Abstract
Recent research has suggested that Staphylococcus epidermidis is a reservoir of genes that, after horizontal transfer, facilitate the potential of Staphylococcus aureus to colonize, survive during infection, or resist antibiotic treatment, traits that are notably manifest in methicillin-resistant S. aureus (MRSA). S. aureus is a dangerous human pathogen and notorious for acquiring antibiotic resistance. MRSA in particular is one of the most frequent causes of morbidity and death in hospitalized patients. S. aureus is an extremely versatile pathogen with a multitude of mechanisms to cause disease and circumvent immune defenses. In contrast, most other staphylococci, such as S. epidermidis, are commonly benign commensals and only occasionally cause disease. Recent findings highlight the key importance of efforts to better understand how genes of staphylococci other than S. aureus contribute to survival in the human host, how they are transferred to S. aureus, and why this exchange appears to be uni-directional.
Keywords: Staphylococcus aureus, MRSA, Staphylococcus epidermidis, horizontal gene transfer, CRISPR
Introduction
Staphylococcus aureus is a frequent and dangerous human pathogen. It is most notorious for causing hospital- and community-acquired skin and lung infections. S. aureus is also a premier cause of endocarditis, osteomyelitis, and septicemia. Importantly, S. aureus infection is a major cause of death in hospital-associated infections, particularly when patients have underlying conditions such as immune deficiencies or primary infections by other pathogens, for example influenza [1].
S. aureus strains may produce a large variety of toxins. S. aureus cytolytic toxins include leukocidins, alpha-toxin, and the phenol-soluble modulins (PSMs) [2,3]. Further important toxins are the S. aureus superantigens, which include toxic shock syndrome toxin and lead to an exacerbated immune response by polyclonal T cell activation and massive cytokine release [4]. S. aureus also produces a large series of toxins that interfere with almost every mechanism of human innate host defense [5].
Notably, toxins are the prime determinants of a specific S. aureus strain's virulence or aggressiveness. Some S. aureus strains may cause particular toxin-related diseases such as toxic shock or scalded skin syndrome [6,7]. These diseases are directly linked to the presence of specific toxin genes in those strains (in these examples, genes encoding toxic shock syndrome toxin and exfoliative toxins, respectively). Similarly, in more common forms of S. aureus diseases, the severity of infection may be related to the toxin repertoire of the infecting strain, which may differ considerably from isolate to isolate. Even for toxin genes that are found in all or almost all S. aureus isolates, differential gene expression may lead to substantially varying aggressiveness [8].
Despite the undisputed importance of aggressive toxins, there are a series of other factors that determine the survival of S. aureus during infection and thus its capacity to cause disease. These include molecular determinants that “shield” S. aureus from the immune system without directly attacking it [9]. Capsular polysaccharides, any substance participating in the formation of extracellular matrix, or the surface protein, protein A, belong to that group of molecules. Importantly, most infections with S. aureus originate from colonizing strains, because at least a considerable subset of the human population is permanently or transiently colonized with S. aureus, mostly in the nose [10,11]. Therefore, any factor promoting asymptomatic colonization may be regarded a risk factor for the subsequent development of infection.
Frequent and often multiple antibiotic resistances considerably narrow therapeutic options to treat S. aureus infections. S. aureus strains may be resistant to virtually all but very recently developed antibiotics [12]. Resistance to some antibiotics, such as penicillin, is so widespread that these antibiotics cannot be considered valuable therapeutics for S. aureus anymore. Because methicillin is a first-choice, efficient drug for S. aureus infections, methicillin-resistant S. aureus (MRSA) is a particular concern for public health. Methicillin resistance rates among infectious S. aureus isolates are high. In many countries, they reach or exceed 50% [13].
Coagulase-negative staphylococci (CoNS) are systematically distinguished from S. aureus by the lack of coagulase. This enzyme promotes blood clotting, and the resulting fibrin coat on the bacterial surface may facilitate immune evasion. CoNS comprise a multitude of species, many of which are opportunistic pathogens [14]. Staphylococcus epidermidis is the most frequently encountered CoNS species on human skin and by far the most frequent source of CoNS infections [15]. S. epidermidis and other CoNS differ from S. aureus in that they are less virulent and typically cause chronic rather than acute infections. In fact, they may have an important role in contributing to immunity of the skin and mucous surfaces toward more harmful pathogens [16]. However, serious complications can arise from chronic CoNS infections, particularly in immune-compromised, hospitalized, and very young or old patients. Notably, S. epidermidis is the most important cause of infections on indwelling medical devices [17].
In accordance with their considerably lower virulence compared to S. aureus, CoNS typically do not produce aggressive toxins [15]. There are rare reports about enterotoxins in CoNS, but it is not clear whether in these cases the toxin genes cause increased virulence. Furthermore, most CoNS have psm genes [18]. The virtual omnipresence of PSM peptides in CoNS may be due to the general functions of those surfactant-like peptides in biofilm formation and surface colonization, while it appears that CoNS – in contrast to S. aureus – do not have genes for strongly cytolytic PSMs or do not express them at considerable amounts [19]. In general, pathogenicity of CoNS appears to stem from molecular determinants that evolved for commensal life on the skin, but may rise to additional use during infection [15,20].
