Abstract
Despite the success of highly active antiretroviral therapy in combating human immunodeficiency virus type 1 (HIV-1) infection, the virus still persists in viral reservoirs, often in a state of transcriptional silence. This review focuses on the HIV-1 protein and regulatory machinery and how expanding knowledge of the function of individual HIV-1-coded proteins has provided valuable insights into understanding HIV transcriptional regulation in selected susceptible cell types. Historically, Tat has been the most studied primary transactivator protein, but emerging knowledge of HIV-1 transcriptional regulation in cells of the monocyte–macrophage lineage has more recently established that a number of the HIV-1 accessory proteins like Vpr may directly or indirectly regulate the transcriptional process. The viral proteins Nef and matrix play important roles in modulating the cellular activation pathways to facilitate viral replication. These observations highlight the cross talk between the HIV-1 transcriptional machinery and cellular activation pathways. The review also discusses the proposed transcriptional regulation mechanisms that intersect with the pathways regulated by microRNAs and how development of the knowledge of chromatin biology has enhanced our understanding of key protein–protein and protein–DNA interactions that form the HIV-1 transcriptome. Finally, we discuss the potential pharmacological approaches to target viral persistence and enhance effective transcription to purge the virus in cellular reservoirs, especially within the central nervous system, and the novel therapeutics that are currently in various stages of development to achieve a much superior prognosis for the HIV-1-infected population.
Introduction
Since the discovery of the human immunodeficiency virus type 1 (HIV-1) more than 28 years ago, the spread of the virus has grown from an epidemic to a severe global pandemic with approximately 33.3 million people living with the virus at the end of year 2010 (UNAIDS; http://www.unaids.org/globalreport/documents/20101123_GlobalReport_full_en.pdf.). The introduction of highly active antiretroviral therapy (HAART) in 1996 has, over time, been a great medical success in terms of increasing survival and improving the quality of life of the infected patient population and has paved the way for an even more effective generation of therapeutic strategies. Successful HAART reduces the plasma viraemia to <50 copies ml−1 of blood, allowing for immune reconstitution and patient improvement and stabilization. However, once the infection has been established, HIV-1 finds its way to privileged sites, likely involving a number of tissues and intracellular environments from which currently available antiretroviral therapies cannot clear the infection. The resting CD4+ T-cells and cells of the monocyte–macrophage lineage are thought to be the major reservoirs of latent HIV-1 infection along with dendritic cells and haematopoietic stem cells in the bone marrow (Alexaki & Wigdahl, 2008; Alexaki et al., 2008; Le Douce et al., 2010; Loré et al., 2005; McNamara & Collins, 2011). These cell lineages are relatively insensitive to therapy, are not detected by immune surveillance, and have relatively longer half-lives. It is well known that latent HIV-1 infection is established early in infection, and reports have identified latent HIV-1 infection in CD4+ T-cells even in patients who are treated with HAART within the first week of infection (Chun et al., 1998). Moreover, HIV-1 infection also occurs in the central nervous system (CNS), bone marrow and gut-associated lymphoid tissue; these sites also likely serve as common sanctuaries and prevent complete eradication of the virus because they serve as pharmacological ‘privileged’ sites where drug penetration is poor (Chun et al., 2008; Davis et al., 1992; Solas et al., 2003). Furthermore, the pharmacokinetics of drugs in these sites is poorly understood.
Therapeutic strategies that target viral latency and/or replication within these privileged reservoirs must be incorporated into current HAART regimens to achieve effective eradication of HIV-1. Another problem requiring attention is how to ensure the penetration and absorption of current and next generation therapeutics into those tissues and cells serving as reservoirs. Strategies designed to intensify HAART by adding candidate drugs like hydroxyurea and cyclophosphamide to activate latent or quiescent HIV-1 provirus did not succeed in decreasing the latently infected reservoirs in otherwise well-controlled patients infected with HIV-1 (Bartlett et al., 2002; Coleman & Wu, 2009; Lisziewicz et al., 2003; Lori et al., 1994). This fact implies that new categories of drugs are needed that can target the virus at pathways that have not previously been considered prime targets. One such process, which we discuss in detail, centres on HIV transcription. Promise is evident from studies that have demonstrated that intervention at the molecular level can lead to HIV-1 reactivation, suggesting that proviral latency is amenable to therapeutic intervention. The challenge is to target expressly the HIV-1-specific processes with minimal effect on the normal physiology of the cell.
In addition to the well-studied Tat protein, another good example of a viral protein that can alter the course of HIV transcription is viral protein R (Vpr). Although Vpr has been deemed an ‘accessory’ protein, it may be an attractive target because its function in nuclear translocation of the preintegration complex (PIC) of HIV-1 in macrophage lineage cells is well established. A big challenge in the development of effective anti-HIV-1 compounds has been the difficulty encountered in evaluating viral proteins in the context of HIV-1 replication in appropriate in vitro surrogate cellular phenotypes. Moreover, the use of non-physiological concentrations of viral proteins and/or associated cellular partners has often attracted criticism, limiting their utility to assess the target efficacy of new antiviral agents. Proteins like Tat and Vpr, besides having a strong intracellular role, are present in the extracellular milieu as well. Studies have shown that Tat released by infected cells can be rapidly internalized through its basic domain and can exert autocrine and paracrine activities that activate various signalling pathways and also induce neurotoxicity in the CNS (Brigati et al., 2003; Kilareski et al., 2009; Strazza et al., 2011). We focus specifically on how the repertoire of HIV-1 proteins functioning in the complex process of HIV-1 transcription can provide valuable clues for more effective therapies. Recent experimental results suggest that HIV-1 replication and the cellular microRNA (miRNA) pathway intersect at several post-transcriptional levels (Houzet & Jeang, 2011; Ouellet et al., 2009; Sánchez-Del Cojo et al., 2011). HIV-1 infection alters the levels of several miRNAs that regulate the levels of HIV-1 proteins involved in the viral life cycle and that regulate the cellular proteins that are required for successful completion of the HIV-1 life cycle within susceptible cell types (Bignami et al., 2012; Sun et al., 2011; Witwer et al., 2012). Not only do HIV-1 proteins like Tat, Vpr and Nef influence the cellular miRNA machinery, but conflicting reports have also suggested that the integrated genome of HIV-1 itself encodes miRNAs (Hayes et al., 2011; Narayanan et al., 2011). The pool of transcription factors available at any time responds according to their native expression profiles in a cell-type-dependent fashion along with the alterations that HIV-1 is able to direct by indirectly modulating the miRNA machinery. All of these studies highlight the importance of understanding the reciprocal interactions between HIV-1 and the miRNA machinery in a manner designed to extract a functional therapeutic tool directed against HIV-1.
This review also discusses the molecular topology of the integrated HIV-1 genome and how the chromatin environment and the epigenetic regulatory events intersect with transcription from the integrated promoter of HIV-1, the long-terminal repeat (LTR). The existence of active APOBEC3G (apolipoprotein B mRNA-editing enzyme catalytic polypeptide 3G) in resting CD4+ T-cells, which causes G-to-A hypermutations during reverse transcription and results in the production of defective viral genomes along with histone modifications like methylation and acetylation, is one of the factors that governs the establishment and maintenance of transcriptional latency in resting CD4+ T-cells (Chiu et al., 2005; Lusic et al., 2003; Van Lint, 2000). A deeper comprehension of the mechanism of HIV-1 latency may lead to the development of better therapeutic strategies, which, when combined with the current HAART regimen, may help eradicate the pool of latently infected cells. We describe current efforts in evaluating still-in-development treatment strategies and concepts based on activation/suppression of HIV-1 transcription, DNA- and histone-methylation, and histone deacetylase (HDAC) inhibitors.
Impact of cellular phenotype on HIV-1 transcription
HIV-1 can infect a variety of immune cells including CD4+ T-cells, macrophages, dendritic cells, bone marrow progenitor cells and brain microglial cells. It has evolved the ability to infect even non-dividing cells. It can infect non-immune cells like astrocytes, which are likely an important target in the CNS, and it has recently been shown that HIV-1-infected astrocytes are involved in disruption of the blood–brain barrier. This detrimental process contributes to HIV-1-associated neurocognitive impairment (Eugenin et al., 2011). Chronic HIV-1 infection has also been shown to dysregulate the gene expression profile of astrocytes, leading to neuropathogenesis (Borjabad et al., 2010). The role of lipid mediators in the process of HIV-1-mediated effects in the CNS is an area that has recently received some attention, but the mechanisms are not yet clear (Bertin et al., 2012).
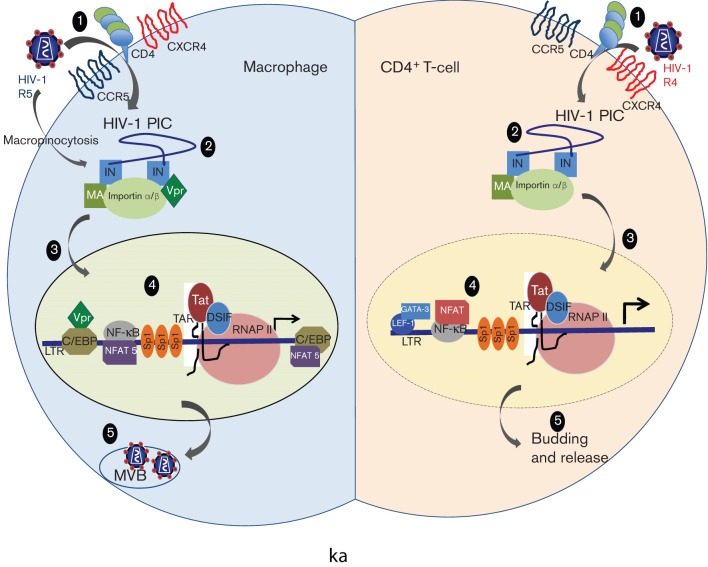
To maximize viral replication, HIV-1 adapts transcription of its integrated proviral genome by exploiting the specific cellular environment (Fig. 1), inducing cellular stimulatory events and impairing transcriptional inhibitory processes. HIV-1 replication in cells of the monocyte–macrophage lineage necessitates a number of specialized events and differs in many respects from that in CD4+ T-cells. Besides exhibiting a different tropism for CD4+ T-lymphocytes and macrophages, productive HIV-1 infection occurs independently of cellular DNA synthesis in macrophages (Fenyö et al., 1988; Weinberg et al., 1991). Because macrophages are non-dividing cells, the HIV-1 PIC is imported through an intact nuclear membrane. In contrast, in T-cells, dissolution of the nuclear membrane during cell division allows the PIC easy access to the host DNA. Moreover, the assembly and budding of viral particles take place in cytoplasmic vacuoles in monocyte–macrophage lineage cells, but not in CD4+ T-cells (Orenstein et al., 1988). It has also been demonstrated that the accessory genes of HIV-1 such as Vpr may have distinct functions in primary macrophages compared with CD4+ T-cells (Sherman et al., 2002; Subbramanian et al., 2002; Swingler et al., 2003). For instance, a Vpr-deficient virus could efficiently infect T-lymphocytes in culture, but was severely compromised in its potential to infect macrophages (Eckstein et al., 2001). Another key step in the viral life cycle, LTR-directed transcription, assumes a different landscape in the two susceptible cell types because the transcription factor expression profile differs in the two cell lineages. For example, some transcription factors (TFs) such as GATA-3, LEF-1, ETS-1 and nuclear factor for activated T-cells (NFATs) are lymphoid or T-cell specific, whereas others, such as the CCAAT/enhancer-binding protein β (C/EBPβ), are necessary for HIV-1 transcription in macrophage lineage cells, but not in CD4+ T-lymphocytes (Henderson & Calame, 1997; Lee et al., 2002; Liu et al., 2010) (Fig. 1). The differential utilization of cellular factors by HIV-1 in T-cells versus macrophages with respect to efficient transcription has in a way stalled efforts to selectively pick a particular set of TF–DNA interactions and use them for therapeutic intervention. This aspect is further complicated by the evolution of genetic variation in the LTR that compromises the binding of cognate TFs and at the same time creates new binding affinities for others (Li et al., 2011). These interactions are relevant in the early phase of HIV-1 transcription, before Tat-directed transcriptional transactivation ensues and also in late-stage disease when the low-level persistent transcription from the LTR continues. Moreover, using brain-derived Tat sequences from demented and non-demented HIV-1-infected patients, it has been shown that alterations in HIV-1 Tat protein, which are independent of its transactivation potential, exert pathogenic effects and contribute to the development of HIV-1-associated dementia (Boven et al., 2007).
Fig. 1.
Differences between the HIV-1 replication cycle in CD4+ T-cells and macrophages. (1) HIV-1 isolates show tropism in the two cell types with CCR5 primarily being utilized on macrophages and CXCR4 primarily used on CD4+ T-cells. Moreover, HIV-1 can alternatively enter macrophages via macropinocytosis. (2) Proteins like Vpr are essential components of the PIC in macrophages, which assist nuclear transport via an intact nuclear membrane (3), whereas in T-cells Vpr is dispensable for integration and the nuclear membrane has increased permeability. (4) A differential transcriptional programme is utilized in the two cell types with the factors like C/EBP and NFAT5 playing a crucial role in macrophages. (5) Viral assembly and budding in multi-vesicular bodies (MVBs) occurs in macrophages, but not in T-cells.
