Abstract
To explore the relationship of gut microbiota with the development of type 2 diabetes (T2DM), we analyzed 121 subjects who were divided into 3 groups based on their glucose intolerance status: normal glucose tolerance (NGT; n = 44), prediabetes (Pre-DM; n = 64), or newly diagnosed T2DM (n = 13). Gut microbiota characterizations were determined with 16S rDNA-based high-throughput sequencing. T2DM-related dysbiosis was observed, including the separation of microbial communities and a change of alpha diversity between the different glucose intolerance statuses. To assess the correlation between metabolic parameters and microbiota diversity, clinical characteristics were also measured and a significant association between metabolic parameters (FPG, CRP) and gut microbiota was found. In addition, a total of 28 operational taxonomic units (OTUs) were found to be related to T2DM status by the Kruskal-Wallis H test, most of which were enriched in the T2DM group. Butyrate-producing bacteria (e.g. Akkermansia muciniphila ATCCBAA-835, and Faecalibacterium prausnitzii L2-6) had a higher abundance in the NGT group than in the pre-DM group. At genus level, the abundance of Bacteroides in the T2DM group was only half that of the NGT and Pre-DM groups. Previously reported T2DM-related markers were also compared with the data in this study, and some inconsistencies were noted. We found that Verrucomicrobiae may be a potential marker of T2DM as it had a significantly lower abundance in both the pre-DM and T2DM groups. In conclusion, this research provides further evidence of the structural modulation of gut microbiota in the pathogenesis of diabetes.
Introduction
Type 2 diabetes mellitus (T2DM) has become one of the fastest growing public health problems in both developed and developing countries. In China, for instance, the prevalence of diabetes and prediabetes have reached 9.7% and 15.5%, respectively, and account for 92.4 million adults and 148.2 million adults, respectively, according to the National Diabetes and Metabolic Disorders Study conducted from 2007 to 2008 [1]. In many countries, type 2 diabetes mellitus has now become the most prevalent type of diabetes in children [2]. Prediabetes is even more prevalent than type 2 diabetes and there is an important demarcation line between the two conditions that indicates whether an individual is going to develop diabetes and cardiovascular disease [1], [3]. Therefore, a more comprehensive understanding of the dynamic development processes of glucose intolerance is fundamentally important for understanding the mechanisms responsible for the development of T2DM and thus strategies to prevent it.
T2DM is thought to involve a complex process involving genetic susceptibility and environmental factors, both of which remain only partially understood. Genome-wide association strategies have offered great opportunities for investigation of the pathogenesis of the disease. However, common genetic variants that are associated with diabetes have only very slightly improved the prediction of future T2DM [4], [5]. Growing evidence that multiple environmental factors contribute to their development was exemplified by emerging evidence of the role of the intestinal microbiota as a potential novel contributor to this epidemic. The metagenomic sequencing of the human microbiome has revealed that there are 3.3 million non-redundant genes, with over 99% of the genes being of bacterial origin [6]. This gene set contains at least100-fold more genes than the complete human genome. With >50 species shared by 90% of the individuals studied, considerable variation occurs in both the types of microbes and in the diversity of microbial functional genes between individuals [6]–[8].
Recent research has shown that gut bacteria play an important role in disorders such as obesity, diabetes, and cardiovascular diseases. Compositional changes of human gut microbiota in response to weight status have been examined in many studies. Gordon and colleagues have reported that human obesity is associated with a low abundance of intestinal Bacteroidetes and a high abundance of Firmicutes, in agreement with results from a weight-loss program involving consumption of restricted diets for a year [9]–[11]. However, Duncan et al. [12] and Jumpertz et al. [13] found no difference in the proportions of fecal microbiota between lean and obese subjects. Relatively few studies have reported the relationship between the gut microbiota composition and T2DM. Larsen et al. [14] showed that men with T2DM had significantly reduced levels of fecal Firmicutes, including Clostridia, compared with non-diabetic control subjects, and that the ratio of Bacteroidetes to Firmicutes, as well as the ratio of Bacteroides-Prevotella to C. coccoides-E. rectale, was correlated positively with the plasma glucose concentration but not with body mass index (BMI). In order to characterize the gut microbiota in diabetic patients, Wu et al. [15] analyzed the diversity and similarity of gut microbiota in 16 T2DM patients and 12 healthy individuals and found that genus Bifidobacterium and Bacteroides vulgatus were less represented in the microbiota of the diabetic group than the non-diabetic group. In our recent metagenome-wide association study of Chinese T2DM patients, we found that patients withT2DM were characterized by a moderate degree of gut microbial dysbiosis, a decrease in the abundance of some universal butyrate-producing bacteria, and an increase in various opportunistic pathogens [16].
Recent population-based studies conducted to evaluate the association between gut microbiota and diabetes have produced conflicting results. Were they limited by the relatively low number of subjects, the diversity of patient enrollment, technical issues, or by racial differences? Probably, bacterial sequences specific for T2DM act as signatures of glucose tolerance status rather than obesity. Moreover, a comprehensive understanding of the microbiota characteristics associated with the natural course of T2DM also remains to be achieved.
In this study, we hypothesized that the proportion and diversity of intestinal microbiota in humans are changed before the period of prediabetes. The aims of the study were to explore the relationships between gut microbiota and different glucose intolerance statuses in a Chinese Han population using 16S rDNA-based high-throughput sequencing.
Results
Clinical characteristics
The anthropometric and metabolic characteristics of the 3 study groups are shown in Table 1. Body mass index (BMI), waist-hip ratio (WHR), fasting plasma glucose (FPG), plasma glucose 2 hours after oral glucose challenge (2HPG), triglyceride (TG), fasting insulin (FINS) concentration, 2-hour insulin (2HINS) concentration, insulin resistance index (IR), and C-reactive protein (CRP) values were all significantly higher in the T2DM group than in the NGT group (P<0.05). In addition, BMI, WHR, FPG, 2HPG, FINS, 2HINS, IR, and CRP values were also higher in the Pre-DM group than in the NGT group (P<0.05). However, there were no significant differences in LDL-C and HDL-C levels between the 3 groups.
Table 1. Characteristics of the subjects with NGT, prediabetes and T2DM.
| Variable | NGT(n = 44) | Pre-DM(n = 64) | T2DM(n = 13) |
| Age (years) | 55 (9) | 54 (7) | 52 (9) |
| BMI (kg/m2) | 23.38 (8.62) | 24.95 (7.18)* | 26.50 (8.61)† |
| Waist-hip ratio (WHR) | 0.85 (3.52) | 0.88 (2.96)* | 0.90 (3.47)† |
| SBP (mmHg) | 122 (15) | 126 (13)* | 129 (11) |
| DBP (mmHg) | 77 (8) | 81 (9)* | 86 (9)† |
| FPG (mmol/L)a | 5.28 (4.91, 5.51) | 5.74 (5.28, 6.29)* | 7.36 (7.09, 8.21)† , ‡ |
| 2HPG (mmol/L)a | 6.81 (6.19, 7.45) | 9.58 (8.61, 10.92)* | 13.52 (12.41, 14.19)† , ‡ |
| FINS (mIU/L)a | 6.6 (4.84, 9.21) | 9.83 (7.1, 13.86)* | 19.15 (11.22, 25.32)† , ‡ |
| 2HINS (mIU/L)a | 35.87 (26.93, 57.03) | 76.2 (53.77, 127.9)* | 80.27 (47.5, 114.1)† |
| IRa | 1.53 (1.08, 2.39) | 2.63 (1.73, 3.67)* | 6.29 (3.69, 8.04)† , ‡ |
| TG (mmol/L)a | 1.35 (0.9, 1.69) | 1.41 (1.04, 1.74) | 1.84 (1.6,0 2.58)† , ‡ |
| TC (mmol/La | 5.0 (4.25, 5.44) | 4.88 (4.47, 5.54) | 4.89 (4.67, 5.47) |
| HDL-C (mmol/L)a | 1.29 (1.02, 1.53) | 1.19 (1.09, 1.32) | 1.12 (0.94, 1.2) |
| LDL-C (mmol/L)a | 2.95 (2.5, 3.54) | 3.10 (2.77, 3.74) | 3.0 (2.69, 3.13) |
| CRP (mg/L)a | 1.02 (0.44, 2.07) | 1.58 (1.03, 2.51)* | 2.12 (1.38, 3.27)† |
Median (interquartile range).
