Abstract
Diffuse large B-cell lymphoma (DLBCL) is the most common lymphoma subtype, and fewer than half of patients are cured with standard front-line therapy. To improve therapeutic options, better animal models that accurately mimic human DLBCL (hDLBCL) are needed. Canine DLBCL (cDLBCL), one of the most common cancers in veterinary oncology, is morphologically similar to hDLBCL and is treated using similar chemotherapeutic protocols. With genomic technologies, it is now possible to molecularly evaluate dogs as a potential large-animal model for hDLBCL. We evaluated canine B-cell lymphomas (cBCLs) using immunohistochemistry and gene expression profiling. Canine B-cell lymphoma expression profiles were similar in many ways to hDLBCLs. For instance, a subset had increased expression of NF-κB pathway genes, mirroring human activated B-cell (ABC)-type DLBCL. Furthermore, immunoglobulin heavy chain (IGH) ongoing mutation status, which is correlated with ABC/germinal center B-cell (GCB) cell of origin in hDLBCL, separated cBCL into two groups with statistically different progression-free and overall survival times. In contrast with hDLBCL, cBCL rarely expressed BCL6 and MUM1/IRF4 by immunohistochemistry. Collectively, these studies identify molecular similarities to hDLBCL that introduce pet dogs as a representative model of hDLBCL for future studies, including therapeutic clinical trials.
Introduction
Non-Hodgkin lymphoma (NHL) is the 7th most common human systemic malignancy, estimated to affect approximately 70,000 people in the U.S. in 2013, and is a leading cause of cancer death (1). Despite improvements in treatments, mortality remains significant (1, 2), necessitating further progress in understanding disease biology and tailoring therapeutic interventions. DLBCL, the most common subtype of NHL, is recognized as a clinically heterogeneous disease (3). The international prognostic index (IPI), a score derived from clinical characteristics, groups patients into risk categories that range widely, from 85% overall survival (OS) at four years in the lowest risk category to 45% OS at four years in the highest-risk group (4, 5). This huge degree of variability within histologically identical DLBCLs prompted attempts to molecularly subcategorize the tumors (6). For example, gene expression profiling (GEP) identified two main subtypes, activated B-cell (ABC) and germinal center B-cell (GCB). These subcategories are thought to be reflective of the cell of origin, with GCBs arising from germinal center cells, and ABC-types arising from post-germinal center cells. In addition, ABC/GCB subtypes can be identified by examining somatic hypermutation (SHM), based on the finding that GCB lymphoma cells are continuously undergoing SHM, while ABC lymphomas arise from cells that have completed SHM and therefore contain static immunoglobulin heavy chain variable region (IGHV) sequences that are different than the germline sequences (7). These two categories of lymphomas have strikingly different survivals, independent of IPI, initially reported as ABC patient survival of 16% at five years and GCB patient survival of 76% at five years. This prognostic disparity was subsequently confirmed in multiple studies, even with the addition of rituximab to the standard cyclophosphamide, adriamycin, vincristine, and prednisone (CHOP) treatment regimen (6). Since it is currently impractical to perform GEP on every patient with DLBCL, a variety of immunohistochemical algorithms have been developed to predict the cell of origin and/or survival of these patients (8-12). Antibodies recognizing antigens expressed by either germinal center or post-germinal center B-cells, such as CD10, MUM1/IRF4, BCL6, GCET1, and FoxP1 are typically included. Concordance with GEP results ranges from 80-93%, and most of these algorithms also predict overall survival OS independent of the IPI, although these results have not yet been consistently reproducible (4, 8-14).
Animal models are useful for better understanding cancer biology and therapeutic responses, with mice being the mainstay of lymphoma model organisms. However, mouse models present inherent properties that can make them less accurate model systems, namely impaired immunity and/or genetic homogeneity, neither of which accurately mimics spontaneously occurring tumors in a diverse population (15, 16). Their small size and metabolic differences can affect pharmacokinetic/pharmacodynamic (PK/PD) parameters as well. These dissimilarities have resulted in therapies that appear promising in laboratory mice being ineffectual when translated to humans. Therefore, more representative animal models that overcome these disadvantages would be a welcome advance.
Pet dogs develop a variety of naturally occurring cancers that are recognized as relevant models of their human counterparts (17-19), similar in clinical presentation, tumor biology, and response to therapy. Dogs provide additional advantages over traditional animal models in that they represent a spectrum of genetic diversity that is not present in most laboratory strains. A recent study showed that T- and B-cell lymphomas in dogs are separable by gene expression profiling (20), paving the way for this technology to be applied with larger sample sizes to further define cBCL. Through a series of studies that were sequentially progressive in resolution, we define aspects of cBCL that are similar to hDLBCL, especially with respect to ABC/GCB subtype. To our knowledge, this is the first study to directly compare canine and human B-cell lymphomas using immunohistochemical algorithms, GEP analysis, and IGHV mutational status. Our data suggest that two subtypes of cBCL can be delineated using IGHV mutational analysis and to a lesser extent, GEP. Interestingly, immunohistochemical algorithms that are used to separate human patients into GCB and ABC subtypes were not useful as prognostic markers in the dog population. This study represents a basis for using canine lymphoma patients to understand both canine and human disease.
