Abstract
Upon studying the diversity of fungal endophytes associated with pine trees in Korea, many species of Penicillium were encountered. In this study, we report two species of Penicillium isolated from the needles of Pinus rigida. Based on ID region analysis, cultural and morphological characteristics, the two species were identified as Penicillium fellutanum and P. toxicarium, both of which are new to Korea.
Keywords: Endophyte, ITS, Lsu-rDNA, Penicillium fellutanum, Penicillium toxicarium, Pinus rigida
The genus Penicillium is large and encompasses many common species that look alike to the uninitiated. However, upon closer study, there is a great deal of variability within the species. The modern taxonomy of Penicillium embraces morphology, colony characters, metabolite profiles and molecular data.
Molecular phylogenetic analyses based on internal transcribed spacer [ITS (Berbee et al., 1995; Skouboe et al., 1996, 1999, 2000; Peterson, 2000)] and recently that of partial β-tublin gene sequences (Seifert and Louis-Seize, 2000; Peterson and Sigler, 2002; Samson et al., 2004) have been used for identification of the Penicillium species. Skouboe et al. (1996, 1999, 2000) analyzed sequences of the ITS region, including the 5.8S region, of several terverticillate Penicillia and reported few sequence differences among the species. Peterson (2000) also found a few differences between terverticillate Penicillium species in the ribosomal DNA region. Seifert and Louis-Seize (2000) as well as Samson et al. (2004) used partial β-tublin sequences to demonstrate a more resolved phylogeny of Penicillium subgenus Penicillium including terverticillate Penicillia. However, for the phylogenetic analysis of Penicillium subgenera Furcatum, Aspergilloides and Geosmithia, ITS and large subunit ribosomal DNA (1surDNA) sequences, but not β-tublin sequences, were used (Peterson, 2000) because of the lack of Penicillium β-tublin sequences in the gene bank.
Endophytes are microorganisms that form symptomless infections within healthy plant tissues (Carroll, 1988). Fungi belonging to this group of microorganisms are increasingly recognized as important mediators of interactions between plants and their competitors, seed dispersers, herbivores, and pathogens (Carroll, 1988; Clay, 1988; Chapela, 1989). Fungal endophytes have also been recognized as a repository of novel secondary metabolites, some of which have beneficial biological activities (Bills and Polishook, 1991).
During the course of our study on the diversity of endophytic fungi from needles of pine trees, many species of Penicillium were encountered. These were identified based on analysis of the ITS and large subunit ribosomal DNA (lsu-rDNA) sequences (ID region) and cultural and morphological characteristics. Here, we report on two endophytic Penicillium species that are new to Korea.
Materials and Methods
Endophyte isolation
Healthy needles of Pinus rigida were collected during June and July 2007, from mountainous areas in Daejeon, Korea. The needles were washed individually in running tap water and moved to a laminar flow hood where sections were cut using a sterile scalpel. These sections were surface-sterilized by dipping in 1% sodium hypochlorite for 3 min, 70% ethanol for 2 min and rinsing in sterile distilled water followed by drying on sterile filter paper (Arnold et al., 2001). The edges of each sampled tissue were cut off and discarded. Sub-samples of the remaining tissue, measuring approximately 10 mm, were placed individually in 9 cm Petri dishes containing potato dextrose agar (PDA; Difco™) supplemented with antibiotic substances such as dichloran, rose Bengal and chloramphenicol (DRBC; Difco™). After incubation at 25℃ for 5, 10 and 25 days, individual hyphal tips of the developing fungal colonies were collected and placed onto PDA media. The media was incubated at 25℃ for 8~10 days and checked for culture purity.