Thus, among extant staphylococcal species most toxins appear to be limited to S. aureus. However, there are several recent findings indicating that S. aureus acquired other factors that facilitate survival during infection - such as antibiotic resistance determinants or molecules promoting immune evasion - from S. epidermidis or other CoNS. Direct evidence for such a process is understandably hard to achieve, but the facts that these factors are more frequently found in CoNS and located on mobile genetic elements indicate a CoNS origin. Here, important examples of such determinants will be discussed to substantiate the overarching hypothesis of a key role of S. epidermidis and other CoNS as a reservoir for genes facilitating the evolution of S. aureus as a successful pathogen. Because MRSA represent the most important S. aureus strains from a clinical point of view and methicillin resistance itself is an important example of a phenotype likely acquired form CoNS, I will focus on MRSA. On the CoNS side, the focus will be on S. epidermidis, as most information is available about this CoNS species, both in general and as a donor of genes to MRSA.
Genes are transferred among and between staphylococcal species by horizontal gene transfer
Many antibiotic resistance and virulence determinants in S. aureus are located on mobile genetic elements (MGEs), such as pathogenicity islands, chromosomal cassettes, transposons, or plasmids [21]. The diversity of S. aureus as a species stems mainly from the different presence of such MGEs, which may be readily lost and acquired, while the remaining genes represent a skeletal “core genome” [22].
As different staphylococcal species live in the same or similar niches on human epithelial surfaces, they are in close enough contact for genetic exchange to occur. In general, genetic material may be exchanged by the use of conjugation, phage transduction, or the uptake of “naked” DNA by natural competence. Competence has not been described in staphylococci [23]. However, it is often assumed that natural competence exists in many bacteria. Direct evidence for competence-mediated horizontal gene transfer and knowledge about competence systems are lacking only because the conditions under which bacteria become competent are hard to established in the laboratory [24]. In contrast, the transfer of virulence and antibiotic resistance genes - carried on conjugative plasmids or transduced by phages - has frequently been described in S. aureus [25]. There is also evidence for transfer between species. For example, a self-transmissible plasmid can transfer between CoNS and S. aureus [26]. However, the mechanism of transfer of MGEs containing methicillin resistance genes, which will be discussed below, is still unclear.
Methicillin resistance is encoded on SCCmec elements, which originated from CoNS
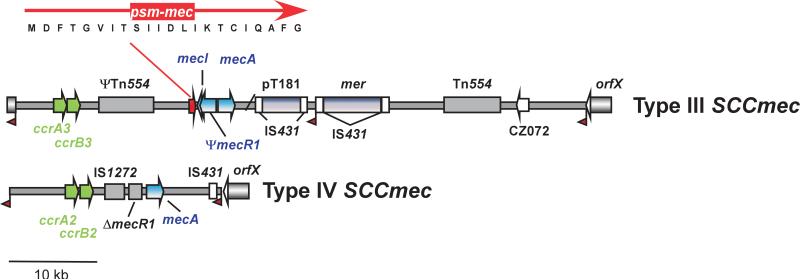
Methicillin resistance in staphylococci is encoded on MGEs called staphylococcal cassette chromosome mec (SCCmec) [27]. SCCmec elements are classified by differences in their key components, the methicillin resistance and immunity systems (mec gene complex) and the recombinase (ccr) genes, and overall genetic composition [28] (Fig. 1). New types of SCCmec elements are frequently reported; the current number of known SCCmec elements is 11.
Fig. 1.
Examples of SCCmec element composition. Shown are SCCmec elements of type III and type IV. Type III elements are common in HA- and type IV elements are characteristic of CA-MRSA. The latter are smaller in total size, which may contribute to increased fitness compared to MRSA strains harboring larger SCCmec elements. Genes of the mec gene complex are shown in blue, recombinases (ccr genes) in green. Some other important, varying components are shown in grey. Red flags depict att recombination sites. The location of the psm-mec cytolysin gene and the amino acid sequence of the PSM-mec protein are shown.
Resistance to methicillin is due to the mecA gene, which encodes an alternative penicillin binding protein with decreased binding affinity to methicillin [29]. Most S. aureus and other staphylococcal strains contain this type of mecA, but homologues of mecA with high similarity (80-90% on the nucleotide level) are found in some S. sciuri and S. vitulinus strains [30,31], now named mecA1 and mecA2 [32]. In addition, homologues of less pronounced similarity to mecA (<70%) were found in S. aureus strain LGA251 and Macrococcus caseolyticus [33,34], which are now named mecB and mecC, respectively [32]. Other parts of the mec gene complex are mecR1 and mecI, which encode a transcriptional repressor and a signal transduction protein involved in regulation of mecA. They are not present on all SCCmec types. The ccr gene complex contains homologues of the ccrA, ccrB, or ccrC recombinase genes. Other genes found in the SCCmec elements vary. Among them are antibiotic resistance genes, insertion elements such as IS431, plasmids such as pT181, or transposons such as Tn554.