HIV-1 transcription is regulated differently in T-cells than in cells of the monocyte–macrophage lineage (Fig. 1) (Herbein & Varin, 2010; Rohr et al., 2003). The expression of different isoforms of the transcription factor C/EBPβ, which are generated by alternative translational initiation, has been shown to regulate HIV-1 LTR-directed transcription in cells of the monocyte–macrophage lineage. The 30–37 kDa activating isoform is required for HIV-1 transcription in cells of the monocyte–macrophage lineage, but not in CD4+ T-cells (Henderson & Calame, 1997). Conversely, the smaller molecular mass isoform, 16–23 kDa, is the dominant-negative inhibitor and reduces HIV-1 transcription in differentiated THP-1 macrophages as a result of induction by beta interferon (IFN-β) (Honda et al., 1998). Besides the two well-characterized binding sites for C/EBP (−116/−103 and −176/−162), unpublished reports (S. Dahiya and B. Wigdahl, unpublished results) indicate the presence of another element downstream of the transcription start site that can interact with C/EBP in the U-937 promonocytic cell line. The same site is competitively occupied by some isoforms of NFAT when present. This observation further extends our understanding of how differential utilization of a particular DNA element in the HIV-1 LTR assumes different binding phenotypes contingent on the cellular phenotype (Fig. 1). In addition to cellular phenotype differences, the sequence variations observed in selected transcription factor-binding sites affect HIV-1 transcription. For instance, we demonstrated previously that sequence variation at both the C/EBP and ATF/CREB sites affects the basal and stimulated LTR activity in monocyte–macrophage lineage cells (Ross et al., 2001). In addition, these TF–DNA interactions respond to extracellular stimuli. For example, treatment with granulocyte-macrophage colony-stimulating factor during differentiation of monocytes to macrophages resulted in decreased viral transcription (R5 strain) in contrast to treatment with macrophage colony-stimulating factor. Viral suppression in this case was related to expression of the inhibitory C/EBPβ isoform (Komuro et al., 2003). These authors also demonstrated the inverse regulation of the Src-like tyrosine kinase Hck and hypothesized that the heterogeneity in macrophage susceptibility to HIV-1 was related to distinct regulation of Hck and the inhibitory isoform of C/EBPβ. These results reinforced the observation that the low level of susceptibility of alveolar macrophages to HIV-1 infection may be dependent on the expression of the dominant-negative inhibitory C/EBPβ isoform (Honda et al., 1998). Activated allogeneic lymphocytes have been shown to reduce the expression of C/EBPβ in differentiated THP-1 cells and alveolar macrophages, increasing their susceptibility to HIV-1 (Hoshino et al., 2002). The differential regulation of LTR-directed transcription via the C/EBP-binding sites was also established using the simian immunodeficiency virus (SIV) model; the authors analysed the functional role of two C/EBPβ-binding sites in activation and IFN-β suppression (Ravimohan et al., 2010). Thus, HIV-1 adaptation to cells of the monocyte–macrophage lineage involves a distinct transcriptional programme, and its regulation varies considerably with the stage of cellular differentiation. Besides exhibiting differential expression of key transcription factors that are responsive to cellular stimulation, these cells were more resistant to the apoptosis induction potential of HIV-1 compared with T-cells (Le Douce et al., 2010). All of these variables make the infected cells of the monocyte–macrophage lineage a major obstacle in the eradication of HIV-1. They may represent a greater portion of the total infected cell population during late-stage disease when CD4+ T-cells have largely been depleted and may contribute significantly at this stage to the circulating levels of virus in vivo (Igarashi et al., 2001; Orenstein et al., 1997). More recently, the role of TCF/LEF family members in regulating the transcriptional activity of HIV-1 in astrocytes has expanded the function of C/EBP isoforms to astrocytes (Narasipura et al., 2012).
Molecular topology of the HIV-1 transcriptome
Integration of the HIV-1 proviral DNA into the host genome is a critical step in the viral life cycle. Thus, inevitably, the chromatin environment at the site of integration plays a crucial role in the regulation of gene expression from the LTR. Regardless of the site of integration, efficient and sustained transcription also requires Tat and other accessory proteins along with the host cellular factors. Various mechanisms restrain viral production. The establishment and maintenance of proviral DNA quiescence are dependent on the chromatin environment and are subject to the same epigenetic mechanisms of regulation as the host as previously reviewed (Hakre et al., 2011). Heterochromatin, as opposed to euchromatin, is highly compact and structured, making the integrated proviral LTR inaccessible to the cellular and viral factors, thereby restricting the initiation of transcription from the integrated proviral DNA template. It is still unclear which integration events yield replication-competent latent proviral DNA genomes because most of the integrants have defective genomes and are replication-incompetent (Chun et al., 1997). This observation may be explained in part by the strong restriction that cellular APOBEC3G exerts on the genome by inducing hypermutations (Malim, 2009). Studies have shown that HIV-1 preferentially integrates into the actively transcribing genes in the host genome during productive infection of cultured T-cells (Schröder et al., 2002). This process is probably the result of interactions of the PIC with host factors that are concentrated in the regions of heavy transcriptional activity. Numerous cellular proteins have been shown to interact with the PIC, thus driving the selection of the site of integration. One such interaction is with the host protein LEDGF/p75, which has been shown to direct the integration of HIV-1 DNA into active sites of the genome (Ciuffi et al., 2005). The presence of LEDGF favours integration into GC-rich regions, whereas its absence results in integration into AT-rich regions. Thus, the level of LEDGF may control integration into a heterochromatin region compared to a euchromatin area. This hypothesis was well supported in studies that showed that the integration sites of quiescent/inducible HIV-1 vectors in T-cell lines could be within the heterochromatin or the actively transcribed genes (Lewinski et al., 2005). It was also shown that HIV-1 PIC interacts with the SET/TAF-Ib complex and that the SET complex acts as a barrier to autointegration of HIV-1 into its own genome (Yan et al., 2009). SET/TAF-Ib has been shown to be associated with transcriptional repression because it binds unacetylated, hypoacetylated or repressively marked histones (Kutney et al., 2004).
It is well established that, regardless of the site of integration, HIV-1 DNA is packaged in such a fashion that two nucleosomes (Nuc-0 and Nuc-1) are positioned strictly with respect to cis-acting regulatory elements on the LTR. Nuc-0 is positioned upstream of the modulatory region, and Nuc-1 is immediately downstream of the transcription start site (+10 to +155). A large body of evidence suggests that Nuc-1 needs to be remodelled for transcriptional activation and that epigenetic silencing at Nuc-1 might assume a state of transcription repression/latency (Colin & Van Lint, 2009).
HIV-1 provirus is in a dynamic state in the cellular genome, and interactions between various host and viral factors with histone acetyltransferases (HATs) and HDACs regulate proviral gene expression. Histones can be methylated as well as acetylated; it is important to note that the histone methyltransferases (HMTs) EZH2 and SUV39H1 have been shown to regulate HIV-1 transcription by inducing histone H3 at lysine 9 (H3K9) methylation. HP1γ binds to methylated H3K9 and further imposes a repressive phenotype on the local chromatin environment (du Chéné et al., 2007). HIV-1 LTR DNA can also be methylated on cytosine residues (CpG islands) and can inhibit viral transcription (Kauder et al., 2009). However, analysis of integrated HIV-1 genomes in resting CD4+ T-cells from HIV-1-infected patients has demonstrated the infrequent methylation of the HIV LTR. Although evidence is accumulating that histone methylation regulates HIV expression, the role of DNA methylation in maintaining HIV-1 latency remains to be explored further as previously reviewed (Choudhary & Margolis, 2011). Recent studies using functional genomic screens (Brass et al., 2008) to identify host proteins crucial for HIV-1 infection and analysing the global landscape of interactions between the HIV-1 and human proteins (Jäger et al., 2012) have identified some key cellular participants that play crucial regulatory roles in the HIV-1 life cycle (König et al., 2008; Yeung et al., 2009a). These studies highlight the importance of studying the molecular architecture of the HIV-1 genome along with the networks that HIV-1 establishes in various stages of its life cycle.
As discussed in the next section, the viral protein Tat plays a key role in allowing generation of full-length transcripts from the integrated genome via its ability to recruit positive transcription elongation factor b (P-TEFb) (Fig. 2). It was recently shown that Tat also recruits elongation factor ELL2 to P-TEFb for more coordinated activation of HIV-1 transcription. Tat also recruits acetyltransferase p300/CBP-associated factor (PCAF), which in turn acetylates Tat and enhances its ability to recruit P-TEFb. In several cell line model systems, Tat has been shown to promote binding of the ATP-dependent remodelling complex SWI/SNF and the histone-modifying enzymes p300 and CREB-binding protein (CBP) as well as the histone chaperone protein nucleosome assembly protein 1 (Vardabasso et al., 2008). These transcription factors further assist in chromatin unfolding and efficient transcription. Thus, the absence or presence of Tat governs the efficiency of LTR-directed transcription in a chromatin environment.
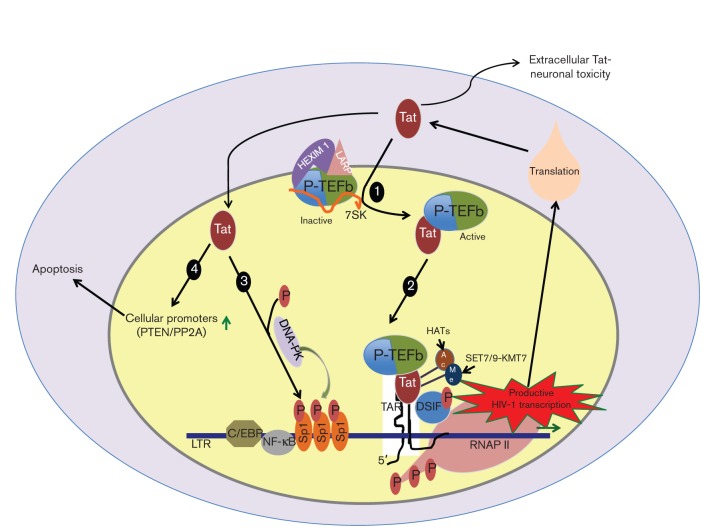
Fig. 2.
HIV-1 Tat interactions and transcriptional regulation. (1) Tat interacts with P-TEFb and triggers its release from the inactive complex formed with 7SK, HEXIM-1 and La ribonucleoprotein domain family member (LARP). (2) The active P-TEFb is recruited to the LTR as a result of the interaction of Tat with the TAR element formed at the 5′ end of the nascent HIV-1 RNA. This event is followed by phosphorylation of the CTD of RNAP II by the CDK9 component of P-TEFb. These phosphorylation events help RNAP II to shift from an initiation mode to an elongation mode, resulting in production of full-length HIV-1 mRNA that exits to the cytosol to be used to make HIV-1 proteins. In addition, the full activity of Tat is associated with its post-translation modifications, namely acetylation and methylation. (3) Tat also regulates the activity of Sp1 in LTR-directed transcription by recruiting DNA-PK to phosphorylate Sp1. (4) Tat activates other cellular promoters such as PTEN and PP2A and is secreted into the extracellular milieu, which has been shown to play a crucial role in apoptosis of CD4+ T-cells and to mediate neuronal toxicity.
HIV-1 protein machinery: from assisting self to combating non-self
In addition to the three structural genes, gag, pol andenv, the compact genome of HIV-1 is equipped with six additional genes. Two of them, tat and rev, encode essential regulatory proteins along with four small accessory factors, Vif, Vpr, Vpu and Nef, which have been shown to be dispensable for viral replication in some cell types, yet they are strongly maintained in the context of natural infections in vivo (Kirchhoff, 2010; Malim & Emerman, 2008). Accumulating evidence, which this review discusses, has identified crucial roles that these accessory proteins, especially Vpr and Nef, serve in the intricate process of viral transcription and replication in counteracting the intracellular proteins that human and other mammals have evolved as a defence against pathogenic viral invasions. In this section, we discuss the viral proteins that play key roles in the sophisticated process of LTR-directed, virus-specific transcription (Figs 2 and 3).
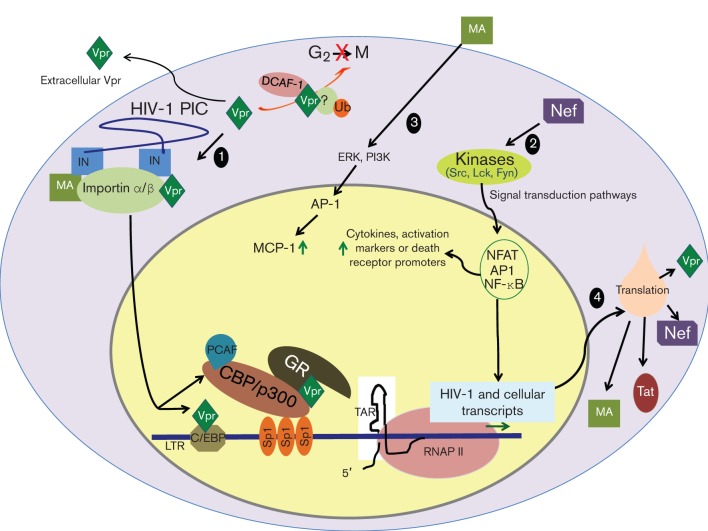
Fig. 3.
HIV-1 protein battery other than Tat and cellular interactions converging on the LTR. (1) The Vpr that is associated with the HIV-1 PIC assists in nuclear importation of the PIC and acts as a transactivator of the LTR. The latter function is achieved via two pathways: (a) association with the LTR in a ternary complex with Sp1 and formation of a complex with GR and HAT-CBP/p300 and (b) formation of a complex with C/EBP at C/EBP site I in the LTR and binding directly to the LTR in the region of C/EBP site I and the NF-κB site. Moreover, Vpr triggers a G2/M arrest in infected cells that involves ubiquitination of an unknown protein, which is recruited to Vpr via interaction with DCAF-1. The G2/M arrest is associated with enhanced transcription from the LTR. Vpr also acts as an extracellular protein that is secreted outside the cell and enters cells from the extracellular milieu. (2) HIV-1 Nef influences the cellular activation pathways that converge on the LTR-directed transcription. It activates important cellular signal transduction pathways that activate key transcription factors like NFAT, AP-1 and NF-κB, which regulate expression from the HIV-1 promoter. Nef also regulates the expression of cytokines and death receptor promoters. (3) MA protein activates key upstream cellular kinases that result in activation of the AP-1 transcription factor and eventual upregulation of AP-1-dependent genes, such as MCP-1, that are crucial in HIV-1 expression. Moreover, MA is part of the PIC and assists in the nuclear transport of the PIC. (4) Once productive transcription from the HIV-1 promoter ensues, the resulting robust translation of HIV-1 proteins ramps up the HIV-1 replication machinery and eventually results in progeny formation.