P<0.05 for Pre-DM vs NGT;
P<0.05 for T2DM vs NGT;
P<0.05 for T2DM vs Pre-DM.
BMI = body mass index; CPR = C-reactive protein; DBP = diastolic blood pressure; FINS = fasting insulin; FPG = fasting plasma glucose; HDL-C = high-density lipoprotein cholesterol; 2HINS = 2-hour insulin concentration; 2HPG = 2-hour plasma glucose concentration; IR = insulin resistance index; LDL-C = low-density lipoprotein cholesterol; NGT = normal glucose tolerance; Pre-DM = prediabetes; SBP = systolic blood pressure; TC = total cholesterol; T2DM = type 2 diabetes mellitus; TG = triglyceride.
Characteristics of gut microbiota at different glucose intolerance levels
A total of 2.2 million sequence reads were generated from the 16S rDNA gene V3–V5 amplicons, with an average of 9474 (±3470 SD) reads per subject. To obtain a detailed structural overview of the microbiome of each subject enrolled in the study, operational taxonomic unit (OTU) analysis was conducted and yielded a total of 8,107 OTUs with a 97% similarity cutoff. Firmicutes and Bacteroidetes were found to be the dominant taxa of the overall structures of the microbiome at phylum level, as were Clostridia and Bacteroidia at class level. However, Clostridia had a higher abundance in the T2DM group compared with the NGT and Pre-DM groups. At genus level, a total of 159 genera were identified but only 31 genera had an abundance more than 0.1% in at least 1 of the 3 groups. The relative abundance of Bacteroides in the T2DM group (∼10%) was lower than that in the NGT and Pre-DM groups, while Dorea, Prevotella and Collinsella had higher relative abundances in the T2DM group than in the NGT group.
Interestingly, the abundance of Streptococcus continued to decrease from the NGT group to the Pre-DM group, and to the T2DM group. Additionally, the average relative abundances of genus Prevotella and genus Megamonas in the Pre-DM group were 1.89-fold and 8.83-fold greater, respectively, than in the NGT group. All of these results indicate that dysbiosis of the gut microbial structure had occurred. To fully explore the relationships between microbes and metabolic disorders, a profile at genus level was generated to perform future analyses, including reads that can only be assigned to a family level. The 20 most abundant clades were then determined (Figure S1).
Analysis of the similarity and diversity of microbial communities among different glucose tolerance statuses
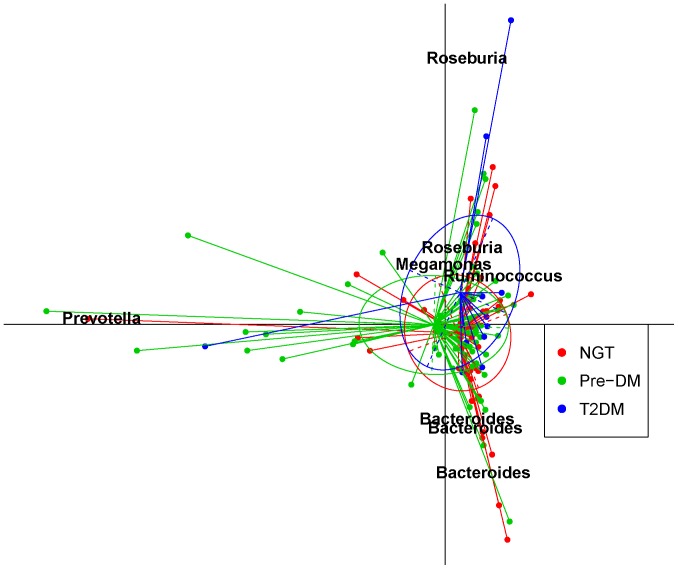
Principal component analysis (PCA) was employed to assess the similarity of microbial communities between the 3 groups. PCA results based on 8,107 OTUs showed a slight separation between the NGT and T2DM groups (Figure 1), which was also observed by PCA based on the taxonomic profile (Figure S2). However, the PERMANOVA test, which was used to assess changes in the overall structure, indicated no notable separation among the 3 groups (P = 0.2626) (Table S1).
Figure 1. Principal component analysis (PCA) analysis of the similarity of microbiota (OTUs) between the NGT, Pre-DM, and T2DM groups.
Data for NGT (n = 44), Pre-DM (n = 64) and T2DM (n = 13) subjects were plotted on the first two principal components of the OTU profiles. The first 2 components (contributing 19.5% of variance) were plotted. Lines connect individuals belonging to the same group and colored circles cover individuals near the center of gravity for each cluster (<1.5σ). The top 7 OTUs (labeled with their annotations) for the main contributors to these groups were determined and plotted by their loadings for these 2 components. PCA was performed with R package ‘ade4’. OTU = operational taxonomic unit.
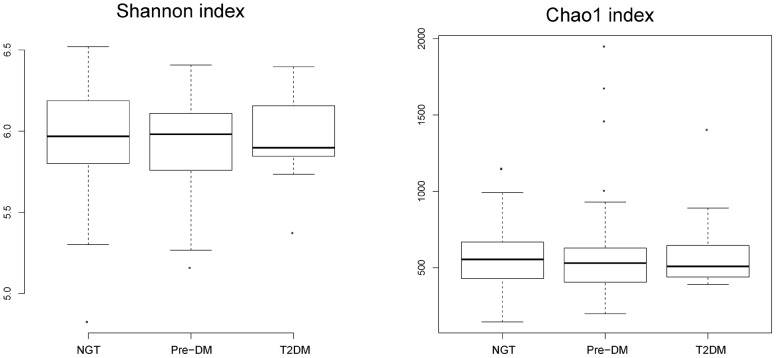
The Chao1 index and Shannon index were calculated to estimate the alpha diversity. No significant differences were found between the 3 groups, but T2DM subjects showed a slightly decreased diversity compared with the NGT group (Figure 2). Using the method documented in the Meta-HIT study [17], we identified 3 enterotypes that were mainly enriched in Bacteroides, Prevotella and Ruminococcus, respectively. Interestingly, this was not associated with T2DM status, and the Fisher exact test was not significant (P = 0.355; Figure S3).
Figure 2. Alpha diversity of the 3 groups.
The Shannon index (left panel) and Chao1 index (right panel) were computed for all 121 subjects. The box depicts the interquartile range (IQR) between the first and third quartiles (25th and 75th percentiles, respectively) and the line inside the box denotes the median.