Recently, the psm-mec gene was identified in SCCmec elements of types II, II, and VIII as conserved part of the class A mec gene complex [35,36] (Fig. 1). PSM-mec is also found frequently among methicillin-resistant S. epidermidis (MRSE) and represents an exception to the rule that S. epidermidis does not transfer toxin genes to S. aureus. This exception may be explained by the fact that the PSM-mec cytolysin, like other PSMs, has additional functions in basic staphylococcal physiology. Furthermore, it only “hitchhikes” on an MGE with a primary function in antibiotic resistance.
Notably, SCCmec elements have only been reported in staphylococci. The abovementioned M. caseolyticus, which is closely related to staphylococci, represents a noticeable exception. While the origin of SCCmec elements is unknown, several lines of evidence indicate that they came from CoNS. The proposed evolutionary scenario is the following: The mecA gene appears to have a common staphylococcal ancestor. This ancestor was originally assumed to be S. sciuri [37], whereas more recent findings point to S. fleuretti [38]. The mec and ccr gene complexes were likely assembled in CoNS, but the source is not known. In CoNS, some alterations occurred, such as mec gene complex deletions, upon which the SCCmec elements were transferred to S. aureus.
The evidence for a transfer of SCCmec elements specifically from S. epidermidis is quite strong. First, the homology of DNA sequences between SCCmec elements of S. aureus and S. epidermidis is very high [39]. For example, S. epidermidis SCCmec type IV shows 98-99% homology to SCCmec IVa of S. aureus (Fig. 1), while the overall similarity of genomic sequences between the two species is much lower [40]. SCCmec type IV was also observed in S. epidermidis much earlier, in the 1970s, than in S. aureus [40]. Second, the insertion element IS1272 is found much more often in S. epidermidis (and S. haemolyticus) than in S. aureus [41]. SCCmec types I and IV in S. aureus and S. epidermidis have identical IS1272-ΔmecR1 junctions [40]. Finally, overall, methicillin resistance is much more frequent among S. epidermidis than S. aureus [42,43]. Altogether, these observations indicate that SCCmec elements were transferred from S. epidermidis to S. aureus, although some parts may have originated from other CoNS before their transfer to S. epidermidis.
As stressed above, the mechanism of SCCmec transfer is unknown. In laboratory experiments, the mec locus could not be transferred by conjugation [44]. Furthermore, there appear to be difficulties introducing SCCmec into staphylococci by common laboratory transformation procedures. Phage-mediated generalized transduction was successful between two S. aureus strains [45], but phage-mediated transfer of genes between different staphylococcal species is rare [27]. It has only been described for the transfer of SaPIbov2, a pathogenicity island encoding the biofilm-associated bap gene, from S. aureus to several CoNS species [46]. In vivo, there is evidence that mecA was transferred from S. epidermidis to S. aureus [47]. Undoubtedly, the SCCmec transfer mechanism represents one of the major unsolved questions in staphylococcal molecular biology. Furthermore, given that there are reports of SCC elements that lack mec, the transfer of such elements may have implications for genetic shuffling between staphylococcal species that is not limited only to antibiotic resistance.
The mobile genetic element ACME represents an excellent example of DNA obtained from S. epidermidis that facilitates MRSA colonization and fitness
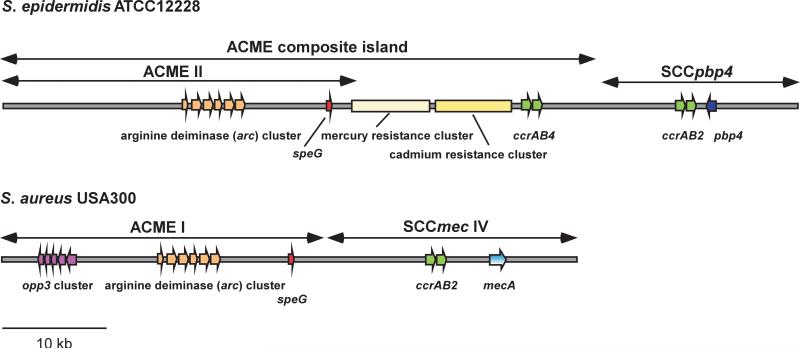
The arginine catabolic mobile element (ACME) is a ~ 31 kb MGE that was found first in the CA-MRSA strain USA300 located next to the SCCmec element of type IV [48] (Fig. 2). ACME encodes an arginine deiminase (arc) gene cluster and an oligopeptide permease (opp3) system. Other arc and opp clusters are found in the S. aureus core genome, which are believed to have basic functions in metabolism, but there is evidence indicating that ACME has a different role. Both ACME and SCCmec of USA300 could be excised using ccrAB recombinases, together and separately [49]. Analysis of these deletion mutants revealed that ACME affected survival of strain USA300 in a rabbit model of bacteremia, in contrast to SCCmec. However, ACME was shown not to impact the capacity of USA300 to cause rat skin abscess and necrotizing pneumonia [50], the two disease manifestations predominantly associated with the USA300 CA-MRSA epidemic [51]. It was concluded that ACME may somehow contribute to competitive fitness of USA300 with the arc or opp gene clusters facilitating colonization or transmissibility capacities of USA300. In fact, it has frequently been reported that opp gene clusters have roles in pathogenesis, such as by facilitating transport of quorum-sensing pheromones or antimicrobial peptides, while the arc gene cluster was speculated possibly to neutralize the acid environment on the skin by producing ammonia [52].
