Abstract
In order to enlarge the substrate binding pocket of the meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum to accommodate larger 2-keto acids, four amino acid residues (Phe146, Thr171, Arg181, and His227) were targeted for site saturation mutagenesis. Among all mutants, the single mutant H227V had a specific activity of 2.39 ± 0.06 U · mg−1, which was 35.1-fold enhancement over the wild-type enzyme.
TEXT
d-Amino acids have attracted great attention because of their increasing importance as chiral building blocks for the synthesis of pharmaceuticals and food ingredients (1–3). Peptides containing d-amino acids show high resistance against proteolytic degradation, which is an important factor in drug design (4). d-p-Hydroxyphenylglycine has been widely used as precursor for the synthesis of some antibiotics and semisynthetic antibiotics, such as amoxicillin, ampicillin, and cefbuparzone (5, 6). d-Phenylalanine is the important chiral component of nateglinide, a drug for the treatment of type 2 diabetes (7). The dipeptide artificial sweeter alitame contains a d-alanine moiety (8). Among the biocatalytic methods for the preparation of d-amino acids, a useful and straightforward approach is the reductive amination of 2-keto acids catalyzed by d-amino acid dehydrogenases (EC1.4.99.1; D-AADH) (9, 10). Unfortunately, the characteristics of membrane-bound native D-AADH have hindered the industrial application of this family of enzymes (11). meso-Diaminopimelate dehydrogenase (EC1.4.1.16; meso-DAPDH) is a special class of D-AADHs which catalyze the reversible oxidative deamination and reductive amination at the d-center of meso-2,6-diaminopimelate (meso-DAP) with high stereoselectivity (12), restricting its application in d-amino acid synthesis. In this context, Vedha-Peters et al. expanded the substrate profile of the meso-DAPDH from Corynebacterium glutamicum by combining site saturation and random mutagenesis, resulting in a few mutants with activity toward a series of 2-keto acids (11). In our recent work, the NADP+-dependent meso-DAPDH from Symbiobacterium thermophilum was found to possess relaxed substrate specificity and catalyze the reductive amination of pyruvic acid, yielding d-alanine with 68% yield and 99% enantiomeric excess (ee). Although this enzyme is less active with other bulky 2-keto acids (e.g., phenylpyruvic acid) (13), it should serve as an excellent starting enzyme to be engineered for expanded substrate specificity.
The three-dimensional structure of the meso-DAPDH from C. glutamicum in a ternary complex showed that the substrate/inhibitor binding residues were Asn253, Gln150, Gly151, Asp90, and Asp120 for the d-center of meso-DAP and His244, Thr171, Arg195, Trp144 for its l-center (14). It was reported that the Cα-hydrogen of the d-center was transferred to the C-4′ of the nicotinamide ring to form an imine intermediate during deamination, and the distal l-center only maintained the correct orientation of meso-DAP (15, 16). In order to enlarge the substrate binding pocket without changing the d-stereospecificity, the amino acid residues interacting with the l-center of meso-DAP in the meso-DAPDH from S. thermophilum were chosen for site saturation mutagenesis. The mutant libraries were screened with phenylpyruvic acid as the substrate in an effort to obtain mutant enzymes capable of accommodating larger 2-keto acids.
By sequence alignment using the BioEdit software package (version 7.0.1) (13), the amino acid residues for the substrate l-center binding in S. thermophilum meso-DAPDH were identified as Phe146, Thr171, Arg181, and His227. The site saturation mutagenesis libraries were created individually for each amino acid residue by using a QuikChange mutagenesis protocol (Stratagene) with NNK as a degenerate codon (17). The recombinant plasmid pET32a (+) with the S. thermophilum meso-DAPDH gene reported previously (13) was used as the template with the primers listed in Table S1 in the supplemental material. The PCR product was transformed into BL21(DE3) competent cells for expression.