OTUs differentially enriched in the NGT and T2DM groups
When we examined a total of 1,640 OTUs that occurred in more than 6 samples, 28 OTUs were found to be associated with a different glucose intolerance status by the Kruskal-Wallis H test (Table 2 and Figure S4; first error rate 1%). Table 2 shows the complete list of OTUs that exhibited different abundances between the 3 groups. OTU4416 (Subdoligranulum) had a relatively higher abundance in the T2DM group than in the NGT and Pre-DM groups, and the average abundance of OTU286 (Clostridiales) in T2DM subjects was twice that in NGT subjects. Seven OTUs annotated to Lachnospiraceae were enriched in the T2DM group compared with the NGT group, and all 3 OTUs annotated to Ruminococcus were enriched in the T2DM group compared with the NGT group. Additionally, OTU5579, OTU2980, OTU7046 and OTU6565 annotated to Eubacterium, Sporobacter, Abiotrophia and Peptostreptococcus, respectively, were enriched significantly in the T2DM group compared with the NGT group, especially OTU7046, the mean abundance of which was 20 times that in the NGT group.
Table 2. Kruskal-Wallis H tests of associations between glucose intolerance status and microbiota composition (OTUs level).
| OTU | P | Occurrence rate | Taxon | Identity | ||
| NGT | Pre-DM | T2DM | ||||
| NGT enrichment: | ||||||
| 6275 | 0.0081626 | 0.18 | 0.02 | 0.08 | Bacteroides | 1 |
| 2101 | 0.0064251 | 0.77 | 0.56 | 0.38 | Blautia | 0.98 |
| 2340 | 0.0087904 | 0.07 | 0.25 | 0.00 | Clostridiales | 1 |
| 3265 | 0.0054547 | 0.27 | 0.52 | 0.08 | Haemophilus | 0.86 |
| 4375 | 0.001644 | 0.32 | 0.59 | 0.23 | Lachnospiraceae | 0.99 |
| 1632 | 0.009626 | 0.11 | 0.30 | 0.00 | Megamonas | 1 |
| 3209a | 0.0024805 | 0.16 | 0.39 | 0.00 | Megamonas | 1 |
| 1900 | 0.003299 | 0.50 | 0.22 | 0.46 | Roseburia | 1 |
| T2DM enrichment: | ||||||
| 7678 | 0.0037128 | 0.05 | 0.31 | 0.23 | Bacteroides | 1 |
| 770 | 0.0057936 | 0.02 | 0.23 | 0.08 | Bacteroides | 1 |
| 6679 | 0.0047138 | 0.84 | 0.63 | 1.00 | Blautia | 1 |
| 286 | 0.0020471 | 0.14 | 0.05 | 0.38 | Clostridiales | 1 |
| 5579 | 0.0071113 | 0.32 | 0.13 | 0.46 | Eubacterium | 0.96 |
| 396 | 0.0010595 | 0.11 | 0.42 | 0.15 | Lachnospiraceae | 0.81 |
| 2508 | 0.0009765 | 0.36 | 0.70 | 0.54 | Lachnospiraceae | 0.84 |
| 3325 | 0.0067516 | 0.30 | 0.56 | 0.62 | Lachnospiraceae | 0.87 |
| 6803 | 0.0017342 | 0.55 | 0.83 | 0.69 | Lachnospiraceae | 0.87 |
| 6718b | 0.0031246 | 0.39 | 0.56 | 0.85 | Lachnospiraceae | 0.95 |
| 4235 | 0.0098852 | 0.20 | 0.50 | 0.38 | Lachnospiraceae | 0.96 |
| 3249 | 0.00129 | 0.07 | 0.06 | 0.38 | Lachnospiraceae | 1 |
| 6565 | 0.0062553 | 0.07 | 0.09 | 0.38 | Peptostreptococcus | 1 |
| 7853 | 0.0005223 | 0.07 | 0.05 | 0.38 | Ruminococcaceae | 0.97 |
| 469 | 0.0022403 | 0.14 | 0.13 | 0.54 | Ruminococcus | 0.97 |
| 4393 | 0.004243 | 0.18 | 0.14 | 0.54 | Ruminococcus | 1 |
| 4508b | 0.0028564 | 0.39 | 0.48 | 0.85 | Ruminococcus | 1 |
| 2980 | 0.0039087 | 0.05 | 0.11 | 0.38 | Sporobacter | 0.92 |
| 4416 | 0.0045742 | 0.50 | 0.39 | 0.85 | Subdoligranulum | 0.97 |
| 7046 | 0.0024899 | 0.02 | 0.06 | 0.31 | Abiotrophia | 0.97 |
The relative abundance of OTUs enriched in the NGT group decreased to 0 with the development of T2DM.
The relative abundance of OTUs increased with the development of T2DM.
NGT = normal glucose tolerance; OTU = operational taxonomic unit; Pre-DM = prediabetes; T2DM = type 2 diabetes mellitus.
Haemophilus (OTU3265) and Megamonas (OTU1632 and OTU3209) showed high relative abundances in the NGT group compared with the T2DM group. Moreover, Roseburia (OTU1900) showed a notably high relative abundance in NGT subjects compared with the other groups.
Association between microbiota diversity and clinical indicators
PERMANOVA tests of associations between microbiota composition and clinical parameters
The PERMANOVA test was used to test the overall effect of gut microbiota on healthy parameters. The model was adjusted for BMI, age and sex as these might be confounding factors. The results showed that fasting plasma glucose (FPG) was significantly associated with the gut microbiota (P = 0.0453) in all samples. In the NGT group, the CRP concentration was significantly associated with microbiota (Table 3).
Table 3. PERMANOVA tests of associations between clinical parameters and microbiota composition (adjusted for age, gender and BMI).
| Phenotype | Df | Sum of squares | Mean squares | F model | R2 | Pr (>F) |
| All groups (95 samples): | ||||||
| FPG | 1 | 0.451807 | 0.451807 | 1.35295 | 0.01423 | 0.0453 |
| NGT group (33 samples): | ||||||
| CRP | 1 | 0.431633 | 0.431633 | 1.320166 | 0.040869 | 0.055 |
| T2DM group (12 samples): | ||||||
| FPG | 1 | 0.428187 | 0.428187 | 1.324532 | 0.113103 | 0.0605 |
BMI = body mass index; CRP = C-reactive protein; FPG = fasting plasma glucose; PERMANOVA = permutational multivariate analysis of variance.
Co-inertia analysis (CIA) of microbiota at the OTU level and clinical parameters
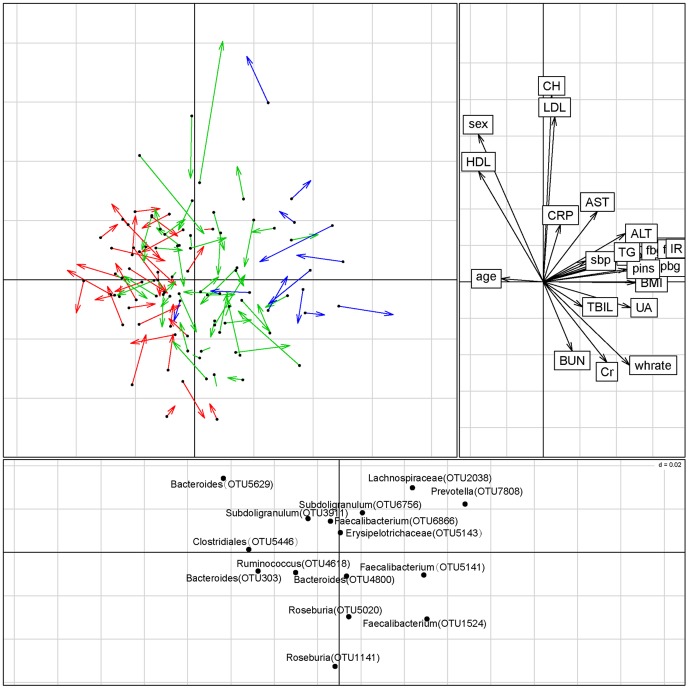
CIA of OTU abundance and clinical parameters revealed a modest relationship (RV coefficient = 0.237, permutation 1000 times, P value = 0.196) between the two datasets (Figure 3). The major genus dedicating to separation was consistent with the Kruskal-Wallis test result referred to above.