Fig. 2.
ACME in S. epidermidis and S. aureus. Important genes on the arginine catabolic mobile element (ACME), and in its vicinity, in the genome-sequenced strains S. epidermidis ATCC12228 and S. aureus USA300 (FPR3757) are shown. Genes of the mec gene complex are shown in blue, recombinases (ccr genes) in green. opp, oligopeptide permease; pbp, penicillin-binding protein; speG, spermidin N-acetyltransferase.
The first mechanistic insight into how ACME may play a role in enhancing survival of USA300 came from a recent study by Joshi et al., who could attribute that role to another part of ACME, a gene termed speG [53]. SpeG is a polyamine N-acetyltransferase that detoxifies polyamines, such as spermin or spermidin, by acetylation. Notably, in contrast to most organisms, S. aureus does not produce polyamines and is hypersensitive to their toxic effects. Enhanced survival characteristics of USA300 on human skin compared to other S. aureus, as proposed previously, may thus at least in part be due to speG. However, in-vivo evidence for such a role of speG still needs to be produced. Furthermore, it remains to be investigated what roles, if any, the other parts of ACME may have for USA300 pathogenesis or transmissibility.
More recently, ACME elements that differ from that originally found in USA300 were identified in S. aureus. ACME type I, the original type, has opp3 and arc, type II has only arc, and type III only opp3. While the USA300 ACME element (type I) was exclusively detected among clones of sequence type (ST) 8, the ST to which USA300 belongs, ACME type II, which lacks opp3, was found in ST5 and ST59 [54,55]. In some S. aureus strains of ST22, the arc gene cluster is up- rather than downstream of SCCmec type IV and opp3 is missing [56]. These findings indicated separate ancestors of S. aureus ACME types.
ACME occurs in S. epidermidis at a high frequency [57]. In a detailed investigation of a large number of S. epidermidis strains, Miragaia et al. found that 52% of global S. epidermidis clones - representing a broad spectrum of the S. epidermidis genetic and geographic diversity - harbored an ACME allotype [58]. Importantly, a close relative of the USA300-type ACME element (now termed ACME I.01), termed ACME I.02, was by far the most predominant among S. epidermidis isolates (27% of all isolates) [58] (Fig. 2). Furthermore, ACME I.02 was particularly frequent in S. epidermidis isolates of clonal complex (CC) 2, a large and widely distributed S. epidermidis strain group associated with hospital infection. Moreover, a large part of the ST22 ACME type has high homology to the ACME composite island of the genome-sequenced S. epidermidis strain ATCC12228 [58].
In conclusion, these findings support the notion that most parts of the ACME elements found in S. aureus originated from S. epidermidis. Only minor parts may have come from other CoNS, such as the opp3 genes that likely originated from S. haemolyticus [59]. Notably, ACME is present in the most successful S. epidermidis lineage and the most widespread clone of S. aureus in the U.S., USA300, supporting the idea that it has an important role in facilitating colonization and/or transmissibility. Mechanistically, this role may be in part due to the recently described speG gene [53], which is present in the widely distributed ACME types I.01 and I.02, but absent from other, less frequent types. SpeG is a polyamine acetyltransferase, which abrogates the unique hypersensitivity of S. aureus to these host-produced molecules.
SasX: Epidemiological evidence for the importance of a colonization and virulence factor obtained from S. epidermidis
The molecular factors underlying the rise of novel HA-MRSA clones are still poorly understood. As with CA-MRSA, the spread of successful clones may not only be linked to virulence sensu stricto, but also non-symptomatic colonization from which infection may arise [10]. While several molecules contributing to S. aureus nasal colonization have been described, no factor has yet been linked to the spread of a particular HA-MRSA clone.
Recently, Li et al. described a novel surface protein, termed SasX, that is located on a prophage and can thus be easily transferred between MRSA strains [60]. SasX was shown in a mouse model to facilitate nasal colonization. Interestingly, SasX also promoted survival of MRSA during experimental skin and lung infection, which likely is a consequence of its impact on bacterial aggregation, which prevents efficient phagocytosis by white blood cells. Notably, the frequency of sasX-positive clones increased significantly among invasive MRSA isolates in eastern China. It was predominantly found in clones of ST239, the most prominent MRSA type in most parts of Asia, and may be in part responsible for the success of that ST in this geographical region. More recently, sasX is spreading to other STs, increasing the recipient clones’ virulence and colonization capacity. Importantly, these findings give first insight into the molecular mechanisms underlying epidemic outbreaks of MRSA [61].
