Abstract
Microcalcifications are a feature of diagnostic significance on a mammogram and a target for stereotactic breast needle biopsy. Here, we report development of a Raman spectroscopy technique to simultaneously identify microcalcification status and diagnose the underlying breast lesion, in real-time, during stereotactic core needle biopsy procedures. Raman spectra were obtained ex vivo from 146 tissue sites from fresh stereotactic breast needle biopsy tissue cores from 33 patients, including 50 normal tissue sites, 77 lesions with microcalcifications, and 19 lesions without microcalcifications, using a compact clinical system. The Raman spectra were modeled based on the breast tissue components and a support vector machine framework was used to develop a single-step diagnostic algorithm to distinguish normal tissue, fibrocystic change (FCC), fibroadenoma (FA) and breast cancer, in the absence and presence of microcalcifications. This algorithm was subjected to leave-one-site-out cross-validation, yielding a positive predictive value, negative predictive value, sensitivity and specificity of 100%, 95.6%, 62.5% and 100% for diagnosis of breast cancer (with or without microcalcifications) and an overall accuracy of 82.2% for classification into specific categories of normal tissue, FCC, FA or breast cancer (with and without microcalcifications). Notably, the majority of breast cancers diagnosed are ductal carcinoma in situ (DCIS), the most common lesion associated with microcalcifications, which could not be diagnosed using previous Raman algorithm(s). Our study demonstrates the potential of Raman spectroscopy to concomitantly detect microcalcifications and diagnose associated lesions, including DCIS, and thus provide real-time feedback to radiologists during such biopsy procedures, reducing non-diagnostic and false negative biopsies.
Keywords: Raman, spectroscopy, breast, cancer, microcalcifications
INTRODUCTION
Breast cancer is the second leading cause of cancer death in women, with one in eight women likely to develop breast cancer in her lifetime. In 2011, 230,480 new cases of breast cancer are estimated to have occurred in the United States alone (1). The most effective approach for preventing breast cancer morbidity and mortality is early detection. X-ray mammography is currently the only accepted routine screening method for early detection (2).
Microcalcifications are localized deposits of calcium species in breast tissue that geographically target the most clinically significant abnormality within the breast and are considered an early mammographic sign of breast cancer (3). However, mammography cannot accurately distinguish microcalcifications associated with benign and malignant breast lesions and even the best mammographic algorithms have limitations arising from dark mammographic backgrounds and densely clustered calcifications (4-6). Therefore, although the type of microcalcification is known to correlate with disease status (e.g. type II microcalcifications co-localize with proliferative lesions (3, 7)), most patients currently undergo vacuum-assisted stereotactic core needle biopsy to determine whether or not the microcalcifications are associated with breast cancer.
In addition, despite stereotactic guidance, core needle biopsy fails to retrieve microcalcifications in up to 15% of patients (8). Failure to retrieve the microcalcifications results in non-diagnostic or false negative biopsies, requiring the patient to undergo repeat, often surgical biopsy. Therefore, there is a clinical need for a tool that can detect microcalcifications in the breast tissue to be biopsied and provide real-time feedback to the radiologist during stereotactic core needle biopsy procedures to ensure that the biopsied tissue core contains the microcalcifications observed during mammography.
Raman spectroscopy is a non-destructive, chemical-specific, inelastic scattering technique (9, 10) that can be performed with fiberoptic probes compatible with vacuum-assisted stereotactic biopsy needles, and so is an ideal choice for detecting microcalcifications during these procedures. Calcium-containing species found in breast microcalcifications, such as calcium hydroxyapatite (CHA) and calcium oxalate (CAO), are strong Raman scatterers. Thus, Raman spectroscopy is sensitive to the presence of microcalcifications and can therefore be used as a clinical tool for guidance of stereotactic breast core needle biopsies for microcalcifications.
Raman spectroscopy is currently being explored by our laboratory (9, 11-18) and others (19-28) for its potential to both detect breast microcalcifications and diagnose breast cancer. Our group has demonstrated the potential of Raman spectroscopy to detect and distinguish type I and II breast microcalcifications and to differentiate type II calcifications associated with benign and malignant breast lesions (12) in Raman microscopy studies of formalin-fixed, paraffin-embedded breast biopsies. We also recently developed the first Raman spectroscopy algorithm to detect microcalcifications in fresh breast needle biopsy tissue cores (18). However, if Raman spectroscopy is to be used as a clinical tool for guidance of stereotactic breast needle biopsies for microcalcifications, it is desirable to not only detect microcalcifications, but also to diagnose the specific breast lesion associated with the microcalcifications. We previously devised a Raman algorithm to diagnose breast cancer and benign breast lesions, including fibrocystic change (FCC) and fibroadenoma (FA). However, the algorithm was developed on tissue sites devoid of microcalcifications (15, 17). This Raman algorithm is, therefore, not applicable to breast lesions associated with microcalcifications because of the large spectral contributions from CHA and/or CAO, which makes it difficult to map the contributions of the other tissue components onto the algorithm domain created without consideration of microcalcifications.
To address this issue, we are developing new Raman algorithms to diagnose breast cancer and benign breast lesions in the presence or absence of microcalcifications. One possible approach is to combine the previous algorithm for microcalcification detection (18) with a new algorithm for diagnosis of breast lesions (irrespective of microcalcification status) (29), thereby constructing a sequential two-step algorithm to identify the breast lesion(s) associated with the microcalcifications. However, this is a laborious and unwieldy process (that also does not provide the required level of accuracy as detailed below), which can be simplified by devising a single algorithm to simultaneously detect microcalcifications and diagnose the associated breast lesion(s).
Here we report the first development of a single-step Raman spectroscopy algorithm to simultaneously determine microcalcification status and diagnose the underlying breast lesion(s), in real-time, during stereotactic breast core needle biopsy procedures. We also compare the diagnostic performance of this single-step algorithm with its two-step counterpart(s) to comprehensively assess the advantages (and/or drawbacks) of pursuing these two approaches.
