, this expanded clade contains all five WSC proteins that were upregulated in the proteome. The WSC-containing proteins outside this clade were highly diverse; they contained a variable number of WSC domains, and many of them had additional Pfam domains.
Fig 3.
Phylogeny of the WSC Pfam family. An unrooted PhyML tree of the first WSC domain in each protein was reconstructed with 1,000 bootstrap replications. Branches with >50% bootstrap support are shown. The phylogeny and Pfam domain architecture of each protein is displayed using iTOL. Proteins marked with a blue star were 2-fold upregulated in the knob proteome compared with the mycelium. Green circles indicate genes that were 2-fold upregulated during infection compared with the knob transcriptome. Red squares show genes that were highly upregulated during infection compared with the knob transcriptome (>10-fold) and highly expressed during infection (10% most expressed transcripts).
Comparisons of protein and mRNA expression levels.
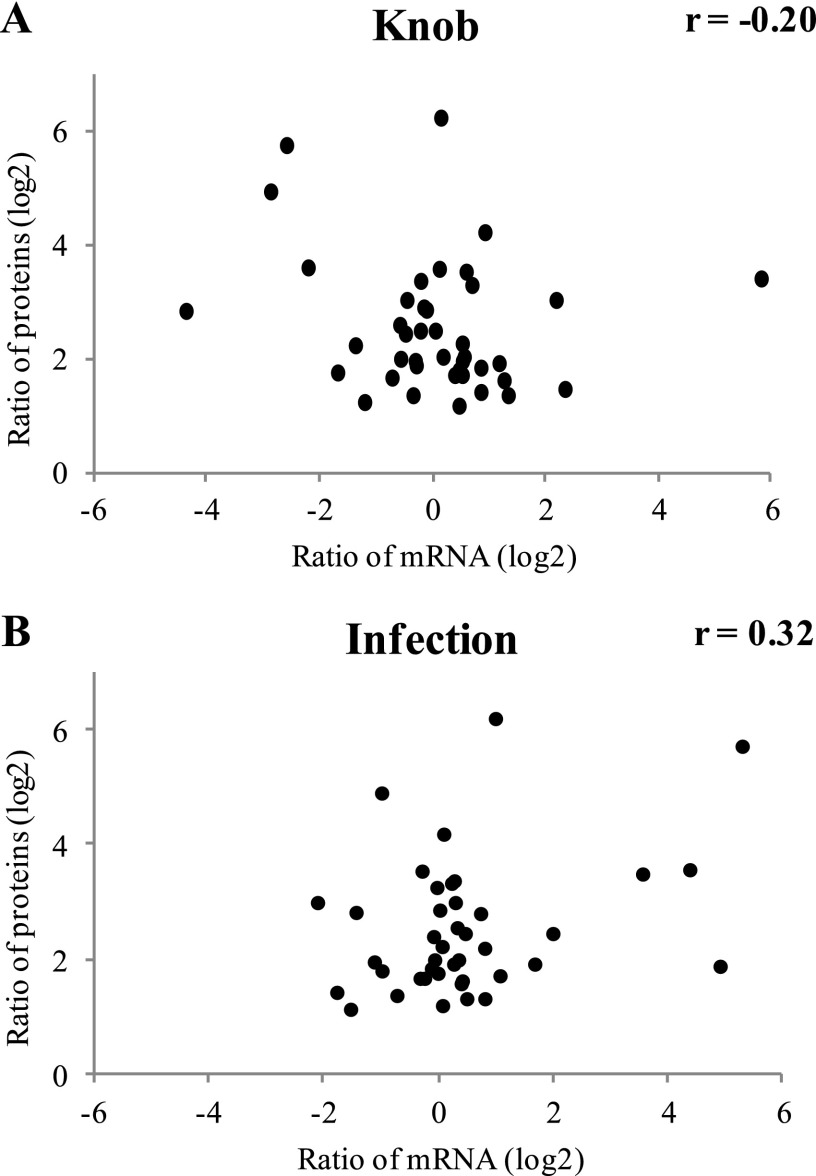
Analysis of the RNA-Seq data showed that 851 gene models (8% of the 10,959 gene models in M. haptotylum) were more than 2-fold upregulated in the knobs compared with the mycelia. The composition of the upregulated knob transcriptome was significantly different from that of the corresponding proteome. Only six of the upregulated knob transcripts were upregulated at the protein level. These proteins included a putative surface protein of the PA14_2/GLEYA family, a glutathione S-transferase, an alcohol dehydrogenase identified in the PHI database, and two hypothetical proteins with predicted secretion signals (Table 1). The lack of correlation between protein and transcript was evident when plotting the fold changes in the abundance of mRNA against its encoded proteins (Fig. 4A). However, in agreement with the upregulated proteome, the upregulated transcriptome was significantly enriched (P = 9.9e−6) with sequences predicted to have a signal peptide (Fig. 2B and C). Indeed, secretion signals were found in 20% of the upregulated knob transcripts, and 87 of the 171 secreted proteins were SSPs. In contrast, the proportion of transcripts from expanded gene families among the upregulated knob transcripts was significantly lower (6%) than that in the upregulated proteome (19%). The most abundant Pfam families in the knob transcriptome included the F-box domain, ankyrin repeat, and CFEM domain, and these were not found among the families detected in the upregulated knob proteome (Table 2).
Fig 4.
Comparisons between protein and transcript changes among the proteins that were significantly more highly expressed in knobs than in mycelia. (A) Log2 values of the ratio of the abundance of proteins in knobs versus mycelia plotted against the ratio of the abundance of the corresponding transcripts. (B) Log2 values of the ratio of the abundance of proteins in knobs versus mycelia plotted against the ratio of the abundance of transcripts in infected hyphae versus knobs. The Pearson correlation coefficients (r) of the comparisons are shown.