Figure 3. Co-inertia analysis (CIA) of the relationship between microbiota at the OTU level, clinical parameters, and disease group.
The upper left panel shows the CIA of the clinical parameter principal component analysis (PCA) and the microbiota PCA; arrows indicate where samples in the clinical parameter dataset are relative to the microbiota dataset. Red lines represent the NGT group, green lines the Pre-DM group, and blue lines the T2DM group. The upper right panel shows clinical parameter loading data; the lower panel displays the associated microbiota at the OTU level (labeled with their annotations). Only OTUs present in at least 1% of the samples were used in the analysis. CIA was performed with R package ‘ade4’. OTU = operational taxonomic unit.
The first two components of the CIA accounted for 47.3% of the variance in the datasets, with component 1 (horizontal) accounting for 31.1% of the variance, and component 2 (vertical) accounting for another 16.1%. NGT and T2DM samples were separated from each other along the primary axis in the analysis. Using a cut-off of 1% abundance, the main OTUs associated with the community metabolome along the first axis were those assigned to Clostridiales, Subdoligranulum, Lachnospiraceae, and Roseburia (Figure 3). Roseburia are known to produce butyrate, while Clostridiales, Subdoligranulum and Lachnospiraceae are opportunistic pathogens.
Correlations between clinical parameters and microbiota diversity
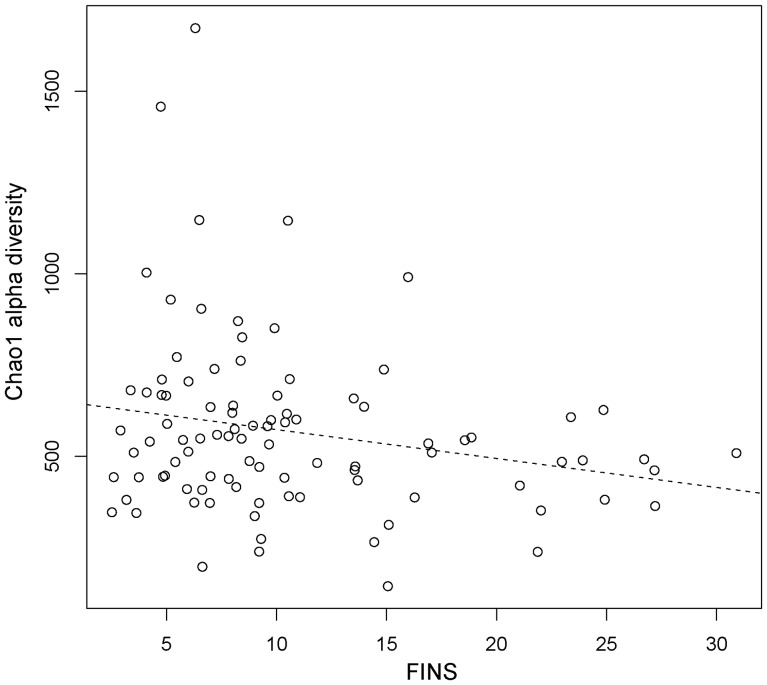
Stepwise multiple regression was conducted to test the relationship between clinical parameters and alpha diversity (Table S2). It was found that the FINS concentration was an independent relevant factor and was negatively correlated with the Chao1 index, which indicates that an insulin-resistant status has a relationship with the diversity of gut microbiota. Simple regression between the FINS and concentration and Chao1 diversity is shown in Figure 4.
Figure 4. Correlations between the fasting insulin (FINS) concentration (mIU/L) and microbiota diversity.
The dashed line was derived from a simple regression model (P-value for the model was 0.034; R = −0.22).
Validation and identification of T2DM markers
Some clades were identified as potential biomarkers for the diagnosis and management of T2DM by Kruskal-Wallis H tests based on changes in the relative abundance of each clade (Tables S3 and S4). In comparison with the NGT group, the relative abundance of Verrucomicrobia and Verrucomicrobiae were significantly decreased in the Pre-DM group. This indicates that Verrucomicrobiae can serve as a signal or a diagnostic biomarker for the progression of glucose intolerance, or it may even be a potentially beneficial microbe for the prevention of T2DM. Inversely, the relative abundance of Betaproteobacteria was significantly increased in the Pre-DM group in comparison with the NGT group, and was even higher in the T2DM group, which may be an indication of a high risk of T2DM. The abundance of Clostridia was also relatively higher in the T2DM group than in Pre-DM subjects, which suggests that it could be useful in the diagnosis of T2DM (Figure S5).
We also checked and compared the T2DM marker taxa reported by Larsen et al. [14] with our data (Tables S3 and S4 and Figure S6). Compared with the NGT group, the proportion of phylum Firmicutes and class Clostridia were a little more abundant in the diabetic group (P value = 0.07), but the ratio of Bacteroidetes to Firmicutes was not correlated with the plasma glucose concentration. Although these findings were not consistent with the study of Larsen et al. [14], similar results were observed in our study for class Betaproteobacteria which was highly enriched in diabetic subjects compared with NGT subjects.
To investigate the T2DM marker species found in the previous metagenomic study [16], we constructed a T2DM marker species profile by BLASTN each OTU's representative reads against 16s rDNA gene (filtered at 97% identity) of the T2DM marker species. The abundance of each species for each sample was calculated and normalized for the corresponding sample's total tags. A Kruskal-Wallis H test was performed between the NGT and T2DM groups based on this normalized profile (Table 4). At a first error of 5%, only 4 species were found to be significantly different among the 3 groups. Faecalibacterium prausnitzii L2-6 and Haemophilus parainfluenzae T3T1 were more abundant in the NGT group than in theT2DM group, which was consistent with the result of Qin et al. [16]. The former taxon is thought to be a butyrate-producing bacteria and healthy for humans. Akkermansia muciniphila ATCCBAA-835 also had a higher abundance in the NGT group than the Pre-DM group, while Clostridiales sp. SS3/4 was less abundant in the NGT group than the Pre-DM and T2DM groups. The results for these 2 species were inconsistent with the findings of Qin et al. [16]. Other markers were not significant, which may have been due to the small sample size.