Tat: a multi-functional transcriptional transactivator
HIV-1 Tat is an early regulatory protein of 101 aa, encoded by two exons. The first exon encodes the first 72 aa and is sufficient for the transactivation function of Tat. However, several studies have suggested that the second exon of Tat is also important for transactivation (Jeang et al., 1993), transrepression (Howcroft et al., 1993) and virus replication (Neuveut & Jeang, 1996). Moreover, functions like modulation of host cell cytoskeleton are exclusively attributable to the second exon (López-Huertas et al., 2010). Besides being an intracellular protein, Tat is also released into the extracellular milieu from productively infected cells; this form can enter target cells and exert its effects. Studies of Tat-derived peptides have demonstrated that residues 48–60 form the basic domain (the protein transduction domain) and account for the functional internalization into cells (Futaki et al., 2001) and that cellular heparan sulfate proteoglycans act as low-affinity cellular receptors for extracellular Tat (Tyagi et al., 2001). Mutational analysis of HIV-1 Tat has identified two important functional domains: an activation domain that mediates its interactions with cellular machinery and an arginine-rich region that is required for binding to the transactivation responsive element (TAR) region of the nascent HIV-1 mRNA (Karn, 2011).
Productive transcription of the HIV-1 genome requires RNA polymerase to work in concert with Tat (Fig. 2). Tat and its team of cofactors function in a highly orchestrated manner to tightly control transcriptional elongation. In the absence of Tat, the basal level of RNA polymerase II (RNAP II) transcription of the HIV-1 provirus is low due to the action of negative factors (NELF and DSIF) that limit elongation. At some point, RNAP II has been shown to be able to transcribe the proviral genome, and the transcript is spliced into an mRNA that encodes Tat. Once it is imported into the nucleus, Tat acts in a positive-feedback loop to strongly activate RNAP II-driven elongation. The level of activation observed in several experimental systems is as large as 100-fold. Tat achieves this by directly binding to P-TEFb (Peterlin & Price, 2006). P-TEFb is a kinase complex consisting of a regulatory cyclin T1 (CycT1) subunit and a catalytic kinase subunit, Cdk-9. Tat binds directly to CycT1; the Tat–P-TEFb complex then binds to the structured TAR element that forms within the first 65 nt of the nascent RNA. Once at this site, the Cdk-9 component of P-TEFb potently stimulates transcriptional elongation by a two-pronged mechanism. First, it phosphorylates the negative elongation factor that removes a powerful block to elongation. Second, it phosphorylates the C-terminal domain (CTD) of RNA polymerase II (Ping & Rana, 2001) (Fig. 2). These two events result in multi-fold activation of the processivity of the polymerase (Karn, 2011). Elucidation of the Tat–P-TEFb crystal structure has clearly shown that Tat forms extensive contacts with both the CycT1 and Cdk-9 subunits of P-TEFb, which results in significant conformational changes and eventual constitutive activation of the enzyme (Tahirov et al., 2010). Tat has also been shown to influence phosphorylation of transcription factors including specificity protein 1 (Sp1), CREB and eIF-2α (Demarchi et al., 1999; Li et al., 2005; Rossi et al., 2006), which regulate basal transcription from the LTR. Landmark studies from several groups have established (i) that stochastic fluctuations in Tat gene expression act as a molecular switch and (ii) how mutations in NF-κB and Sp1-binding sites, which mimic changes in initiation rates, are sufficient to restrict Tat production and eventually result in increased rates of viral entry into latency (Pearson et al., 2008; Weinberger et al., 2005, 2008). Other investigators have shown that mutations in Tat that attenuate its activity lead to enhanced viral entry into latency. Also, it was recently established that latently infected CD4+ T-cells are enriched for Tat variants with impaired transactivation potential (Yukl et al., 2009). All of the aforementioned studies reinforce the importance of Tat as a switch between active transcription and latency. Adding to the complexity of Tat function, it is now well established that Tat is heavily regulated by the post-translational modifications it undergoes, which include phosphorylation, methylation and acetylation (Fig. 2). Specifically, HATs can acetylate Tat at lysines 28, 50 and/or 51, all of which impart different functions. For instance, acetylation at lysine 50 and 51 by CBP/p300 and GCN5 promotes the dissociation of Tat from the TAR-binding domain, indicating the start of transcriptional elongation as previously reviewed (Easley et al., 2010). In addition, Tat has been shown to be autoacetylated at lysines 41 and 71, which is important for acetylation of nucleosome H4 (Deng et al., 2001). The association of Tat with HMTs and the ability of Tat to be methylated at lysines 50 and 51 have been shown to recruit remodelling proteins to the LTR (Van Duyne et al., 2008). All of the aforementioned actions of Tat in boosting transcription from the LTR are compromised initially in undifferentiated monocytes due to poor expression of CycT1; however, on differentiation, increased availability of CycT1 results in robust transactivation by Tat (Dong et al., 2009). The monocyte component of HIV-1 infection responds to cues emanating from its own differentiation programme because transient expression of CycT1 in undifferentiated monocytes could not rescue Tat-mediated transactivation of the LTR (Dong et al., 2009). Thus, it is important to evaluate the differences between the transcriptional programmes adopted by HIV-1 in the two compartments, CD4+ T-cells and monocyte–macrophage cells.
Besides being a prominent intracellular molecule in the HIV-1 transcriptional programme, Tat is released by infected macrophages, microglia and astrocytes. Extracellular Tat can enter neurons via endocytosis through interaction with the low-density lipoprotein receptor-related protein present on the neuronal surface. This interaction induces apoptosis in neurons, thus explaining the neurotoxic effects of Tat (Kim et al., 2008a, b; Wong et al., 2005). Tat increases the intracellular Ca2+ in neurons, which is taken up by mitochondria. This activity results in the generation of reactive oxygen species, activation of caspases, and, eventually, initiation of apoptosis. The neurotoxic function of Tat is further supported by the observation that the mRNA levels for Tat are elevated in brain extracts of patients with HIV-1-associated dementia (Boven et al., 2007). More recently, it was demonstrated that Tat can associate with the promoters of phosphatase and tensin homologue (PTEN) and protein phosphatase 2A (PP2A) subunits and that these interactions result in transcriptional activation of apoptotic pathways in HIV-1-infected CD4+ T-cells (Kim et al., 2010). This study and others have strengthened the belief that Tat, besides regulating the HIV-1 promoter, affects cellular promoters that result in activation of transcriptional programmes and end in apoptosis of infected cells and uninfected bystander cells. This process contributes to neuronal cell death and explains Tat-mediated neurotoxicity (Le Douce et al., 2010). Whether the toxic functions of Tat are also operative in the periphery with equally detrimental effects is unknown. If Tat is not toxic in the periphery, it could be due to the inherent differences in the environments of compartments such as the brain and bone marrow. These areas may have higher local concentrations of extracellular Tat compared with the periphery, thereby causing cellular toxicity that may not occur in the peripheral blood. In addition, it may not be possible to achieve highly localized concentrations of Tat in the peripheral blood because of the absence of discrete physical boundaries to facilitate the development of elevated localized concentrations of Tat.
Establishment of Tat as a central regulator of HIV-1 transcription along with its ability to modulate the transcription of numerous cellular genes relevant to HIV-1 infection has provoked a number of proposals to use Tat as an important target for antiretroviral therapy (discussed below). However, cell type-specific differences in Tat function and Tat’s interactions in the CNS need to be addressed in a more holistic sense. One criticism of the studies involving Tat is that the concentration used in the experimental system is usually much higher than that achievable in localized microenvironments (difficult if not impossible to assess experimentally). Thus, it is essential to concentrate efforts on the development of experimental approaches to assess Tat function in systems in which the biological functions of Tat can be examined at a concentration closer to what one might predict to be present in an in vivo setting. A number of questions remain unanswered. How is Tat shuttled to the neighbouring cells? Does this process have any specificity or does it involve simple diffusion? How variable are the effects of Tat in different susceptible cell types? Addressing these questions can extend our understanding of Tat so that it can be exploited as a potential therapeutic target.
Vpr: an accessory protein with a versatile repertoire of functions
Vpr is a 96 aa protein encoded in the HIV-1 genome that is also packaged into the mature virion. The exact functions of Vpr have been difficult to elucidate. Many studies have implicated this protein in a multitude of activities dependent on the cellular context and on the in vitro culture system used. Although various studies have demonstrated that Vpr is dispensable for HIV-1 replication in CD4+ T-cells, the relevance of this protein in the overall regulation of HIV-1 pathogenesis is highlighted by the fact that the transactivation ability of Vpr is conserved in the different strains of HIV and SIV (HIV-1, HIV-2, SIV-mac and SIV-agm) (Kogan & Rappaport, 2011; Malim & Emerman, 2008). It is also supported by the observation that Vpr is a necessary positive regulator of viral transcription and infectivity in cells of the monocyte–macrophage lineage where it has been implicated in assisting the importation of PIC into the nucleus (Herbein et al., 2010). Vpr enhances HIV-1 cytopathogenicity by promoting viral transcription through interactions with the viral promoter and host cell factors. Vpr assists HIV-1 replication through its moderate transactivation effect on the LTR, which in turn results in increased transcription and production of progeny virus. Since Vpr is packaged into virions, it is present immediately following HIV-1 infection and considerably before the early gene transcription that results in the production of the regulatory proteins Tat and Rev (Seelamgari et al., 2004). Although the transactivation effect of Vpr is modest compared with that of Tat, Vpr-induced transactivation is considered to be synergistic as well as a prerequisite for optimal Tat transactivation.
An important, unambiguous attribute of Vpr is its ability to delay or arrest cells in the G2 phase of the cell cycle (Fig. 3). Studies performed by a number of investigators have suggested that the cell death phenotype induced by Vpr is linked to the pathway that leads to G2 arrest. The efficacy of Vpr-induced transactivation of the LTR has been shown to correlate with the induction of G2 arrest in host cells (Andersen et al., 2006). It was also demonstrated that LTR activity is increased four- to sixfold in the G2 phase of the cell cycle. The observation that infected cells in individuals with HIV-1 infection appear to be enriched for cells in G2 highlights the importance of the role of Vpr in vivo (Ardon et al., 2006; Zimmerman et al., 2006). Vpr has also been shown to interact with transcription factor Sp1 in a ternary complex with the GC-box array in the HIV-1 LTR (Fig. 3) (Kino & Pavlakis, 2004). Vpr has been shown to bind the LTR in a sequence-specific manner to the sequences that span the C/EBP site I and the promoter-distal NF-κB site in the viral LTR (Hogan et al., 2003). It was also shown that with HIV disease progression and in patients with HIV-1-associated dementia, LTR SNPs (particularly, a C-to-T change at position 3 in a consensus B C/EBP site I background) emerge, become prevalent, and show reduced C/EBP binding and enhanced Vpr binding to the HIV-1 LTR (Burdo et al., 2004; Hogan et al., 2003). The ability of Vpr to bind directly to the HIV-1 LTR C/EBP site I, combined with the indispensable role of C/EBP factors in HIV-1 replication in the cells of the monocyte–macrophage lineage, has raised the interesting possibility that both of these factors may function either coordinately or, perhaps, in opposition during HIV-1 replication in this cell population. Consistent with these observations, we have demonstrated that Vpr and C/EBP form a complex at C/EBP site I and that this interaction may be integrally involved in the regulation of transcription in monocytic cell populations (S. Dahiya and B. Wigdahl, data not shown).
In addition to being associated with the virion, where it may be involved in nucleic acid condensation during assembly of new progeny virus, and endogenously expressed, extracellular Vpr is found in the plasma and the CSF (Piller et al., 1998) and has been shown to enter cells of the monocyte–macrophage lineage and function as an endogenously expressed protein (Levy et al., 1995; Varin et al., 2005). In addition, Vpr has been shown to modulate the host glucocorticoid receptor (GR) to affect transcription from the LTR as well as that of a number of host genes (Refaeli et al., 1995). The underlying mechanism involves Vpr induction of R-interacting protein 1 nuclear translocation in a GR-dependent manner. Vpr was also shown to transactivate promoters containing glucocorticoid-responsive elements. This modulation is most likely via direct interaction with the GR, with Vpr acting as a co-activator of GR (Kino et al., 1999). The role of glucocorticoids in HIV-1 replication remains controversial. Several researchers have reported that glucocorticoids suppress the HIV-1 LTR, whereas others have suggested that the role of glucocorticoid signalling in HIV-1 infection is not influenced by Vpr (Kogan & Rappaport, 2011). Vpr has been shown (i) to induce oxidative stress through the hypoxia-inducible factor pathway in microglia cells (Deshmane et al., 2009); (ii) to be implicated in the induction of apoptosis via both intrinsic (mitochondria-mediated) and extrinsic pathways; and (iii) to interact with proteins including ANT, PTPC, PP2A and HAX-1 that regulate the process of apoptosis and eventually neurodegeneration [for detailed mechanisms refer to (Na et al., 2011; Zhao et al., 2011)].
The results of more than two decades of research have established that Vpr is a crucial accessory protein in HIV-1 pathogenesis and is involved at each step of the viral life cycle in proliferating and non-proliferating cells. Future strategies to counter the ability of Vpr to inhibit host-mediated restriction could potentially be used to alleviate the impact of viral infection.