The large differences between the knob proteome and knob transcriptome were also displayed when comparing the distributions of proteins and transcripts upregulated in knobs in different eukaryotic orthologous groups (KOGs) (see Fig. S3 in the supplemental material). In the proteome, the KOG classes “signal transduction mechanisms”; “replication, recombination, and repair”; and “transcription” were more enriched in the knobs than in the mycelia. However, in the transcriptome, the KOG classes “coenzyme transport and metabolism”; “translation, ribosomal structure, and biogenesis”; and “amino acid transport and metabolism” were more enriched in the knobs than in the mycelia.
Distinct differences were also found between the knob proteome and the transcriptome that was upregulated during infection of the nematode C. briggsae (Meerupati et al., submitted). Only 11 upregulated knob proteins were found among the 842 gene models that were more than 2-fold upregulated in the penetrating hyphae (in a pairwise comparison with the knob transcriptome) (Table 1). Furthermore, there was weak correlation between the changes in proteins in the knob and the changes in the corresponding transcripts in the infected mycelium (Fig. 4B). Notably, 5 of the 11 upregulated protein-transcript pairs encoded proteins with the WSC domain. Accordingly, all upregulated WSC domain proteins present in the knob proteome were significantly upregulated at the transcriptional level during infection. In addition, two proteins presumably involved in stress response (displaying sequence similarity to thioredoxin and HSP70, respectively)—an actin binding protein and two SSPs—were upregulated in both the knob proteome and the infection transcriptome (Table 1).
DISCUSSION
This is the first study where the proteome of the trapping cells of a nematophagous fungus has been characterized in detail. Previous proteome studies of nematode-trapping fungi have involved species such as Monacrosporium lysipagum and A. oligospora, where the trap cells cannot be isolated from the mycelium (10, 38). Given that the biomass of trap cells is considerably lower than that of the mycelium, it is difficult to infer the trap proteome by comparing the proteomes of a mycelium with and without traps. M. haptotylum offers a unique opportunity in this context, because functionally intact trap cells can be isolated from the mycelium, meaning that the proteomes of the two tissues can be analyzed separately (8).
The upregulated knob proteome of M. haptotylum was characterized by an overrepresentation of secreted proteins (including SSPs) and a number of extracellular proteins, including proteins with WSC or GLEYA domains, peptidases, and proteins involved in stress response. Of note, the knob proteome was significantly different from the transcriptomes expressed in the mycelium, knob, or infecting hypha. Actively growing yeast cells generally show concordance between mRNA and protein expression profiles (39–41). However, the knob is a specialized cell that develops at the apex of a hypha, meaning that growth has been arrested. There are at least two possible reasons for poor correlation between mRNA and protein levels: faster protein synthesis and slower degradation (42). Both explanations are feasible in this context. Early experiments in M. haptotylum showed that treatment with the protein synthesis inhibitor cycloheximide does not affect the infection process (43). Thus, the traps of M. haptotylum appear to contain many of the proteins needed in the early stages of infection, and it is reasonable to assume that trap cells can tightly control the translation and degradation of these proteins to minimize the cost of protein synthesis.
Cell surface proteins. (i) Proteins containing the WSC domain.
Several of the most highly upregulated proteins in the trap proteome contained the WSC Pfam domain. WSC is one of the most expanded gene families in M. haptotylum, with 33 members. This number is very high: other ascomycetes have 0 to 18 proteins containing the domain (Meerupati et al., submitted). The closely related fungus, A. oligospora, has 16 proteins with this domain. The only fungus, to our knowledge, that has a number of WSC-containing proteins similar to that of M. haptotylum is the root-colonizing endophyte Piriformospora indica, with 36 genes (44).
The phylogenetic tree shown in Fig. 3 revealed rapid expansion of a clade containing 15 M. haptotylum paralogs. This clade has only one ortholog in A. oligospora, indicating that the expansion of the M. haptotylum paralogs in the clade occurred after the speciation of M. haptotylum and A. oligospora. Another phylogenetic analysis using all WSC domains (instead of the first domain in each protein, as in Fig. 3) revealed that the first WSC domains in each protein tend to cluster together (see Fig. S4 in the supplemental material). Accordingly, the two domains were found in the ancestral protein of the expanded clade containing the 15 WSC paralogs.
Gene duplication and protein family expansion are important genomic mechanisms shaping the evolution of pathogenic fungi (45). Many gene family expansions are species specific (46, 47). Interestingly, the clade of 15 expanded WSC proteins in M. haptotylum contains all 5 WSC proteins that were upregulated in the knob proteome and all 13 transcripts encoding the WSC domain that were upregulated in the transcriptome during infection. This indicates that these proteins have an important role during pathogenicity. Two of the five upregulated WSC proteins are found in a gene cluster of secreted proteins that are adjacent in the M. haptotylum genome (cluster 74) (Meerupati et al., submitted). All five proteins in this cluster are highly (>10-fold) upregulated during infection. In addition to the two WSC proteins, the cluster includes two SSPs and one protein with the DUF3129 domain. Taking the data together, it is likely that this gene cluster is associated with pathogenicity and that its deletion affects pathogenicity, similar to what has been shown in Ustilago maydis (48).