Materials and Methods
Subjects, sample acquisition
After owner consent, neoplastic lymph nodes (LN) were collected from 49 dogs presenting to the North Carolina State Veterinary Health Complex (NCSU VHC), or in some cases, referral veterinary clinics for evaluation and management between June 2008 and August 2010 (Table 1). LN from referral clinics were shipped overnight in chilled physiologic saline to NCSU VHC for processing. In cases where clients preferred not to have LN resection performed, cells were collected via fine needle aspiration (FNA) of enlarged peripheral LN. These were obtained from 19 additional dogs diagnosed with B-cell lymphoma via flow cytometry (68 total dogs in this study). FNAs were placed immediately into chilled RPMI/2% FBS, stored at 4°C and processed within 24 hours. These samples combined are referred to as canine B-cell lymphoma (cBCL) throughout the text.
Table 1.
Characteristics of the patients and samples used in this study.
| Signalment | |
| Number of dogs | 68 |
| Breeds | 31 (11 LR, 7 GR, 6 GSD) |
| Spay/Neuter | (3 IF, 31 SF, 3 IM, 31 NM) |
| Age (years) | mean, 7.1 (range, 2-14) |
| B-cell lymphoma | |
| Diagnosis | 20 LN FNA, 48 LN biopsy |
| Stage | 1 II, 14 III, 18 IV, 28 V, 7 unknown |
| Substage | 46 a, 17 b, 5 unknown |
LR = Labrador retriever, GR = golden retriever, GSD = German shephard dog IF = intact female, SF = spayed female, IM = intact male, NM = neutered male, LN = lymph node, FNA = fine needle aspirate
LN morphology and immunohistochemistry
LN morphology was evaluated using H&E staining of 35 available formalin fixed and paraffin embedded samples (NCSU VHC). Lymphoma was diagnosed and subtyped in alignment with the 4th edition of the World Health Organization (WHO) classification of hematopoietic and lymphoid tissues (21). B-cell phenotype was determined by immunohistochemistry and/or flow cytometry using CD79a and CD3 antibodies (AbD Serotec, Raleigh, NC). MUM1/IRF4 (Dako, Glostrup, Denmark) and, PAX5, CD10, and BCL6 antibodies (Leica Microsystems, Wetzlar, Germany) were used to both confirm B-cell phenotype (PAX5) and determine the cell of origin. External positive and negative control tissue was available for all antibodies. For all antibodies, a sample was called positive if >30% of the neoplastic cells were definitively staining (9).
RNA preparation and array processing
Excised LNs were manually homogenized in chilled RPMI/2% fetal bovine serum (FBS, Mediatech, Manassas, VA) and passed through a 70 μm filter. FNAs were washed twice in ice-cold phosphate-buffered saline, pH 7.4 (PBS, Mediatech) followed by red blood cell (RBC) lysis (Roche Applied Science, Indianapolis, IN). After counting, 1 × 107 cells were placed into Trizol (Sigma-Aldrich, St. Louis, MO), then frozen at −80°C. RNA was prepared according to the manufacturer’s protocol, and then purified over an RNeasy column (Qiagen, Valencia, CA). RNA quality was verified on a bioanalyzer (Agilent Technologies, Santa Clara, CA); all samples had an RNA Integrity Number (RIN) >9. Microarray gene expression data were obtained by the Lineberger Functional Genomics Core Facility using Affymetrix Canine Genome 2.0 Arrays (Santa Clara, CA). One microgram of total RNA was processed for microarray hybridization to Affymetrix GeneChips using the MessageAmp II-Biotin Enhanced Kit from Ambion (Life Technologies, Grand Island, NY). Briefly the RNA was reverse transcribed into cDNA using T7 Oligo(dT) primer. Following the first and second strand cDNA synthesis, in vitro transcription was carried out at 37°C for 14 hours to generate biotin-labeled aRNA. The aRNA was fragmented at 94°C for 35 min. and prepared for hybridization according to the Affymetrix Expession Analysis Technical Manual. The hybridization cocktail was hybridized to the arrays at 45°C for 16 hours. Following hybridization the arrays were washed with the Affymetrix Fluidics Station 450 wash stations. The arrays were scanned with the Affymetrix GeneChip Scanner 3000 7G Plus with Autoloader. GeneChip Command Console Software was used for control of all of the instrumentation. Basic data analysis and quality of the arrays was carried out using Affymetrix Expression Console software. Data have been annotated and deposited according to MIAME guidelines with Gene Expression Omnibus (GEO) accession number GSE 43664.
Statistical analysis
Microarray data were normalized using Robust Multi-array Average (RMA) software implemented by the Bioconductor package in R (22), version 2.12.1. Identification of differentially expressed genes (> three samples varying by at least four fold from the median) and average linkage hierarchical clustering was done with Cluster 2.11 (23), and visualized with Java Treeview (24). The annotation file Canine_2.na31 was used to identify genes from probeset information.
To match probe sets between the dog and human arrays we used the Homologene (build 64), and RefSeq (release 38, vertebrate mammalian sequences) databases from NCBI.
Overall survival was defined as the time from diagnosis until death from any cause. Progression-free survival was defined as the time from diagnosis until progression of disease or death from any cause. Outcomes were censored as of April 30, 2011. One dog was euthanized the day of diagnosis, and one was lost to follow up. These two dogs were not included in the survival analyses. Differences in survival by subgroup were tested with Cox proportional hazards regression, after testing that proportional hazards assumptions were met (25). Survival curves were estimated using the Kaplan-Meier estimator (26). Association between the hypermutation status and group categories was performed using a Chi Square test of association. Association analysis was performed in Stata v10 (www.stata.com) and R (22).
Human DLBCL Data Sets
CEL files from GEO accession number GSE11318 contain expression data from 203 human DLBCL samples analyzed on Affymetrix Human Genome U133 Plus 2.0 expression arrays (27). These were downloaded and normalized with RMA. The systematic data biases between human gene expression data and dog gene expression were detected and removed by Distance Weighted Discrimination (DWD) before the two datasets were combined (28). Twenty-five genes were then retrieved from the combined datasets and a two-way average linkage hierarchical co-cluster was generated.