Nef: inducer of cellular transcription factors
HIV-1 Nef is a 27 kDa, myristoylated viral protein that is associated with cytoplasmic membranes. It is expressed early during the viral life cycle. Nef is required for maintenance of high viral loads and has been shown to accelerate HIV-1 disease (Baur, 2011), but its functions are not completely understood. Nef serves a number of functions including control of cellular trafficking, gene and receptor surface expression, cell migration, antigen presentation and signal transduction. Both Nef and Tat are involved in the enhancement of interleukin (IL)-2 promoter activity. Nef contributes to the transcriptional activation of the HIV-1 promoter through induction of cellular transcription factors, such as NF-AT, NF-κB and AP-1, which elevate transcription from the LTR, often in a cell-type-dependent manner, and participates in the activation of cellular genes including those encoding inflammatory cytokines, activation markers and death receptors (Fenard et al., 2005; Fortin et al., 2004) (Fig. 3). Recently, it was shown using primary quiescent T-cells that Nef superinduces NF-AT and IL-2 production, bypassing early T-cell antigen receptor (TCR) effector molecules (Neri et al., 2011). The same study also established that early phosphorylation of PLC-γ1, the induction of NFAT, and the expression of IL-2 are impaired by Nef in suboptimally activated or resting CD4+ T-cells, suggesting that Nef plays a dual role in the modulation of TCR signalling, thereby enhancing HIV-1 replication in both quiescent and metabolically active CD4+ T-cells. Nef is known to interact with a multi-protein signalling complex that includes kinases of the Src family, Lck and Fyn, which are proximal signalling kinases activated immediately on TCR stimulation (Arold et al., 1997; Trible et al., 2006). Nef has also been shown to modulate the activation of downstream effectors essential for activation-induced cytoskeletal rearrangement including PAK2, CDC42 and Vav (Rauch et al., 2008; Simmons et al., 2005). Nef associates with PAK2 in a multi-protein complex found in detergent-insoluble lipid rafts (Krautkrämer et al., 2004). It was recently established that Nef-mediated enhancement of cellular activation and viral replication in primary T-cells was dependent on PAK2 and on the potency of the activating stimuli. This enhancement correlated with the ability of Nef to associate with PAK2 (Olivieri et al., 2011). Thus, Nef serves as an adaptor protein to divert host cell proteins like cellular kinases to aberrant functions that enhance the amplification of viral replication. Besides cellular activation, four other functions of Nef have been established in various in vitro studies: (i) downregulation of CD4; (ii) downregulation of cell surface levels of major histocompatibility class I molecules; (iii) enhancement of viral particle infectivity by CD4-independent mechanisms (Kirchhoff, 2010); and (iv) downregulation of surface expression of CXCR4 and CCR5 (Sloan et al., 2010).
Recent studies have suggested that Nef may not only manipulate host cells infected with HIV-1, but may also cause remarkable changes in the cellular environment. These changes include its ability to induce secretion of factors from infected macrophages that attract T-cells and render them more susceptible to HIV-1 infection. Moreover, Nef has been shown to regulate the intracellular trafficking of a number of cell surface proteins of helper T-cells and macrophages with central roles in immunity and the viral life cycle (Roeth & Collins, 2006). The functions of Nef related to viral replication and pathogenesis in vivo are not yet completely understood, but it appears that HIV-1 and SIV have evolved Nef as a tool to manipulate the key cell types of the acquired immune system and to modulate various cellular mechanisms/pathways for the immunological restriction of HIV-1.
Matrix (MA): creator of a favourable environment for HIV-1 transcription
HIV-1 MA protein or p17 is a structural protein that plays a key role in regulating the early and late steps in viral morphogenesis. Increasing evidence suggests that p17 may also be active extracellularly in deregulating the biological activities of immune cells that are either directly or indirectly involved in AIDS pathogenesis as previously reviewed (Mascarenhas & Musier-Forsyth, 2009). MA has two nuclear localization signals and was among the first HIV-1 proteins to be implicated in the nuclear importation of PIC. Earlier studies demonstrated that mutations of the nuclear localization signals perturbed HIV-1 nuclear importation, especially in non-dividing macrophages. However, later studies questioned the role of p17 in nuclear transport (Mascarenhas & Musier-Forsyth, 2009). p17 is also found in extracellular spaces through utilization of alternative secretion pathways, where it dysregulates the biological activities of many different immune cells that are directly or indirectly involved in HIV-1-associated pathological effects (Fiorentini et al., 2010). It was established that monocytes treated with p17 selectively produce monocyte chemotactic protein-1 (MCP-1) (Marini et al., 2008). The effect of p17 on MCP-1 expression was at the transcriptional level and was primarily dependent on the activation of AP-1. Many studies have shown a tight link between biologically deregulated monocytes, AP-1 activation, MCP-1 release, and HIV-1 pathogenesis (Bukrinskaya, 2007). There is mounting evidence that p17 stimulation modulates the ERK and PI3K/Akt signalling cascades, which may represent upstream events in activation of AP-1, which eventually influences transcription from the LTR (Fig. 3) (Fiorentini et al., 2010; Giagulli et al., 2011). Further understanding of the mechanisms underlying p17 signalling may increase our understanding of how p17 modulates both innate and adaptive immune responses in favour of HIV-1 transcription (Fiorentini et al., 2010) and how the interactions with various signalling components change with the cellular context.
miRNAs and HIV-1: regulatory intersections
Several previous studies have strongly indicated that HIV infection induces a dramatic change in the gene expression profile of infected cells. Recent studies have attempted to elucidate the complex interaction between HIV-1 and host miRNA silencing machinery (Zamore & Haley, 2005). The host RNA interference (RNAi) machinery along with the recent idea that HIV-1 may encode miRNAs has triggered a new interest in determining the extent to which these two interfaces influence viral replication, latency and the efficiency of host defences (Narayanan et al., 2011). In this section, we summarize our current state of knowledge with respect to aspects of multi-faceted interactions between HIV-1 and miRNA-guided silencing machinery.
Algorithms using thermodynamically favourable miRNA in target pairing studies have suggested that candidate HIV-1 genes could be controlled by host miRNAs (Chable-Bessia et al., 2009) (Table 1). It was shown that virus replication kinetics are enhanced in Drosha- or Dicer-depleted peripheral blood mononuclear cells from HIV-1-infected patients using small interfering RNAs (siRNAs) to Drosha and Dicer (Triboulet et al., 2007). Recent studies demonstrated that the 3′ UTR of almost all HIV-1 mRNA produced during latency in resting primary CD4+ T-lymphocytes contain a 1.2 kb fragment that can be recognized by cellular miRNAs (miR-28, -125b, -150, -223 and -382), resulting in a negative impact on viral protein production (Huang et al., 2007). Combined with the relatively inefficient synthesis of Tat and Rev, miRNAs harboured by resting CD4+ T-cells may participate in post-transcriptional regulation of HIV-1 mRNA and contribute to the state of viral latency, as observed in patients with suppressive HAART (Ouellet et al., 2009). These new elements contribute to our understanding of the molecular basis of viral latency and will aid in the design of therapeutic strategies aimed at purging HIV-1-infected patients of quiescent virus. To this end, it has been shown that neutralizing these cellular miRNAs by transfecting specific antagonists into non-activated CD4+ T-cells from patients treated with HAART resulted in a 10-fold increase in the in vitro efficiency of virus isolation (Corbeau, 2008).
Table 1. miRNAs and siRNAs and their proposed regulation in HIV-1.
| miRNA name | Proposed function | Reference(s) |
| miR-17/92 cluster (miR-17-5p and miR-20a) | Indirectly affects HIV-1 replication by targeting cellular proteins like histone acetyltransferase (HAT) Tat cofactor PCAF | Triboulet et al. (2007) |
| miR-N367 | Suppresses Nef expression | Omoto & Fujii (2005); Omoto et al. (2004) |
| hsa-miR-29a | Interferes with Nef protein expression and HIV-1 replication; directs HIV-1 transcripts to P bodies | Ahluwalia et al. (2008); Nathans et al. (2009) |
| miR-28, miR-125b, miR-150, miR-223, miR-382 | Target sequences in the 3′ end of HIV-1 RNA | Huang et al. (2007) |
| miR-TAR-5p, miR-TAR-3p | Chromatin remodelling of the HIV-1 LTR; inhibits apoptosis by downregulating ERCC1 and IER3 | Klase et al. (2007, 2009); Ouellet et al. (2009) |
| vsiRNA1 | Rescues Env mRNA expression | Bennasser et al. (2005) |
| miR-122, miR-370, miR-373, miR-297 | Unclear in HIV-1 infection but were shown to be upregulated during HIV-1 infection of Jurkat T-cells | Triboulet et al. (2007) |
| miR-29c, miR-26a, miR-21 | Unclear, shown to be downregulated in HIV-1 patient samples | Houzet et al. (2008) |
| miR-146a | Targets CCL8/MCP-2 in HIV-1-infected microglia for maintenance of HIV-mediated chronic inflammation of the brain | Rom et al. (2010) |
| miR-34a | Targets transcription factor CREB (levels deregulated by Vpr resulting in neuronal dysfunction) | Mukerjee et al. (2011) |
| Predicted in silico | ||
| hsa-miR-29a and -29b | Targets Nef gene | Hariharan et al. (2005) |
| hsa-miR-149 | Targets Vpr gene | Hariharan et al. (2005) |
| hsa-miR-378 | Targets Env | Hariharan et al. (2005) |
| hsa-miR-324-5p | Targets Vif gene | Hariharan et al. (2005) |
Several studies have proposed a regulation paradigm whereby cellular miRNAs target mRNAs that encode cellular proteins involved in virus replication (Table 1). For example, the miR-17/92 cluster, which encodes seven miRNAs, among which miR-17-5p and miR-20 may target HATs and HIV-1 Tat cofactor PCAF, was substantially decreased during HIV-1 infection. PCAF has been shown to be a host cofactor for Tat transactivation of the HIV-1 LTR and is also recruited by Tat to remodel the histone architecture in the vicinity of the LTR (Triboulet et al., 2007). This knowledge can enhance our understanding of how latent reservoirs, which are resistant to HAART, can be activated and eventually flushed out of the system.
The studies reviewed above suggest that most of the time, the host machinery works to curtail the spread of HIV-1. Thus, it is intuitive to investigate the reverse question: does HIV-1 adopt strategies to subvert the RNAi machinery? In this regard, downregulation of a large pool of miRNAs in HeLa cells transfected with the pNL4-3 infectious HIV-1 molecular clone was observed (Yeung et al., 2005). How does the virus achieve this downregulation to facilitate its own transcription and replication? Adenoviral protein VA1 was shown to inhibit the nucleocytoplasmic transport of pre-miRNA by binding to exportin 5, which is required for this transport (Lu & Cullen, 2004). HIV-1 has been hypothesized to act against the host RNAi machinery by targeting two of its key players – Dicer and TRBP. Purified HIV-1 Tat protein was shown to inhibit the capacity of Dicer to process dsRNA to siRNA in vitro (Bennasser et al., 2005). Also, the TAR and RRE sequences of HIV-1 RNA bind to TRBP and thus competitively sequester it away from its normal function of pre-miRNA processing, impacting miRNA biogenesis and function (Christensen et al., 2007; Gatignol et al., 2005). HIV-1 Tat RNAi suppressor functions have been reported (Qian et al., 2009; Schnettler et al., 2009). Other studies have reported otherwise and suggested that HIV-1 Tat and TAR do not reduce the efficacy of cellular RNA silencing mechanisms (Sanghvi & Steel, 2011). Thus, the impact on HIV-1 infection of these inhibitory processes is yet to be established unequivocally. The argument that HIV-1 needs to counter the endogenous miRNA machinery is supported by the observation that a point mutation in Tat that compromises its function to inhibit Dicer, but not its transactivation effect on the LTR, results in a reduction in viral replication (Bennasser et al., 2005). Similarly, it was shown that efficient viral production requires Tat to block Dicer activity (Haasnoot et al., 2007). In addition to having a globally negative impact on miRNA production, HIV-1 has been shown specifically to regulate, positively or negatively, the level of expression of some miRNAs (Table 1). The mechanism behind this selective regulation of miRNAs by HIV-1 is unclear. Other examples of selective regulation include a study showing that miRNA-198 regulated HIV-1 replication in cells of the monocyte–macrophage lineage through regulation of cyclin T1 expression (Chiang et al., 2012; Sung & Rice, 2009). Cell-type-specific regulation of HIV-1 has been shown to correlate with differential miRNA expression. For instance, monocytes express high levels of the anti-HIV miRNAs (specifically miRNA-28, -150, -223 and -382). As differentiation from monocytes to macrophages ensues, the expression profile of this series of miRNAs decreases, and these decreases inversely correlate with susceptibility to HIV-1 infection (Wang et al., 2009).
Recent studies utilizing deep sequencing (Schopman et al., 2012) and pyrosequencing (Yeung et al., 2009b) of small non-coding RNAs in virus-infected cells have alluded to the existence of HIV-encoded small RNAs. However, their function in cells is yet to be established. Recently, it was shown that TAR miRNA protects the infected cell from undergoing apoptosis by targeting the cellular proteins ERCC1 and IER3 (Cobos-Jiménez et al., 2011).
HIV-1 proteins Vpr and Nef have also been shown to modulate host RNAi pathways to promote viral latency or to counteract anti-HIV-1 immune responses. It was recently shown, using primary human neurons and neuronal cell lines, that Vpr plays a major role in HIV-1-associated neuronal dysfunction. This role can be attributed, in part, to its ability to deregulate miR-34 and its target gene, CREB (Mukerjee et al., 2011). On the other hand, Nef, along with Tat, has been shown to downregulate Dicer expression, thus inhibiting the RNAi machinery. In addition, HIV-1 has been shown to induce the expression of miRNA-146a that targets MCP-2, a potent inhibitor of CD4/CCR5-mediated HIV-1 entry (Rom et al., 2010).