MATERIALS AND METHODS
Patient population
This study was performed on the Raman spectroscopy data set used previously to develop our decision algorithms to detect breast microcalcifications (18). Raman spectroscopy was performed ex vivo on fresh breast tissue cores obtained from 33 female patients (ages 38-79) undergoing vacuum-assisted stereotactic core needle breast biopsy procedures in the Breast Health Center at University Hospitals-Case Medical Center. All studies were approved by the Case Cancer Institutional Review Board and the Massachusetts Institute of Technology Committee On the Use of Humans as Experimental Subjects, in accordance with assurances filed with and approved by the U.S. Department of Health and Human Services. Informed consent was obtained from all subjects prior to their biopsy procedures.
Raman spectral acquisition
The Raman spectra were obtained using a portable clinical Raman spectroscopy system, previously described in detail (18). The instrument delivers 830nm NIR excitation light to the tissue via an optical fiber probe. The fiber probe, which is in the form of a flexible catheter of outer diameter 2mm, consists of a central excitation fiber surrounded by nine acquisition fibers, each of 200μm diameter. The acquisition fibers are coupled to an f/1.8i spectrograph for dispersion onto a thermoelectrically-cooled CCD detector. Laser power at the probe-tissue interface was 98-105 mW. 10 spectral acquisitions were obtained from each tissue site and these were summed to provide the final spectrum corresponding to the tissue site. Each of the 10 spectral acquisitions was performed in 0.25s, for a total collection time of 2.5s per tissue site. Analysis of the spectra was performed in real-time, that is, in a few tens of milliseconds as detailed previously (30). In other words, each spectrum was acquired, model fit, analyzed and the diagnostic classification for the tissue site displayed on a computer interface in just over 2.5 seconds.
Raman spectra were collected from several tissue sites of interest on each tissue core (typically normal tissue, lesions (grossly abnormal tissue) without microcalcifications and lesions with microcalcifications) identified by gross inspection and comparison with the accompanying specimen radiograph. The tissue cores were roughly cylindrical and measured ca. 1.7 mm by 2 cm, as determined by the size of the needle biopsy port. Spectra were also collected from different tissue cores in each biopsy, so the number of spectra varied from patient to patient. The number of tissue sites studied per core varied from 1-8 and the number of cores per patient from 1-6. All spectra were obtained within 30 minutes of excision.
Histopathology
After spectral acquisition, the tissue sites from which Raman spectra were obtained were uniquely identified with multicolored colloidal inks. The tissue was then fixed in 10% neutral buffered formalin and paraffin embedded, and sections cut and hematoxylin and eosin (H&E) stained for microscopic examination by an experienced breast pathologist. Histopathological evaluation, in conjunction with the radiographic assessment, was used as the gold standard for comparison with the Raman spectral diagnosis.
Raman data analysis
The Raman system was wavenumber calibrated and the Raman spectra corrected for the system wavelength response and background subtracted, as previously described (18). The Raman spectra were then fit with a previously developed breast model (13), in which the Raman spectrum is considered as a linear combination of the basis spectra of 10 breast tissue constituents, including epithelial cell nuclei (ECN) and cytoplasm (ECC), fat, cholesterol-like deposits (CHOL), β-carotene (β-CAR), collagen (COLL), oxy-hemoglobin (oxy-HB), CHA, CAO and water; and 2 fiberoptic probe materials, epoxy and sapphire. Ordinary least squares (OLS) fitting was used to determine the contribution of each basis spectrum to the tissue spectrum, yielding fit coefficients (FC) that provide information about the morphological and chemical composition of the tissue. The goodness of the model fit was qualitatively estimated by visual inspection of the residual (spectrum minus fit) and quantitatively from the standard deviation of the residual. The extracted FCs were used to develop the Raman algorithms outlined below.
Raman algorithm development
In this study, support vector machines (SVM) were used to construct single-step and two-step algorithms to concomitantly detect microcalcifications and diagnose the associated breast lesion(s) based on the Raman FCs. SVMs are a relatively new class of non-linear classification techniques (31-33), whose robust nature with respect to sparse and noisy data has enabled its extensive usage, especially in bioinformatics and chemometrics.
First, a single-step SVM Raman algorithm was constructed to simultaneously detect microcalcifications and diagnose the associated breast lesion(s). In this single-step algorithm, we considered the positive class to be cancer (with or without microcalcifications), unless otherwise specified. All other classes including normal, fibroadenoma and fibrocystic change, irrespective of microcalcification status, were considered to be negative. This single-step SVM Raman algorithm was then compared to two-step Raman algorithms. Initially, a “naïve” two-step algorithm was built by combining the logistic regression (LR) algorithm for the detection of microcalcifications described in Saha et al. (18) with an SVM algorithm developed for diagnosis of lesions irrespective of microcalcification status described in Dingari et al. (29). In the first step of this two-step naïve algorithm, LR was performed to first discriminate the entire dataset into three classes: normal breast tissue, lesions without microcalcifications and lesions with microcalcifications. For the LR step, a likelihood ratio test was used to estimate the FCs most critical for diagnosis, namely COLL, fat and the combined contribution of CAH and CAO. In the second step of the naïve algorithm, a breast model FC-based SVM algorithm was employed for all tissue sites to further diagnose the specific tissue type, namely normal, FCC, FA or cancer.