Table 4. T2DM markers in the studya.
| Species | P-value | Relative abundance mean | Occurrence rate | ||||
| NGT | Pre-DM | T2DM | NGT | Pre-DM | T2DM | ||
| Akkermansiamuciniphila ATCCBAA-835 | 0.025 | 9.17E-05 | 1.16E-05 | 1.72E-05 | 0.227 | 0.063 | 0.077 |
| Bacteroides 20-3 | 0.142 | 0.003859 | 0.003822 | 0.001607 | 0.909 | 0.906 | 0.769 |
| Bacteroides intestinalis DSM17393 | 0.434 | 0.008022 | 0.003493 | 0.004447 | 0.841 | 0.766 | 0.769 |
| Clostridiales sp. SS3/4 | 0.007 | 0.000846 | 0.001873 | 0.001716 | 0.886 | 0.953 | 1.000 |
| Clostridium HGF2 | 0.271 | 0.000423 | 0.000115 | 0.000198 | 0.455 | 0.359 | 0.462 |
| Clostridium SS2-1 | 0.209 | 0.012028 | 0.011949 | 0.018385 | 0.977 | 0.984 | 1.000 |
| Clostridium bolteae str16351 | 0.590 | 0.001972 | 0.001052 | 0.001313 | 0.795 | 0.922 | 1.000 |
| Clostridium symbiosum WAL14673 | 0.547 | 0.00055 | 0.000904 | 0.000954 | 0.614 | 0.734 | 0.769 |
| Eggerthella lenta DSM2243 | 0.750 | 0.000678 | 0.00043 | 0.000421 | 0.591 | 0.516 | 0.538 |
| Escherichia coli ED1a | 0.315 | 0.004086 | 0.022505 | 0.007521 | 0.841 | 0.859 | 0.769 |
| Eubacterium eligens ATCC27750 | 0.813 | 0.006233 | 0.009685 | 0.004943 | 0.773 | 0.813 | 0.769 |
| Eubacterium rectale ATCC33656 | 0.915 | 0.021439 | 0.016453 | 0.032689 | 0.841 | 0.922 | 0.846 |
| Faecalibacterium prausnitzii L2-6 | 0.031 | 0.054632 | 0.074787 | 0.04992 | 0.977 | 1.000 | 1.000 |
| Haemophilus parainfluenzae T3T1 | 0.011 | 0.00018 | 0.000319 | 3.30E-05 | 0.364 | 0.531 | 0.077 |
| Roseburia intestinalis XB6B4 | 0.862 | 0.040707 | 0.044163 | 0.060625 | 0.909 | 1.000 | 1.000 |
| Roseburia inulinivorans DSM16841 | 0.942 | 0.007569 | 0.008261 | 0.005562 | 0.818 | 0.875 | 0.923 |
| Clostridium ramosum DSM1402 | 0.658 | 0.001806 | 0.000649 | 0.002932 | 0.523 | 0.516 | 0.308 |
These markers were identified in the study of Qin et al. [16].
The data in this table show the average relative abundance and occurrence rates in the three groups.
Discussion
Pre-DM is a key stage in the progression from normal glucose metabolism to diabetes. Our data have shown that the relative abundances of Bacteroides and Clostridium undergo marked undulations with the progression of glucose intolerance. The relative abundances of Verrucomicrobiae and Betaproteobacteria from NGT to Pre-DM and T2DM showed an opposite trend, while the relative abundance of Streptococcus continued to decrease from NGT to Pre-DM and then to T2DM. Thus, our results support the hypothesis that dysbiosis in gut microbiota occurs early before the prediabetes stage, which suggests that the proportion and diversity of microbiota can serve as potential makers for a high risk of diabetes. Therefore, retaining the balance of gut microbiota in the prediabetic stage may be a unique intervention to delay the development of diabetes.
It was, however, surprising to find an inconsistency with regard to the enrichment of Akkermansia muciniphila when we compared data derived from this study using 16S rDNA gene amplicon sequencing with that derived from metagenomics [16]. There may be many reasons for this. Firstly, different target genes were used to calculate the abundances. 16S rDNA gene is a ubiquitous and extraordinarily conservative biomarker in terms of the key role it plays in the translational apparatus, and rare horizontal gene transfer has been reported [18]. Not all other genes are suitable for taxonomic assignment and therefore for abundance determination, especially as only a few genes were detected for some species. This study was based on 16S rDNA amplicon sequencing as it is theoretically representative of the microbial community structure. A better method for microbial characterization in metagenomics may be the clade-specific marker gene identified by Segata et al. [19]. This indicates the need for a combination of biomarker genes (amplicon sequencing and qPCR) and metagenomic shotgun sequencing.
Secondly, many other confounding factors such as dietary or pharmaceutical treatments may also affect gut flora [20], [21]. In the present study, no subjects were taking antibiotics, hormones, or other drugs such as glucose- or lipid-lowering agents so they could not significantly change the gut microbiota composition. Dietary factors are complex, and we did not quantitate each subject's dietary composition as is usually done in animal models. It has frequently been reported that the fat and fiber content of diets can affect gut microbiota composition [22]–[24]. As no dietary information was provided in the study of Qin et al. [16], a further study is needed to explore this.
As a mucin-degrading bacterium, Akkermansia muciniphila has been widely identified in the intestinal tract and considerable efforts have been made to explore its relationship with human health [25]. Recently, more and more reports have pointed out that this species plays a key role in maintaining the integrity of the mucin layer and reducing inflammation, and it also protects against obesity and T2DM via the same mechanism [26]. The strain Akkermansia muciniphila ATCCBAA-835 used by Everard et al. [26] was also identified in the present study and a higher abundance of this organism was observed in the NGT group. In addition, a negative correlation between A. muciniphila and body weight, fat mass and metabolic disorders associated with obesity has been observed [26]–[29]. Taken together, these findings and the decrease of Verrucomicrobiae in pre-DM indicate that the abundance of Verrucomicrobiae is meaningful for the diagnosis and risk estimation of T2DM. A similar conclusion was also reached by Liou et al. [30].
Interestingly, a higher abundance of genus Collinsella in T2DM compared with NGT and pre-DM was detected, and it has been reported that this genus shows a positive correlation with serum cholesterol, and is also enriched in patients with symptomatic atherosclerosis [31], [32]. This may indicate an association between Collinsella, T2DM and atherosclerosis.
Our data showed that among the 28 OTUs enriched in the NGT and T2DM groups, OTUs identified as Haemophilus, Roseburia and Megamonas were enriched in the NGT group, while OTUs identified as Lachnospiraceae, Clostridiales, Ruminococcus, Eubacterium, Sporobacter, Abiotrophia, Peptostreptococcus and Subdoligranulum had relatively high abundances in the T2DM group. It has been reported that almost all species of Roseburia are buryrate-producing bacteria and some species of Megamonas can ferment glucose into acetate and propionate [33], [34], and that all of these short-chain fatty acids (SCFAs) are beneficial for health. These findings indicate that T2DM is associated with compositional changes in the gut microbiota, especially a decrease of probiotics. Some of our findings are consistent with study of Larsen et al. [14] in 36 male adults, 18 of whom had T2DM. This study also found and validated the T2DM-associated microbiota markers that were identified in previous metagenome-wide study [16]. Some of these were opportunistic pathogens such as Clostridiales, Subdoligranulum and Lachnospiraceae. Roseburia which are known to be butyrate-producing bacteria, seem to have a protective role, as has been recognized in previous reports [35], [36]. The finding that Roseburia (OTU1900) had a notably high relative abundance in NGT subjects compared with the other study groups was in agreement with previously reported findings.
Growing evidence from animal studies has suggested that changes in gut microbiota composition in response to a high-fat diet results in endotoxemia and inflammation, which in turn triggers the development of obesity, insulin resistance and even diabetes [11], [37], [38]. Furet et al. [39] analyzed the gut microbiota composition in 13 lean individuals and 30 obese subjects, including 7 T2DM patients who underwent a Roux-en-Y gastric bypass. They found that there was a negative correlation between F. prausnitzii and insulin resistance status, and that the proportion of F. prausnitzii was directly linked to the reduction of low-grade inflammation upon therapeutic intervention in T2DM. F. prausnitzii was one of the 2 major butyrate-producing bacterial groups and a lower abundance was detected in pre-DM compared with NGT. Using quantitative real-time PCR, Fujimoto et al. [40] found an inverse correlation between the abundance of F. prausnitzii and disease activity in a study of 47 Japanese patients with Crohn's disease and 20 healthy subjects, and this decrease was independent of genetic background. As fermentative organisms, F. prausnitzii and Roseburia and Eubacterium spp. are thought to supply butyrate and other SCFAs to the colonic epithelium, which could reduce oxidative stress and inflammation [41]. The observation that inflammation occurs in association with a decrease of these benefical microbes, especially F. prausnitzii, indicates that this species plays key role in maintaining colonic epithelial cells.