WSC is a carbohydrate-binding domain that is found in proteins with many different functions (49). Notably, 28 of the 33 WSC proteins in M. haptotylum—including the five proteins that were upregulated in the trap proteome—have structural features that are found in fungal adhesins (50). These features include a signal peptide, tandem repeats, and O-glycosylation sites (see Table S2 in the supplemental material). Three of the putative adhesins have glycosylphosphatidylinositol (GPI) anchor signals. Based on these features, we suggest that a large proportion of the WSC domain proteins in M. haptotylum are extracellular and located on the surfaces of knob cells.
None of the five upregulated WSC proteins in M. haptotylum displayed significant sequence similarity to proteins found in the UniProt database. However, several other WSC domain proteins in M. haptotylum displayed significant sequence similarity to other proteins. Protein H072_1433 displayed sequence similarity (2e−33) to WscB in Aspergillus nidulans, which is involved in the cell wall integrity (CWI) pathway (51). WscB and other so-called cell surface sensors have been extensively characterized in yeast, where they monitor cell stress, including internal turgor pressure, through the CWI pathway (52–54). Interestingly, the CWI pathway is essential for appressorium penetration and invasive growth in Magnaporthe oryzae (55). In addition, four WSC proteins in M. haptotylum (H072_10165, H072_2046, H072_6644, and H072_8090) displayed sequence similarity (≤1e−124) to a legume lectin beta-domain-containing protein (H1UW68) identified in the plant-pathogenic fungus Colletotrichum higginsianum.
(ii) GLEYA proteins and other cell surface proteins.
Another cell surface protein that was upregulated in the knobs was the cupin_1 protein, which showed sequence similarity to spherulin 1a, a cell wall glycoprotein in the slime mold Physarum polycephalum (56). Furthermore, another upregulated knob protein had a PA14_2 (also known as GLEYA) domain, which is a carbohydrate-binding domain that is found in fungal adhesins (57, 58). GLEYA is one of the most expanded gene families in M. haptotylum, with 28 members. Notably, three GLEYA proteins also had a WSC domain, but none of them was found in the clade containing the upregulated WSC knob proteins (Fig. 3).
(iii) Lectins.
The adhesion of nematodes to nematode-trapping fungi might be mediated by an interaction between lectins present on the trap surface and carbohydrate ligands found on the nematode surface, as indicated by sugar inhibition experiments published in 1979 (59). Later, a gene encoding such a lectin was characterized from A. oligospora (60). However, deletion of the gene did not affect the ability to infect nematodes (61), which suggests that the fungus might express other lectins mediating attachment to nematodes. Indeed, in the recently published genome of A. oligospora, seven lectins with different sugar specificities were identified (10). The expression levels of the mRNAs and the corresponding proteins of these lectins did not change during trap formation, and their roles in the trapping mechanism remain unclear (10). In the genome of M. haptotylum, we identified homologs of six of the seven lectins in A. oligospora. None of the lectins identified in M. haptotylum were upregulated in the knob proteome. However, the transcripts of two lectins were upregulated in the knobs during nematode infection. One of the lectins in A. oligospora has a WSC domain. This sequence is homologous to the WSC gene model H072_8090 in M. haptotylum, but H072_8090 was not found in the rapidly evolving clade of WSC knob proteins.
Peptidases. (i) Subtilisins.
Several peptidases were upregulated in the knob proteome of M. haptotylum, including a subtilisin (H072_11420) with the peptidase_S8 domain. In A. oligospora, extracellular subtilisins have a key role in the early stages of infection, including immobilization of the captured nematodes (10, 62, 63). Subtilisins are an expanded gene family in M. haptotylum, and the genome contains 59 gene models with a peptidase_S8 domain (Meerupati et al., submitted). Two subtilisin genes (spr1 and spr2) in M. haptotylum are significantly upregulated during infection of Caenorhabditis elegans (64). In the genome, spr1 corresponds to H072_6551 and spr2 to H072_5672; thus, the subtilisin that was upregulated in the proteome was not identical to SPR1 or SPR2. The transcriptome data (Table 2) showed that three genes encoding a peptidase_S8 domain were upregulated in knobs compared with mycelia and that eight genes were upregulated during the infection of nematodes. One of the three genes that were upregulated in knobs was spr2, but neither spr1 nor spr2 was significantly upregulated during infection. spr2 did show the second highest expression level among all the transcripts upregulated during infection, but the P value was too high for the result to be significant.
(ii) Carboxypeptidases and other peptidases.
In addition to subtilisins, two carboxypeptidases were upregulated in knobs. One of them (H072_11104) shows sequence similarity to a carboxypeptidase that is produced during root colonization by the egg-parasitic nematophagous fungus Pochonia chlamydosporia (65). The other upregulated carboxypeptidase (H072_4030) shows sequence similarity to carboxypeptidase Y in M. grisea, which is expressed during appressorium formation (66).
Other upregulated peptidases included aminopeptidases (H072_4639 and H072_6217), an aspartic endopeptidase (H072_3605), and a metalloendopeptidase (H072_8764) belonging to the M36 fungalysin family. Notably, the M36 peptidase is a species-specific protein that displayed no sequence similarity to other fungal peptidases.
Fungal pathogenicity factors.
The trapping cells of nematode-trapping fungi and the appressoria formed by plant-pathogenic fungi have several structural and functional similarities (9). Both are specialized infection structures that develop from hyphae. The structures contain an adhesive layer that binds to the host surface. Following attachment, both traps and appressoria form a hypha that penetrates the host using a combination of physical force and extracellular enzyme activities (3, 67). The plasticity of infection structures in nematode-trapping fungi has been extensively studied in A. oligospora (68). Hence, this fungus has the capacity to colonize plant roots by the aid of appressorium-like structures (69). Furthermore, it was observed that the infected plant cells contained structures similar to the infection bulbs that the fungus forms inside penetrated nematodes (3).