For 95% coverage of each library, 95 colonies were picked randomly and transferred into a deep 96-well plate containing 600 μl of LB medium with ampicillin (100 mg · liter−1) per well and incubated for 16 h at 37°C. After being replicated into a new deep 96-well plate, the colonies were incubated at 37°C in a shaker for microtiter plates (INFORS, Switzerland) at 400 rpm for 6 h. After isopropyl-β-d-thiogalactopyranoside (IPTG; 0.1 mM) was added, the plate was incubated at 30°C for 16 h. Cell pellets were harvested by centrifugation, suspended into 1.5 ml of phosphate buffer (100 mM, pH 7.0), and then disrupted by using an ultrasonic liquid processor (Q700; Qsonica, USA) with 96-tip horns. The resulting supernatants were used for activity screening by monitoring the NADPH consumption at room temperature in a reaction system of phosphate buffer (100 mM, pH 7.0) containing 240 μl of crude enzyme extract, 50 μl of substrate mixture (20 mM phenylpyruvic acid and 200 mM NH4Cl), and 10 μl of NADPH (0.26 mM) (13). Variants with 2-fold higher activity toward phenylpyruvic acid than wild-type enzyme were selected. No positive variant was obtained from the library of Phe146. A series of variants from the libraries of the other three sites showed up to about 35-fold of activity enhancement over wild-type enzyme (data not shown).
Among all mutants, five variants with the highest activities, T171P, T171S, R181F, H227C, and H227V, were purified for verification. Expression of the genes was induced using IPTG (0.1 mM) for 6 h at 37°C when the optical density at 600 nm (OD600) was 0.6 to 0.8. Cells were harvested by centrifugation and disrupted with a low-temperature ultrahigh-pressure continuous flow cell disrupter (JNBIO, China). Wild-type and all mutant enzymes were purified on an ÄKTA Avant 25 system (GE Healthcare, USA) with a His trap high-performance (HP) column (GE Healthcare, USA) as described in our previous work (13). The mutant enzymes were soluble and could be purified with yields similar to the yield of the wild-type enzyme. The specific activities for the reductive amination of phenylpyruvic acid were measured as described before (13). As shown in Table 1, residue His227 exerted a greater effect on the enzyme activity than residues Thr171 and Arg181. The mutants at position 171 (T171S and T171P) showed about 2.5-fold enhancement over wild-type enzyme in specific activity toward phenylpyruvic acid, while activity was increased 6.4-fold when Arg181 was mutated into Phe. The variants at H227 (H227C and H227V) showed much more improvement in activity toward phenylpyruvic acid over the wild type, with increases of 15.1- and 35.1-fold, or about 1.03 and 2.39 U · mg−1, respectively.
Table 1.
Specific activities, conversions, and kinetic parameters of the wild type and all variants toward phenylpyruvic acida
| Enzyme | Sp act (U · mg−1)b | Enhancement of activity | Conversion (%)c | Km (mM) | kcat (s−1) |
|---|---|---|---|---|---|
| WT | 0.07 ± 0.01 | 5.0 | 12.5 ± 2.1 | 0.11 ± 0.01 | |
| T171P | 0.15 ± 0.01 | 2.2 | 30.9 | 15.8 ± 1.0 | 0.53 ± 0.01 |
| T171S | 0.19 ± 0 | 2.8 | 27.5 | 19.6 ± 2.6 | 0.88 ± 0.04 |
| R181F | 0.44 ± 0.02 | 6.4 | 85.8 | 11.1 ± 1.6 | 1.07 ± 0.05 |
| H227C | 1.03 ± 0.07 | 15.1 | 88.7 | 15.5 ± 2.3 | 2.48 ± 0.15 |
| H227V | 2.39 ± 0.06 | 35.1 | 96.9 | 24.3 ± 3.2 | 3.98 ± 0.24 |
| T171S H227V | 0.74 ± 0.02 | 10.6 | 40.7 | 13.9 ± 2.5 | 2.37 ± 0.1 |
| R181F H227V | 1.35 ± 0.02 | 19.3 | 97.7 | 20.8 ± 1.4 | 3.43 ± 0.09 |
| T171S R181F | 0.28 ± 0.01 | 4.0 | 81.3 | 13.9 ± 1.8 | 0.70 ± 0.04 |
| T171S R181F H227V | 1.02 ± 0.01 | 14.6 | 74.2 | 11.0 ± 2.1 | 2.09 ± 0.17 |
Kinetic parameters of mutant and wild-type enzymes toward phenylpyruvic acid were measured in 100 mM Na2CO3-NaHCO3 buffer (pH 8.5) at 30°C. The concentrations of NH4Cl and of NADPH were 200 mM and 0.5 mM, respectively; the concentration of phenylpyruvic acid varied from 5 to 50 mM.