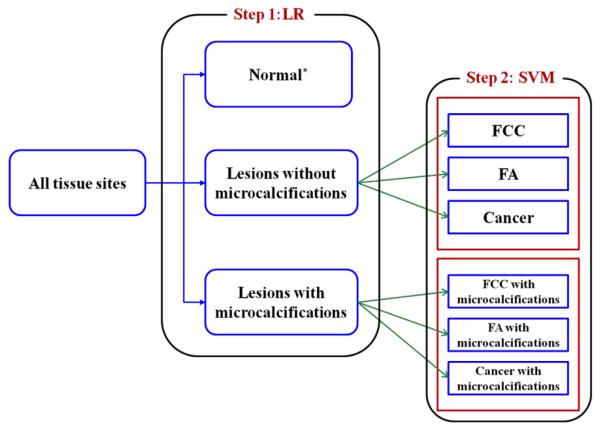
Additionally, in order to comprehensively assess the two-step algorithm, we sub-divided the second-step into two new SVM Raman algorithms (as diagrammed in Fig. 1), one optimized for classification of breast lesions without microcalcifications and one optimized for classification of breast lesions with microcalcifications. We call this implementation of the two-step algorithm the “optimized” version. It is worth noting that the first-step (the LR algorithm for detection of microcalcifications) is identical in both the naïve and optimized versions of the two-step algorithms. All three scenarios (namely, single-step, two-step (naïve) and two-step (optimized) algorithms) result in classification of the tissue sites into one of eight categories, based on the tissue type (normal breast tissue, FCC, FA or cancer) and microcalcification status (with and without microcalcifications).
Figure 1.
Schematic diagram of sequential two-step optimized Raman algorithm using a logistic regression algorithm for Step 1 and two separate support vector machine classification models (naïve and optimized) for Step 2. Both these algorithms were used in a leave-one-out cross-validation protocol. (*Normal tissue sites with microcalcifications were not considered for algorithm development as they represent a discordance between radiographic assessment (lesion with microcalcifications) and histopathological evaluation (normal))
Here, LR was performed using in-house code (MATLAB R2010b, Math Works, Natick, MA). The SVM classification analysis was performed using Orange (http://www.ailab.si/orange), an open-source data mining suite featuring Python scripting and a graphic interface (34). Specifically, a radial basis function (RBF) kernel with a Gaussian profile K(xi,xj) = exp(-g∥xi-xj∥2) was used for non-linear SVM classification, where g represents the RBF kernel parameter. The optimal model parameters C (cost parameter) and g that give minimum error in cross-validation were determined by performing a grid search over appropriate ranges (30).
The performance of the single-step and two-step SVM Raman algorithms were assessed using the following metrics: sensitivity (SE), specificity (SP), positive predictive value (PPV) and negative predictive value (NPV) for the diagnosis of cancer (with or without microcalcifications) and overall accuracy (OA) for the diagnosis of all categories: normal tissue, FCC, FA and cancer, with and without microcalcifications. These metrics were evaluated based on the rates of true and false positive and negative results per standard definitions (35). Algorithm performance was validated using a leave-one-out cross validation technique (LOOCV) (15). In this technique, the data from a particular tissue site is eliminated, and a decision algorithm developed that classifies all of the remaining tissue sites in the dataset (including ones from the same tissue core and patient) optimizing agreement with the histopathology diagnoses. The resulting decision algorithm is then used to classify the excluded site. This process is successively applied to each site.
RESULTS
The data set initially included Raman spectra obtained from 158 tissue sites. Five tissue sites with miscellaneous tissue histopathological diagnoses (fat necrosis, healing biopsy site, etc.) were excluded from analysis during algorithm development. Seven additional tissue sites were excluded during LOOCV as unallocated, based on their relatively low probability of belonging to any class, including: 1 ductal carcinoma in situ (DCIS) with microcalcifications; 4 FCC with microcalcifications; 1 FCC without microcalcifications; and 1 normal breast tissue site. (Specifically, these 7 tissue sites yielded low probability of classification on application of the single-step Raman algorithm and were excluded from all ensuing analysis for the sake of consistency. In clinical practice, tissue sites for which there is no definitive spectral diagnosis would be designated as unclassified or equivocal and additional Raman measurements made in the hope of obtaining more definitive results.) Analysis was performed on the remaining 146 tissue sites, whose reference classifications are as follows: 50 normal; 3 normal with microcalcifications; 17 FA all with microcalcifications; 60 FCC, 43 with and 17 without microcalcifications; 16 cancers, 14 with microcalcifications (13 DCIS and 1 invasive ductal carcinoma (IDC)) and 2 without microcalcifications (1 DCIS and 1 lobular carcinoma in situ (LCIS)). Breast lesions were classified as having microcalcifications if microcalcifications were seen at that tissue site on either the specimen radiograph or the H&E stained tissue sections. In particular, there were 13 microcalcifications seen on the H&E stained sections that were not observed on the specimen radiograph. In addition, 3 normal tissue sites were classified as normal with microcalcifications since microcalcifications were seen on radiography that were not seen on histopathology. Of the 77 tissue sites with microcalcifications, 75 had type II (CHA-derived) and only 2 had type I (CAO-derived) microcalcifications.
Raman spectra
Figure 2 shows the histopathology and Raman spectrum (blue) with model fit (red) and residual (black) for a typical breast lesion (FCC) with type II microcalcifications. The microcalcifications are visible as dark blue concretions (arrow) in the photomicrograph in Figure 2(a) (H&E; 10X). The corresponding Raman spectrum in Fig. 2(b) shows a prominent band at 960 cm−1 due to CHA (arrow) (arising from the ν1(PO4) totally symmetric stretching mode of the “free” tetrahedral phosphate ion) present in the microcalcifications.
Figure 2.
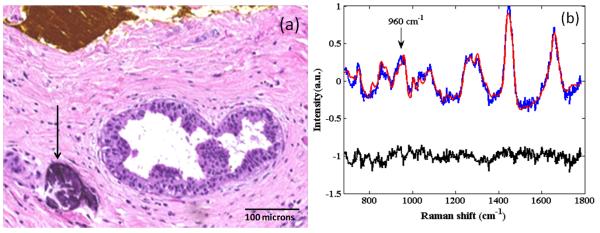
Histopathology and Raman spectrum (blue) with model fit (red) and residual (black) for a typical breast lesion (FCC) with type II microcalcifications. The microcalcifications are visible as dark blue concretions (arrow) in the photomicrograph in Figure 2(a) (H&E; 10X). Note the yellow ink on the breast tissue surface at the top in Figure 2(a), marking the site for spectral correlation. The corresponding Raman spectrum in Figure 2(b) shows a prominent band at 960 cm−1 due to CHA (arrow), which is a major constituent of type II microcalcifications.