Phenotypic analysis
Penicillium isolates were then inoculated onto Czapek yeast extract agar (CYA; K2HPO4 1.0 g, Czapek concentrate 10 ml, yeast extract 5 g, sucrose 30 g, agar 15 g, distilled water 1 l), malt extract agar (MEA; malt extract 20 g, peptone 1.0 g, glucose 20 g, agar 20 g, distilled water 1 l) and yeast extract sucrose (YES) agar (yeast extract 20 g, sucrose 15 g, MgSO4·7H2O 0.05 g, CuSO4·5H2O 0.0005 g, ZnSO4·7H2O 0.001 g, agar 20 g, distilled water 1 l) under the conditions recommended by Pitt (1979). Colony appearance, exudate production, pigmentation and reverse coloration were assessed. Colony diameters were measured and recorded after one week of growth at 25℃. Colony color names were based on the Rayner (1970) nomenclature.
DNA isolation
The isolates were grown in liquid shake culture in potato dextrose broth medium for 3~4 days at 25℃. Mycelia were collected from the cultures by filtration and transferred to 1.5 ml tubes. These samples were frozen at -70℃. DNA was extracted by method of Cubero et al. (1999).
PCR amplification and sequencing
For amplification of the ID region, primers ITS1 (5'-TCC GTA GGT GAA CCT GCG G-3') and D2R (5'-AAC CAG GCA CAA AGT TCT GC-3') (White et al., 1990; Peterson, 1993) were used. The PCR mixture contained 0.5 pmol of each prsimer, 0.25 mM of dNTP's, 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, 2.5 U Taq polymerase and 15 ng of template DNA. PCR cycling conditions were as follows: an initial denaturation step of 96℃ for 7 min followed by 30 cycle of 96℃ for 30 sec, 51℃ for 30 sec and 72℃ for 150 sec. A final elongation step of 72℃ for 10 min was performed.
The PCR product was purified using a Wizard PCR prep kit (Promega, Madison, WI, U.S.A.). Purified double stranded PCR fragments were directly sequenced with BigDye terminator cycle sequencing kits (Applied Biosystems, Forster City, CA, U.S.A.) following the manufacturer's instructions. Same primer sets with PCR amplification were used to sequence both DNA strands. Agarose gel electrophoresis and data collection were performed on an ABI Prism 310 Genetic Analyzer (Applied Biosystems, Forster City, CA, U.S.A). Sequences of the two isolates were deposited in Genbank under the accession numbers FJ557247 and FJ557248.
Phylogenetic analysis