Western blotting
Ten randomly chosen cDLBCLs were used for this experiment; five were “GCB-like”, i.e. from group 1 in Fig. 5 (Dogs 6, 8, 14, 28, 44), and five were “ABC-like”, i.e. from group 2 in Fig. 5 (Dogs 2, 3, 7, 21, 49). Western blotting was performed as previously described (29) except cell pellets stored at −80°C were used as the starting material. Antibodies directed against phospho-NF-κB p65 (Ser536) and NF-κB p65 (Cell Signaling, Boston, MA) and β-actin as a load control (Sigma-Aldrich, St. Louis, MO) were used. Bands were visualized with ECL Prime Western Blotting Detection Reagent (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). Densitometry was performed using NIH ImageJ 1.46.
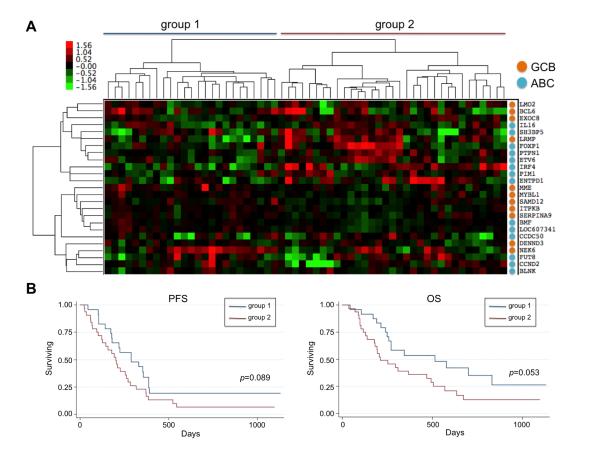
Figure 5.
Clustering of cBCLs using “dog specific” ABC/GCB classifier genes. (A) Hierarchical clustering with 1180 “dog specific” ABC/GCB genes. Differentially expressed genes categorized as being in the “B-cell receptor pathway”, are listed on the left. (B) Kaplan-Meier survival curves and Cox regression were performed on the two groups.
RNA isolation and qPCR
Total cellular RNA was extracted from TRIzol, and RNA (1μg) was then reverse-transcribed into cDNA using Transcriptor First Strand cDNA Synthesis kit (Roche, Indianapolis, IN) according to manufacturer instructions. Quantitative PCR (qPCR) was performed in a 25 μl reaction volume using SsoFast™ EvaGreen® (Bio-Rad, Hercules, CA) Mastermix. Reactions used 400 nM each of forward and reverse primers prepared by the Nucleic Acids Core Facility (University of North Carolina, Chapel Hill, NC). The following primers were used: IRAK1BP1, 5′-GAGGCCAAGAAGAGCGTTTG-3′ and 5′-GCTTGGCGTCGAAGATTCTC-3′; STAT4, 5′-GCAGAGAAGCTTACAGCCCA-3′ and 5′-TGCGGGACTCAGGTTTTCTC-3′; and GAPDH, 5′-CGGAGTCAACGGATTTGGCCGTA-3′ and 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′. Annealing temperature was 61.5°C for all reactions. The ten cDLBCL samples used were the same as those used in the Western blotting experiment.
IGHV sequencing
The dog IGHV gene was sequenced according to the method previously described for the human IGHV gene (30). However, since 39/41 functional members of the canine IGHV repertoire are in a single family, only a single set of primers was used, rather than the seven family-specific sets used in humans. Each canine sample was amplified successfully with these primers: Forward 5′-CCATGGAGTCTGTGCTCTGC-3′ containing the ATG at the start of the IGHV open reading frame, and Reverse 5′-CTGAGGAGACGGTGACCAGG-3′, located in the JH region, producing a 432bp amplicon. RNA from each lymphoma sample was converted to cDNA using the Transcriptor First Strand cDNA synthesis kit (Roche Applied Science, Indianapolis, IN). The PCR conditions for cDNA amplification were 94°C for 3 min.; followed by 35 cycles of (94°C for 1 min.; 65°C for 45 sec.; and 72°C for 1 min.) with a final elongation step of 72°C for 10 min. The PCR product was then cloned using the Zero Blunt TOPO PCR cloning kit (Invitrogen, Carlsbad, CA) or the Topo II cloning kit. Colonies were screened by PCR or EcoRI restriction digest, and at least 12 clones from two separate PCR/cloning reactions were sequenced for each subject. S6 universal primer was used. Conventional dideoxynucleotide sequencing was performed by the DNA sequencing core at UNC-CH Genomic Analysis Facility, University of North Carolina at Chapel Hill. IGHV mutation status was then defined as “static”, i.e. greater than 50% of the subclones identical or nearly identical, or “ongoing”, i.e. less than or equal to 50% of the subclones identical or nearly identical.
Results
LN phenotyping and morphology
Thirty-five canine lymph nodes that were CD79a+ and CD3−, designated as large B-cell lymphomas, were selected for additional immunophenotyping. 28/35 (80%) of the dogs were morphologically characterized as DLBCL and 7/35 (20%) were morphologically characterized as late marginal zone lymphoma (MZL). Similar to hDLBCL, cDLBCL is characterized by a complete loss of LN architecture via the expansion of a population of large neoplastic B-cells (Fig. 1A). Canine late MZL, in contrast to human MZL, is an aggressive lymphoma characterized by the expansion of large neoplastic B-cells apparently arising from the marginal zone which, in the later stages of the disease, leads to the typical finding of “fading germinal centers” (Fig. 1B). Given that these two types of B-cell lymphomas have similar cellular morphology and clinical behavior, and that they account for the vast majority (>90%) of canine large B-cell lymphomas (31), we analyzed them together in this study.