Single-step SVM Raman algorithm
We devised a single-step Raman spectral algorithm to diagnose normal breast tissue, FCC, FA and breast cancer with and without microcalcifications. This algorithm uses SVM to classify the tissue sites into the different lesion categories based on the FCs extracted using the OLS model. In particular, the FCs corresponding to CAH, ECC, fat, oxy-HB, COLL and CHOL were selected as input parameters to the SVM model as they provided the optimal diagnostic performance on LOOCV. The inclusion of the other parameters (e.g. epoxy and water FCs) had a slightly detrimental effect on the classification capability of the algorithm, which can be attributed to lack of correlation between the presence of these components and the lesion type.
The LOOCV results for this single-step SVM Raman algorithm are shown in the confusion matrix in Table 1. This algorithm, which takes into account microcalcification status for the first time, has a SE of 62.5%, SP of 100%, PPV of 100% and NPV of 95.6% for the diagnosis of breast cancer (with or without microcalcifications) and an OA of 82.2% for the classification into the specific categories of normal breast tissue, FCC, FA or breast cancer (with and without microcalcifications). Here, the area under the curve (AUC) is 0.92, and indicates the robustness of the algorithm (with respect to a maximum AUC of 1.00 for a perfect algorithm).
Table 1.
Confusion matrix for leave-one-out cross validation (LOOCV) of single-step SVM Raman decision algorithm for all 7 diagnostic categories
| Pathology Diagnosis |
Raman Diagnosis | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Normal | Normal with Microcalcs |
FCC | FCC with Microcalcs |
FA with Microcalcs |
Cancer | Cancer with Microcalcs |
Total | |
| Normal | 50 | 0 | 0 | 0 | 0 | 0 | 0 | 50 |
|
Normal
with Microcalcs |
0 | 0 | 1 | 1 | 1 | 0 | 0 | 3 |
| FCC | 2 | 0 | 3 | 12 | 0 | 0 | 0 | 17 |
|
FCC with
Microcalcs |
1 | 0 | 6 | 34 | 2 | 0 | 0 | 43 |
|
FA with
Microcalcs Note |
0 | 0 | 1 | 11 | 5 | 0 | 0 | 17 |
| Cancer | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 2 |
|
Cancer with
Microcalcs |
1 | 0 | 0 | 4 | 0 | 0 | 9 | 14 |
| Total | 55 | 0 | 11 | 62 | 8 | 0 | 10 | 146 |
There were no tissue sites with fibroadenoma without microcalcifications.
Abbreviation: Microcalcs = microcalcifications
In these studies, we chose the decision line whose operating point results in maximal PPV for the diagnosis of breast cancer, as is typically done in a clinical situation where the disease to be diagnosed is serious, should not be missed and is treatable (35). However, the situation is more complex in the case of Raman spectroscopy guidance of stereotactic breast biopsies for microcalcifications. In this instance, the radiologist’s goal is to diagnosis breast cancer if present or, failing that, to retrieve the targeted microcalcifications. So, the radiologist needs to know whether or not there are microcalcifications present in the tissue to be biopsied and whether or not the associated breast lesion is cancer. In order to take this into account, we also carefully considered the OA of the diagnostic algorithm, the only metric that fully takes into account the combined diagnosis of microcalcification status and the underlying breast lesion.
The performance of the single-step SVM algorithm for simultaneous detection of microcalcification status and diagnosis of the underlying breast lesion is virtually identical to that of a previously developed SVM Raman algorithm that ignored microcalcification status, which has a SE of 62.5%, SP of 100%, PPV of 100% and NPV of 95.6% for the diagnosis of breast cancer (ROC AUC=0.92) and overall accuracy of 81.5%, in this same data set (29), as shown in Table 2. (This is not surprising as, even though the new algorithm simultaneously detects microcalcification status and diagnoses the underlying lesion, here we are assessing algorithm performance for the diagnosis of breast cancer irrespective of microcalcification status for the sake of comparison. We have also assessed its performance for an instance of a combined diagnosis of microcalcification status and lesion diagnosis (See Concomitant diagnosis of microcalcifications and underlying breast lesion below.)) Further, the performance of this single-step SVM algorithm is markedly superior to that obtained with our previously developed LR Raman algorithm (in a prospective ex vivo validation study), which has a SE of 83%, SP of 93%, PPV of only 36% and NPV of 99% for the diagnosis of breast cancer in the absence of microcalcifications (17). Significantly, the majority of breast cancers in the current study are DCIS, a lesion that could not be diagnosed with the previous LR Raman algorithm, as the latter algorithm was developed in a data set that consisted primarily of samples with IDC (15).
Table 2.
Comparison of diagnostic performance of single-step, two-step and previous Raman algorithms for detection of breast cancer, with and without microcalcifications
| Raman Algorithm | Classification Scheme | SE | SP | PPV | NPV | OA |
|---|---|---|---|---|---|---|
| Single-step SVM | All tissue sites | 62.5% | 100% | 100% | 95.6% | 82.2% |
| Tissue sites with microcalcifications |
64.3% | 100% | 100% | 92.7% | 70.1% | |
| Tissue sites without microcalcifications |
50% | 100% | 100% | 98.5% | 95.7% | |
|
| ||||||
| Two-step LR-SVM (naive) |
All tissue sites | 53.8% | 94% | 95% | 81.3% | 73.4% |
|
| ||||||
| Two-step LR-SVM (optimized) |
All tissue sites | 56.3% | 100% | 100% | 94.9% | 80% |
|
| ||||||
| Previous SVM algorithma |
All tissue sites | 62.5% | 100% | 100% | 95.6% | 81.5% |
|
| ||||||
| Previous LR algorithmb |
83% | 93% | 36% | 99% | 78.3% | |
Impact of microcalcifications on single-step SVM Raman algorithm performance
In order to specifically assess the impact of the presence of microcalcifications on the performance of the single-step SVM Raman algorithm, the 7 specific diagnostic categories were collapsed into 2 sub-categories: tissue sites (normal, FCC, FA and cancer) with and without microcalcifications, as shown in Table 3. This resulted in a PPV of 100%, NPV of 92.7% and an OA of 70.1% for lesions with microcalcifications, compared to a PPV of 100%, NPV of 98.5% and OA of 95.7% for lesions without microcalcifications. These results indicate that the SVM Raman algorithm is more robust for classification of tissue sites without microcalcifications than for tissue sites with microcalcifications, especially when one considers classification of all classes (i.e. the OA metric) as compared to discrimination of cancer sites only (i.e. the PPV and NPV metrics). Nevertheless, the SVM algorithm provides comparable accuracy for the critical metrics of PPV and, to a slightly lesser extent, NPV. The confusion matrices for these two sub-classification schemes are strikingly similar to that of the 7-category classification scheme (Table 1), and are shown in the Supplementary Information (Tables S1 and S2). Table S3 also provides a detailed breakdown of the tissue sites by pathology and the corresponding diagnoses based on the single-step SVM Raman decision algorithm. Overall, these results indicate that performance of the single-step SVM Raman algorithm is remarkably consistent, irrespective of the presence of microcalcifications.