The sequences obtained from the present study were compared to the ID region available in the Genbank database using the BLAST search. Sequences generated from materials in this study and retrieved from GenBank were initially aligned using the program CLUSTAL X (Thompson et al., 1997), and then the alignment was manually refined using the PHYDIT program version 3.0 (Chun 1995; available at http://plaza.snu.ac.kr/~jchun/phydit). Maximum parsimony (MP) analysis was estimated using heuristic searches consisting of random addition order and tree bisection-reconnection (TBR) branch swapping. Bootstrap analysis using 1000 replications was performed to assess the relative stability of the branches.
Results and Discussion
Phylogenetic analysis
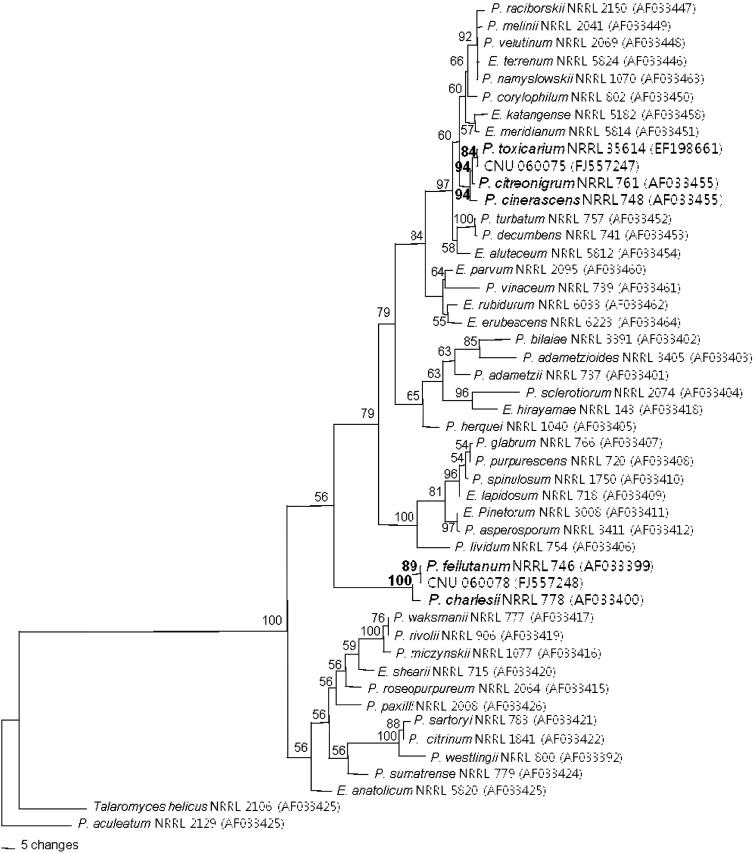
The ITS and lsu-rDNA (ID) from both Penicillium isolates from Pinus rigida in Korea were amplified. Amplification of the ID region with primers ITS1 and D2R yielded a fragment of approximately 1120 bp and alignment of the sequences resulted in a dataset of 1156 characters (27.1% variable and 20.9% parsimony informative characters). Parsimony analysis yielded sixth-seven parsimonious trees of 668 steps (CI = 0.6189, RI = 0.8293) (Fig. 1). In the maximum parsimony analysis, CNU 060078 isolate, P. fellutanum NRRL 746 and P. charlesii NRRL 778 were clustered together in a group, which was supported by a bootstrap value of 89%. The ID region sequence of CNU 060078 was identical to that of neotype of P. fellutanum NRRL 746 (Fig. 1).
Fig. 1.
One of sixth-seven parsimonious trees inferred from sequence of ID region. The numbers above each branch indicate bootstrap values obtained after a bootstrap test with 1000 replications. The bar represents 0.01 substitutions per site. Numbers in parentheses are GenBank accession numbers.
The CNU 060075 isolate was most closely related to P. cinerascens NRRL 748, P. citreonigrum NRRL 761 and P. toxicarium NRRL 35614. The ID region sequence of CNU 060075 was identical to that of P. toxicarium NRRL 35614. P. toxicarium and P. citreonigrum were considered nonspecific. Both species were thus morphologically and culturally similar (Samson et al., 2000). Serra et al. (2008) reported that P. toxicarium was closely related to P. cinerascens, but was a distinct species based on multilocus sequences. In the present study, P. toxicarium was clearly distinguished from P. cinerascens and the previously reported Penicillium. The phylogenetic tree inferred from the ID region showed good correlation with species that are defined by cultural and morphological characteristics.
Taxonomic description
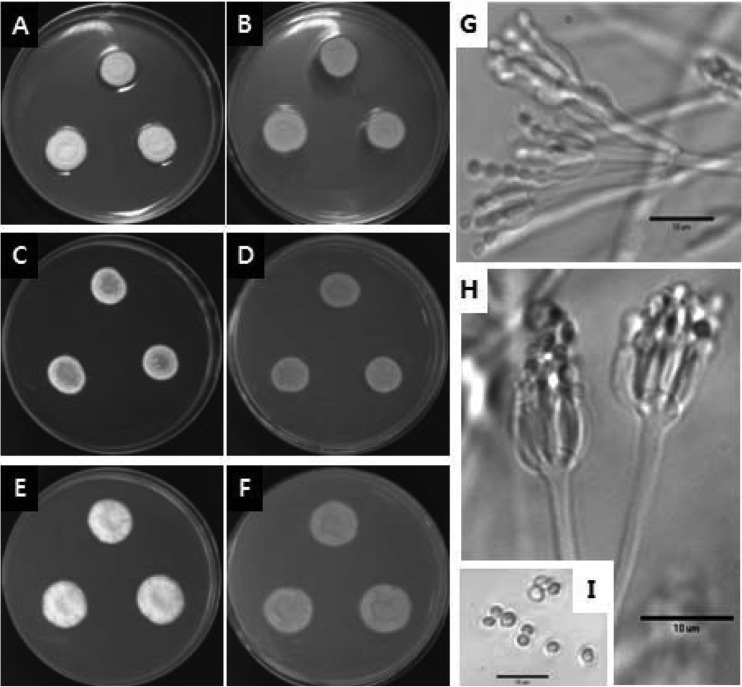
Penicillium fellutanum Biourge Fig. 2
Fig. 2.