According to the Meta-HIT study [17], individual gut microbiota can be assigned to 1 of 3 enterotypes driven by Bacteroides, Prevotella and Ruminococcus, respectively. In agreement with this, all 3 enterotypes were also observed in our study. No significant relationship was found between enterotypes and Pre-DM or T2DM status which was consistent with the results of a previous study [16].
In the present study, after adjustment for BMI, age and sex (which could be confounding factors), PERMANOVA tests showed that FPG and CRP concentrations were significantly associated with the gut microbiota composition. Another study in overweight adults found significantly lower concentrations of CRP and changes in gut flora composition after treatment with galacto-oligosaccharides, prebiotics that can positively modify the gut microbiota and immune system. The different clades included increased Bifidobacteria and decreased Bacteroides spp. and C. histolyticum [42]. This may be due to an increase in CRP in response to inflammation caused by bacterial infection or other conditions, and their role is to bind to phosphocholine on microbes and clear necrotic and apoptotic cells. Thus, in healthy subjects when gut flora are modulated by prebiotics such as galacto-oligosaccharides and fructo-oligosaccharides, beneficial microbes (including butyrate-producing bacteria and Bifidobacteria spp.) will become more abundant or have higher activity levels, and help to maintain epithelial cells and prevent inflammation and destruction of the mucus layer – which is important for normal intestinal tract function. This was confirmed by Lapthorne et al. [43] who found that inflammation is a consequence of the dysbiosis of gut microbiota. Interestingly, stepwise multiple regression showed that the FINS concentration was an independent relevant factor and was negatively correlated with the diversity of gut microbiota. Furthermore, in comparison with subjects with NGT, the FPG and FINS concentration and IR index were significantly higher in the Pre-DM group. These data are in line with the hypothesis that the structure of gut microbiota may have a relationship with the inflammation system and with an insulin-resistant status.
Over last 20 years, a series of diabetes prevention trials have indicated that lifestyle improvement, weight reduction, and medications such as metformin, acarbose and rosiglitazone can significantly reduce the risk of progression from prediabetes to diabetes and improve risk factors for cardiovascular disease [44]–[49]. However, most of the controlled clinical trials that have been conducted have focused on changes in glycemia as the primary outcome. Accumulating evidence indicates that the gut microbiota is involved in host metabolism by increased energy extraction, immune system modulation, and altered lipid metabolism, all which have been demonstrated to contribute to progression to T2DM [13], [50]–[52]. The association of gut microbiota and prediabetes not only provides a clue to monitor the natural course of diabetes, but also raises the possibility of manipulating the microbiotic environment to prevent or treat the onset of T2DM.
In conclusion, this is the first study to show that the proportion and diversity of intestinal microbiota are changed early in the period of prediabetes. Although the relatively low number of subjects was a limitation of the study, these findings enhance our understanding of the influence of changes in human gut microbiota on the pathogenesis of diabetes. Additional research is needed to explore the biologic plausibility and the dynamic interrelation between gut microbiota and diabetes.
Design and Methods
Subjects
A total of 121 adult subjects were enrolled in the study in Beijing, China and were divided into 3 groups according to WHO criteria for the diagnosis of diabetes [53]. The 3 groups were those with normal glucose tolerance (NGT group; n = 44, M/F ratio = 12/32, mean age 55±9 years); prediabetes subjects including those with impaired fasting glucose and impaired glucose tolerance (Pre-DM group; n = 64, M/F ratio = 23/41, mean age 54±7 years), and subjects with newly diagnosed T2DM (T2DM group; n = 13, M/F ratio = 7/6, mean age 52±9 years). Exclusion criteria were clinical existence of renal and/or hepatic diseases, gastrointestinal tract diseases, psychiatric disorders, neoplasia, and any other disease that could interfere with the conduct of the study. None of the participants had taken pharmacological doses of antibiotics, hormones, antiobesity agents, lipid-lowering drugs, antihyperglycemic agents, and probiotics at least 1 month before the study. The study complied with the principles expressed in the Declaration of Helsinki and was approved by the Ethics Committee of Peking University People's Hospital. Written informed consent was obtained from each participant of the study.
Sample collection and processing
Venous blood samples were taken from study participants after an overnight fast for at least 10 hours, and serum samples were obtained in the normal manner. An oral glucose tolerance test (OGTT) was performed in each participant by measuring plasma glucose concentrations at 0 and 2 hours after oral ingestion of 75 g glucose. Aliquots of serum samples after fasting and 2 hours after glucose challenge were snap frozen in liquid nitrogen, and stored at −80°C until analyzed. Two fecal samples were also collected from each participant using sterile cups after defecation. One sample was used to perform routine tests to exclude subjects with bloody stools, pus or parasitic infections. Another fecal sample was brought to the laboratory within 2 hours and stored at −80°C for microbiota analysis.
Clinical chemistry measurements
Plasma glucose concentrations were measured by the glucose oxidase method. Serum total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), triglycerides (TG), C-reactive protein (CRP), and liver function and renal function indicators were measured using an automatic biochemical analyzer. Fasting insulin (FINS) and 2-hour insulin (2HINS) concentrations were determined by an electrochemiluminescence immunoassay (Elecsys 2010 system, Roche Diagnostics Ltd, Basel, Switzerland). Insulin resistance index (IR) was calculated from the fasting plasma glucose (FPG, mmol/L) and insulin (FINS, µU/ml) concentrations as: homeostasis model of assessment–insulin resistance (HOMA-IR) = FPG • FINS/22.5 [54].
DNA extraction and pyrosequencing
DNA extraction and PCR amplification were performed as described previously [55]. The V3–V5 region of the 16S ribosomal RNA (rRNA) gene from each DNA sample was amplified using the bacterial universal forward primer 341F (5′-CCTACGGGNGGCWGCAG-3′) and the reverse primer 926R (5′- CCGTCAATTCMTTTRAGT-3′), with the following parameters: initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 45 s, 56°C for 1 min, and 72°C for 1 min with a final extension at 72°C for 10 min. Polymerase chain reaction (PCR) products were run on 1.5% agarose gel electrophoresis and the DNA band with the correct size was excised and purified using Wizard® SV Gel and PCR Clean-Up System (Promega, St. Louis, MO, USA). Sample-unique 10-base barcodes were added to the 5′ end of the primer 341F and used for sorting of PCR amplicons into different samples. Emulsion PCR was set up according to Roche's protocols for the three methods. Sequencing was performed on a 454 GS FLX titanium pyrosequencer (454 Life Sciences, Branford, CT, USA) at BGI-Shenzhen. All sequences used in this study are available from GenBank, Accession No. SRA068175.