Table 3.
Performance of single-step SVM Raman decision algorithm for tissue sites with and without microcalcifications
| Pathology Diagnosis |
Raman Diagnosis | ||
|---|---|---|---|
|
| |||
| Correct Lesion Classification |
Incorrect Lesion Classification |
Total | |
|
Tissue sites
with Microcalcifications |
54 (70.1%) | 23 (29.9%) | 77 (100%) |
|
Tissue sites
without Microcalcifications |
66 (95.7%) | 3 (4.3%) | 69 (100%) |
| Total | 120 (82.2%) | 26 (17.8%) | 146 (100%) |
Misdiagnoses using single-step SVM Raman algorithm
As might be suspected from the results above, a significant number of tissue site misdiagnoses using the single-step SVM Raman algorithm involved misdiagnosis of the underlying lesion in tissue sites with microcalcifications. All 3 normal breast tissue sites with microcalcifications were misclassified, one as FCC, one as FCC with microcalcifications and one as FA with microcalcifications. Presumably, the spectroscopy results for these tissue sites (which showed normal breast tissue on histopathology) are correct as they agree with the radiographic assessment of the presence of lesions with microcalcifications at these sites, and the apparent misclassifications arise due to spectroscopy-histopathology registration errors. The majority of tissue sites with FA with microcalcifications (11 of 17) were also misclassified as FCC with microcalcifications. For these tissue sites, microcalcifications are the dominant spectral contributors, and the spectral contributions from the other tissue components (such as CHOL, fat and COLL) are fairly comparable for FA and FCC. In other words, we suspect that the SVM class allocation probability for these tissue sites was reasonably similar for both the FA and FCC categories, and the resulting misclassification can be attributed to the combination of an imperfect decision plane (between these two classes) and the uncertainty inherent in spectral assignment (from shot noise etc.). It is worth noting that the substantially larger number of samples designated as FCC with microcalcification (43) in relation to FA with microcalcification (17) in this dataset may have also artificially skewed the decision plane in favor of the former. Fortunately, this problem can be remedied by adequately increasing the sample size, e.g. by performing larger clinical studies. These misclassifications are likely not clinically significant in the context of Raman guidance of stereotactic breast biopsies, viewed in terms of overall biopsy (or patient) results, and not individual tissue site results, as the overall assessment that these biopsies harbor benign breast lesions with microcalcifications that would be retrieved at biopsy is correct.
Further, 5 of 14 tissue sites with cancer with microcalcifications were misclassified by the single-step SVM Raman algorithm as normal (1) or FCC with microcalcifications (4). The first of these was classified as normal due to a high FC of fat. The spectroscopy result for this tissue site is most likely correct, as it appeared normal (largely fat) on gross inspection. Thus, the apparent misclassification of this site is likely due to a spectroscopy-histopathology registration error. Also, 3 out of the 4 tissue site misclassifications as FCC with microcalcifications belonged to an individual patient, the spectral data from whom exhibited tissue features that were not accounted for by the breast constituent model. The misclassification of these 4 cancer with microcalcifications sites, while potentially clinically significant, are not unexpected due to the relatively high FC of CHA in both of these categories (FCC and cancer with microcalcifications). There were only two tissue sites with cancer without microcalcifications, and these were both misclassified as normal (1) and cancer with microcalcifications (1). Again, the misclassification for cancer without microcalcifications is not likely clinically significant, as the biopsy also contained tissue sites correctly classified by the Raman algorithm as cancer with microcalcifications. Thus the overall assessment that the biopsy harbors cancer with microcalcifications that would be retrieved at biopsy is correct.
Other misdiagnoses resulted from misclassification of microcalcification status. The majority of tissue sites with FCC without microcalcifications (12 of 17) were misclassified as FCC with microcalcifications, most likely because the corresponding decision plane is dominated by the FC of COLL as opposed to the FC of CHA. Even so, these misclassifications are again likely not clinically significant, as these biopsies also contained tissue sites correctly classified by the Raman algorithm as FCC with microcalcifications. Thus the overall assessment that these biopsies harbor benign breast lesions with microcalcifications that would be retrieved at biopsy is again correct. As alluded to above in the case of misclassification of FA sites as FCC with microcalcifications, the SVM classification algorithm is likely to perform with greater accuracy for more extensive clinical datasets, which have sufficiently large number of FCC without microcalcifications sites.
Two-step SVM algorithm
Performance of the single-step SVM Raman algorithm was compared to that of the naïve and optimized two-step algorithms. The naïve two-step algorithm, which sequentially applies our LR algorithm (for microcalcification detection) and a SVM algorithm (for lesion discrimination), yields a SE of 53.8%, SP of 94%, PPV of 95% and NPV of 81.3% and an OA of 73.4%. Clearly, the performance of this algorithm in LOOCV is considerably inferior to that of the single-step SVM algorithm across the board (Table 2). This reveals the underlying deficiencies of employing a two-step method where the multistage or hierarchical decision scheme amplifies errors emanating from the imperfect decision planes in each of the steps.