Thus, the reciprocal interactions between cellular miRNAs, viral miRNA and HIV-1 add a complex level of regulation to HIV-1 gene expression. This expression is not only contingent on the cellular phenotype, but is also responsive to the extracellular stimuli that HIV-1-susceptible cells receive when they traffic to different end organs like brain, lung, kidney and bone marrow, and to a number of mucosal compartments. Currently, there is no general agreement in the field concerning whether the HIV-1 genome encodes miRNAs in an in vivo setting and how this new arm of regulation fits into the existing knowledge of miRNA-based gene regulation. It was once assumed that miRNAs exerted their influence only at the translational level, but recent studies have altered that paradigm and demonstrated that miRNAs can also regulate gene expression at the epigenetic level. In particular, new evidence indicates that miRNAs can initiate gene silencing either by specifically inducing methylation along promoter sequences or by directly remodelling the surrounding chromatin by modifying the promoter-associated histones (Morris, 2005). How this plays at the HIV-1 promoter or LTR remains to be determined. However, understanding how various viral proteins regulate the expression of cellular genes via miRNA along with their ability to regulate and be regulated by the RNAi machinery will enable investigators to use this aspect of HIV-1 gene expression regulation within the context of therapeutic intervention strategies.
Therapeutic interventions to target HIV-1 transcription
Current antiretroviral therapeutic strategies have reduced HIV-1 RNA levels to below 50 copies ml−1. However, any interruption in the therapy results in a rebound of viral replication from the latently infected cellular reservoirs, which include resting CD4+ T-cells and cells of the monocyte–macrophage lineage. Two important characteristics of these cellular compartments make them effective drug-resistant reservoirs: (i) their quiescent nature and (ii) their presence/residence in tissue sanctuary sites, like the brain, that are protected more from drug penetration. Recent developments concerning the molecular mechanisms of HIV-1 latency (Choudhary & Margolis, 2011; Margolis, 2011), combined with the efforts to elucidate targets for pharmacological intervention to induce viral transcription utilizing in vitro cell systems, have not yet translated into successful eradication of latent HIV from patients. Current approaches to disrupt latency are studied in chronically infected cell lines or primary cells ex vivo. Both of these systems are still deficient in being able to uniformly mimic an in vivo scenario because they depend on mutations in viral genes or on biological effects contingent on specific integration sites. We believe that newer strategies that can specifically target reservoirs, along with elimination by enhanced ART, will yield promising results in purging the infection. Another important method for controlling HIV-1 replication, pathogenesis and disease will centre on the suppression of viral transcription with the goal of maintaining the state of latency (discussed below).
One difficulty associated with the development of an effective anti-HIV-1 therapy is the lack of a suitable animal model. An animal model system is required not only to fully understand the dynamics of HIV-1 latency in vivo, but also to validate the efficacy of drug candidates. The available animal models for the evaluation of candidate anti-HIV-1 therapeutics have been recently reviewed in detail (Clements et al., 2011; Sato & Koyanagi, 2011).
Transcriptional inhibitors: a novel strategy to control HIV-1
An important consideration in controlling HIV-1 replication is to reinforce the state of latency by using transcriptional inhibitors that effectively target unique, selective features of HIV-1 transcription (Fig. 4a). It is envisioned that these inhibitors would be used as adjuncts to the current HAART combinations. Tat, for example, a critical regulator of transcription, constitutes a major therapeutic target in the HIV-1 life cycle. Additionally, drugs can be designed to target crucial cellular cofactors involved in transcriptional activation. In this intervention paradigm, it is important to consider proteins that are used selectively to enhance HIV-1 transcription with minimal impact on the normal physiological processes. Although it will be difficult to find a cellular target that is used only in HIV-1 transcription, there are candidate cellular proteins or protein domains that do not greatly impact cellular transcription but are crucial for HIV-1, as discussed below, that can serve as good starting points for the development of novel therapeutics. Several studies have characterized and demonstrated the efficacy of transcriptional inhibitors such as C-terminally truncated STAT5, Staf-50, prothymosin α and thioredoxin reductase in controlling HIV-1 transcription in human macrophages. Another important factor, C/EBP, which is dispensable for viral transcription in T-cells but critical for LTR-mediated transcription in monocyte–macrophage cells, can be evaluated for devising strategies for controlling HIV-1 expression. It would be of interest to determine how induction of cellular inhibitors of C/EBP, CHOP (C/EBP homologous protein, GADD153) and SHARP1/DEC2 would impact HIV-1 transcription in the monocyte–macrophage cell lines. In addition, inhibiting the interaction of nuclear factor for activated T-cells 5 (NFAT5) with the LTR using RNAi may also provide a promising target because it has been shown to suppress HIV-1 transcription in primary macrophages (Ranjbar et al., 2006). The observation that IL-10 inhibits HIV-1 LTR-directed transcription in human macrophages by a two-pronged mechanism involving the induction of (i) cyclin T1 proteolysis and (ii) inhibitory C/EBPβ through STAT-3 can be seen as a way to control viral expression in these cells (Tanaka et al., 2005). Recent observations that have involved O-linked N-acetylglucosaminylation of Sp1 inhibition of LTR-mediated transcription have demonstrated the potential for novel ways to control HIV-1. Several reports have indicated that OTK18, a zinc finger protein, was able to suppress Tat-induced HIV-1 LTR activity in the macrophage lineage cells (Horiba et al., 2007). Studies highlighting the importance of zinc finger proteins, Sp1 and OTK18, in sequence-specific LTR transcriptional regulation, have triggered a new interest in the field to use engineered transcription factors with zinc fingers as novel candidates for repressing LTR-directed transcription. This interest is further fuelled by the observation that zinc finger proteins can also influence chromatin packing and nuclear events via regulation of proteins that are involved in epigenetic regulation of the HIV-1 LTR (Verschure et al., 2006). Such experimental strategies need first to have a relatively comprehensive understanding of the protein–protein and protein–DNA interactions that occur at the LTR and then use this knowledge to target crucial interactions between transcription factors rather than single transcription factors. This approach centres on disruption of the transcriptional circuits employed by HIV-1 that will have minimal impact on the host compared with current therapies, which are still toxic. Combining these new approaches to an already established antiretroviral therapy with improving the ability of any class of drugs to penetrate the blood–brain barrier will go a long way to fully capitalize on our understanding of HIV-1 transcriptional regulation.
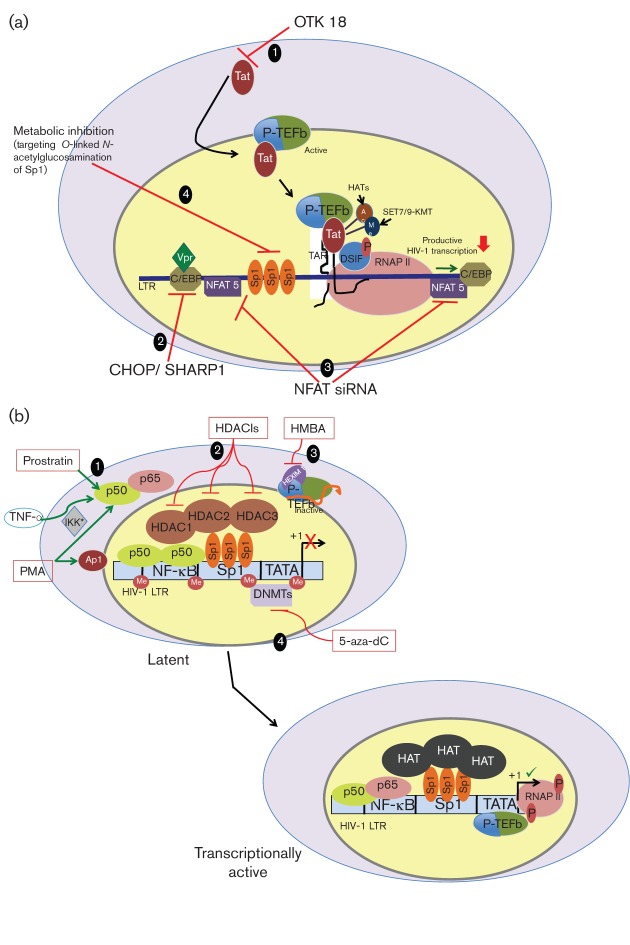
Fig. 4.
Therapeutic strategies for HIV-1. (a) Strategies to suppress HIV-1 transcription: (1) inhibit Tat-mediated transactivation using the C2H2 zinc finger protein OTK18, resulting in overall inhibition of transcription from the LTR. (2) Target the C/EBP protein, a crucial transcriptional regulator in cells of the monocyte–macrophage lineage, by using overexpression of cellular inhibitors like CHOP/SHARP1 to suppress LTR-directed transcription. (3) Evaluate inhibition of the NFAT family of proteins C as a potential therapy to block HIV-1 expression. (4) Regulate the function of the Sp1 transcription factor through post-translational modifications including O-linked N-acetylglucosamination; evaluate strategies to inhibit the relevant enzymes to maintain HIV-1 in an inactive state. (b) Strategies to purge HIV-1: (1) activate NF-κB to achieve a stimulated expression from the latent integrated HIV-1 genome. The homodimeric p50 form of NF-κB recruits HDACs during latency. In resting CD4+ T-cells, the activating p50–p65 heterodimer of NF-κB is sequestered in the cytosol; use of activators like phorbol 12-myristate 13-acetate, TNF-α and prostratin, which activate IκB kinase and induce nuclear translocation of p50–p65 and recruitment of HATs to the NF-κB sites, can result in activation of HIV-1 transcription. (2) Use class I selective HDAC inhibitors that inactivate HDACs 1, 2 and 3 at the LTR and result in unrestricted action of HATs that are recruited to the LTR by various transcription factors, resulting in acetylation of histones at nucleosomes near the site of initiation of LTR-directed transcription. (3) Use HMBA to release P-TEFb from inactive complexes with HEXIM 1, resulting in phosphorylation of RNAP II CTD and subsequent shift to processive transcription of the integrated HIV-1. (4) Reverse transcriptional silencing imposed by the methylation of cytosine residues at the LTR by DNA methyltransferases by inhibiting the enzyme using 5-aza-2′ deoxycytidine, resulting in stimulation of HIV-1 transcription.
Therapeutic interventions to purge latent HIV-1
The long-standing idea for eradicating HIV-1 from patients is to activate and purge latent HIV-1 from the reservoirs, which would then be taken care of either by the host immune system or by antiretroviral therapy. Because we understand that viral latency is multi-factorial, there is probably a need to combine several targeting strategies to achieve a successful outcome. To date, many eradication protocols have passed preclinical trials, but none has made it through clinical trials. Here, we discuss and suggest targets that can be utilized for therapeutic intervention to clear HIV-1 from the system (Fig. 4b).
P-TEFb
As discussed before, HIV-1 Tat recruits active P-TEFb to the LTR and efficiently relieves the block at transcriptional elongation. Restriction in the nuclear levels of active P-TEFb is a key factor in contributing to proviral latency. P-TEFb is tightly regulated in active and inactive complexes by tethering to HMBA-induced protein 1/2 (HEXIM1/2). Investigations have established that P-TEFb can activate HIV-1 transcription in the absence of Tat (Klichko et al., 2006). Hexamethylene bisacetamide (HMBA) can trigger the release of P-TEFb from HEXIM1 and enhance HIV-1 LTR-directed transcription independently of Tat and NF-κB. It has also been suggested that P-TEFb can be recruited to the LTR by interactions with several transcription factors such as NF-κB or Brd4 and Sp1 in the absence of Tat (Klichko et al., 2006). This observation is important because molecules like HMBA may be efficient in inducing the expression of quiescent HIV-1 lingering in latent pools of resting CD4+ T-cells and macrophages.
The cellular protein phosphatase PP1 is involved in the release of P-TEFb in both Tat-dependent and -independent manners. We also know that CDK9 kinase activity itself is regulated by phosphorylation of residues at the C terminus (Ser347, Ser354, Ser357, Thr350, Thr354 and Thr186). Phosphorylation of CDK9 at Thr29 is related to the inhibition of kinase activity at the LTR. Recently, PP1 was shown to activate CDK9 by dephosphorylating Ser175 (Ammosova et al., 2011a); expression of the PP1 inhibitor, cdNIPP1, increased Thr186 phosphorylation and inhibited HIV-1 transcription (Ammosova et al., 2011b). These studies indicate that full activation of CDK9 requires a complex series of phosphorylation and dephosphorylation events at the LTR. Although they are not fully understood, coordinated actions of PP1 and PP2A are thought to control this process. Our lack of complete understanding of P-TEFb regulation of HIV-1 transcription is reflected in the mixed results investigators have obtained in global inhibition of PP2A using pharmacological inhibitors like okadaic acid. Some studies have demonstrated induction of HIV-1 expression in chronically infected cell lines, whereas others have shown inhibition (Epie et al., 2006; Vlach et al., 1995). Thus, P-TEFb still presents itself as a promising target; a complete understanding of its role in HIV-1 transcription is required before it can be used as a targeted therapy.
Signalling pathways: the protein kinase-C (PKC) and IL-6 pathways
The HIV-1 promoter or LTR contains binding sites for transcription factors like NFAT, NF-κB and activating protein 1 (AP-1) that stimulate the transcription of the integrated genome of HIV-1. All of these factors are themselves regulated by PKC signalling. The NF-κB signalling pathway plays several key roles in HIV-1 transcription. On activation, the p50–p65 heterodimer replace the chromatin repressive p50–p50 homodimer with a concomitant displacement of HDAC (Williams & Greene, 2007). This step is followed by recruitment of HATs such as p300 and CBP by the p50–p65 heterodimer to the LTR that results in acetylation of histones and Tat. It has also been suggested that NF-κB assists in recruitment of P-TEFb. All of these crucial downstream functions of PKC signalling in the context of HIV-1 transcription have inspired the evaluation of PKC agonists to flush out latent HIV-1 from reservoirs. Indeed, pharmacological PKC agonists like prostratin, jatrophane diterpene (SJ23B), and 13-phenylacetate have been shown to activate transcription in latently infected CD4+ T-cells (Bedoya et al., 2009; Bocklandt et al., 2003; Kulkosky et al., 2001). In fact, it was recently shown that prostratin in combination with HDAC inhibitors resulted in the synergistic activation of HIV-1 transcription from latently infected patient cells and also from chronically infected cell lines (Burnett et al., 2010). In addition to activating transcription of the HIV-1 promoter, prostratin also downregulates CD4 and CXCR4 receptors on cells, thus resulting in a decrease in HIV-1 susceptibility. However, the toxicity issues related to prostratin have plagued its development as a drug. To utilize this aspect of the HIV-1 transcriptional intervention paradigm in a pharmacological setting, newer methods are required to synthetically design prostratin-like compounds having favourable toxicological parameters.