One unit (U) was defined as the amount of enzyme consuming 1 μmol of NADPH per minute.
The biotransformation systems contained 0.2 mg of meso-DAPDH, wild-type meso-DAPDH, or the variants.
To investigate the synergistic effects of His227, Thr171, and Arg181, double mutants T171S H227V, R181F H227V, and T171S R181F and the triple mutant T171S R181F H227V were created by site-directed mutagenesis using the primers listed in Table S1 in the supplemental material. The specific activities of purified double and triple mutants (Table 1) showed that H227V coupling with T171S and/or R181F showed lower specific activities than the single mutant H227V. Moreover, the double mutant T171S R181F was also less active than the single mutant R181F.
Vedha-Peters et al. succeeded in creating a highly stereoselective D-AADH with a broad substrate profile from C. glutamicum meso-DAPDH by three rounds of site saturation mutagenesis and error-prone PCR techniques. This variant (BC621) exhibited a specific activity of 0.11 U · mg−1 toward phenylpyruvic acid (11). Interestingly, the variant BC621 showed a 625-fold enhancement in specific activity toward cyclohexyl pyruvate (11), which has a similar bulkiness in the side chain as phenylpyruvic acid. However, variant BC621 had five mutations including Gln151Leu and Asp155Gly, in addition to the mutations Arg196Met, Thr170Ile, and His245Asn, at the binding sites for the l-center of the native substrate. A thermostable NADP+-dependent D-AADH was also obtained by site-directed mutagenesis of Ureibacillus thermosphaericus meso-DAPDH at the equivalent residues described by Vedha-Peters et al., but the enzyme activity toward phenylpyruvic acid was not reported (18).
Kinetic parameters of the variants and wild type toward phenylpyruvic acid were measured as reported in Gao et al. (13). The Km and kcat values were calculated by nonlinear fitting and are summarized in Table 1. There were slight effects on Km observed for all mutants, but kcat values increased greatly from 0.11 ± 0.01 s−1 to 3.98 ± 0.24 s−1, which was the major contributor to improve catalytic efficiency.
In order to determine the enantiomeric excess (ee) and conversions, biotransformation of phenylpyruvic acid was performed. The procedure was as follows. Phenylpyruvic acid (0.1 mmol), NH4Cl (0.3 mmol), glucose (80 mg), glucose dehydrogenase (GDH; 0.2 mg), NADP+ (0.5 mg) and meso-DAPDH (mutant/wild type, 0.2 mg) were added into 1 ml of Na2CO3-NaHCO3 buffer (200 mM, pH 8.5). The reaction mixture was incubated at 37°C for 24 h, and the reaction was stopped by addition of 30 μl of HClO4 solution. After centrifugation the supernatants were filtered through a 0.22-μm-pore-size filter and analyzed by high-performance liquid chromatography (HPLC) [CrownPak CR(+) column (Daicel) with a detection wavelength of 200 nm, mobile phase of HClO4 (pH 2.0), and flow rate of 0.5 ml · min−1]. The mutant enzymes catalyzed the reduction amination of phenylpyruvic acid to give d-phenylalanine with up to 97.7% conversion (Table 1). The ee values for all variants were >99%, the same as for the wild-type enzyme, demonstrating that the distal l-center amino acid residues do not affect stereospecificity of the enzyme.
To shed some insight on the enzyme activity improvement of variant H227V, molecular modeling and 10-ns molecular dynamic (MD) simulations of the wild type and H227V were performed.