(i) Homologs of proteins in plant-pathogenic fungi.
Three of the upregulated knob proteins in M. haptotylum were homologs of proteins in plant-pathogenic fungi that are involved in the development and function of appressoria. Alpha-glucosidase (H072_7564) shows sequence similarity to GAS1 in U. maydis, which is induced during pathogenesis. Mutants lacking gas1 were able to form an appressorium but were arrested in growth after penetration of the plant surface (70). The protein H072_9515 shows sequence similarity to a short-chain dehydrogenase/reductase that is involved in M. oryzae pathogenicity. Mutants lacking this gene showed reduced appressorium development (71). Reduced appressorium development was also seen in mutants of M. grisea lacking a nonhistone protein 6 gene that shows sequence similarity to H072_4754 in M. haptotylum (72).
(ii) Proteins with peroxisomal target signals.
Microscopic studies have shown that traps contain a large number of peroxisome-like organelles called microbodies (73–75). The number of these microbodies rapidly decreases during penetration and development of the infection hyphae (3). Organelles with peroxisomal function are also found in the appressoria of plant-pathogenic fungi, including Colletotrichum orbiculare (76). Similar to microbodies, these organelles are selectively degraded at the stage of plant invasion (77). The organelles might have a role in the recycling of cellular components that are needed for the synthesis of compounds required for host invasion (77). Notably, we identified only two proteins with peroxisomal PTS1 target signals in the upregulated knob proteome of M. haptotylum. H072_1845 is an SSP with unknown function, while H072_10514 is a member of the YjgF/Yer057p/UK114 family. Even though the primary sequence is highly conserved among members of this family, their biological functions vary (78–81). For the predicted protein H072_10514, the highest sequence similarity was found with the L-PSP endoribonuclease family protein Brt1. A BLAST search of sequences in the UniProt database revealed that there are other fungal L-PSP endoribonuclease proteins that have PTS1 target signals. The fact that that only two of the upregulated knob proteins had peroxisomal PTS1 target signals raises the question of whether the microbodies are peroxisomes. Alternatively, the membranes of the microbodies were not disrupted during sample preparation.
(iii) Tyrosinases.
One of the most highly upregulated (18-fold) knob proteins (compared with mycelium proteins) in M. haptotylum was a tyrosinase. In addition, eight other tyrosinase genes were upregulated in the transcriptome during infection. Tyrosinases are involved in the synthesis of melanin (82). Several fungi require melanization for pathogenic processes, for example, appressorial penetration by Colletotrichum lagenarium (83). Tyrosinases also oxidize protein- and peptide-bound tyrosyl residues, resulting in the formation of inter- and intramolecular cross-links between peptides, proteins, and carbohydrates (84). Cross-linking between proteins and polymers present in the extracellular layer might have a role in mediating binding between the trap of nematode-trapping fungi and the nematode cuticle (85).
(iv) Cell wall-degrading enzymes.
The upregulated knob proteins also included two putative plant cell wall-degrading enzymes. One of these was a feruloyl esterase (EC 3.1.1.73), an enzyme that hydrolyzes ester bonds between hydroxycinnamic acids and sugars present in plant cell walls (86). The other putative cell wall-degrading enzyme (H072_5927) displayed significant sequence similarity to exo-β-1,3 glucanases (EC 3.2.1.58). Further annotation showed that it has the same catalytic sites as other enzymes with β-1,3 glucan-hydrolyzing activity belonging to glycoside hydrolase family 55 (87). β-1,3 Glucans are building blocks of callose, which is found in plant cell walls (88), but β-1,3 glucans are also major constituents of the fungal cell wall (89).
(v) SSPs.
Many plant-pathogenic fungi are thought to employ small secreted proteins during infection (90). Entomopathogenic fungi also have a large battery of SSPs that might be involved in pathogenesis (91). Typically, such SSPs have a limited phylogenetic distribution, possible because of rapid evolution. Notably, we identified three SSPs with unknown functions among the upregulated knob proteins in M. haptotylum. The transcripts of two of the SSPs were highly upregulated during infection. The three SSPs showed no sequence similarity to genes in other species, and they lack Pfam domains, i.e., they are orphans (35). In the genome of M. haptotylum, we have recently identified 695 SSPs (Meerupati et al., submitted). The sequences of these SSPs are highly divergent. A homology-based clustering of secreted proteins using an all-against-all BLASTP similarity search showed that only a few of the SSPs belonged to clusters containing three or more paralogs (Meerupati et al., submitted). One of the three SSPs upregulated in the knob was found in a cluster with two other SSPs in the genome. The sequences of the paralogs in this cluster share 12 conserved cysteine residues. The DNA sequences of the other two SSPs that are upregulated in the knob proteome show evidence of repeat-induced point (RIP) mutations. RIP mutation has been suggested to be a major mechanism generating sequence divergence of SSPs in the genome of M. haptotylum (Meerupati et al., submitted).
Conclusion.
We have shown that there are large differences between the protein content of the trapping knob and that of the vegetative mycelia. The knob proteome was overrepresented in secreted proteins, including SSPs, peptidases, and carbohydrate-binding proteins of the WSC family. Previous studies have shown that transcripts encoding such proteins are highly upregulated in M. haptotylum during early stages of infection (Meerupati et al., submitted). Hence, the trap proteome contains several of the proteins needed during the initial stages of infection, i.e., adhesion and penetration. Phylogenetic analysis shows that the WSC proteins that were upregulated in the knob proteome are members of a large clade of rapidly evolving WSC genes. Further studies will show if they have a role during the adhesion of nematodes or what function they have during pathogenicity.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Swedish Research Council (VR) and the Crafoord Foundation.