One potential problem of altering NF-κB signalling to control HIV-1 transcription is the off-target effects including T-cell activation, which can be a huge detriment in moving forward with this line of investigation. It was recently shown that the compound 5-hydroxynaphthalene-1,4-dione was able to activate latent infection in primary cells without activating T-cells (Yang et al., 2009a, b).
Similarly, activating the IL-6 pathway is an important axis that can be used to activate LTR-directed transcription. The IL-6 pathway converges on the LTR in the form of NF-IL-6/C/EBPβ, which has been shown by us and others to be indispensable in activating transcription from the cells of the monocyte–macrophage lineage (Kilareski et al., 2009). However, none of these aforementioned strategies has moved forward because of the difficulties inherent in activating HIV-1 without globally activating cellular signalling.
Combination therapies and epigenetic regulation
The integrated HIV-1 LTR has been shown to have consistent nucleosome packaging with Nuc-1, which is present immediately downstream from the transcriptional start site. Remodelling of the chromatin at this location is highly regulated and crucial for transcriptional activation. Also, deacetylation of histones at Nuc-1 by class I HDACs has been shown to be critical in maintaining latency in chronically infected cell lines (Quivy et al., 2007; Ylisastigui et al., 2004). It was shown that viral expression in resting CD4+ T-cells could be activated by inhibition of HDAC enzymic activity (Archin et al., 2009). These investigators also showed that treating resting CD4+ T-cells with HDAC inhibitor (HDACI) did not result in activation of T-cells. Another important feature of some HDACIs currently under investigation is their lipophilic nature, which can also serve to efficiently target latent HIV-1 in anatomical reservoirs like brain microglia.
In addition to being used as a stand-alone therapy, HDACIs have been combined with the glutathione synthesis inhibitor, buthionine sulfoximine (BSO), which created a pro-oxidant environment and resulted in stimulation of HIV-1 transcription (Savarino et al., 2009). The ability of HDACIs to induce HIV-1 transcription was enhanced by the addition of BSO; the doses of both drugs were lowered to levels that were not toxic for uninfected cells. Moreover, some of the cells shown to be insensitive to HDACIs were rendered sensitive when HDACIs were combined with BSO and added to the population of responding cells. This study, although in a cell line model of HIV-1 quiescence, shows potential to be extrapolated to different models of latency and in general has indicated that combinations of targeted approaches may serve as a way to achieve efficacious therapies with low toxicity. Similarly, the results of another study indicated the strong synergistic activation of HIV-1 transcription when the HDACI trichostatin A was combined with the NF-κB inducer tumour necrosis factor alpha (TNF-α) in a latent U1 promonocytic cell line model (Quivy et al., 2007). Again, the biggest hindrance to translating these therapeutic strategies to HIV-1-infected humans is their toxicity. Combinations of several HDACIs in human clinical therapies including prostratin have also been examined (Reuse et al., 2009).
A drug approved to treat epilepsy, valproic acid (VPA), has been shown to induce latent infection in ex vivo primary cells. Its efficacy was synergized by use with the potent class I HDACI, SAHA, utilizing latently infected cell lines, promonocytic U1 and J-Lat T-cells (Reuse et al., 2009). The observed synergistic activation resulted in the remodelling of the nucleosome Nuc-1 also, using indirect end-labelling experiments. These results constitute a proof of concept for co-administration of at least two categories of therapeutically promising drugs that can activate latent HIV-1. Coupling these to currently available antiretroviral therapeutic strategies to decrease the pool of latent HIV-1 reservoirs in preclinical and clinical trials may be the next step. However, the issue of toxicity of such therapeutics needs to be addressed by coupling refinement of the pharmacophore design with continuous evaluation of new combinations using in vitro and ex vivo studies.
One more aspect of the epigenetic regulation of the HIV-1 LTR to consider in the development of new therapeutic approaches for HIV-1 eradication centres on the histone methylation of the integrated HIV-1 genome. Factors that stimulate HIV-1 transcription result in demethylation of H3K9 and H3K2 and are associated with a restrictive chromatin environment at the LTR (Tyagi et al., 2010). Targeting enzymes, namely HMTs, that maintain these repressive alterations can be an important target in the arsenal of novel therapeutics. The HMT inhibitors like chaetocin, 3-deazaneplanocin A and BIX01294, which have already found utility in cancer chemotherapy, have just begun to be studied for the purpose of antiretroviral therapy. These studies have used chronically infected cell line models to point to a strong interplay between acetylation and methylation mechanisms (Blazkova et al., 2009; Kauder et al., 2009). For instance, it was demonstrated that an HDACI, depsipeptide, resulted in activation of HIV-1 transcription, which was followed by a decrease in both CpG methylation and H3K9 methylation in the LTR region (Wu et al., 2008). However, not all studies have confirmed a synergistic interaction between these two classes of drugs. A cytosine methylation inhibitor (5-aza-2′-deoxycytidine) combined with VPA failed to synergistically activate HIV-1 transcription in latently infected cell lines (Kauder et al., 2009). Novel combinations like HMT inhibitors with a PKC agonist could represent a new direction in the effort to purge HIV-1 completely or at least further reduce the levels from infected patients, but this approach still warrants an in-depth analysis using primary cells and other model systems.
Conclusions and future perspectives
More than 25 years of research have delineated several regulatory pathways that play crucial roles in the HIV-1 life cycle. However, our understanding of HIV-1 pathogenesis remains limited due to the lack of suitable animal model systems. Various steps of the HIV-1 cell cycle, including transcription, are potential targets for therapeutic intervention. Greater specificities are urgently needed to target key viral players like Tat and Vpr because of their multi-faceted effects on host cellular machinery and their role in HIV-1 pathogenesis. The field of epigenetic regulation of HIV-1 transcription is yet to be evaluated fully in ex vivo primary cells. Research in these areas is slowed by the lack of availability of appropriate infected blood and tissue samples. The ideal combination of therapeutic agents should be orally available, active but not toxic to various cell types, and able to target and eliminate HIV from its sanctuaries like the CNS. The molecular mechanisms of latency established to date need to be evaluated rigorously in multiple in vitro model systems for possible translation into therapy for HIV-1-infected individuals. Perhaps the greatest therapeutic challenge ahead will involve the irreversible silencing of proviral DNA in all reservoirs and/or the identification of latent proviral DNA and complete excision of latent integrated proviral DNA in the absence of genomic activation strategies or cellular and tissue cure of HIV-1 infection.
Acknowledgements
This work was supported in part by funds from the Public Health Service, National Institutes of Health, through grants from the National Institute of Neurological Disorders and Stroke (NS32092 to B. W.) and the National Institute of Drug Abuse (DA19807 to B. W.). Dr Michael Nonnemacher was also supported by faculty development funds provided by the Department of Microbiology and Immunology and the Institute for Molecular Medicine and Infectious Disease. The authors would also like to acknowledge the editorial assistance of Pamela Fried in Academic Publishing Services of the Drexel University College of Medicine.
References
- Ahluwalia J. K., Khan S. Z., Soni K., Rawat P., Gupta A., Hariharan M., Scaria V., Lalwani M., Pillai B. & other authors (2008). Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology 5, 117 10.1186/1742-4690-5-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexaki A., Wigdahl B. (2008). HIV-1 infection of bone marrow hematopoietic progenitor cells and their role in trafficking and viral dissemination. PLoS Pathog 4, e1000215 10.1371/journal.ppat.1000215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexaki A., Liu Y., Wigdahl B. (2008). Cellular reservoirs of HIV-1 and their role in viral persistence. Curr HIV Res 6, 388–400 10.2174/157016208785861195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammosova T., Obukhov Y., Kotelkin A., Breuer D., Beullens M., Gordeuk V. R., Bollen M., Nekhai S. (2011a). Protein phosphatase-1 activates CDK9 by dephosphorylating Ser175. PLoS ONE 6, e18985 10.1371/journal.pone.0018985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammosova T., Yedavalli V. R., Niu X., Jerebtsova M., Van Eynde A., Beullens M., Bollen M., Jeang K. T., Nekhai S. (2011b). Expression of a protein phosphatase 1 inhibitor, cdNIPP1, increases CDK9 threonine 186 phosphorylation and inhibits HIV-1 transcription. J Biol Chem 286, 3798–3804 10.1074/jbc.M110.196493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J. L., DeHart J. L., Zimmerman E. S., Ardon O., Kim B., Jacquot G., Benichou S., Planelles V. (2006). HIV-1 Vpr-induced apoptosis is cell cycle dependent and requires Bax but not ANT. PLoS Pathog 2, e127 10.1371/journal.ppat.0020127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin N. M., Espeseth A., Parker D., Cheema M., Hazuda D., Margolis D. M. (2009). Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses 25, 207–212 10.1089/aid.2008.0191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardon O., Zimmerman E. S., Andersen J. L., DeHart J. L., Blackett J., Planelles V. (2006). Induction of G2 arrest and binding to cyclophilin A are independent phenotypes of human immunodeficiency virus type 1 Vpr. J Virol 80, 3694–3700 10.1128/JVI.80.8.3694-3700.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arold S., Franken P., Strub M. P., Hoh F., Benichou S., Benarous R., Dumas C. (1997). The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling. Structure 5, 1361–1372 10.1016/S0969-2126(97)00286-4 [DOI] [PubMed] [Google Scholar]
- Bartlett J. A., Miralles G. D., Sevin A. D., Silberman M., Pruitt S. K., Ottinger J., Gryszowska V., Fiscus S. A., Bucy R. P., ACTG 380 Study Team (2002). Addition of cyclophosphamide to antiretroviral therapy does not diminish the cellular reservoir in HIV-infected persons. AIDS Res Hum Retroviruses 18, 535–543 10.1089/088922202753747888 [DOI] [PubMed] [Google Scholar]
- Baur A. S. (2011). HIV-Nef and AIDS pathogenesis: are we barking up the wrong tree? Trends Microbiol 19, 435–440 10.1016/j.tim.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Bedoya L. M., Márquez N., Martínez N., Gutiérrez-Eisman S., Alvarez A., Calzado M. A., Rojas J. M., Appendino G., Muñoz E., Alcamí J. (2009). SJ23B, a jatrophane diterpene activates classical PKCs and displays strong activity against HIV in vitro. Biochem Pharmacol 77, 965–978 10.1016/j.bcp.2008.11.025 [DOI] [PubMed] [Google Scholar]
- Bennasser Y., Le S. Y., Benkirane M., Jeang K. T. (2005). Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22, 607–619 10.1016/j.immuni.2005.03.010 [DOI] [PubMed] [Google Scholar]
- Bertin J., Barat C., Méthot S., Tremblay M. J. (2012). Interactions between prostaglandins, leukotrienes and HIV-1: possible implications for the central nervous system. Retrovirology 9, 4 10.1186/1742-4690-9-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignami F., Pilotti E., Bertoncelli L., Ronzi P., Gulli M., Marmiroli N., Magnani G., Pinti M., Lopalco L. & other authors (2012). Stable changes in CD4+ T-lymphocyte microRNA expression following exposure to HIV-1. Blood. 10.1182/blood-2011-09-379503. 10.1182/blood-2011-09-379503 [DOI] [PubMed] [Google Scholar]
- Blazkova J., Trejbalova K., Gondois-Rey F., Halfon P., Philibert P., Guiguen A., Verdin E., Olive D., Van Lint C. & other authors (2009). CpG methylation controls reactivation of HIV from latency. PLoS Pathog 5, e1000554 10.1371/journal.ppat.1000554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocklandt S., Blumberg P. M., Hamer D. H. (2003). Activation of latent HIV-1 expression by the potent anti-tumor promoter 12-deoxyphorbol 13-phenylacetate. Antiviral Res 59, 89–98 10.1016/S0166-3542(03)00034-2 [DOI] [PubMed] [Google Scholar]
- Borjabad A., Brooks A. I., Volsky D. J. (2010). Gene expression profiles of HIV-1-infected glia and brain: toward better understanding of the role of astrocytes in HIV-1-associated neurocognitive disorders. J Neuroimmune Pharmacol 5, 44–62 10.1007/s11481-009-9167-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boven L. A., Noorbakhsh F., Bouma G., van der Zee R., Vargas D. L., Pardo C., McArthur J. C., Nottet H. S., Power C. (2007). Brain-derived human immunodeficiency virus-1 Tat exerts differential effects on LTR transactivation and neuroimmune activation. J Neurovirol 13, 173–184 10.1080/13550280701258399 [DOI] [PubMed] [Google Scholar]
- Brass A. L., Dykxhoorn D. M., Benita Y., Yan N., Engelman A., Xavier R. J., Lieberman J., Elledge S. J. (2008). Identification of host proteins required for HIV infection through a functional genomic screen. Science 319, 921–926 10.1126/science.1152725 [DOI] [PubMed] [Google Scholar]