The initial structure of the meso-DAPDH from S. thermophilum was created by homology modeling based on the structures of the meso-DAPDHs from C. glutamicum (Protein Data Bank [PDB] code 1f06) and Porphyromonas gingivalis W83 (PDB code 3BIO) using Discovery Studio, version 3.5 (Accelrys, USA). The structure of the H227V variant was obtained in silico upon mutation of the wild type in the targeted position with the same software. During the modeling, ligand NADPH was copied from the templates. Because docking the high-energy intermediate rather than the ground state of substrate or product significantly improved the prediction of the potential substrates of the enzymes (19–21), the imine intermediate (12) was docked into the binding pocket with Autodock, version 4.0, for MD simulations. All MD simulations were performed using the AMBER 11 simulation package (22). The standard AMBER force field (ff03.r1) and the general AMBER force field (gaff) were used to establish the topologies and parameters of enzyme and substrate, respectively. The program Antechamber was used to handle the force field parameters of the substrate, and NADPH parameters reported by Ryde were used (23). Five sodium cations were placed around the complex to maintain the system's neutrality. The whole system was solvated in an octahedral periodic box of TIP3P water molecules with a minimum solute-wall distance of 9 Å. Prior to the MD simulations, minimizations were carried out via three steps. First, waters/ions were optimized by restraining the protein. Second, the side chains of the protein were relaxed by restraining the backbone atoms. Third, the whole system was optimized without any restraints. Following minimizations, the systems were gradually heated from 0 to 303 K over 60 ps, and then 10-ns MD simulations with a 2.0-fs time step were performed under a constant temperature of 303 K. The SHAKE algorithm was employed to constrain bonds involving hydrogen atoms. Particle mesh Ewald (PME) summation was employed to deal with the long-range electrostatic interactions (24).
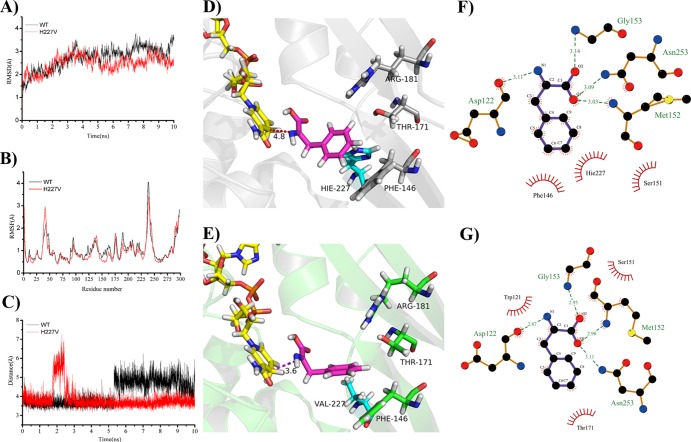
The root mean square displacement (RMSD) data of the skeleton during the full simulation showed that the wild-type complex and variant complex became dynamically equilibrated after 4 ns of simulation (Fig. 1A). The root mean square fluctuation (RMSF) of protein assay during 6- to 10-ns simulations (Fig. 1B) showed that there were no major changes in the stability in either the mutated position or the adjacent amino acids between the wild-type and mutant enzymes. Superimpositions of the average structures of the wild-type and H227V complex between 6 and 8 ns were used to analyze the hydrogen binding, hydrophobic interactions, and so on. As shown in Fig. 1C to E, the distance between the Cα of the imine intermediate and the C-4′ of the NADPH nicotinamide ring of the wild type was longer than that of the variant H227V, with values of 4.8 Å and 3.6 Å, respectively. In addition, there were slight differences in the orientations of the phenyl ring. Two-dimensional representations were visualized using Ligplot+ software (version 1.4.4 [http://www.ebi.ac.uk/thornton-srv/software/LigPlus/]) (25). As shown in Fig. 1F and G, Asp122, Met152, Gly153, and Asn253 interacted with the amino and carboxyl groups of the substrate via hydrogen bonds in both wild-type and mutant enzymes. These residues were the equivalent to the ones interacting with the d-center of meso-DAP in the meso-DAPDH from C. glutamicum (14). However, there were great differences around the phenyl ring of the substrate. In H227V, hydrophobic interactions were found between Thr171 and the phenyl ring, while the phenyl ring interacted with three amino residues (Phe146, His227, and Ser151) in the wild-type protein. Thus, the substrate was pulled closer to NADPH in the variant than that in the wild-type enzyme because of the weaker hydrophobic interactions in the mutant.