We thank Björn Canbäck for valuable advice on bioinformatics analysis and Bioinformatics Infrastructure for Life Sciences (BILS) for bioinformatics support. We also thank Mats Mågård for performing the mass spectrometry runs.
Footnotes
Published ahead of print 14 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01390-13.
REFERENCES
- 1. Barron GL. 1977. The nematode-destroying fungi. Canadian Biological Publications, Guelph, Canada [Google Scholar]
- 2. Tunlid A, Ahrén D. 2011. Molecular mechanisms of the interaction between nematode-trapping fungi and nematodes: lessons from genomics, p 145–169 In Davies K, Spiegel Y. (ed), Biological control of plant-parasitic nematodes, vol 11 Springer, Dordrecht, Netherlands [Google Scholar]
- 3. Dijksterhuis J, Veenhuis M, Harder W, Nordbring-Hertz B. 1994. Nematophagous fungi: physiological aspects and structure-function relationships. Adv. Microb. Physiol. 36:111–143 [PubMed] [Google Scholar]
- 4. Liou GY, Tzean SS. 1997. Phylogeny of the genus Arthrobotrys and allied nematode-trapping fungi based on rDNA sequences. Mycologia 89:876–884 [Google Scholar]
- 5. Ahrén D, Ursing BM, Tunlid A. 1998. Phylogeny of nematode-trapping fungi based on 18S rDNA sequences. FEMS Microbiol. Lett. 158:179–184 [DOI] [PubMed] [Google Scholar]
- 6. Yang Y, Yang E, An ZQ, Liu XZ. 2007. Evolution of nematode-trapping cells of predatory fungi of the Orbiliaceae based on evidence from rRNA-encoding DNA and multiprotein sequences. Proc. Natl. Acad. Sci. U. S. A. 104:8379–8384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tunlid A, Johansson T, Nordbring-Hertz B. 1991. Surface polymers of the nematode-trapping fungus Arthrobotrys oligospora. J. Gen. Microbiol. 137:1231–1240 [DOI] [PubMed] [Google Scholar]
- 8. Friman E. 1993. Isolation of trap cells from the nematode-trapping fungus Dactylaria candida. Exp. Mycol. 17:368–370 [Google Scholar]
- 9. Ahrén D, Tholander M, Fekete C, Rajashekar B, Friman E, Johansson T, Tunlid A. 2005. Comparison of gene expression in trap cells and vegetative hyphae of the nematophagous fungus Monacrosporium haptotylum. Microbiology 151:789–803 [DOI] [PubMed] [Google Scholar]
- 10. Yang JK, Wang L, Ji XL, Feng Y, Li XM, Zou CG, Xu JP, Ren Y, Mi QL, Wu JL, Liu SQ, Liu Y, Huang XW, Wang HY, Niu XM, Li J, Liang LM, Luo YL, Ji KF, Zhou W, Yu ZF, Li GH, Liu YJ, Li L, Qiao M, Feng L, Zhang KQ. 2011. Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 7:e1002179. 10.1371/journal.ppat.1002179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bradford MM. 1976. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 12. Kessner D, Chambers M, Burke R, Agusand D, Mallick P. 2008. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24:2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hakkinen J, Vincic G, Mansson O, Warell K, Levander F. 2009. The Proteios Software Environment: an extensible multiuser platform for management and analysis of proteomics data. J. Proteome Res. 8:3037–3043 [DOI] [PubMed] [Google Scholar]
- 14. Craig R, Beavis RC. 2004. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20:1466–1467 [DOI] [PubMed] [Google Scholar]
- 15. Sandin M, Ali A, Hansson K, Mansson O, Andreasson E, Resjo S, Levander F. 2013. An adaptive alignment algorithm for quality-controlled label-free LC-MS. Mol. Cell. Proteomics 12:1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bellew M, Coram M, Fitzgibbon M, Igra M, Randolph T, Wang P, May D, Eng J, Fang RH, Lin CW, Chen JZ, Goodlett D, Whiteaker J, Paulovich A, McIntosh M. 2006. A suite of algorithms for the comprehensive analysis of complex protein mixtures using high-resolution LC-MS. Bioinformatics 22:1902–1909 [DOI] [PubMed] [Google Scholar]
- 17. Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang HZ, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh LSL. 2004. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 32:D115–D119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 19. Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer ELL, Eddy SR, Bateman A. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Winnenburg R, Urban M, Beacham A, Baldwin TK, Holland S, Lindeberg M, Hansen H, Rawlings C, Hammond-Kosack KE, Kohler J. 2008. PHI-base update: additions to the pathogen-host interaction database. Nucleic Acids Res. 36:D572–D576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 23. Neuberger G, Maurer-Stroh S, Eisenhaber B, Hartig A, Eisenhaber F. 2003. Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J. Mol. Biol. 328:581–592 [DOI] [PubMed] [Google Scholar]
- 24. Gao QA, Jin K, Ying SH, Zhang YJ, Xiao GH, Shang YF, Duan ZB, Hu XA, Xie XQ, Zhou G, Peng GX, Luo ZB, Huang W, Wang B, Fang WG, Wang SB, Zhong Y, Ma LJ, St Leger RJ, Zhao GP, Pei Y, Feng MG, Xia YX, Wang CS. 2011. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 7:e1001264. 10.1371/journal.pgen.1001264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, Garcia JL, Garcia MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jimenez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafton A, Latge JP, Li WX, Lord A, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O'Neil S, Paulsen I, Penalva MA, Pertea M, Price C, Pritchard BL, Quail MA, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, de Cordoba SR, Rodriguez-Pena JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sanchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, de Aldana CRV, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156 [DOI] [PubMed] [Google Scholar]