Figure 1.
Histologic appearance and immunophenotyping of canine BCL. (A) cDLBCL is characterized by lymph node effacement with diffuse sheets of large B-cells (H&E stain, first panel) that are CD79a-positive (second panel) and CD3-negative (third panel) by immunohistochemistry. (B) Canine MZL is characterized by nodules of large B-cells with “fading germinal centers” (H&E stain, first panel) that are CD79a-positive (second panel) and CD3-negative (third panel) by immunohistochemistry. Insets show the cell size similarity between the two morphologic types.
Canine B-cell lymphomas are not separable by immunohistochemical stains that differentiate human DLBCLs
We studied immunohistochemical (IHC) staining of PAX5 to corroborate our CD79a/CD3 data. This was supplemented with evaluation of three additional antibodies, CD10, BCL6, and MUM1/IRF4, which are used in hDLBCL as a surrogate for gene expression profiling to separate ABC and GCB subtypes. In our canine samples, 34/35 samples were PAX5 positive, confirming a B-cell phenotype, consistent with nearly universal PAX5-positivity in hDLBCL (Supplemental Fig. 1A) (32). Eight samples of 35 (23%) were positive for CD10 staining, suggesting a GCB subtype using human algorithms, where CD10-positivity ranges from 26-40% (Supplemental Fig. 1B) (9, 11). Notably 2/7 (28%) MZLs were CD10-positive, reinforcing the distinction from human MZL, which is almost never CD10-positive (33, 34). Interestingly, only a single sample tested positive for BCL6 (Supplemental Fig. 1C) and a second single sample tested positive for MUM1/IRF4 (Supplemental Fig. 1D). This is in contrast to human DLBCL, in which 70-80% of cases are positive for BCL6 and 20-40% are positive for MUM1 (8, 12).
Gene expression profiling does not distinguish MZL from DLBCL in cBCL
A total of 58 cBCLs, 39 with a histologic diagnosis and 19 obtained by FNA and diagnosed only as cBCL, were collected for transcriptional profiling. Genes that were differentially expressed were identified, and unsupervised hierarchical clustering using these 1079 probe sets did not identify robust separation into subtypes, as has been reported with hDLBCL (Supplemental Fig. 2) (6). Given that a subset of our samples were obtained by FNA and therefore did not have histologic diagnosis available, we first analyzed whether MZL could be distinguished from DLBCL by expression profiling. Based on population averages, >95% of our cBCL samples are expected to be either MZL or DLBCL, divided approximately as 25% MZL and 75% DLBCL (31). We analyzed the 39 samples with known histology, 7 MZL and 32 DLBCL, to see whether any genes were differentially expressed. Using two-class unpaired Significance Analysis of Microarrays (SAM) analysis (35), no genes were statistically significantly differentially expressed. We also performed principal component analysis with these 39 samples and there was no separation between MZL and DLBCL (Fig. 2), in agreement with previous results (20).
Figure 2.
Principal component analysis does not distinguish cMZL and cDLBCL. The 1079 genes that were most differentially expressed (Supplemental Fig. 2) among cBCL were used for principal component analysis. There is no division between histologic subtypes of cBCL by gene expression data.
Expression of genes homologous to ABC/GCB signature genes distinguish two subtypes of canine B-cell lymphoma
We hypothesized, based on similar morphology and veterinary pathology classification standards, that the majority of cBCLs would be molecularly similar to hDLBCL (31). Therefore, we were interested in determining whether the two major subclasses of hDLBCL could be identified in cBCL. We analyzed all 58 cBCLs as a single group, given that there were no detectable differences in their gene expression profiles based on MZL/DLBCL distinction (possibly because late MZLs in dogs are similar to DLBCL). We used a 27-gene classifier that separates hDLBCL into ABC/GCB subtypes (36) and analyzed both human and canine lymphomas together. Two genes were omitted from this analysis: TBC1D27 did not have a known canine homolog and IGHM was so strongly expressed in some cBCL cases that it dominated the clustering when included. The hDLBCL samples clustered into two clear groups, corresponding to human ABC and GCB DLBCL, while the cBCL samples were interspersed among the human samples in both groups (Fig. 3). Next, we used this set of 25 human ABC/GCB classifier genes and performed unsupervised hierarchical clustering with cBCL samples. Quantitative PCR (qPCR) was performed on five of these genes and showed good correlation with gene expression array data (Pearson’s r=0.842), (data not shown). There was no distinct separation into two subgroups, although the two subgroups had differences in progression-free and overall survival times that approached statistical significance (p=0.089 and p=0.053, respectively; Fig. 4B).
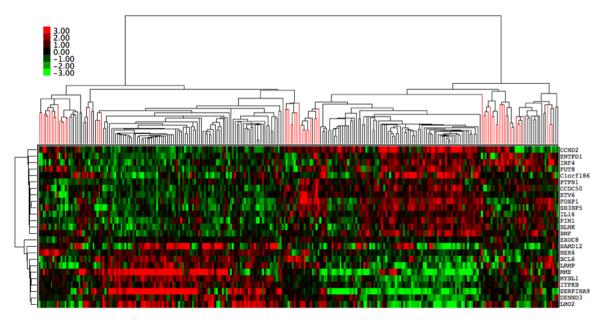
Figure 3.
cBCL and hDLCBL co-cluster using human ABC/GCB classifier genes. Expression data from 203 hDLBCLs was combined with 58 cBCL expression profiles. Distance Weighted Discrimination (DWD) was used to remove systematic biases between the two groups, and unsupervised hierarchical clustering was performed using the listed genes. Red branches are cBCL samples and black branches are hDLBCL samples.