The 127-kb ΦSPβ-like prophage that harbors the sasX gene at its end is very similar to a prophage found in the genome of S. epidermidis strain RP62A [62]. The striking similarity suggests that the prophage was acquired from S. epidermidis by horizontal gene transfer, which is supported by earlier findings reporting high frequency of the S. epidermidis sasX homologue, sesI, among S. epidermidis strains [63]. However, a more exhaustive analysis of the frequency of the phage in S. epidermidis strains will be needed to substantiate that hypothesis.
CRISPR: A mechanism directing the gene flow from S. epidermidis to S. aureus?
While these findings indicate that S. aureus frequently takes up genetic material from CoNS, especially S. epidermidis, and stably includes it in its own genome, S. epidermidis does not appear to acquire genes from S. aureus. If S. epidermidis were to do so, one would expect that many of the multiple toxins and other virulence determinants of S. aureus be transferred to S. epidermidis over time. However, S. epidermidis clearly lacks the vast repertoire of aggressive virulence factors known from S. aureus [14,15], a situation that has been hypothesized to fit perfectly to the distinctly different lifestyles of these two closely related bacteria [64].
What is the mechanism underlying this virtually unidirectional gene transfer? The answer may lie in the recently discovered CRISPR (clustered, regularly interspaced, short palindromic repeat loci) [65], a bacterial system of self versus non-self discrimination [66] (Fig. 3). CRISPR loci contain sequences that, when transcribed to RNA, bind non-host DNA specifically, and target it for degradation. That process protects bacteria against phages, for example, but it could also be the mechanism preventing foreign DNA from a related bacterial species being incorporated into the S. epidermidis genome. Interestingly, much of the work on CRISPR has been performed using S. epidermidis. S. epidermidis strain RP62A, several other S. epidermidis strains [67], as well as ~40% to ~90% of sequenced bacterial genomes contain CRISPR sequences [68]. However, CRISPR sequences are much less frequent in S. aureus. Therefore, CRISPR sequences may be involved in preventing uptake of foreign DNA in S. epidermidis. In contrast, S. aureus as a species appears to take the risk of incorporating harmful DNA, such as viruses, to benefit from the possibility to accumulate virulence factors from other bacteria.
Fig. 3.
Directionality of gene transfer between S. epidermidis and S. aureus explained by CRISPR interference. S. aureus is known to collect a large variety of toxin and other virulence determinant genes from mobile genetic elements – in clear contrast to S. epidermidis, which virtually lacks such genes. This may be explained at least in part by the presence of CRISPR loci in S. epidermidis and their absence in S. aureus. CRISPR loci (clustered, regularly interspaced, short palindromic repeat loci) consist of repeats, between which DNA sequences are found that hybridize with components of foreign DNA. CRISPR RNA, which arises from CRISPR loci after transcription and processing, hybridizes to such foreign sequences and the foreign DNA is cleaved by products of the cas (CRISPR-associated genes) complex.
Certainly, the role of CRISPR in directing the gene flow from S. epidermidis to S. aureus is still hypothetical. First, CRISPR is only found in some S. epidermidis genomes. Second, some S. aureus strains contain CRISPR sequences but also MGEs. However, in those cases, MGEs may have been present before the acquisition of CRISPR. Third, one would expect characteristic nucleotide sequences of MGEs found in S. aureus to be present in S. epidermidis CRISPR spacer regions. Substantiating the role of CRISPR in limiting gene transfer from S. aureus to S. epidermidis will require exhaustive further investigation. For example, genome sequencing of more S. epidermidis strains should be performed to show whether CRISPR loci are more frequently present in S. epidermidis than S. aureus. Draft sequences obtained by pan-genome analysis do not necessarily allow a rigorous analysis of CRISPR presence, in part because CRISPR heterogeneity complicates identification. Furthermore, analysis of CRISPR spacer sequences may reveal sequences homologous to bacteriophages involved in gene transfer between staphylococci. So far, spacer analysis of S. epidermidis CRISPR sequences has not revealed such sequences; however, this may be related to our general lack of knowledge on interspecies phage-dependent gene transfer between staphylococci, which also requires more emphasis. Finally, retrospective analysis of historical strains may allow insight into whether CRISPR sequences were acquired by CRISPR-positive S. aureus strains before or after acquisition of MGEs.
Conclusions
Recent studies have provided additional examples supporting the hypothesis that S. epidermidis and other CoNS have an important function as providers of genes that contribute to the survival of S. aureus during infection and as a commensal. This hypothesis is based on the higher prevalence and, where information is available, earlier appearance of transferred determinants in S. epidermidis compared to S. aureus. The opposite scenario with S. aureus as the source appears much less likely because of (i) the general paucity of characteristic S. aureus genes that are found in S. epidermidis, and (ii) the fact that the transferred determinants are not restricted to a specific S. epidermidis clone, which would suggest more recent acquisition from S. aureus. However, more genetic data will be needed to confirm this scenario.