- 26. Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D'Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115 [DOI] [PubMed] [Google Scholar]
- 27. Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu JR, Pan HQ, Read ND, Lee YH, Carbone I, Brown D, Oh YY, Donofrio N, Jeong JS, Soanes DM, Djonovic S, Kolomiets E, Rehmeyer C, Li WX, Harding M, Kim S, Lebrun MH, Bohnert H, Coughlan S, Butler J, Calvo S, Ma LJ, Nicol R, Purcell S, Nusbaum C, Galagan JE, Birren BW. 2005. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434:980–986 [DOI] [PubMed] [Google Scholar]
- 28. Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 29. Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guindon S, Lethiec F, Duroux P, Gascuel O. 2005. PHYML Online: a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557–W559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Letunic I, Bork P. 2011. Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39:W475–W478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo SJ, McCurdy S, Foy M, Ewan M, Roth R, George D, Eletr S, Albrecht G, Vermaas E, Williams SR, Moon K, Burcham T, Pallas M, DuBridge RB, Kirchner J, Fearon K, Mao J, Corcoran K. 2000. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 18:630–634 [DOI] [PubMed] [Google Scholar]
- 33. Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tautz D, Domazet-Loso T. 2011. The evolutionary origin of orphan genes. Nat. Rev. Genet. 12:692–702 [DOI] [PubMed] [Google Scholar]
- 36. Heinisch JJ, Dupres V, Wilk S, Jendretzki A, Dufrene YF. 2010. Single-molecule atomic force microscopy reveals clustering of the yeast plasma-membrane sensor Wsc1. PLoS One 5:e11104. 10.1371/journal.pone.0011104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dupres V, Heinisch JJ, Dufrene YF. 2011. Atomic force microscopy demonstrates that disulfide bridges are required for clustering of the yeast cell wall integrity sensor Wsc1. Langmuir 27:15129–15134 [DOI] [PubMed] [Google Scholar]
- 38. Khan A, Williams KL, Soon J, Nevalainen HKM. 2008. Proteomic analysis of the knob-producing nematode-trapping fungus Monacrosporium lysipagum. Mycol. Res. 112:1447–1452 [DOI] [PubMed] [Google Scholar]
- 39. Gygi SP, Rochon Y, Franza BR, Aebersold R. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19:1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lackner DH, Schmidt MW, Wu SD, Wolf DA, Bahler J. 2012. Regulation of transcriptome, translation, and proteome in response to environmental stress in fission yeast. Genome Biol. 13:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee MV, Topper SE, Hubler SL, Hose J, Wenger CD, Coon JJ, Gasch AP. 2011. A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol. Syst. Biol. 7:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Greenbaum D, Colangelo C, Williams K, Gerstein M. 2003. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 4:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Friman E. 1993. Uv inhibition of the nematode infection process in Arthrobotrys oligospora and Dactylaria candida. J. Gen. Microbiol. 139:2841–2847 [Google Scholar]
- 44. Zuccaro A, Lahrmann U, Guldener U, Langen G, Pfiffi S, Biedenkopf D, Wong P, Samans B, Grimm C, Basiewicz M, Murat C, Martin F, Kogel KH. 2011. Endophytic life strategies decoded by genome and transcriptome analyses of the mutualistic root symbiont Piriformospora indica. PLoS Pathog. 7:e1002290. 10.1371/journal.ppat.1002290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Powell AJ, Conant GC, Brown DE, Carbone I, Dean RA. 2008. Altered patterns of gene duplication and differential gene gain and loss in fungal pathogens. BMC Genomics 9:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Soanes DM, Alam I, Cornell M, Wong HM, Hedeler C, Paton NW, Rattray M, Hubbard SJ, Oliver SG, Talbot NJ. 2008. Comparative genome analysis of filamentous fungi reveals gene family expansions associated with fungal pathogenesis. PLoS One 3:e2300. 10.1371/journal.pone.0002300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abramyan J, Stajich JE. 2012. Species-specific chitin-binding module 18 expansion in the amphibian pathogen Batrachochytrium dendrobatidis. mBio 3:e00150–12. 10.1128/mBio.00150-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kamper J, Kahmann R, Bolker M, Ma LJ, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Muller O, Perlin MH, Wosten HAB, de Vries R, Ruiz-Herrera J, Reynaga-Pena CG, Snetselaar K, McCann M, Perez-Martin J, Feldbrugge M, Basse CW, Steinberg G, Ibeas JI, Holloman W, Guzman P, Farman M, Stajich JE, Sentandreu R, Gonzalez-Prieto JM, Kennell JC, Molina L, Schirawski J, Mendoza-Mendoza A, Greilinger D, Munch K, Rossel N, Scherer M, Vranes M, Ladendorf O, Vincon V, Fuchs U, Sandrock B, Meng S, Ho ECH, Cahill MJ, Boyce KJ, Klose J, Klosterman SJ, Deelstra HJ, Ortiz-Castellanos L, Li WX, Sanchez-Alonso P, Schreier PH, Hauser-Hahn I, Vaupel M, Koopmann E, Friedrich G, Voss H, Schluter T, Margolis J, Platt D, Swimmer C, Gnirke A, Chen F, Vysotskaia V, Mannhaupt G, Guldener U, Munsterkotter M, Haase D, Oesterheld M, Mewes HW, Mauceli EW, DeCaprio D, Wade CM, Butler J, Young S, Jaffe DB, Calvo S, Nusbaum C, Galagan J, Birren BW. 2006. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444:97–101 [DOI] [PubMed] [Google Scholar]