Figure 4.
Clustering of cBCLs using human ABC/GCB classifier genes. (A) Hierarchical clustering of gene expression data reveal two groups of cBCLs that are not robustly separated. Also, genes more highly expressed in ABC lymphomas (teal) do not cluster distinctly from the genes more highly expressed in GCB lymphomas (orange) on the vertical axis. (B) Kaplan-Meier survival curves and Cox regression analysis were performed on the two groups. Overall and progression-free survival by ABC/GCB grouping approaches, but does not reach, statistical significance.
Canine lymphomas differentiated by human ABC/GCB distinguishing genes can be better separated with a canine-specific set of genes
Hypothesizing that cBCL subtypes might be better distinguished by a canine-specific gene signature, we attempted to use the two groups of canine lymphomas defined by the human ABC/GCB signature genes (Fig. 4) to develop a more robust canine gene signature. Two-class unpaired SAM analysis was used to find genes that were differentially expressed between the “human ABC-like” and “human GCB-like” groups, and 1180 probe sets were discovered using a cutoff false-discovery rate (FDR) of 5%. We used these to perform hierarchical clustering of cBCL, which resulted in the separation into two distinct groups (Fig. 5A). There was a statistically significant progression-free survival difference between the groups, and a trend towards an overall survival difference (Fig. 5B). It should be noted that overall survival in pet dogs is not as robust an endpoint as it is in humans, because owners choose when to euthanize their pets, and factors other than extent of disease therefore affect this endpoint.
ABC/GCB genes are not conserved between dogs and humans, but pathways and biological processes that distinguish germinal and post-germinal center groups are shared across species
We analyzed the 1180 genes (Supplemental Table 1) that were statistically differentially expressed between cBCL groups when separated using two-class unpaired SAM analysis. While individual genes were not uniformly conserved across species, pathways and processes were strikingly conserved. Ingenuity Pathway Analysis (Ingenuity® Systems, www.ingenuity.com) was used to determine the top biologic processes and pathways enriched for members of the canine ABC/GCB gene list. Similar to the case in hDLBCL, B-cell activation, B-cell receptor signaling, and the NF-κB pathway were among the most statistically significantly associated processes. Two genes, IRAK1BP1 and STAT4, which were more highly expressed in group 1, the "GCB-like" group, were confirmed with qPCR in ten cDLBCL samples (five ABC-like, five GCB-like). Similar to the expression array data, the GCB-like group had higher expression of both genes, with an average 10.46-fold higher expression of IRAK1BP1 and 7.44-fold higher expression of STAT4. To further confirm biologic differences, we also performed immunoblotting for p65 (RelA). This protein has previously been shown to be active in a majority of cDLBCLs, with activation of downstream targets in approximately half of cDLBCLs (37). In our samples, we observed higher total cellular expression of p65, concordant with the mRNA expression data. Phospho-p65 was also present in the majority of samples (Supplemental Fig. 3). One of the top two canonical pathways found in our pathway analysis was the B-cell receptor signaling pathway, with a p-value of 2.79 × 10−6 (Fig. 5A). To ensure that inclusion of MZLs was not obscuring our ability to define subgroups, we repeated the entire gene expression analysis using only known DLBCLs (n=32), and found no substantial differences, other than loss of statistical significance in the survival analyses, presumably due to smaller sample size (data not shown). Collectively, these data indicate that B-cell survival pathways are conserved as differentiators of cBCL subtypes, albeit in some cases with different pathway members differentially expressed than those in humans.
Canine lymphomas have all undergone somatic hypermutation, and a subgroup exhibits ongoing somatic hypermutation
To further explore a potential ABC/GCB distinction in cBCL, we attempted to assess other phenotypes associated with ABC/GCB subgroups. We obtained the IGHV sequence from 53 of the 58 dogs and compared each with published parental germline sequences (38). There was an obvious difference in the degree of ongoing SHM in the canine samples (Supplemental Table 3). We divided the dogs into two groups based on this phenotype; there were no genes significantly differentially expressed between the two groups when analyzed by SAM analysis. However, there was a significant difference in the progression-free survival (p=0.018) between dogs with ongoing vs. static IGHV hypermutation (Fig. 6A). The same trend was true when the 32 DLBCLs were analyzed separately, although the p-value was no longer signficant with the smaller sample size (p=0.10 for PFS). Notably, MZLs were also found in both ongoing and static IGHV groups, but the sample size was too small for survival analysis (n=5). It has been reported in a small number of hDLBCL patients that ABC/GCB phenotype is well correlated with IGHV status (ongoing vs. static) (7). However, the two phenotypes were not well correlated in cBCL patients, as shown in Fig. 6B. Based on hDLBCL, static IGHV mutation status should be correlated with the group of dogs that is more similar to ABC hDLBCL. However, the X2 test showed no statistically significant difference, though it was trending in that direction (p=0.097). In addition, a multivariate analysis using both IGHV status and GEP failed to show that either was independently predictive of survival.
Figure 6.
Immunoglobulin heavy chain mutation status predicts survival in cBCL. The IGHV gene was subcloned and sequenced in 53 cBCLS, which were then categorized as “ongoing” (many different subclones, indicating ongoing SHM) or “static” (subclones identical or nearly identical, indicating completed SHM) (A) Kaplan-Meier survival curves and Cox regression were performed on the two groups. (B) Overlap between groups defined by canine “ABC/GCB” genes (Fig. 5) and IGHV status is shown. p=0.097 for the X2 test.