In comparison, the optimized two-step algorithm, which uses two separate SVM algorithms to sub-categorize lesions with and without microcalcifications as FCC, FA and breast cancer in step two, yields a SE of 56.3%, SP of 100%, PPV of 100% and NPV of 94.9% for the diagnosis of breast cancer and an OA of 80.2% for classification into the specific categories of normal breast tissue, FCC, FA or breast cancer (with and without microcalcifications). Thus, when optimized, performance of the two-step Raman algorithm is comparable to that of the single-step SVM Raman algorithm (Table 2). Nevertheless, it should be noted that application of two separate SVM algorithms for lesion discrimination in our limited dataset may run the risk of overtraining (overfitting) due to the concomitant reduction in number of samples available for each algorithm (i.e. data sparsity). For example, 12 tissue sites were classified by the LR algorithm as lesions without microcalcifications and, as all of these 12 were designated as FCC by histopathology, the corresponding SVM algorithm was able to readily identify all these tissue sites as FCC. For larger datasets, one would anticipate that lesion discrimination would be more difficult due to the presence of other types of lesions without microcalcifications. Thus, viewed from the perspective of diagnostic accuracy (better PPV, NPV and OA) as well as robustness (lower chance of overfitting), use of the single-step SVM Raman algorithm appears more promising. Nevertheless, the two-step algorithm has certain intrinsic advantages in terms of interpretability, as it decomposes the overall classification into two independent steps to determine microcalcification status and diagnose the underlying lesion. Furthermore, in specific cases, only one step of the decomposition (for example, microcalcification status) may be of interest to the radiologist.
Concomitant diagnosis of microcalcifications and underlying breast lesions
Finally, to further validate the capability of the single-step SVM algorithm to concomitantly diagnose both microcalcification status and the underlying lesion, performance metrics were also calculated for the diagnosis of breast cancer with microcalcifications. In this case, we considered the positive class to be cancer with microcalcifications. All other classes, including cancer without microcalcifications, were considered to be negative. In this case, our single-step SVM algorithm exhibits a SE of 64.3%, SP of 100%, PPV of 100% and NPV of 96.4%, a slight improvement in SE and NPV over those for the diagnosis of breast cancer with or without microcalcifications (62.5% and 95.6%, respectively). An ROC curve illustrating the performance of this single-step Raman algorithm is shown in Fig. 3. The AUC for the diagnosis of breast cancer with microcalcifications was 0.92, indicating the robustness of the algorithm.
Figure 3.
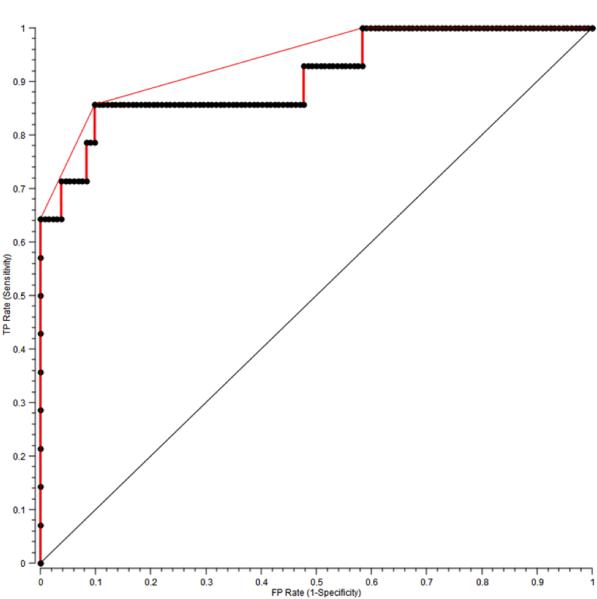
ROC curve for the single-step SVM Raman decision algorithm for the diagnosis of breast cancer with microcalcifications. The x- and y-axis represent the false positive (FP) rate and the true positive (TP) rate, respectively. The ROC curve of two indistinguishable populations, represented by the dashed line, is included for comparison. The area under the curve (AUC) is 0.92, the AUC for a perfect algorithm is 1.
DISCUSSION
Our research focuses on development of Raman spectroscopy as a clinical tool for the real-time diagnosis of breast cancer, motivated by its exquisite chemical specificity especially for detection of microcalcifications. Here we report on the development of a novel single-step Raman spectroscopy algorithm to simultaneously determine microcalcification status and diagnose the underlying breast lesion, in real time, at stereotactic breast needle core biopsy. Our SVM-derived algorithm yielded a PPV and NPV of 100% and 95%, respectively, for the diagnosis of breast cancer, with or without microcalcifications. The single-step algorithm had an OA of 82% for the specific diagnosis of microcalcification status and the underlying breast lesion. Significantly, the algorithm is able to classify DCIS cases, the most common type of breast cancer associated with microcalcifications, which could not be done with our previous algorithms (15, 17). This is a vital step in the development of Raman spectroscopy as a viable biopsy guidance tool. Specifically, we see Raman spectroscopy as a clinically viable adjunct to stereotactic breast needle biopsy procedures. Real time analysis of the Raman spectra using our single-step algorithm would reveal whether or not the tissue harbors the targeted microcalcifications and diagnose any breast lesions present, helping the radiologist to decide how many cores to take for submission for pathology examination. This should improve the likelihood of an adequate, diagnostic biopsy that contains the targeted microcalcifications and reduce the need for repeat, surgical biopsy.
The findings in this study suggest that the new single-step SVM Raman algorithm is more robust and accurate than two-step Raman algorithms utilizing previously developed or newly constructed SVM algorithms. In fact, due to the potential error propagation in a multistage algorithm, it is expected that a single-step algorithm simultaneously considering all the variables will do a superior job. An analogy can be drawn to the relatively poor performance of multistage decision tree algorithms (where the dataset is repetitively split based on the criterion that maximizes the separation of the tissue type) in relation to support vector machines in classifying large and complex datasets (29, 31-33, 36, 37).