Bioinformatics analysis
All reads were filtered and trimmed as outlined by Pat Schloss using Mothur (v.1.21.1). Briefly, all reads shorter than 200 bp or with more than 8 homopolymers were removed. Two mismatches and no mismatches were allowed for the primer and barcode, respectively. The PyroNoise algorithm in Mothur (v.1.21.1) was implemented to remove noise from pyrosequenced amplicons with the default parameter [56]. Subsequently, all sequences were clustered using the program CD-HIT and both the similarity and the coverage for shorter sequences were set at 99% [57]. To determine the taxa for the samples at the OTU level, the most abundant sequence of each cluster generated by CD-HIT was selected as a representative and used to cluster into the OTU using QIIME with default parameters [58]. Each OTU's most abundant sequence was selected to determine the phylogeny of the OTU using the RDP classfier [59]. A taxonomic profile was generated based on the OTU annotation using PERL script.
Statistical analysis
Clinical data were expressed as means ± standard deviation (SD) or medians (interquartile range), and differences between groups were compared by ANOVA (one-way analysis of variance). Relative abundances of gut microbiota were compared between the NGT, Pre-DM, and T2DM groups using the Kruskal-Wallis rank sum test. Richness and diversity estimations used the Shannon diversity and Chao1 index. Principal component analysis (PCA) was used to analyze the beta diversity between the 3 groups. PERMANOVA (permutational multivariate analysis of variance), Pearson correlations, and co-inertia analysis (CIA) were used to test the relationship between microbiota and clinical parameters.
Supporting Information
Relative abundance boxplot (121 samples) of the 20 most abundant clades.
(TIF)
Principal component analysis (PCA) results for the genus profile.
(TIF)
Enterotypes of the human gut microbiome. Principal component analysis (PCA) figures were generated for the genus profiles. The 3 enterotypes [determined by the method of Arumugam et al. [17]] were labeled and grouped in the PCA figures. Fisher's exact test for the association between T2DM status and enterotypes was performed and was not significant (P = 0.3546).
(TIF)
Changes in abundance for operational taxonomic units (OTUs,) that exhibited a significant difference in the Wilcoxon rank sum test between the NGT, Pre-DM, and T2DM groups. All OTUs found to be changed significantly are labeled in different colors. The annotation for each OTU is listed in the corresponding color. The vertical arrows indicate that the corresponding OTUs have a higher or lower abundance.
(TIF)
Average abundances of each clade in the 3 groups. Taxons less than 1% and all tags assigned no genus in the 3 groups were counted as others. An average of 27.82%, 24.73% and 27.50% of the total tags could not be assigned to any known genus for the NGT, pre-DM and T2DM groups, respectively.
(TIF)
Relationship between FBG (fasting blood glucose), PBG (postprandial blood glucose) and the ratio of Bacteroidetes to Firmicutes . The lines in the figures were derived from a simple regression model (the model was not significant at alpha 0.05).
(TIF)
PERMANOVA test of the significance of the shift in gut microbiota structure.
(DOCX)
Correlations between the Chao1 alpha diversity index and healthy parameters. Stepwise regression was performed between the sample Chao1 alpha diversity index and healthy parameters by the “both” stepwise method. The final model Akaike information criterion (AIC) value was 1039.48, R2 value was 0.1119, and the P-value was 0.02898. The final model included SBP, FINS, TBIL, and Cr but only FINS was significant at the P<0.05 level. This result is consistent with the PERMANOVA test result, with every 8 mIU/L increase in FINS resulting in a 1 point decrease in the Chao1 alpha diversity index. *P<0.05; **P<0.001.
(DOCX)
Phylum level Kruskal-Wallis test. aThe Firmicutes result was not significant at a first error rate of 5%. bThe Verrucomicrobia result was significant.
(DOCX)
Class level test. Relative abundance mean = average relative abundance of each group; occurrence rate = ratio of samples that detected corresponding clades for each group.
(DOCX)
Acknowledgments
We thank all the participants for agreeing to join this study. We are grateful to the staffs at PKU Diabetes Centre for their assistance with subject recruitment, conducting the clinical protocol and laboratory test. We are indebted to the technicians of BGI-Shenzhen for their practical work in this study.
Funding Statement
Financial support from the National High Technology Research and Development Program (2006AA02A409). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Yang W, Lu J, Weng J, Jia W, Ji L, et al. (2010) China National Diabetes and Metabolic Disorders Study Group. Prevalence of diabetes among men and women in China. N Engl J Med 362: 1090–1101. [DOI] [PubMed] [Google Scholar]
- 2. Diamond Project Group (2006) Incidence and trends of childhood type 1 diabetes worldwide 1990–1999. Diabet Med 23: 857–866. [DOI] [PubMed] [Google Scholar]
- 3. Coutinho M, Gerstein HC, Wang Y, Yusuf S (1999) The relationship between glucose and incident cardiovascular events. A metaregression analysis of published data from 20 studies of 95,783 individuals followed for 12.4 years. Diabetes Care 22: 233–240. [DOI] [PubMed] [Google Scholar]
- 4. Meigs JB, Shrader P, Sullivan LM, McAteer JB, Fox CS, et al. (2008) Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med 359: 2208–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, et al. (2008) Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med 359: 2220–2232. [DOI] [PubMed] [Google Scholar]
- 6. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, et al. (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, et al. (2005) Diversity of the human intestinal microbial flora. Science 308: 1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, et al. (2006) Metagenomic analysis of the human distal gut microbiome. Science 312: 1355–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ley RE, Turnbaugh PJ, Klein S, Gordon JI (2006) Microbial ecology: human gut microbes associated with obesity. Nature 444: 1022–1023. [DOI] [PubMed] [Google Scholar]
- 10. Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, et al. (2005) Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 102: 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, et al. (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031. [DOI] [PubMed] [Google Scholar]
- 12. Duncan SH, Lobley GE, Holtrop G, Ince J, Johnstone AM, et al. (2008) Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes (Lond) 32: 1720–1724. [DOI] [PubMed] [Google Scholar]
- 13. Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, et al. (2011) Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr 94: 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, et al. (2010) Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 5: e9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu X, Ma C, Han L, Nawaz M, Gao F, et al. (2010) Molecular characterisation of the faecal microbiota in patients with type II diabetes. Curr Microbiol 61: 69–78. [DOI] [PubMed] [Google Scholar]
- 16. Qin J, Li Y, Cai Z, Li S, Zhu J, et al. (2012) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490: 55–60. [DOI] [PubMed] [Google Scholar]
- 17. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, et al. (2011) Enterotypes of the human gut microbiome. Nature 473: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Woese CR, Fox GE (1977) Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A 74: 5088–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, et al. (2012) Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 9: 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yan H, Potu R, Lu H, Vezzoni de Almeida V, Stewart T, et al. (2013) Dietary fat content and fiber type modulate hind gut microbial community and metabolic markers in the pig. PLoS One 8: e59581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davis LM, Martínez I, Walter J, Goin C, Hutkins RW (2011) Barcoded pyrosequencing reveals that consumption of galactooligosaccharides results in a highly specific bifidogenic response in humans. PLoS One 6: e25200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang C, Zhang M, Pang X, Zhao Y, Wang L, et al. (2012) Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J 6: 1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lam YY, Ha CW, Campbell CR, Mitchell AJ, Dinudom A, et al. (2012) Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS One 7: e34233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, et al. (2012) Gut microbiota composition correlates with diet and health in the elderly. Nature 488: 178–184. [DOI] [PubMed] [Google Scholar]