Possibly, CRISPR-mediated immunity of CoNS strains to DNA uptake may play a role in directing the gene flow between S. aureus and S. epidermidis. However, other yet unidentified mechanisms probably exist that resulted in the divergent evolution of S. aureus as a virulent pathogen on the one hand and CoNS as usually harmless commensals and only occasionally invasive pathogens on the other. Notably, these findings highlight the importance of research providing molecular insight into the function of genes facilitating survival of CoNS species in the human host, because they may ultimately contribute to the virulence potential of a dangerous pathogen.
Acknowledgements
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), U. S. National Institutes of Health (NIH).
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–32. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Prevost G, Mourey L, Colin DA, Menestrina G. Staphylococcal pore-forming toxins. Curr Top Microbiol Immunol. 2001;257:53–83. doi: 10.1007/978-3-642-56508-3_4. [DOI] [PubMed] [Google Scholar]
- 3.Wang R, Braughton KR, Kretschmer D, Bach TH, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–4. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 4.McCormick JK, Yarwood JM, Schlievert PM. Toxic shock syndrome and bacterial superantigens: an update. Annu Rev Microbiol. 2001;55:77–104. doi: 10.1146/annurev.micro.55.1.77. [DOI] [PubMed] [Google Scholar]
- 5.Rooijakkers SH, van Kessel KP, van Strijp JA. Staphylococcal innate immune evasion. Trends Microbiol. 2005;13:596–601. doi: 10.1016/j.tim.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Schlievert PM. Role of superantigens in human disease. J Infect Dis. 1993;167:997–1002. doi: 10.1093/infdis/167.5.997. [DOI] [PubMed] [Google Scholar]
- 7.Iwatsuki K, Yamasaki O, Morizane S, Oono T. Staphylococcal cutaneous infections: invasion, evasion and aggression. J Dermatol Sci. 2006;42:203–14. doi: 10.1016/j.jdermsci.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 8.Li M, Diep BA, Villaruz AE, Braughton KR, et al. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2009;106:5883–8. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster TJ. Immune evasion by staphylococci. Nat Rev Microbiol. 2005;3:948–58. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 10.von Eiff C, Becker K, Machka K, Stammer H, et al. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N Engl J Med. 2001;344:11–6. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 11.Wertheim HF, Melles DC, Vos MC, van Leeuwen W, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–62. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 12.Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest. 2003;111:1265–73. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diekema DJ, Pfaller MA, Schmitz FJ, Smayevsky J, et al. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin Infect Dis. 2001;32(Suppl 2):S114–32. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- 14.Otto M. Virulence factors of the coagulase-negative staphylococci. Front Biosci. 2004;9:841–63. doi: 10.2741/1295. [DOI] [PubMed] [Google Scholar]
- 15.Otto M. Staphylococcus epidermidis - the ‘accidental’ pathogen. Nat Rev Microbiol. 2009;7:555–67. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–9. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207–28. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rautenberg M, Joo HS, Otto M, Peschel A. Neutrophil responses to staphylococcal pathogens and commensals via the formyl peptide receptor 2 relates to phenol-soluble modulin release and virulence. Faseb J. 2011;25:1254–63. doi: 10.1096/fj.10-175208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheung GY, Rigby K, Wang R, Queck SY, et al. Staphylococcus epidermidis strategies to avoid killing by human neutrophils. PLoS Pathog. 2010;6:e1001133. doi: 10.1371/journal.ppat.1001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otto M. Molecular basis of Staphylococcus epidermidis infections. Semin Immunopathol. 2012;34:201–14. doi: 10.1007/s00281-011-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novick RP, Schlievert P, Ruzin A. Pathogenicity and resistance islands of staphylococci. Microbes Infect. 2001;3:585–94. doi: 10.1016/s1286-4579(01)01414-9. [DOI] [PubMed] [Google Scholar]
- 22.Holden MT, Feil EJ, Lindsay JA, Peacock SJ, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A. 2004;101:9786–91. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morikawa K, Inose Y, Okamura H, Maruyama A, et al. A new staphylococcal sigma factor in the conserved gene cassette: functional significance and implication for the evolutionary processes. Genes Cells. 2003;8:699–712. doi: 10.1046/j.1365-2443.2003.00668.x. [DOI] [PubMed] [Google Scholar]
- 24.Claverys JP, Martin B. Bacterial “competence” genes: signatures of active transformation, or only remnants? Trends Microbiol. 2003;11:161–5. doi: 10.1016/s0966-842x(03)00064-7. [DOI] [PubMed] [Google Scholar]