Further, these preliminary results are not representative of the best classification performance that is likely to be obtainable after further optimization of the probe hardware (optical excitation and collection) and algorithm selection procedures. Specifically, we are designing and fabricating customized fiber probes that integrate non-imaging optical elements and bifocal lenses to enhance the efficiency of collection of Raman photons. In terms of algorithm performance enhancement, we anticipate that hybrid multivariate classification schemes that tailor to individual categories of lesion types and microcalcification status will be developed with the incorporation of larger clinical datasets. More extensive clinical studies will also enable us to expand our diagnostic algorithm to encompass breast lesions (such as epithelial hyperplasia, sclerosing adenosis and Monckeberg’s arteriosclerosis), which were not observed in the current dataset. Furthermore, such datasets would enable us to perform cross-validation by leaving out a larger number of tissue sites (e.g. 25% of the total dataset), or alternately by leaving out the sites corresponding to a single patient. The final milestone in algorithm validation would be in prospective application of the developed algorithm in patients undergoing stereotactic breast needle biopsy.
ACKNOWLEDGEMENTS
The authors thank the entire Breast Center, radiology and pathology staff at the University Hospitals Case Medical Center for their assistance in the research. In addition, the authors would like to acknowledge the women who participated in the study.
GRANT SUPPORT This research was supported by the National Institute of Biomedical Imaging and Bioengineering (9P41EB015871-26A1) and the National Cancer Institute (R01-CA140288).
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interests were disclosed.
REFERENCES
- 1.American Cancer Society . Breast Cancer Facts & Figures 2011-2012. American Cancer Society, Inc.; Atlanta: [Google Scholar]
- 2.Rim A, Chellman-Jeffers M. Trends in breast cancer screening and diagnosis. Clev Clin J Med. 2008;75:S2–9. doi: 10.3949/ccjm.75.suppl_1.s2. [DOI] [PubMed] [Google Scholar]
- 3.Johnson JM, Dalton RR, Wester SM, Landercasper J, Lambert PJ. Histological correlation of microcalcifications in breast biopsy specimens. Arch Surg. 1999;134:712–715. doi: 10.1001/archsurg.134.7.712. [DOI] [PubMed] [Google Scholar]
- 4.Markopoulos C, Kouskos E, Koufopoulos K, Kyriakou V, Gogas J. Use of artificial neural networks (computer analysis) in the diagnosis of microcalcifications on mammography. Eur J Radiol. 2001;39:60–5. doi: 10.1016/s0720-048x(00)00281-3. [DOI] [PubMed] [Google Scholar]
- 5.Betal D, Roberts N, Whitehouse GH. Segmentation and numerical analysis of microcalcifications on mammograms using mathematical morphology. Br J Radiol. 1997;70:903–17. doi: 10.1259/bjr.70.837.9486066. [DOI] [PubMed] [Google Scholar]
- 6.Shen L, Rangayyan RM, Desautels JEL. Application of shape-analysis to mammographic calcifications. IEEE T Med Imaging. 1994;13:263–74. doi: 10.1109/42.293919. [DOI] [PubMed] [Google Scholar]
- 7.Radi MJ. Calcium oxalate crystals in breast biopsies. An overlooked form of microcalcification associated with benign breast disease. Arch Pathol Lab Med. 1989;113:1367–9. [PubMed] [Google Scholar]
- 8.Jackman RJ, Rodriguez-Soto J. Breast microcalcifications: retrieval failure at prone stereotactic core and vacuum breast biopsy--frequency, causes, and outcome. Radiology. 2006;239:61–70. doi: 10.1148/radiol.2383041953. [DOI] [PubMed] [Google Scholar]
- 9.Haka AS, Fitzmaurice M. Raman spectroscopy diagnosis of breast cancer and atherosclerosis: a primer. In: Tunnel JW, editor. Biophotonics: in vivo clinical imaging and diagnosis. McGraw-Hill; New York: 2011. pp. 317–50. [Google Scholar]
- 10.Raman CV, Krishnan KS. A new type of secondary radiation. Nature. 1928;121:501–2. [Google Scholar]
- 11.Manoharan R, Shafer K, Perelman L, Wu J, Chen K, Deinum G, et al. Raman spectroscopy and fluorescence photon migration for breast cancer diagnosis and imaging. Photochem Photobio. 1998;67:15–22. [PubMed] [Google Scholar]
- 12.Haka AS, Shafer-Peltier KE, Fitzmaurice M, Crowe J, Dasari RR, Feld MS. Identifying microcalcifications in benign and malignant breast lesions by probing differences in their chemical composition using Raman spectroscopy. Cancer Res. 2002;62:5375–80. [PubMed] [Google Scholar]
- 13.Shafer-Peltier KE, Haka AS, Fitzmaurice M, Crowe J, Myles J, Dasari RR, et al. Raman microspectroscopic model of human breast tissue: implications for breast cancer diagnosis in vivo. J Raman Spectrosc. 2002;33:552–63. [Google Scholar]
- 14.Shafer-Peltier KE, Haka AS, Motz JT, Fitzmaurice M, Dasari RR, Feld MS. Model-based biological Raman spectral imaging. J Cell Biochem Suppl. 2002;87:125–37. doi: 10.1002/jcb.10418. [DOI] [PubMed] [Google Scholar]
- 15.Haka AS, Shafer-Peltier KE, Fitzmaurice M, Crowe J, Dasari RR, Feld MS. Diagnosing breast cancer by using Raman spectroscopy. Proc Nat Acad Sci. 2005;102:12371–6. doi: 10.1073/pnas.0501390102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haka AS, Volynskaya Z, Gardecki JA, Nazemi J, Lyons J, Hicks D, et al. In vivo margin assessment during partial mastectomy breast surgery using Raman spectroscopy. Cancer Res. 2006;66:3317–22. doi: 10.1158/0008-5472.CAN-05-2815. [DOI] [PubMed] [Google Scholar]