Fig 1.
MD simulation analysis of wild-type and variant H227V enzyme with the imine intermediate as the substrate. (A) RMSD of the backbone. (B) RMSF averaged between 6 ns and 10 ns. (C) Distance between the C-4′ of the nicotinamide ring and the carbon atom of the C=N bond of the imine intermediate. (D) Superimposition of the average structure of the wild type with the imine intermediate (purple) and NADPH (yellow). (E) Superimposition of the average structure of H227V complex. (F) Two-dimensional representation of the interactions of the substrate against the wild type. (G) Two-dimensional representation of the interactions of the substrate against H227V.
Since the mutations did not affect stereoselectivity but improved activity, we assumed that the hydrogen bonds involving the amino and carboxyl groups of the substrate with the protein residues were stronger than the hydrophobic interactions between the residues and the phenyl ring of the substrate. This view is consistent with the observation that the mutation did not significantly affect Km values.
In conclusion, site saturation mutagenesis of the amino acid residues interacting with the l-center of the substrate in meso-DAPDH from S. thermophilum resulted in mutant enzymes with improved activity for the reductive amination of phenylpyruvic acid. The mutant H227V catalyzed the conversion of phenylpyruvic acid to d-phenylalanine, with 96.9% conversion and 99% ee, demonstrating its potential for the direct synthesis of d-phenylalanine. Furthermore, it was affirmed that amino acid residues interacting with the l-center of the substrate have no relationship with the stereoselectivity of meso-DAPDH. Therefore, it is possible that efficient d-amino acid dehydrogenases for different substrates could be tailor-made by engineering of the meso-DAPDH from S. thermophilum while keeping its excellent stereospecificity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Neotrident Technology, Ltd., for supporting us in the molecular modeling. This work was financially supported by the National Natural Science Foundation of China (grant 21072151) and the National Basic Research Program of China (973 Program, grant 2011CB710801).
We thank Ph.D. candidate Hao Zhou for helpful discussions and constructive suggestions on MD simulations. We also thank Jon Stewart at the University of Florida for proofreading the manuscript and for his suggestions.
Footnotes
Published ahead of print 31 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01049-13.
REFERENCES
- 1. Ma JS. 2003. Unnatural amino acids in drug discovery. Chim. Oggi 21:65–68 [Google Scholar]
- 2. Friedman M, Levin C. 2012. Nutritional and medicinal aspects of d-amino acids. Amino Acids 42:1553–1582 [DOI] [PubMed] [Google Scholar]
- 3. Friedman M. 2010. Origin, microbiology, nutrition, and pharmacology of d-amino acids. Chem. Biodivers. 7:1491–1530 [DOI] [PubMed] [Google Scholar]
- 4. Tugyi R, Uray K, Iván D, Fellinger E, Perkins A, Hudecz F. 2005. Partial d-amino acid substitution: improved enzymatic stability and preserved Ab recognition of a MUC2 epitope peptide. Proc. Natl. Acad. Sci. U. S. A. 102:413–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Louwrier A, Knowles CJ. 1996. The purification and characterization of a novel d(−)-specific carbamoylase enzyme from an Agrobacterium sp. Enzyme Microb. Technol. 19:562–571 [Google Scholar]
- 6. Lee D-C, Lee S-G, Kim H-S. 1996. Production of d-p-hydroxyphenylglycine from d,l-5-(4-hydroxyphenyl)hydantoin using immobilized thermostable d-hydantoinase from Bacillus stearothermophilus SD-1. Enzyme Microb. Technol. 18:35–40 [Google Scholar]
- 7. White JR, Campbell RK. 2001. Recent developments in the pharmacological reduction of blood glucose in patients with type 2 diabetes. Clin. Diabetes 19:153–159 [Google Scholar]