- 25.Lindsay JA, Holden MT. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 2004;12:378–85. doi: 10.1016/j.tim.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Archer GL, Johnston JL. Self-transmissible plasmids in staphylococci that encode resistance to aminoglycosides. Antimicrob Agents Chemother. 1983;24:70–7. doi: 10.1128/aac.24.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–55. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito T, Katayama Y, Asada K, Mori N, et al. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:1323–36. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambers HF, Hartman BJ, Tomasz A. Increased amounts of a novel penicillin-binding protein in a strain of methicillin-resistant Staphylococcus aureus exposed to nafcillin. J Clin Invest. 1985;76:325–31. doi: 10.1172/JCI111965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Couto I, de Lencastre H, Severina E, Kloos W, et al. Ubiquitous presence of a mecA homologue in natural isolates of Staphylococcus sciuri. Microb Drug Resist. 1996;2:377–91. doi: 10.1089/mdr.1996.2.377. [DOI] [PubMed] [Google Scholar]
- 31.Schnellmann C, Gerber V, Rossano A, Jaquier V, et al. Presence of new mecA and mph(C) variants conferring antibiotic resistance in Staphylococcus spp. isolated from the skin of horses before and after clinic admission. J Clin Microbiol. 2006;44:4444–54. doi: 10.1128/JCM.00868-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito T, Hiramatsu K, Tomasz A, de Lencastre H, et al. Guidelines for reporting novel mecA gene homologues. Antimicrob Agents Chemother. 2012;56:4997–9. doi: 10.1128/AAC.01199-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsubakishita S, Kuwahara-Arai K, Baba T, Hiramatsu K. Staphylococcal cassette chromosome mec-like element in Macrococcus caseolyticus. Antimicrob Agents Chemother. 2010;54:1469–75. doi: 10.1128/AAC.00575-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Alvarez L, Holden MT, Lindsay H, Webb CR, et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chatterjee SS, Chen L, Joo HS, Cheung GY, et al. Distribution and regulation of the mobile genetic element-encoded phenol-soluble modulin PSM-mec in methicillin-resistant Staphylococcus aureus. PLoS ONE. 2011;6:e28781. doi: 10.1371/journal.pone.0028781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Queck SY, Khan BA, Wang R, Bach TH, et al. Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathog. 2009;5:e1000533. doi: 10.1371/journal.ppat.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu S, Piscitelli C, de Lencastre H, Tomasz A. Tracking the evolutionary origin of the methicillin resistance gene: cloning and sequencing of a homologue of mecA from a methicillin susceptible strain of Staphylococcus sciuri. Microb Drug Resist. 1996;2:435–41. doi: 10.1089/mdr.1996.2.435. [DOI] [PubMed] [Google Scholar]
- 38.Tsubakishita S, Kuwahara-Arai K, Sasaki T, Hiramatsu K. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob Agents Chemother. 2010;54:4352–9. doi: 10.1128/AAC.00356-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbier F, Ruppe E, Hernandez D, Lebeaux D, et al. Methicillin-resistant coagulase-negative staphylococci in the community: high homology of SCCmec IVa between Staphylococcus epidermidis and major clones of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2010;202:270–81. doi: 10.1086/653483. [DOI] [PubMed] [Google Scholar]
- 40.Wisplinghoff H, Rosato AE, Enright MC, Noto M, et al. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob Agents Chemother. 2003;47:3574–9. doi: 10.1128/AAC.47.11.3574-3579.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Archer GL, Thanassi JA, Niemeyer DM, Pucci MJ. Characterization of IS1272, an insertion sequence-like element from Staphylococcus haemolyticus. Antimicrob Agents Chemother. 1996;40:924–9. doi: 10.1128/aac.40.4.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones RN, Barry AL, Gardiner RV, Packer RR. The prevalence of staphylococcal resistance to penicillinase-resistant penicillins. A retrospective and prospective national surveillance trial of isolates from 40 medical centers. Diagn Microbiol Infect Dis. 1989;12:385–94. doi: 10.1016/0732-8893(89)90108-9. [DOI] [PubMed] [Google Scholar]
- 43.Schmitz FJ, Verhoef J, Fluit AC. Prevalence of resistance to MLS antibiotics in 20 European university hospitals participating in the European SENTRY surveillance programme. Sentry Participants Group. J Antimicrob Chemother. 1999;43:783–92. doi: 10.1093/jac/43.6.783. [DOI] [PubMed] [Google Scholar]
- 44.Lacey RW. Transfer of chromosomal genes between staphylococci in mixed cultures. J Gen Microbiol. 1972;71:399–401. doi: 10.1099/00221287-71-2-399. [DOI] [PubMed] [Google Scholar]
- 45.Cohen S, Sweeney HM. Effect of the prophage and penicillinase plasmid of the recipient strain upon the transduction and the stability of methicillin resistance in Staphylococcus aureus. J Bacteriol. 1973;116:803–11. doi: 10.1128/jb.116.2.803-811.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tormo MA, Knecht E, Gotz F, Lasa I, et al. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology. 2005;151:2465–75. doi: 10.1099/mic.0.27865-0. [DOI] [PubMed] [Google Scholar]