- Brigati C., Giacca M., Noonan D. M., Albini A. (2003). HIV Tat, its TARgets and the control of viral gene expression. FEMS Microbiol Lett 220, 57–65 10.1016/S0378-1097(03)00067-3 [DOI] [PubMed] [Google Scholar]
- Bukrinskaya A. (2007). HIV-1 matrix protein: a mysterious regulator of the viral life cycle. Virus Res 124, 1–11 10.1016/j.virusres.2006.07.001 [DOI] [PubMed] [Google Scholar]
- Burdo T. H., Nonnemacher M., Irish B. P., Choi C. H., Krebs F. C., Gartner S., Wigdahl B. (2004). High-affinity interaction between HIV-1 Vpr and specific sequences that span the C/EBP and adjacent NF-κB sites within the HIV-1 LTR correlate with HIV-1-associated dementia. DNA Cell Biol 23, 261–269 10.1089/104454904773819842 [DOI] [PubMed] [Google Scholar]
- Burnett J. C., Lim K. I., Calafi A., Rossi J. J., Schaffer D. V., Arkin A. P. (2010). Combinatorial latency reactivation for HIV-1 subtypes and variants. J Virol 84, 5958–5974 10.1128/JVI.00161-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chable-Bessia C., Meziane O., Latreille D., Triboulet R., Zamborlini A., Wagschal A., Jacquet J. M., Reynes J., Levy Y. & other authors (2009). Suppression of HIV-1 replication by microRNA effectors. Retrovirology 6, 26 10.1186/1742-4690-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang K., Sung T. L., Rice A. P. (2012). Regulation of cyclin T1 and HIV-1 replication by microRNAs in resting CD4+ T lymphocytes. J Virol 86, 3244–3252 10.1128/JVI.05065-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y. L., Soros V. B., Kreisberg J. F., Stopak K., Yonemoto W., Greene W. C. (2005). Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435, 108–114 10.1038/nature03493 [DOI] [PubMed] [Google Scholar]
- Choudhary S. K., Margolis D. M. (2011). Curing HIV: pharmacologic approaches to target HIV-1 latency. Annu Rev Pharmacol Toxicol 51, 397–418 10.1146/annurev-pharmtox-010510-100237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen H. S., Daher A., Soye K. J., Frankel L. B., Alexander M. R., Lainé S., Bannwarth S., Ong C. L., Chung S. W. & other authors (2007). Small interfering RNAs against the TAR RNA binding protein, TRBP, a Dicer cofactor, inhibit human immunodeficiency virus type 1 long terminal repeat expression and viral production. J Virol 81, 5121–5131 10.1128/JVI.01511-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T. W., Carruth L., Finzi D., Shen X., DiGiuseppe J. A., Taylor H., Hermankova M., Chadwick K., Margolick J. & other authors (1997). Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387, 183–188 10.1038/387183a0 [DOI] [PubMed] [Google Scholar]
- Chun T. W., Engel D., Berrey M. M., Shea T., Corey L., Fauci A. S. (1998). Early establishment of a pool of latently infected, resting CD4+ T cells during primary HIV-1 infection. Proc Natl Acad Sci U S A 95, 8869–8873 10.1073/pnas.95.15.8869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T. W., Nickle D. C., Justement J. S., Meyers J. H., Roby G., Hallahan C. W., Kottilil S., Moir S., Mican J. M. & other authors (2008). Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J Infect Dis 197, 714–720 10.1086/527324 [DOI] [PubMed] [Google Scholar]
- Ciuffi A., Llano M., Poeschla E., Hoffmann C., Leipzig J., Shinn P., Ecker J. R., Bushman F. (2005). A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med 11, 1287–1289 10.1038/nm1329 [DOI] [PubMed] [Google Scholar]
- Clements J. E., Gama L., Graham D. R., Mankowski J. L., Zink M. C. (2011). A simian immunodeficiency virus macaque model of highly active antiretroviral treatment: viral latency in the periphery and the central nervous system. Curr Opin HIV AIDS 6, 37–42 10.1097/COH.0b013e3283412413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos-Jiménez V., Booiman T., Hamann J., Kootstra N. A. (2011). Macrophages and HIV-1. Curr Opin HIV AIDS 6, 385–390 10.1097/COH.0b013e3283497203 [DOI] [PubMed] [Google Scholar]
- Coleman C. M., Wu L. (2009). HIV interactions with monocytes and dendritic cells: viral latency and reservoirs. Retrovirology 6, 51 10.1186/1742-4690-6-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin L., Van Lint C. (2009). Molecular control of HIV-1 postintegration latency: implications for the development of new therapeutic strategies. Retrovirology 6, 111 10.1186/1742-4690-6-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbeau P. (2008). Interfering RNA and HIV: reciprocal interferences. PLoS Pathog 4, e1000162 10.1371/journal.ppat.1000162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L. E., Hjelle B. L., Miller V. E., Palmer D. L., Llewellyn A. L., Merlin T. L., Young S. A., Mills R. G., Wachsman W., Wiley C. A. (1992). Early viral brain invasion in iatrogenic human immunodeficiency virus infection. Neurology 42, 1736–1739 [DOI] [PubMed] [Google Scholar]
- Demarchi F., Gutierrez M. I., Giacca M. (1999). Human immunodeficiency virus type 1 Tat protein activates transcription factor NF-κB through the cellular interferon-inducible, double-stranded RNA-dependent protein kinase, PKR. J Virol 73, 7080–7086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Wang D., de la Fuente C., Wang L., Li H., Lee C. G., Donnelly R., Wade J. D., Lambert P., Kashanchi F. (2001). Enhancement of the p300 HAT activity by HIV-1 Tat on chromatin DNA. Virology 289, 312–326 10.1006/viro.2001.1129 [DOI] [PubMed] [Google Scholar]
- Deshmane S. L., Mukerjee R., Fan S., Del Valle L., Michiels C., Sweet T., Rom I., Khalili K., Rappaport J. & other authors (2009). Activation of the oxidative stress pathway by HIV-1 Vpr leads to induction of hypoxia-inducible factor 1α expression. J Biol Chem 284, 11364–11373 10.1074/jbc.M809266200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Kwas C., Wu L. (2009). Transcriptional restriction of human immunodeficiency virus type 1 gene expression in undifferentiated primary monocytes. J Virol 83, 3518–3527 10.1128/JVI.02665-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Chéné I., Basyuk E., Lin Y. L., Triboulet R., Knezevich A., Chable-Bessia C., Mettling C., Baillat V., Reynes J. & other authors (2007). Suv39H1 and HP1γ are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J 26, 424–435 10.1038/sj.emboj.7601517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easley R., Van Duyne R., Coley W., Guendel I., Dadgar S., Kehn-Hall K., Kashanchi F. (2010). Chromatin dynamics associated with HIV-1 Tat-activated transcription. Biochim Biophys Acta 1799, 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein D. A., Sherman M. P., Penn M. L., Chin P. S., De Noronha C. M., Greene W. C., Goldsmith M. A. (2001). HIV-1 Vpr enhances viral burden by facilitating infection of tissue macrophages but not nondividing CD4+ T cells. J Exp Med 194, 1407–1419 10.1084/jem.194.10.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epie N., Ammosova T., Turner W., Nekhai S. (2006). Inhibition of PP2A by LIS1 increases HIV-1 gene expression. Retrovirology 3, 65 10.1186/1742-4690-3-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugenin E. A., Clements J. E., Zink M. C., Berman J. W. (2011). Human immunodeficiency virus infection of human astrocytes disrupts blood-brain barrier integrity by a gap junction-dependent mechanism. J Neurosci 31, 9456–9465 10.1523/JNEUROSCI.1460-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenard D., Yonemoto W., de Noronha C., Cavrois M., Williams S. A., Greene W. C. (2005). Nef is physically recruited into the immunological synapse and potentiates T cell activation early after TCR engagement. J Immunol 175, 6050–6057 [DOI] [PubMed] [Google Scholar]
- Fenyö E. M., Morfeldt-Månson L., Chiodi F., Lind B., von Gegerfelt A., Albert J., Olausson E., Asjö B. (1988). Distinct replicative and cytopathic characteristics of human immunodeficiency virus isolates. J Virol 62, 4414–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentini S., Giagulli C., Caccuri F., Magiera A. K., Caruso A. (2010). HIV-1 matrix protein p17: a candidate antigen for therapeutic vaccines against AIDS. Pharmacol Ther 128, 433–444 10.1016/j.pharmthera.2010.08.005 [DOI] [PubMed] [Google Scholar]
- Fortin J. F., Barat C., Beauséjour Y., Barbeau B., Tremblay M. J. (2004). Hyper-responsiveness to stimulation of human immunodeficiency virus-infected CD4+ T cells requires Nef and Tat virus gene products and results from higher NFAT, NF-κB, and AP-1 induction. J Biol Chem 279, 39520–39531 10.1074/jbc.M407477200 [DOI] [PubMed] [Google Scholar]
- Futaki S., Suzuki T., Ohashi W., Yagami T., Tanaka S., Ueda K., Sugiura Y. (2001). Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J Biol Chem 276, 5836–5840 10.1074/jbc.M007540200 [DOI] [PubMed] [Google Scholar]
- Gatignol A., Lainé S., Clerzius G. (2005). Dual role of TRBP in HIV replication and RNA interference: viral diversion of a cellular pathway or evasion from antiviral immunity? Retrovirology 2, 65 10.1186/1742-4690-2-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giagulli C., Marsico S., Magiera A. K., Bruno R., Caccuri F., Barone I., Fiorentini S., Andò S., Caruso A. (2011). Opposite effects of HIV-1 p17 variants on PTEN activation and cell growth in B cells. PLoS ONE 6, e17831 10.1371/journal.pone.0017831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot J., de Vries W., Geutjes E. J., Prins M., de Haan P., Berkhout B. (2007). The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog 3, e86 10.1371/journal.ppat.0030086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakre S., Chavez L., Shirakawa K., Verdin E. (2011). Epigenetic regulation of HIV latency. Curr Opin HIV AIDS 6, 19–24 10.1097/COH.0b013e3283412384 [DOI] [PubMed] [Google Scholar]
- Hariharan M., Scaria V., Pillai B., Brahmachari S. K. (2005). Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun 337, 1214–1218 10.1016/j.bbrc.2005.09.183 [DOI] [PubMed] [Google Scholar]
- Hayes A. M., Qian S., Yu L., Boris-Lawrie K. (2011). Tat RNA silencing suppressor activity contributes to perturbation of lymphocyte miRNA by HIV-1. Retrovirology 8, 36 10.1186/1742-4690-8-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A. J., Calame K. L. (1997). CCAAT/enhancer binding protein (C/EBP) sites are required for HIV-1 replication in primary macrophages but not CD4+ T cells. Proc Natl Acad Sci U S A 94, 8714–8719 10.1073/pnas.94.16.8714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbein G., Varin A. (2010). The macrophage in HIV-1 infection: from activation to deactivation? Retrovirology 7, 33 10.1186/1742-4690-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbein G., Gras G., Khan K. A., Abbas W. (2010). Macrophage signaling in HIV-1 infection. Retrovirology 7, 34 10.1186/1742-4690-7-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan T. H., Nonnemacher M. R., Krebs F. C., Henderson A., Wigdahl B. (2003). HIV-1 Vpr binding to HIV-1 LTR C/EBP cis-acting elements and adjacent regions is sequence-specific. Biomed Pharmacother 57, 41–48 10.1016/S0753-3322(02)00333-5 [DOI] [PubMed] [Google Scholar]
- Honda Y., Rogers L., Nakata K., Zhao B. Y., Pine R., Nakai Y., Kurosu K., Rom W. N., Weiden M. (1998). Type I interferon induces inhibitory 16-kD CCAAT/enhancer binding protein (C/EBP)β, repressing the HIV-1 long terminal repeat in macrophages: pulmonary tuberculosis alters C/EBP expression, enhancing HIV-1 replication. J Exp Med 188, 1255–1265 10.1084/jem.188.7.1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiba M., Martinez L. B., Buescher J. L., Sato S., Limoges J., Jiang Y., Jones C., Ikezu T. (2007). OTK18, a zinc-finger protein, regulates human immunodeficiency virus type 1 long terminal repeat through two distinct regulatory regions. J Gen Virol 88, 236–241 10.1099/vir.0.82066-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino Y., Nakata K., Hoshino S., Honda Y., Tse D. B., Shioda T., Rom W. N., Weiden M. (2002). Maximal HIV-1 replication in alveolar macrophages during tuberculosis requires both lymphocyte contact and cytokines. J Exp Med 195, 495–505 10.1084/jem.20011614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzet L., Jeang K. T. (2011). MicroRNAs and human retroviruses. Biochim Biophys Acta 1809, 686–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzet L., Morichaud Z., Didierlaurent L., Muriaux D., Darlix J. L., Mougel M. (2008). Nucleocapsid mutations turn HIV-1 into a DNA-containing virus. Nucleic Acids Res 36, 2311–2319 10.1093/nar/gkn069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howcroft T. K., Strebel K., Martin M. A., Singer D. S. (1993). Repression of MHC class I gene promoter activity by two-exon Tat of HIV. Science 260, 1320–1322 10.1126/science.8493575 [DOI] [PubMed] [Google Scholar]
- Huang J., Wang F., Argyris E., Chen K., Liang Z., Tian H., Huang W., Squires K., Verlinghieri G., Zhang H. (2007). Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med 13, 1241–1247 10.1038/nm1639 [DOI] [PubMed] [Google Scholar]
- Igarashi T., Brown C. R., Endo Y., Buckler-White A., Plishka R., Bischofberger N., Hirsch V., Martin M. A. (2001). Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): implications for HIV-1 infections of humans. Proc Natl Acad Sci U S A 98, 658–663 10.1073/pnas.021551798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S., Cimermancic P., Gulbahce N., Johnson J. R., McGovern K. E., Clarke S. C., Shales M., Mercenne G., Pache L. & other authors (2012). Global landscape of HIV-human protein complexes. Nature 481, 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeang K. T., Berkhout B., Dropulic B. (1993). Effects of integration and replication on transcription of the HIV-1 long terminal repeat. J Biol Chem 268, 24940–24949 [PubMed] [Google Scholar]