Penicillium fellutanum. (A~B) 7-day-old colonies on CYA, (C~D) 7-day-old colonies on MEA, (E~F) 7-day-old colonies on YES agar, (A, C, E) obverse, (B, D, F) reverse; (G~H) conidiophores; (I) conidia. Scale bar = 10 µm.
Cellule 33: 262, 1923
Colonies on CYA were 17~23 mm in diameter, very dense, velutinous, radially sulcate, of narrow margins, of white mycelium; of light to heavy conidiogenesis, pale greenish grey if conidia sparse, but more commonly dark green or lavender grey, and sometimes having a clear exudate; the colonies showed reverse coloration of pale to occasionally yellow brown. Colonies on MEA were 15~21 mm in diameter, low and dense, velutinous with a floccose central area, radially sulcate; the margins were usually very narrow, less than 1 mm wide, of white mycelium, of typically heavy conidiogenesis, lighter in floccose isolates, showed colour similarity to CYA and no exudates were seen; the colonies showed reverse coloration of pale to olive brown. Colonies on YES were 19~25 mm in diameter, showed slow and dense growth patterns, were radially or annularly sulcate, often centrally umbonate; showing sparse and peripheral conidiogenesis that were dark grey green; no exudates were present and the colonies showed reverse coloration of pale grey to white.
The conidiophores were borne from aerial hyphae, characteristically of indeterminate form, terminating in well defined penicilli, bearing metulae in a random manner, with or without solitary phialides, less commonly of well defined monoverticillate penicilli borne perpendicular to fertile hyphae always with at least two terminal metulae; stipes smooth walled, of irregular and often indeterminate length; metulae 10~25 µm long, thin walled vesicles, 4~6 µm in diameter; phialides ampulliform, borne parallel in verticils of 8~12, 5~10 µm long; conidia ellipsoidal, 2.5~3.5 × 2.0~2.5 µm, with surface roughened, borne in long irregular columns.
Isolate examined: CNU 060078
Note: Colony characteristics and micro-morphology of the species agreed well with the description of P. fellutanum (Pitt, 1979). Microscopically it was distinguished by conidiophores with irregularly located metulae of variable length, terminating in definite vesicles; and macroscopically by close textured colonies and dark green conidia (Pitt, 1979). Besides, growth on MEA was slower than on CYA and YES agar. This is the first record of P. fellutanum in Korea.
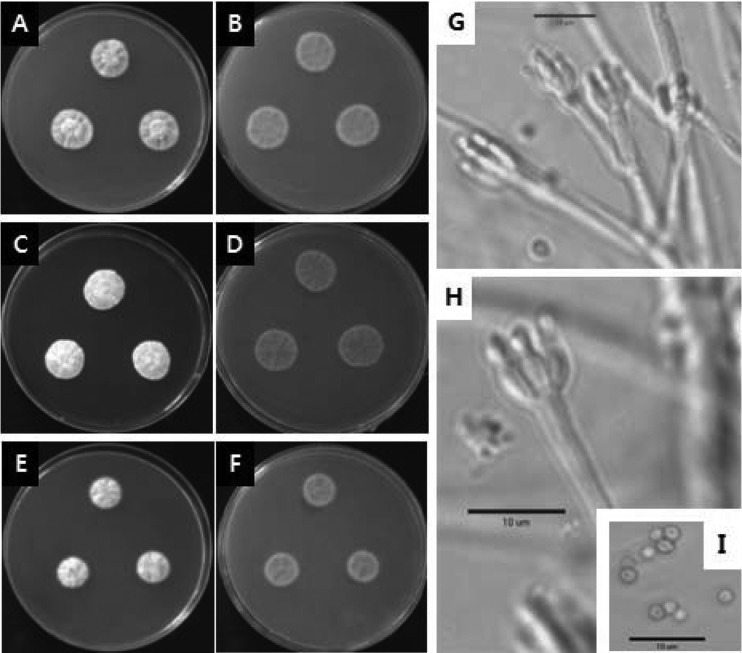
Penicillium toxicarium Miyake ex C. Ramirez Fig. 3
Fig. 3.