- 47.Wielders CL, Vriens MR, Brisse S, de Graaf-Miltenburg LA, et al. In-vivo transfer of mecA DNA to Staphylococcus aureus. Lancet. 2001;357:1674–5. doi: 10.1016/s0140-6736(00)04832-7. [DOI] [PubMed] [Google Scholar]
- 48.Diep BA, Gill SR, Chang RF, Phan TH, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–9. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 49.Diep BA, Stone GG, Basuino L, Graber CJ, et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2008;197:1523–30. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- 50.Montgomery CP, Boyle-Vavra S, Daum RS. The arginine catabolic mobile element is not associated with enhanced virulence in experimental invasive disease caused by the community-associated methicillin-resistant Staphylococcus aureus USA300 genetic background. Infect Immun. 2009;77:2650–6. doi: 10.1128/IAI.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–68. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diep BA, Otto M. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 2008;16:361–9. doi: 10.1016/j.tim.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joshi GS, Spontak JS, Klapper DG, Richardson AR. Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol Microbiol. 2011;82:9–20. doi: 10.1111/j.1365-2958.2011.07809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Urushibara N, Kawaguchiya M, Kobayashi N. Two novel arginine catabolic mobile elements and staphylococcal chromosome cassette mec composite islands in community-acquired methicillin-resistant Staphylococcus aureus genotypes ST5-MRSA-V and ST5-MRSA-II. J Antimicrob Chemother. 2012;67:1828–34. doi: 10.1093/jac/dks157. [DOI] [PubMed] [Google Scholar]
- 55.Bartels MD, Hansen LH, Boye K, Sorensen SJ, et al. An unexpected location of the arginine catabolic mobile element (ACME) in a USA300-related MRSA strain. PLoS ONE. 2011;6:e16193. doi: 10.1371/journal.pone.0016193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shore AC, Rossney AS, Brennan OM, Kinnevey PM, et al. Characterization of a novel arginine catabolic mobile element (ACME) and staphylococcal chromosomal cassette mec composite island with significant homology to Staphylococcus epidermidis ACME type II in methicillin-resistant Staphylococcus aureus genotype ST22-MRSA-IV. Antimicrob Agents Chemother. 2011;55:1896–905. doi: 10.1128/AAC.01756-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barbier F, Lebeaux D, Hernandez D, Delannoy AS, et al. High prevalence of the arginine catabolic mobile element in carriage isolates of methicillin-resistant Staphylococcus epidermidis. J Antimicrob Chemother. 2011;66:29–36. doi: 10.1093/jac/dkq410. [DOI] [PubMed] [Google Scholar]
- 58.Miragaia M, de Lencastre H, Perdreau-Remington F, Chambers HF, et al. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PLoS ONE. 2009;4:e7722. doi: 10.1371/journal.pone.0007722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pi B, Yu M, Chen Y, Yu Y, et al. Distribution of the ACME-arcA gene among meticillin-resistant Staphylococcus haemolyticus and identification of a novel ccr allotype in ACME-arcA-positive isolates. J Med Microbiol. 2009;58:731–6. doi: 10.1099/jmm.0.007351-0. [DOI] [PubMed] [Google Scholar]
- 60.Li M, Du X, Villaruz AE, Diep BA, et al. MRSA epidemic linked to a quickly spreading colonization and virulence determinant. Nat Med. 2012;18:816–9. doi: 10.1038/nm.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kriegeskorte A, Peters G. Horizontal gene transfer boosts MRSA spreading. Nat Med. 2012;18:662–3. doi: 10.1038/nm.2765. [DOI] [PubMed] [Google Scholar]
- 62.Holden MT, Lindsay JA, Corton C, Quail MA, et al. Genome sequence of a recently emerged, highly transmissible, multi-antibiotic- and antiseptic-resistant variant of methicillin-resistant Staphylococcus aureus, sequence type 239 (TW). J Bacteriol. 2010;192:888–92. doi: 10.1128/JB.01255-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soderquist B, Andersson M, Nilsson M, Nilsdotter-Augustinsson A, et al. Staphylococcus epidermidis surface protein I (SesI): a marker of the invasive capacity of S. epidermidis? J Med Microbiol. 2009;58:1395–7. doi: 10.1099/jmm.0.008771-0. [DOI] [PubMed] [Google Scholar]
- 64.Massey RC, Horsburgh MJ, Lina G, Hook M, et al. The evolution and maintenance of virulence in Staphylococcus aureus: a role for host-to-host transmission? Nat Rev Microbiol. 2006;4:953–8. doi: 10.1038/nrmicro1551. [DOI] [PubMed] [Google Scholar]
- 65.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–75. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 66.Bikard D, Marraffini LA. Innate and adaptive immunity in bacteria: mechanisms of programmed genetic variation to fight bacteriophages. Curr Opin Immunol. 2012;24:15–20. doi: 10.1016/j.coi.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 67.Conlan S, Mijares LA, Comp Seq Program N, Becker J, et al. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol. 2012;13:R64. doi: 10.1186/gb-2012-13-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–71. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]