- 25. Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S (2007) Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol 73: 7767–7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, et al. (2013) Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 110: 9066–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Santacruz A, Collado MC, García-Valdés L, Segura MT, Martín-Lagos JA, et al. (2010) Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr 104: 83–92. [DOI] [PubMed] [Google Scholar]
- 28. Karlsson CL, Onnerfält J, Xu J, Molin G, Ahrné S, et al. (2012) The microbiota of the gut in preschool children with normal and excessive body weight. Obesity (Silver Spring) 20: 2257–2261. [DOI] [PubMed] [Google Scholar]
- 29. Everard A, Lazarevic V, Derrien M, Girard M, Muccioli GG, et al. (2011) Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes 60: 2775–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liou AP, Paziuk M, Luevano JM Jr, Machineni S, Turnbaugh PJ, et al. (2013) Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med 5: 178ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lahti L, Salonen A, Kekkonen RA, Salojärvi J, Jalanka-Tuovinen J, et al. (2013) Associations between the human intestinal microbiota, Lactobacillus rhamnosus GG and serum lipids indicated by integrated analysis of high-throughput profiling data. PeerJ 1: e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karlsson FH, Fåk F, Nookaew I, Tremaroli V, Fagerberg B, et al. (2012) Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun 3: 1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chevrot R, Carlotti A, Sopena V, Marchand P, Rosenfeld E (2008) Megamonas rupellensis sp. nov., an anaerobe isolated from the caecum of a duck. Int J Syst Evol Microbiol 58: 2921–2914. [DOI] [PubMed] [Google Scholar]
- 34. Sakon H, Nagai F, Morotomi M, Tanaka R (2008) Sutterella parvirubra sp. nov. and Megamonas funiformis sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 58: 970–975. [DOI] [PubMed] [Google Scholar]
- 35. Biagi E, Nylund L, Candela M, Ostan R, Bucci L, et al. (2010) Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One 5: e10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang T, Cai G, Qiu Y, Fei N, Zhang M, et al. (2012) Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J 6: 320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, et al. (2007) Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56: 1761–1772. [DOI] [PubMed] [Google Scholar]
- 38. Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, et al. (2010) Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328: 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, et al. (2010) Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes 59: 3049–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fujimoto T, Imaeda H, Takahashi K, Kasumi E, Bamba S, et al. (2013) Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn's disease. J Gastroenterol Hepatol 28: 613–619. [DOI] [PubMed] [Google Scholar]
- 41. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, et al. (2008) Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 105: 16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vulevic J, Juric A, Tzortzis G, Gibson GR (2013) A mixture of trans-galactooligosaccharides reduces markers of metabolic syndrome and modulates the fecal microbiota and immune function of overweight adults. J Nutr 143: 324–331. [DOI] [PubMed] [Google Scholar]
- 43. Lapthorne S, Pereira-Fantini PM, Fouhy F, Wilson G, Thomas SL, et al. (2013) Gut microbial diversity is reduced and is associated with colonic inflammation in a piglet model of short bowel syndrome. Gut Microbes 4: 212–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pan XR, Li GW, Hu YH, Wang JX, Yang WY, et al. (1997) Effect of diet and exercise in preventing NIDDM in people with impaired glucose tolerance. The Da Qing IGT and Diabetes Study. Diabetes Care 20: 537–544. [DOI] [PubMed] [Google Scholar]
- 45. Tuomilehto J, Lindström J, Eriksson JG, Valle TT, Hämäläinen H, et al. (2001) Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med 344: 1343–1350. [DOI] [PubMed] [Google Scholar]
- 46. Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, et al. (2002) Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 346: 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chiasson JL, Josse RG, Gormis R, Hanefeld M, Karasik A, et al. (2002) STOP-NIDDM Trial Research Group. Acarbose for prevention of type 2 diabetes mellitus: the STOP-NIDDM randomized trial. Lancet 359: 2072–2077. [DOI] [PubMed] [Google Scholar]
- 48. Chiasson JL, Josse RG, Gormis R, Hanefeld M, Karasik A, et al. (2003) STOP-NIDDM Trial Research Group. Acarbose treatment and the risk of cardiovascular disease and hypertension in patients with impaired glucose tolerance: the STOP-NIDDM randomized trial. JAMA 290: 486–494. [DOI] [PubMed] [Google Scholar]
- 49. Gerstein HC, Yusuf S, Bosch J, Pogue J, Sheridan P, et al. (2006) Effect of rosiglitazone on the frequency of diabetes in patients with impaired glucose tolerance or impaired fasting glucose: a randomized controlled trial. Lancet 368: 1096–1105. [DOI] [PubMed] [Google Scholar]
- 50. Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, et al. (2008) Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57: 1470–1481. [DOI] [PubMed] [Google Scholar]
- 51. Dumas ME, Barton RH, Toye A, Cloarec O, Blancher C, et al. (2006) Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci U S A 103: 12511–12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, et al. (2013) Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17: 225–235. [DOI] [PubMed] [Google Scholar]
- 53. Alberti KG, Zimmet PZ (1998) Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med 15: 539–553. [DOI] [PubMed] [Google Scholar]
- 54. Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, et al. (1985) Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28: 412–419. [DOI] [PubMed] [Google Scholar]
- 55. Chun J, Kim KY, Lee JH, Choi Y (2010) The analysis of oral microbial communities of wild-type and Toll-like receptor 2-deficient mice using a 454 GS FLX titanium pyrosequencer. BMC Microbiol 10: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ (2011) Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. [DOI] [PubMed] [Google Scholar]
- 58. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative abundance boxplot (121 samples) of the 20 most abundant clades.
(TIF)
Principal component analysis (PCA) results for the genus profile.
(TIF)
Enterotypes of the human gut microbiome. Principal component analysis (PCA) figures were generated for the genus profiles. The 3 enterotypes [determined by the method of Arumugam et al. [17]] were labeled and grouped in the PCA figures. Fisher's exact test for the association between T2DM status and enterotypes was performed and was not significant (P = 0.3546).
(TIF)
Changes in abundance for operational taxonomic units (OTUs,) that exhibited a significant difference in the Wilcoxon rank sum test between the NGT, Pre-DM, and T2DM groups. All OTUs found to be changed significantly are labeled in different colors. The annotation for each OTU is listed in the corresponding color. The vertical arrows indicate that the corresponding OTUs have a higher or lower abundance.
(TIF)
Average abundances of each clade in the 3 groups. Taxons less than 1% and all tags assigned no genus in the 3 groups were counted as others. An average of 27.82%, 24.73% and 27.50% of the total tags could not be assigned to any known genus for the NGT, pre-DM and T2DM groups, respectively.
(TIF)
Relationship between FBG (fasting blood glucose), PBG (postprandial blood glucose) and the ratio of Bacteroidetes to Firmicutes . The lines in the figures were derived from a simple regression model (the model was not significant at alpha 0.05).
(TIF)
PERMANOVA test of the significance of the shift in gut microbiota structure.
(DOCX)
Correlations between the Chao1 alpha diversity index and healthy parameters. Stepwise regression was performed between the sample Chao1 alpha diversity index and healthy parameters by the “both” stepwise method. The final model Akaike information criterion (AIC) value was 1039.48, R2 value was 0.1119, and the P-value was 0.02898. The final model included SBP, FINS, TBIL, and Cr but only FINS was significant at the P<0.05 level. This result is consistent with the PERMANOVA test result, with every 8 mIU/L increase in FINS resulting in a 1 point decrease in the Chao1 alpha diversity index. *P<0.05; **P<0.001.
(DOCX)
Phylum level Kruskal-Wallis test. aThe Firmicutes result was not significant at a first error rate of 5%. bThe Verrucomicrobia result was significant.
(DOCX)
Class level test. Relative abundance mean = average relative abundance of each group; occurrence rate = ratio of samples that detected corresponding clades for each group.
(DOCX)