Penicillium toxicarium. (A~B) 7-day-old colonies on CYA, (C~D) 7-day-old colonies on MEA, (E~F) 7-day-old colonies on YES agar, (A, C, E) obverse, (B, D, F) reverse; (G~H) conidiophores; (I) conidia. Scale bar = 10 µm.
Rep. Res. Inst. Rice Improvement 1: 1, 1940 (nom. inval., Art. 36)
Colonies on CYA were 20~26 mm in diameter, moderately deep, dense and velutinous, radially sulcate and often centrally wrinkled; the margins were narrow with mycelium that were white to bright yellow; the conidiogenesis was sparse to moderate and greenish grey in colour; clear exudates produced rarely; yellow soluble pigment typically produced; a reverse coloration of typically brilliant yellow to less commonly yellow brown was reported. Colonies on MEA were 22~25 mm in diameter, low, dense and velutinous, plane to lightly sulcate; margins, white, entire or irregular; mycelium white, becoming yellow or buff centrally; the conidiogenesis was moderate and similar in color to those on CYA; exudates were rarely produced, and the colonies were clear to yellow; a brown soluble pigment was sometimes produced; reverse coloration of pale to brown or deep reddish brown. Colonies on YES were 15~21 mm in diameter, plane, sulcate or convolute, low to deep, velutinous to floccose; conidiogenesis was light, lavender greyish; exudates were absent; yellow soluble pigment produced; reverse colouration of pale to straw or brown.
Conidiophores borne from aerial hyphae, stipes delicate, 60~100 × 2.0~2.5 µm, smooth walled, nonvesiculate; monoverticillate, occasionally bimetulate; phialides, ampulliform, in verticils of 5~8, length varing with isolate, 5.2~13.0 µm long; conidia spheroidal to sub-spheroidal, 1.8~2.5 µm in diameter, with walls smooth to finely roughened, borne in short disordered chains.
Isolate examined: CNU 060075
Note: Colony characteristics and micro-morphology of the fungus agreed well with the description of P. toxicarium (Pitt, 1979). P. toxicarium produced compact yellow colonies; stipes were slender, conidia were tiny and smooth walled (Pitt, 1979). This is the first record of P. toxicarium in Korea.
Acknowledgements
This study was supported in part by a grant from the BioGreen 21 program of Rural Development Administration, and in part by a grant from National Institute of Biological Resources, Korea.