- 49. Ponting CP, Hofmann K, Bork P. 1999. A latrophilin/CL-1-like GPS domain in polycystin-1. Curr. Biol. 9:R585–R588 [DOI] [PubMed] [Google Scholar]
- 50. Linder T, Gustafsson CM. 2008. Molecular phylogenetics of ascomycotal adhesins: a novel family of putative cell-surface adhesive proteins in fission yeasts. Fungal Genet. Biol. 45:485–497 [DOI] [PubMed] [Google Scholar]
- 51. Futagami T, Nakao S, Kido Y, Oka T, Kajiwara Y, Takashita H, Omori T, Furukawa K, Goto M. 2011. Putative stress sensors WscA and WscB are involved in hypo-osmotic and acidic pH stress tolerance in Aspergillus nidulans. Eukaryot. Cell 10:1504–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Verna J, Lodder A, Lee K, Vagts A, Ballester R. 1997. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 94:13804–13809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lodder AL, Lee TK, Ballester R. 1999. Characterization of the wsc1 protein, a putative receptor in the stress response of Saccharomyces cerevisiae. Genetics 152:1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Merchan S, Bernal D, Serrano R, Yenush L. 2004. Response of the Saccharomyces cerevisiae Mpk1 mitogen-activated protein kinase pathway to increases in internal turgor pressure caused by loss of Ppz protein phosphatases. Eukaryot. Cell 3:100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li GT, Zhou XY, Xu JR. 2012. Genetic control of infection-related development in Magnaporthe oryzae. Curr. Opin. Microbiol. 15:678–684 [DOI] [PubMed] [Google Scholar]
- 56. Bernier F, Lemieux G, Pallotta D. 1987. Gene families encode the major encystment-specific proteins of Physarum polycephalum Plasmodia. Gene 59:265–277 [DOI] [PubMed] [Google Scholar]
- 57. Rigden DJ, Mello LV, Galperin MY. 2004. The PA14 domain, a conserved all-beta domain in bacterial toxins, enzymes, adhesins and signaling molecules. Trends Biochem. Sci. 29:335–339 [DOI] [PubMed] [Google Scholar]
- 58. Frieman MB, McCaffery JM, Cormack BP. 2002. Modular domain structure in the Candida glabrata adhesin Epa1p, a beta 1,6 glucan-cross-linked cell wall protein. Mol. Microbiol. 46:479–492 [DOI] [PubMed] [Google Scholar]
- 59. Nordbring-Hertz B, Mattiasson B. 1979. Action of a nematode-trapping fungus shows lectin-mediated host-microorganism interaction. Nature 281:477–479 [Google Scholar]
- 60. Rosén S, Kata M, Persson Y, Lipniunas PH, Wikstrom M, Van Den Hondel CAMJ, Van Den Brink JM, Rask L, Heden LO, Tunlid A. 1996. Molecular characterization of a saline-soluble lectin from a parasitic fungus: extensive sequence similarities between fungal lectins. Eur. J. Biochem. 238:822–829 [DOI] [PubMed] [Google Scholar]
- 61. Balogh J, Tunlid A, Rosén S. 2003. Deletion of a lectin gene does not affect the phenotype of the nematode-trapping fungus Arthrobotrys oligospora. Fungal Genet. Biol. 39:128–135 [DOI] [PubMed] [Google Scholar]
- 62. Tunlid A, Jansson S. 1991. Proteases and their involvement in the infection and immobilization of nematodes by the nematophagous fungus Arthrobotrys oligospora. Appl. Environ. Microbiol. 57:2868–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Åhman J, Johansson T, Olsson M, Punt PJ, van den Hondel CA, Tunlid A. 2002. Improving the pathogenicity of a nematode-trapping fungus by genetic engineering of a subtilisin with nematotoxic activity. Appl. Environ. Microbiol. 68:3408–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fekete C, Tholander M, Rajashekar B, Ahrén D, Friman E, Johansson T, Tunlid A. 2008. Paralysis of nematodes: shifts in the transcriptome of the nematode-trapping fungus Monacrosporium haptotylum during infection of Caenorhabditis elegans. Environ. Microbiol. 10:364–375 [DOI] [PubMed] [Google Scholar]
- 65. Lopez-Llorca LV, Gomez-Vidal S, Monfort E, Larriba E, Casado-Vela J, Elortza F, Jansson HB, Salinas J, Martin-Nieto J. 2010. Expression of serine proteases in egg-parasitic nematophagous fungi during barley root colonization. Fungal Genet. Biol. 47:342–351 [DOI] [PubMed] [Google Scholar]
- 66. Kim ST, Yu S, Kim SG, Kim HJ, Kang SY, Hwang DH, Jang YS, Kang KY. 2004. Proteome analysis of rice blast fungus (Magnaporthe grisea) proteome during appressorium formation. Proteomics 4:3579–3587 [DOI] [PubMed] [Google Scholar]
- 67. Tucker SL, Talbot NJ. 2001. Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 39:385–417 [DOI] [PubMed] [Google Scholar]