We also analyzed the IGHV parental gene used in each lymphoma. Humans have 123 IGHV genes, including 79 pseudogenes, that fall into seven gene families (39). Dogs have a total of 80 IGHV genes, including 39 pseudogenes, that fall into three gene families, but the majority (76) are in VH1, the human VH3 family ortholog (38). The majority (57%) of cBCLs used VH1-44. This is the second most commonly used parental gene in normal canine B-cells (23%) (38). Parental gene usage in cBCL was significantly different from usage in normal B-cells (p<0.0002), indicating a bias in IGHV usage in cBCL. When looking at MZL (2/5, or 40%, used VH1-44) and DLBCL (29/52, or 56% used VH1-44) separately, results were similar, although the number of MZLs was too small for statistical comparisons.
Discussion
In contrast to other available models, the population of pet dogs in the United States represents a diverse and abundant source of spontaneously occurring lymphomas (18, 19, 40). Recently, with improvements in genomic technologies and the advent of subspecialty veterinary care, including oncology, this readily available resource is now primed for use to augment clinic research. This study represents the first molecular analysis of cBCL combining modalities (gene expression profiling, immunohistochemistry, and IGHV status) specifically aimed at defining molecular similarities to hDLBCL. We initially used antibodies against known human antigens that are expressed by either germinal center or post-germinal center B-cells in an effort to determine if cBCL can also be separated into these subtypes. Although the anti-human CD10, BCL6, and MUM1/IRF4 antibodies all cross-reacted with the canine antigens, the rare positivity for BCL6 and MUM1/IRF4 make human immunohistochemical algorithms less useful. This reinforces the notion that different proteins may be the hallmarks of cBCL subtypes, as compared with hDLBCL. Even if GCB and ABC correlates are not strictly conserved in dogs, discovering immunohistochemical stains to distinguish the germinal center and post-germinal center subtypes we characterized in this report will be important for prognostic purposes.
Our canine B-cell lymphomas were separable into two major histologic subtypes by LN morphology, which corroborates previous studies (21). The majority (32/39, or 82%) were cDLBCL, and 7/39 (18%) were canine marginal zone lymphomas. Our analyses revealed no distinctions that reliably separated these two histologic subtypes, similar to a recently published analysis with a smaller number of cBCLs (10 DLBCL, 5 MZL) (20). Gene expression profiles were similar, with no genes differentially expressed between the two groups in our samples (albeit with only seven MZLs). Franz et al also found similar gene expression profiles, but did find genes that were differentially expressed between cMZL and cDLBCL. However, it is possible that the smaller number of cBCLs in that study could account for this. In our study, MZLs and DLBCLs also were not distinguishable by IHC, IGHV mutation status, or by ABC/GCB subtyping. Furthermore, we repeated our entire analysis using only DLBCLs (n=32), and found no substantial differences from our findings presented here, other than loss of statistical significance in the survival analyses, presumably due to smaller sample size (data not shown). Theoretically, canine MZL could begin as an indolent disease that is not detected early in its course and then progresses to a more aggressive form in the later stages when enlarged peripheral lymph nodes become apparent. In any case, it appears that by the time canine MZL is clinically detectable, the disease is as biologically aggressive as DLBCL and molecularly similar to it. Therefore, LN morphology in canine B-cell LSA may not be prognostic or clinically relevant at this time. More canine MZL cases will need to be studied before making any final conclusions.
Similar to hDLBCL, we found heterogeneity in gene expression using microarray analysis. Genes that distinguish ABC and GCB subtypes in hDLBCL separate cBCL into two groups. Using this distinction to select a canine-specific set of differentially expressed genes yields two distinct groups with statistically different survival times. Furthermore, these canine-specific “ABC/GCB” discriminating genes, while different from the human ABC/GCB gene list, are involved in the same pathways and processes (e.g. NF-κB signaling and B-cell receptor signaling). The importance of B-cell receptor signaling in canine lymphoma has been demonstrated previously by responses to ibrutinib, an inhibitor of B-cell receptor signaling, in a subset of cBCL patients (41). NF-κB signaling in cBCL is complex, showing both similarities and differences to human DLBCL in both our results and in other reports (37). Our protein studies of NF-κB support differential expression of pathway members found by GEP, although future studies using nuclear extracts/staining will be needed to definitively show activation of the pathway in canine ABC-like DLBCL. These and other studies, including generation of canine lymphoma cell lines and more detailed study of molecular aberrations in these lymphomas, will be necessary to definitively determine whether both ABC and GCB subtypes exist in cBCL.
Like ABC/GCB gene expression pattern, immunoglobulin heavy chain status (ongoing SHM vs. completed SHM) also identified two cBCL groups with statistically different survival times. In hDLBCL, these two phenotypes (ABC/GCB gene expression pattern and completed/ongoing SHM) are reported to be overlapping in most cases. However, this observation is based on a limited number of samples (n=14) and may not be as perfectly correlated as previously reported (7). If that is the case, ongoing SHM may be a better predictor of survival in hDLBCL, as it is in cBCL. Further study of SHM in hDLBCL is therefore warranted.
Several limitations of our study should be highlighted. The enrolled dogs were not part of a clinical trial, so while they were generally treated with a standard front-line CHOP-based protocol, variability in treatment did occur. Uniform treatment should increase the statistical power of future results. Another source of heterogeneity is the type of biopsy (39 excisional biopsies and 19 FNAs for the gene expression studies). Both types of biopsies were equally represented in both classes of canine lymphomas in both Fig. 5 and 6 (chi-squared p-value >0.25 in both cases), making bias caused by type of biopsy unlikely in this study. Additional excisional biopsies will be needed to adequately power studies of whether stromal expression signatures are relevant in cBCL. Finally, since many different breeds were included, our study does not address the existence of breed-specific lymphoma subtypes, which could be of interest. Our data create a reference group against which future breed-specific cohorts can be compared.