References
- 1.Arnold AE, Maynard Z, Gilbert GS. Fungal endophytes in dicotyledonous neotropical trees: parrterns of abundance and diversity. Mycol Res. 2001;105:1502–1507. [Google Scholar]
- 2.Berbee ML, Yoshimura A, Sugiyama J, Taylor JW. Is Penicillium monophyletic? An evaluation of phylogeny in the family Trichocomaceae from 18S, 5.8S and ITS ribosomal DNA sequence data. Mycologia. 1995;87:210–222. [Google Scholar]
- 3.Bills GF, Polishook JD. Microfungi from Carpinus caroliniana. Can J Bot. 1991;69:1477–1482. [Google Scholar]
- 4.Biourge P. Les moisissures du groupe Penicillium. Link Cellule. 1923;33:7–331. [Google Scholar]
- 5.Carroll GC. Fungal endophytes in stems and leaves: from latent pathogens to mutualistic symbiont. Ecology. 1988;62:2–9. [Google Scholar]
- 6.Chapela IH. Fungi in healthy stems and branches of American beech and aspen: a comparative study. New Phytologist. 1989;113:65–75. [Google Scholar]
- 7.Chun J. Computer-assisted classification and identification of actinomycetes. Newcastle upon Tyne, UK: University of Newcastle; 1995. Ph.D. Thesis. [Google Scholar]
- 8.Clay K. Fungal endophytes of grasses: a defensive mutualism between plants and fungi. Ecololgy. 1988;69:10–16. [Google Scholar]
- 9.Cubero OF, Crespo ANA, Fatehi F, Bridge PD. DNA extraction and PCR amplication method suitable for fresh, herbarium-stored, lichenized, and other fungi. Pl Syst Evol. 1999;216:243–249. [Google Scholar]
- 10.Peterson SW. Molecular genetic assessment of relatedness of Penicillium subgenus Penicillim. In: Reynolds DR, Taylor JW, editors. The Fungal Holomorph: Mitotic, Meiotic and Pleomorhic Speciation in Fungal Systematic. United Kingdom: Wallingford, CAB International; 1993. pp. 121–128. [Google Scholar]
- 11.Peterson SW. Phylogenetic analysis of Penicillium species based on ITS and LSU-rDNA nucleotide sequences. In: Samson RA, Pitt JI, editors. Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification. Amsterdam: Harwood Academic Publishers; 2000. pp. 163–178. [Google Scholar]
- 12.Pitt JI. The Genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. Academic press inc. (London) Ltd; 1979. [Google Scholar]
- 13.Pitt JI, Samson RA, Frisvad JC. List of accepted species and their synonyms in the family Trichocomaceae. In: Samson RA, Pitt JI, editors. Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification. Amsterdam: Harwood Academic Publishers; 2000. pp. 9–49. [Google Scholar]
- 14.Rayner RW. A mycological colour chart. Commonwealth Agricultural Bureaux; 1970. [Google Scholar]
- 15.Serra R, Peterson S, Venâncio A CTCOR. Multilocus sequence identification of Penicillium species in cork bark during plank preparation for the manufacture of stoppers. Res Microbiol. 2008;159:178–186. doi: 10.1016/j.resmic.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Seifert KA, Louis-Seize G. Phylogeny and species concept in the Penicillium aurantiogriseum complex as inferred from β-tublin gene DNA sequences. In: Samson RA, Pitt JI, editors. Integration of Modern Taxonomy Method for Penicillium and Aspergillus Classification. Amsterdam: Harwood Academic Publishers; 2000. pp. 189–198. [Google Scholar]
- 17.Skouboe P, Boysen M, Pedersen LH, Frisvad JC, Rossen L. Identification of Penicillium species using the internal transcribed spacer (ITS) regions. In: Rossen L, Rubio V, Dawson MT, Frisvad JC, editors. Fungal Identification Techniques. Brussels: European Commission; 1996. pp. 160–164. [Google Scholar]
- 18.Skouboe P, Frisvad JC, Taylor JW, Lauritsen D, Boysen M, Rossen L. Phylogenetic analysis of nucleotide sequences from the ITS region of terverticillate Penicillium species. Mycol Res. 1999;103:837–881. [Google Scholar]
- 19.Skouboe P, Taylor JW, Frisvad JC, Rossen L. Molecular methods for differentiation of closely related Penicillium species. In: Samson RA, Pitt JI, editors. Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification. Amsterdam: Harwood Academic Publishers; 2000. pp. 171–180. [Google Scholar]
- 20.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (* and other methods). Versin 4. Sunderland, Massachusetts: Sinauer Associates; 2003. [Google Scholar]
- 21.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. ClustalX: windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;25:4876–4878. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal DNA for phylogenetics. In: Innes MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: A guide to the Methods and Applications. New York: Academic Press; 1990. p. 315. [Google Scholar]