- 68. Nordbring-Hertz B. 2004. Morphogenesis in the nematode-trapping fungus Arthrobotrys oligospora: an extensive plasticity of infection structures. Mycologist 18:125–133 [Google Scholar]
- 69. Bordallo JJ, Lopez-Llorca LV, Jansson HB, Salinas J, Persmark L, Asensio L. 2002. Colonization of plant roots by egg-parasitic and nematode-trapping fungi. New Phytol. 154:491–499 [DOI] [PubMed] [Google Scholar]
- 70. Schirawski J, Bohnert HU, Steinberg G, Snetselaar K, Adamikowa L, Kahmann R. 2005. Endoplasmic reticulum glucosidase II is required for pathogenicity of Ustilago maydis. Plant Cell 17:3532–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kwon M, Kim KS, Lee YH. 2010. A short-chain dehydrogenase/reductase gene is required for infection-related development and pathogenicity in Magnaporthe oryzae. Plant Pathol. J. 26:8–16 [Google Scholar]
- 72. Lu J, Feng X, Liu X, Lu Q, Wang H, Lin F. 2007. Mnh6, a nonhistone protein, is required for fungal development and pathogenicity of Magnaporthe grisea. Fungal Genet. Biol. 44:819–829 [DOI] [PubMed] [Google Scholar]
- 73. Veenhuis M, Nordbring-Hertz B, Harder W. 1984. Occurrence, characterization and development of two different types of microbodies in the nematophagous fungus Arthrobotrys oligospora. FEMS Microbiol. Lett. 24:31–38 [Google Scholar]
- 74. Veenhuis M, Vanwijk C, Wyss U, Nordbring-Hertz B, Harder W. 1989. Significance of electron dense microbodies in trap cells of the nematophagous fungus Arthrobotrys oligospora. Antonie Van Leeuwenhoek 56:251–261 [DOI] [PubMed] [Google Scholar]
- 75. Veenhuis M, Harder W, Nordbring-Hertz B. 1989. Occurrence and metabolic significance of microbodies in trophic hyphae of the nematophagous fungus Arthrobotrys oligospora. Antonie Van Leeuwenhoek 56:241–249 [DOI] [PubMed] [Google Scholar]
- 76. Kimura A, Takano Y, Furusawa I, Okuno T. 2001. Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell 13:1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Asakura M, Ninomiya S, Sugimoto M, Oku M, Yamashita S, Okuno T, Sakai Y, Takano Y. 2009. Atg26-mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbiculare. Plant Cell 21:1291–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Melloni E, Michetti M, Salamino F, Pontremoli S. 1998. Molecular and functional properties of a calpain activator protein specific for mu-isoforms. J. Biol. Chem. 273:12827–12831 [DOI] [PubMed] [Google Scholar]
- 79. Morishita R, Kawagoshi A, Sawasaki T, Madin K, Ogasawara T, Oka T, Endo Y. 1999. Ribonuclease activity of rat liver perchloric acid-soluble protein, a potent inhibitor of protein synthesis. J. Biol. Chem. 274:20688–20692 [DOI] [PubMed] [Google Scholar]
- 80. Sinha S, Rappu P, Lange SC, Mantsala P, Zalkin H, Smith JL. 1999. Crystal structure of Bacillus subtilis YabJ, a purine regulatory protein and member of the highly conserved YjgF family. Proc. Natl. Acad. Sci. U. S. A. 96:13074–13079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kim JM, Yoshikawa H, Shirahige K. 2001. A member of the YER057c/yjgf/Uk114 family links isoleucine biosynthesis and intact mitochondria maintenance in Saccharomyces cerevisiae. Genes Cells 6:507–517 [DOI] [PubMed] [Google Scholar]
- 82. Bell AA, Wheeler MH. 1986. Biosynthesis and functions of fungal melanins. Annu. Rev. Phytopathol. 24:411–451 [Google Scholar]
- 83. Perpetua NS, Kubo Y, Takano Y, Furusawa I. 1996. Cloning and characterization of a melanin biosynthetic THR1 reductase gene essential for appressorial penetration of Colletotrichum lagenarium. Mol. Plant Microbe Interact. 9:323–329 [DOI] [PubMed] [Google Scholar]
- 84. Selinheimo E, NiEidhin D, Steffensen C, Nielsen J, Lomascolo A, Halaouli S, Record E, O'Beirne D, Buchert J, Kruus K. 2007. Comparison of the characteristics of fungal and plant tyrosinases. J. Biotechnol. 130:471–480 [DOI] [PubMed] [Google Scholar]
- 85. Tunlid A, Jansson HB, Nordbring-Hertz B. 1992. Fungal attachment to nematodes. Mycol. Res. 96:401–412 [Google Scholar]
- 86. Williamson G, Kroon PA, Faulds CB. 1998. Hairy plant polysaccharides: a close shave with microbial esterases. Microbiology 144:2011–2023 [DOI] [PubMed] [Google Scholar]
- 87. Kawai R, Igarashi K, Samejima M. 2006. Gene cloning and heterologous expression of glycoside hydrolase family 55 beta-1,3-glucanase from the basidiomycete Phanerochaete chrysosporium. Biotechnol. Lett. 28:365–371 [DOI] [PubMed] [Google Scholar]
- 88. Chen XY, Kim JY. 2009. Callose synthesis in higher plants. Plant Signal. Behav. 4:489–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bowman SM, Free SJ. 2006. The structure and synthesis of the fungal cell wall. Bioessays 28:799–808 [DOI] [PubMed] [Google Scholar]
- 90. Rep M. 2005. Small proteins of plant-pathogenic fungi secreted during host colonization. FEMS Microbiol. Lett. 253:19–27 [DOI] [PubMed] [Google Scholar]
- 91. Xiao GH, Ying SH, Zheng P, Wang ZL, Zhang SW, Xie XQ, Shang YF, St Leger RJ, Zhao GP, Wang CS, Feng MG. 2012. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci. Rep. 2:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.