In conclusion, our work represents a first combination approach using detailed immunohistochemical and molecular characterization of cBCL, thus providing a pathway for this widely available resource to be further developed as a large-animal model for the study of hDLBCL. Our data are consistent with a germinal center and post-germinal center phenotype in cBCL, although how closely these mimic human GCB and ABC subtypes clinically remains to be determined. As molecular similarities are better defined, pet dogs will be useful in clinical trials with new agents that target particular molecular subtypes of lymphoma with aberrations that are shared between canines and humans. Given the lack of shared specific genes that are aberrantly expressed, comparative translational oncology will likely focus more on conserved pathways that are deranged rather than specific gene products. Targeting these shared pathways in canine lymphoma patients will allow the rapid development of new therapies by gathering PK/PD and efficacy data from the same model organism. This study provides an important step toward the development of a more faithful and representative animal model for the development of hDLBCL therapeutics.
Supplementary Material
Acknowledgements
We would like to thank Alexis Brown for assistance with immunohistochemical analysis, and Mike Vernon for gene expression profiling at the UNC functional genomics core facility. We thank Avenelle Turner and Holly Mills for critical reading of the manuscript and helpful discussions. This work was supported by a developmental grant from the North Carolina University Cancer Research Fund administered by the Lineberger Comprehensive Cancer Center to KLR, AMR, MB, and SES.
Footnotes
Disclosure of Conflicts of Interest The authors declare no conflicts of interest for this study
Authorship Contributions SES, KLR, AMR, and MB conceived and wrote the grant to fund this study. SES collected the cBCL samples, isolated DNA and RNA, and wrote the manuscript. KLR prepared DNA and RNA, performed the IGHV analysis, and wrote the manuscript. AMR, DMN, and CF performed GEP statistical analysis. CS provided bioinformatic support. HC performed the IGHV analysis and analyzed the VH family usage. GS prepared RNA and performed the qPCR, Western, and IGHV analysis. YF and LB performed the lymph node immunohistochemical staining and analysis. MB, RT, SSD, and CP provided significant intellectual input, and all authors reviewed the manuscript.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Edwards BK, Brown ML, Wingo PA, Howe HL, Ward E, Ries LA, et al. Annual report to the nation on the status of cancer, 1975-2002, featuring population-based trends in cancer treatment. J Natl Cancer Inst. 2005;97:1407–27. doi: 10.1093/jnci/dji289. [DOI] [PubMed] [Google Scholar]
- 3.Nastoupil LJ, Rose AC, Flowers CR. Diffuse large B-cell lymphoma: current treatment approaches. Oncology (Williston Park) 2012;26:488–95. [PubMed] [Google Scholar]
- 4.Salles G, de Jong D, Xie W, Rosenwald A, Chhanabhai M, Gaulard P, et al. Prognostic significance of immunohistochemical biomarkers in diffuse large B-cell lymphoma: a study from the Lunenburg Lymphoma Biomarker Consortium. Blood. 2011;117:7070–8. doi: 10.1182/blood-2011-04-345256. [DOI] [PubMed] [Google Scholar]
- 5.A predictive model for aggressive non-Hodgkin’s lymphoma. The International Non-Hodgkin’s Lymphoma Prognostic Factors Project. N Engl J Med. 1993;329:987–94. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 6.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 7.Lossos IS, Alizadeh AA, Eisen MB, Chan WC, Brown PO, Botstein D, et al. Ongoing immunoglobulin somatic mutation in germinal center B cell-like but not in activated B cell-like diffuse large cell lymphomas. Proc Natl Acad Sci U S A. 2000;97:10209–13. doi: 10.1073/pnas.180316097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi WW, Weisenburger DD, Greiner TC, Piris MA, Banham AH, Delabie J, et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res. 2009;15:5494–502. doi: 10.1158/1078-0432.CCR-09-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–82. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 10.Meyer PN, Fu K, Greiner TC, Smith LM, Delabie J, Gascoyne RD, et al. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J Clin Oncol. 2011;29:200–7. doi: 10.1200/JCO.2010.30.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muris JJ, Meijer CJ, Vos W, van Krieken JH, Jiwa NM, Ossenkoppele GJ, et al. Immunohistochemical profiling based on Bcl-2, CD10 and MUM1 expression improves risk stratification in patients with primary nodal diffuse large B cell lymphoma. J Pathol. 2006;208:714–23. doi: 10.1002/path.1924. [DOI] [PubMed] [Google Scholar]
- 12.Visco C, Li Y, Xu-Monette ZY, Miranda RN, Green TM, Tzankov A, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia. 2012;26:2103–13. doi: 10.1038/leu.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez-Garcia G, Cardesa-Salzmann T, Climent F, Gonzalez-Barca E, Mercadal S, Mate JL, et al. Gene-expression profiling and not immunophenotypic algorithms predicts prognosis in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Blood. 2011;117:4836–43. doi: 10.1182/blood-2010-12-322362. [DOI] [PubMed] [Google Scholar]