- 17.Haka AS, Volynskaya Z, Gardecki JA, Nazemi J, Shenk R, Wang N, et al. Diagnosing breast cancer using Raman spectroscopy: prospective analysis. J Biomed Opt. 2009;14:054023. doi: 10.1117/1.3247154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saha A, Barman I, Dingari NC, McGee S, Volynskaya Z, Galindo LH, et al. Raman spectroscopy: a real-time tool for identifying microcalcifications during stereotactic breast core needle biopsies. Biomed Opt Exp. 2011;2:2792–803. doi: 10.1364/BOE.2.002792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bitar RA, Martinho Hda S, Tierra-Criollo CJ, Zambelli Ramalho LN, Netto MM, Martin AA. Biochemical analysis of human breast tissues using Fourier-transform Raman spectroscopy. J Biomed Opt. 2006;11:054001. doi: 10.1117/1.2363362. [DOI] [PubMed] [Google Scholar]
- 20.Chowdary MV, Kumar KK, Kurien J, Mathew S, Krishna CM. Discrimination of normal, benign, and malignant breast tissues by Raman spectroscopy. Biopolymers. 2006;83:556–69. doi: 10.1002/bip.20586. [DOI] [PubMed] [Google Scholar]
- 21.Baker R, Matousek P, Ronayne KL, Parker AW, Rogers K, Stone N. Depth profiling of calcifications in breast tissue using picosecond Kerr-gated Raman spectroscopy. The Analyst. 2007;132:48–53. doi: 10.1039/b614388a. [DOI] [PubMed] [Google Scholar]
- 22.Matousek P, Stone N. Prospects for the diagnosis of breast cancer by noninvasive probing of calcifications using transmission Raman spectroscopy. J Biomed Opt. 2007;12:024008. doi: 10.1117/1.2718934. [DOI] [PubMed] [Google Scholar]
- 23.Stone N, Baker R, Rogers K, Parker AW, Matousek P. Subsurface probing of calcifications with spatially offset Raman spectroscopy (SORS): future possibilities for the diagnosis of breast cancer. Analyst. 2007;132:899–905. doi: 10.1039/b705029a. [DOI] [PubMed] [Google Scholar]
- 24.Krishna CM, Kurien J, Mathew S, Rao L, Maheedhar K, Kumar KK, et al. Raman spectroscopy of breast tissues. Expert Rev Mol Diagn. 2008;8:149–66. doi: 10.1586/14737159.8.2.149. [DOI] [PubMed] [Google Scholar]
- 25.Majumder SK, Keller MD, Boulos FI, Kelley MC, Mahadevan-Jansen A. Comparison of autofluorescence, diffuse reflectance, and Raman spectroscopy for breast tissue discrimination. J Biomed Opt. 2008;13:054009. doi: 10.1117/1.2975962. [DOI] [PubMed] [Google Scholar]
- 26.Chowdary MV, Kalyan Kumar K, Mathew S, Rao L, Krishna CM, Kurien J. Biochemical correlation of Raman spectra of normal, benign and malignant breast tissues: a spectral deconvolution study. Biopolymers. 2009;91:539–46. doi: 10.1002/bip.21171. [DOI] [PubMed] [Google Scholar]
- 27.Abramczyk H, Brozek-Pluska B, Surmacki J, Jablonska-Gajewicz J, Kordek R. Raman ‘optical biopsy’ of human breast cancer. Prog Biophys Mol Biol. 2012;108:74–81. doi: 10.1016/j.pbiomolbio.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Keller MD, Vargis E, de Matos Granja N, Wilson RH, Mycek MA, Kelley MC, et al. Development of a spatially offset Raman spectroscopy probe for breast tumor surgical margin evaluation. J Biomed Opt. 2011;16:077006. doi: 10.1117/1.3600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dingari NC, Barman I, Saha A, McGee S, Galindo LH, Liu W, et al. Development and comparative assessment of Raman spectroscopic classification algorithms for lesion discrimination in stereotactic breast biopsies with microcalcifications. J Biophotonics. 2012 doi: 10.1002/jbio.201200098. DOI: 10.1002/jbio.201200098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barman I, Dingari NC, Rajaram N, Tunnell JW, Dasari RR, Feld MS. Rapid and accurate determination of tissue optical properties using least-squares support vector machines. Biomed Opt Exp. 2011;2:592–9. doi: 10.1364/BOE.2.000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cortez C, Vapnik V. Support-Vector Networks. Machine Learning. In: Vapnik VN, editor. The Nature of Statistical Learning Theory. Springer-Verlag; Berlin: 1995. pp. 273–97. [Google Scholar]
- 32.Dingari NC, Barman I, Myakalwar AK, Tewari SP, Gundawar MK. Incorporation of support vector machines in the LIBS toolbox for sensitive and robust classification amidst unexpected sample and system variability. Anal Chem. 2012;84:2686–94. doi: 10.1021/ac202755e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brereton RG, Lloyd GR. Support vector machines for classification and regression. Analyst. 2010;135:230–67. doi: 10.1039/b918972f. [DOI] [PubMed] [Google Scholar]
- 34.Demsar J, Zupan B, Leban G, Curk T. Orange: from experimental machine learning to interactive data mining. European Conference of Machine Learning; Pisa, Italy. Springer Verlag; 2004. pp. 537–9. [Google Scholar]
- 35.Fitzmaurice M. Principles and pitfalls of diagnostic test development: implications for spectroscopic tissue diagnosis. J Biomed Opt. 2000;5:119–30. doi: 10.1117/1.429978. [DOI] [PubMed] [Google Scholar]
- 36.Foody GM, Mathur A. A relative evaluation of multiclass image classification by support vector machines. IEEE Trans Geosci Remote Sensing. 2004;42:1335–43. [Google Scholar]
- 37.He J, Hu HJ, Harrison R, Tai PC, Pan Y. Rule generation for protein secondary structure prediction with support vector machines and decision tree. IEEE Trans Nanobiosci. 2006;5:46–53. doi: 10.1109/tnb.2005.864021. [DOI] [PubMed] [Google Scholar]