- 8. Walters DE. 1995. Using models to understand and design sweeteners. J. Chem. Educ. 72:680–683 [Google Scholar]
- 9. Zhu D, Hua L. 2009. Biocatalytic asymmetric amination of carbonyl functional groups—a synthetic biology approach to organic chemistry. Biotechnol. J. 4:1420–1431 [DOI] [PubMed] [Google Scholar]
- 10. Höhne M, Bornscheuer UT. 2009. Biocatalytic routes to optically active amines. ChemCatChem 1:42–51 [Google Scholar]
- 11. Vedha-Peters K, Gunawardana M, Rozzell JD, Novick SJ. 2006. Creation of a broad-range and highly stereoselective d-amino acid dehydrogenase for the one-step synthesis of d-amino acids. J. Am. Chem. Soc. 128:10923–10929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Misono H, Togawa H, Yamamoto T, Soda K. 1979. Meso-alpha, epsilon-diaminopimelate d-dehydrogenase: distribution and the reaction product. J. Bacteriol. 137:22–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao X, Chen X, Liu W, Feng J, Wu Q, Hua L, Zhu D. 2012. A novel meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum: overexpression, characterization, and potential for d-amino acid synthesis. Appl. Environ. Microbiol. 78:8595–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cirilli M, Scapin G, Sutherland A, Vederas JC, Blanchard JS. 2000. The three-dimensional structure of the ternary complex of Corynebacterium glutamicum diaminopimelate dehydrogenase-NADPH-l-2-amino-6-methylene-pimelate. Protein Sci. 9:2034–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Olsiewski PJ, Kaczorowski G, Walsh C. 1980. Purification and properties of d-amino acid dehydrogenase, an inducible membrane-bound iron-sulfur flavoenzyme from Escherichia coli B. J. Biol. Chem. 255:4487–4494 [PubMed] [Google Scholar]
- 16. Scapin G, Cirilli M, Reddy SG, Gao Y, Vederas JC, Blanchard JS. 1998. Substrate and inhibitor binding sites in Corynebacterium glutamicum diaminopimelate dehydrogenase. Biochemistry 37:3278–3285 [DOI] [PubMed] [Google Scholar]
- 17. Zheng L, Baumann U, Reymond J-L. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Akita H, Doi K, Kawarabayasi Y, Ohshima T. 2012. Creation of a thermostable NADP+-dependent d-amino acid dehydrogenase from Ureibacillus thermosphaericus strain A1 meso-diaminopimelate dehydrogenase by site-directed mutagenesis. Biotechnol. Lett. 34:1693–1699 [DOI] [PubMed] [Google Scholar]
- 19. Tyagi S, Pleiss J. 2006. Biochemical profiling in silico-predicting substrate specificities of large enzyme families. J. Biotechnol. 124:108–116 [DOI] [PubMed] [Google Scholar]
- 20. Hermann JC, Ghanem E, Li Y, Raushel FM, Irwin JJ, Shoichet BK. 2006. Predicting substrates by docking high-energy intermediates to enzyme structures. J. Am. Chem. Soc. 128:15882–15891 [DOI] [PubMed] [Google Scholar]
- 21. Juhl PB, Trodler P, Tyagi S, Pleiss J. 2009. Modelling substrate specificity and enantioselectivity for lipases and esterases by substrate-imprinted docking. BMC Struct. Biol. 9:39–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Case D, Darden T, Cheatham T, III, Simmerling C, Wang J, Duke R, Luo R, Walker R, Zhang W, Merz K. 2010. AMBER 11. User's manual. University of California, San Francisco, CA [Google Scholar]
- 23. Holmberg N, Ryde U, Bülow L. 1999. Redesign of the coenzyme specificity in L-lactate dehydrogenase from Bacillus stearothermophilus using site-directed mutagenesis and media engineering. Protein Eng. 12:851–856 [DOI] [PubMed] [Google Scholar]
- 24. Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577–8593 [Google Scholar]
- 25. Laskowski RA, Swindells MB. 2011. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model 51:2778–2786 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.