- 14.Morton LM, Cerhan JR, Hartge P, Vasef MA, Neppalli VT, Natkunam Y, et al. Immunostaining to identify molecular subtypes of diffuse large B-cell lymphoma in a population-based epidemiologic study in the pre-rituximab era. Int J Mol Epidemiol Genet. 2011;2:245–52. [PMC free article] [PubMed] [Google Scholar]
- 15.Maddison K, Clarke AR. New approaches for modelling cancer mechanisms in the mouse. The Journal of pathology. 2005;205:181–93. doi: 10.1002/path.1698. [DOI] [PubMed] [Google Scholar]
- 16.O’Connor OA, Toner LE, Vrhovac R, Budak-Alpdogan T, Smith EA, Bergman P. Comparative animal models for the study of lymphohematopoietic tumors: strengths and limitations of present approaches. Leukemia & lymphoma. 2005;46:973–92. doi: 10.1080/10428190500083193. [DOI] [PubMed] [Google Scholar]
- 17.Breen M, Modiano JF. Evolutionarily conserved cytogenetic changes in hematological malignancies of dogs and humans--man and his best friend share more than companionship. Chromosome Res. 2008;16:145–54. doi: 10.1007/s10577-007-1212-4. [DOI] [PubMed] [Google Scholar]
- 18.Khanna C, Lindblad-Toh K, Vail D, London C, Bergman P, Barber L, et al. The dog as a cancer model. Nat Biotechnol. 2006;24:1065–6. doi: 10.1038/nbt0906-1065b. [DOI] [PubMed] [Google Scholar]
- 19.Paoloni M, Khanna C. Translation of new cancer treatments from pet dogs to humans. Nat Rev Cancer. 2008;8:147–56. doi: 10.1038/nrc2273. [DOI] [PubMed] [Google Scholar]
- 20.Frantz AM, Sarver AL, Ito D, Phang TL, Karimpour-Fard A, Scott MC, et al. Molecular Profiling Reveals Prognostically Significant Subtypes of Canine Lymphoma. Vet Pathol. 2013 doi: 10.1177/0300985812465325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swerdlow SH, editor. WHO classification of tumours of haematopoietic and lymphoid tissues. Vol. 2008. International Agency for Research on Cancer; Lyon, France: 2008. [Google Scholar]
- 22.Team RDC. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2010. [Google Scholar]
- 23.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 25.Breslow NE. Analysis of Survival Data under the Proportional Hazards Model. International Statistical Review / Revue Internationale de Statistique. 1975;43:45–57. [Google Scholar]
- 26.Kaplan EL, Meier P. Nonparametric Estimation from Incomplete Observations. Journal of the American Statistical Association. 1958;53:457–81. [Google Scholar]
- 27.Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A. 2008;105:13520–5. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benito M, Parker J, Du Q, Wu J, Xiang D, Perou CM, et al. Adjustment of systematic microarray data biases. Bioinformatics. 2004;20:105–14. doi: 10.1093/bioinformatics/btg385. [DOI] [PubMed] [Google Scholar]
- 29.Suter SE, Small GW, Seiser EL, Thomas R, Breen M, Richards KL. FLT3 mutations in canine acute lymphocytic leukemia. BMC Cancer. 2011;11:38. doi: 10.1186/1471-2407-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell MJ, Zelenetz AD, Levy S, Levy R. Use of family specific leader region primers for PCR amplification of the human heavy chain variable region gene repertoire. Mol Immunol. 1992;29:193–203. doi: 10.1016/0161-5890(92)90100-c. [DOI] [PubMed] [Google Scholar]
- 31.Ponce F, Marchal T, Magnol JP, Turinelli V, Ledieu D, Bonnefont C, et al. A morphological study of 608 cases of canine malignant lymphoma in France with a focus on comparative similarities between canine and human lymphoma morphology. Vet Pathol. 2010;47:414–33. doi: 10.1177/0300985810363902. [DOI] [PubMed] [Google Scholar]
- 32.Desouki MM, Post GR, Cherry D, Lazarchick J. PAX-5: a valuable immunohistochemical marker in the differential diagnosis of lymphoid neoplasms. Clin Med Res. 2010;8:84–8. doi: 10.3121/cmr.2010.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kost CB, Holden JT, Mann KP. Marginal zone B-cell lymphoma: a retrospective immunophenotypic analysis. Cytometry B Clin Cytom. 2008;74:282–6. doi: 10.1002/cyto.b.20426. [DOI] [PubMed] [Google Scholar]
- 34.Petit B, Chaury MP, Le Clorennec C, Jaccard A, Gachard N, Moalic-Judge S, et al. Indolent lymphoplasmacytic and marginal zone B-cell lymphomas: absence of both IRF4 and Ki67 expression identifies a better prognosis subgroup. Haematologica. 2005;90:200–6. [PubMed] [Google Scholar]
- 35.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991–6. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaurnier-Hausser A, Patel R, Baldwin AS, May MJ, Mason NJ. NEMO-binding domain peptide inhibits constitutive NF-kappaB activity and reduces tumor burden in a canine model of relapsed, refractory diffuse large B-cell lymphoma. Clin Cancer Res. 2011;17:4661–71. doi: 10.1158/1078-0432.CCR-10-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bao Y, Guo Y, Xiao S, Zhao Z. Molecular characterization of the VH repertoire in Canis familiaris. Vet Immunol Immunopathol. 2010 doi: 10.1016/j.vetimm.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 39.Matsuda F, Ishii K, Bourvagnet P, Kuma K, Hayashida H, Miyata T, et al. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J Exp Med. 1998;188:2151–62. doi: 10.1084/jem.188.11.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paoloni MC, Khanna C. Comparative oncology today. Vet Clin North Am Small Anim Pract. 2007;37:1023–32. v. doi: 10.1016/j.cvsm.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Honigberg LA, Smith AM, Sirisawad M, Verner E, Loury D, Chang B, et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc Natl Acad Sci U S A. 2010;107:13075–80. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.