- Karn J. (2011). The molecular biology of HIV latency: breaking and restoring the Tat-dependent transcriptional circuit. Curr Opin HIV AIDS 6, 4–11 10.1097/COH.0b013e328340ffbb [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauder S. E., Bosque A., Lindqvist A., Planelles V., Verdin E. (2009). Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog 5, e1000495 10.1371/journal.ppat.1000495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilareski E. M., Shah S., Nonnemacher M. R., Wigdahl B. (2009). Regulation of HIV-1 transcription in cells of the monocyte-macrophage lineage. Retrovirology 6, 118 10.1186/1742-4690-6-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Martemyanov K. A., Thayer S. A. (2008a). Human immunodeficiency virus protein Tat induces synapse loss via a reversible process that is distinct from cell death. J Neurosci 28, 12604–12613 10.1523/JNEUROSCI.2958-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. A., Chang S., Yoon J. H., Ahn S. G. (2008b). TAT-Hsp40 inhibits oxidative stress-mediated cytotoxicity via the inhibition of Hsp70 ubiquitination. FEBS Lett 582, 734–740 10.1016/j.febslet.2008.01.053 [DOI] [PubMed] [Google Scholar]
- Kim N., Kukkonen S., Gupta S., Aldovini A. (2010). Association of Tat with promoters of PTEN and PP2A subunits is key to transcriptional activation of apoptotic pathways in HIV-infected CD4+ T cells. PLoS Pathog 6, e1001103 10.1371/journal.ppat.1001103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T., Pavlakis G. N. (2004). Partner molecules of accessory protein Vpr of the human immunodeficiency virus type 1. DNA Cell Biol 23, 193–205 10.1089/104454904773819789 [DOI] [PubMed] [Google Scholar]
- Kino T., Gragerov A., Kopp J. B., Stauber R. H., Pavlakis G. N., Chrousos G. P. (1999). The HIV-1 virion-associated protein Vpr is a coactivator of the human glucocorticoid receptor. J Exp Med 189, 51–62 10.1084/jem.189.1.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff F. (2010). Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8, 55–67 10.1016/j.chom.2010.06.004 [DOI] [PubMed] [Google Scholar]
- Klase Z., Kale P., Winograd R., Gupta M. V., Heydarian M., Berro R., McCaffrey T., Kashanchi F. (2007). HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol 8, 63 10.1186/1471-2199-8-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klase Z., Winograd R., Davis J., Carpio L., Hildreth R., Heydarian M., Fu S., McCaffrey T., Meiri E. & other authors (2009). HIV-1 TAR miRNA protects against apoptosis by altering cellular gene expression. Retrovirology 6, 18 10.1186/1742-4690-6-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klichko V., Archin N., Kaur R., Lehrman G., Margolis D. (2006). Hexamethylbisacetamide remodels the human immunodeficiency virus type 1 (HIV-1) promoter and induces Tat-independent HIV-1 expression but blunts cell activation. J Virol 80, 4570–4579 10.1128/JVI.80.9.4570-4579.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan M., Rappaport J. (2011). HIV-1 accessory protein Vpr: relevance in the pathogenesis of HIV and potential for therapeutic intervention. Retrovirology 8, 25 10.1186/1742-4690-8-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro I., Yokota Y., Yasuda S., Iwamoto A., Kagawa K. S. (2003). CSF-induced and HIV-1-mediated distinct regulation of Hck and C/EBPβ represent a heterogeneous susceptibility of monocyte-derived macrophages to M-tropic HIV-1 infection. J Exp Med 198, 443–453 10.1084/jem.20022018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König R., Zhou Y., Elleder D., Diamond T. L., Bonamy G. M., Irelan J. T., Chiang C. Y., Tu B. P., De Jesus P. D. & other authors (2008). Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135, 49–60 10.1016/j.cell.2008.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautkrämer E., Giese S. I., Gasteier J. E., Muranyi W., Fackler O. T. (2004). Human immunodeficiency virus type 1 Nef activates p21-activated kinase via recruitment into lipid rafts. J Virol 78, 4085–4097 10.1128/JVI.78.8.4085-4097.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J., Culnan D. M., Roman J., Dornadula G., Schnell M., Boyd M. R., Pomerantz R. J. (2001). Prostratin: activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood 98, 3006–3015 10.1182/blood.V98.10.3006 [DOI] [PubMed] [Google Scholar]
- Kutney S. N., Hong R., Macfarlan T., Chakravarti D. (2004). A signaling role of histone-binding proteins and INHAT subunits pp32 and Set/TAF-Iβ in integrating chromatin hypoacetylation and transcriptional repression. J Biol Chem 279, 30850–30855 10.1074/jbc.M404969200 [DOI] [PubMed] [Google Scholar]
- Le Douce V., Herbein G., Rohr O., Schwartz C. (2010). Molecular mechanisms of HIV-1 persistence in the monocyte-macrophage lineage. Retrovirology 7, 32 10.1186/1742-4690-7-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. S., Sarma D., Zhou H., Henderson A. J. (2002). CCAAT/enhancer binding proteins are not required for HIV-1 entry but regulate proviral transcription by recruiting coactivators to the long-terminal repeat in monocytic cells. Virology 299, 20–31 10.1006/viro.2002.1500 [DOI] [PubMed] [Google Scholar]
- Levy D. N., Refaeli Y., Weiner D. B. (1995). Extracellular Vpr protein increases cellular permissiveness to human immunodeficiency virus replication and reactivates virus from latency. J Virol 69, 1243–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski M. K., Bisgrove D., Shinn P., Chen H., Hoffmann C., Hannenhalli S., Verdin E., Berry C. C., Ecker J. R., Bushman F. D. (2005). Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J Virol 79, 6610–6619 10.1128/JVI.79.11.6610-6619.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. C., Lee D. C., Cheung B. K., Lau A. S. (2005). Mechanisms for HIV Tat upregulation of IL-10 and other cytokine expression: kinase signaling and PKR-mediated immune response. FEBS Lett 579, 3055–3062 10.1016/j.febslet.2005.04.060 [DOI] [PubMed] [Google Scholar]
- Li L., Aiamkitsumrit B., Pirrone V., Nonnemacher M. R., Wojno A., Passic S., Flaig K., Kilareski E., Blakey B. & other authors (2011). Development of co-selected single nucleotide polymorphisms in the viral promoter precedes the onset of human immunodeficiency virus type 1-associated neurocognitive impairment. J Neurovirol 17, 92–109 10.1007/s13365-010-0014-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisziewicz J., Foli A., Wainberg M., Lori F. (2003). Hydroxyurea in the treatment of HIV infection: clinical efficacy and safety concerns. Drug Saf 26, 605–624 10.2165/00002018-200326090-00002 [DOI] [PubMed] [Google Scholar]
- Liu Y., Nonnemacher M. R., Stauff D. L., Li L., Banerjee A., Irish B., Kilareski E., Rajagopalan N., Suchitra J. B. & other authors (2010). Structural and functional studies of CCAAT/enhancer binding sites within the human immunodeficiency virus type 1 subtype C LTR. Biomed Pharmacother 64, 672–680 10.1016/j.biopha.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Huertas M. R., Callejas S., Abia D., Mateos E., Dopazo A., Alcamí J., Coiras M. (2010). Modifications in host cell cytoskeleton structure and function mediated by intracellular HIV-1 Tat protein are greatly dependent on the second coding exon. Nucleic Acids Res 38, 3287–3307 10.1093/nar/gkq037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loré K., Smed-Sörensen A., Vasudevan J., Mascola J. R., Koup R. A. (2005). Myeloid and plasmacytoid dendritic cells transfer HIV-1 preferentially to antigen-specific CD4+ T cells. J Exp Med 201, 2023–2033 10.1084/jem.20042413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lori F., Malykh A., Cara A., Sun D., Weinstein J. N., Lisziewicz J., Gallo R. C. (1994). Hydroxyurea as an inhibitor of human immunodeficiency virus-type 1 replication. Science 266, 801–805 10.1126/science.7973634 [DOI] [PubMed] [Google Scholar]
- Lu S., Cullen B. R. (2004). Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and microRNA biogenesis. J Virol 78, 12868–12876 10.1128/JVI.78.23.12868-12876.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusic M., Marcello A., Cereseto A., Giacca M. (2003). Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J 22, 6550–6561 10.1093/emboj/cdg631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim M. H. (2009). APOBEC proteins and intrinsic resistance to HIV-1 infection. Philos Trans R Soc Lond B Biol Sci 364, 675–687 10.1098/rstb.2008.0185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim M. H., Emerman M. (2008). HIV-1 accessory proteins–ensuring viral survival in a hostile environment. Cell Host Microbe 3, 388–398 10.1016/j.chom.2008.04.008 [DOI] [PubMed] [Google Scholar]
- Margolis D. M. (2011). Histone deacetylase inhibitors and HIV latency. Curr Opin HIV AIDS 6, 25–29 10.1097/COH.0b013e328341242d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini E., Tiberio L., Caracciolo S., Tosti G., Guzman C. A., Schiaffonati L., Fiorentini S., Caruso A. (2008). HIV-1 matrix protein p17 binds to monocytes and selectively stimulates MCP-1 secretion: role of transcriptional factor AP-1. Cell Microbiol 10, 655–666 10.1111/j.1462-5822.2007.01073.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas A. P., Musier-Forsyth K. (2009). The capsid protein of human immunodeficiency virus: interactions of HIV-1 capsid with host protein factors. FEBS J 276, 6118–6127 10.1111/j.1742-4658.2009.07315.x [DOI] [PubMed] [Google Scholar]
- McNamara L. A., Collins K. L. (2011). Hematopoietic stem/precursor cells as HIV reservoirs. Curr Opin HIV AIDS 6, 43–48 10.1097/COH.0b013e32834086b3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K. V. (2005). siRNA-mediated transcriptional gene silencing: the potential mechanism and a possible role in the histone code. Cell Mol Life Sci 62, 3057–3066 10.1007/s00018-005-5182-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukerjee R., Chang J. R., Del Valle L., Bagashev A., Gayed M. M., Lyde R. B., Hawkins B. J., Brailoiu E., Cohen E. & other authors (2011). Deregulation of microRNAs by HIV-1 Vpr protein leads to the development of neurocognitive disorders. J Biol Chem 286, 34976–34985 10.1074/jbc.M111.241547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na H., Acharjee S., Jones G., Vivithanaporn P., Noorbakhsh F., McFarlane N., Maingat F., Ballanyi K., Pardo C. A. & other authors (2011). Interactions between human immunodeficiency virus (HIV)-1 Vpr expression and innate immunity influence neurovirulence. Retrovirology 8, 44 10.1186/1742-4690-8-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasipura S. D., Henderson L. J., Fu S. W., Chen L., Kashanchi F., Al-Harthi L. (2012). Role of β-catenin and TCF/LEF family members in transcriptional activity of HIV in astrocytes. J Virol 86, 1911–1921 10.1128/JVI.06266-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A., Kehn-Hall K., Bailey C., Kashanchi F. (2011). Analysis of the roles of HIV-derived microRNAs. Expert Opin Biol Ther 11, 17–29 10.1517/14712598.2011.540564 [DOI] [PubMed] [Google Scholar]
- Nathans R., Chu C. Y., Serquina A. K., Lu C. C., Cao H., Rana T. M. (2009). Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell 34, 696–709 10.1016/j.molcel.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri F., Giolo G., Potestà M., Petrini S., Doria M. (2011). The HIV-1 Nef protein has a dual role in T cell receptor signaling in infected CD4+ T lymphocytes. Virology 410, 316–326 10.1016/j.virol.2010.11.018 [DOI] [PubMed] [Google Scholar]
- Neuveut C., Jeang K. T. (1996). Recombinant human immunodeficiency virus type 1 genomes with Tat unconstrained by overlapping reading frames reveal residues in Tat important for replication in tissue culture. J Virol 70, 5572–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri K. C., Mukerji J., Gabuzda D. (2011). Nef-mediated enhancement of cellular activation and human immunodeficiency virus type 1 replication in primary T cells is dependent on association with p21-activated kinase 2. Retrovirology 8, 64 10.1186/1742-4690-8-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto S., Fujii Y. R. (2005). Regulation of human immunodeficiency virus 1 transcription by nef microRNA. J Gen Virol 86, 751–755 10.1099/vir.0.80449-0 [DOI] [PubMed] [Google Scholar]
- Omoto S., Ito M., Tsutsumi Y., Ichikawa Y., Okuyama H., Brisibe E. A., Saksena N. K., Fujii Y. R. (2004). HIV-1 nef suppression by virally encoded microRNA. Retrovirology 1, 44 10.1186/1742-4690-1-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein J. M., Meltzer M. S., Phipps T., Gendelman H. E. (1988). Cytoplasmic assembly and accumulation of human immunodeficiency virus types 1 and 2 in recombinant human colony-stimulating factor-1-treated human monocytes: an ultrastructural study. J Virol 62, 2578–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein J. M., Fox C., Wahl S. M. (1997). Macrophages as a source of HIV during opportunistic infections. Science 276, 1857–1861 10.1126/science.276.5320.1857 [DOI] [PubMed] [Google Scholar]
- Ouellet D. L., Plante I., Barat C., Tremblay M. J., Provost P. (2009). Emergence of a complex relationship between HIV-1 and the microRNA pathway. Methods Mol Biol 487, 415–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R., Kim Y. K., Hokello J., Lassen K., Friedman J., Tyagi M., Karn J. (2008). Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J Virol 82, 12291–12303 10.1128/JVI.01383-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin B. M., Price D. H. (2006). Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23, 297–305 10.1016/j.molcel.2006.06.014 [DOI] [PubMed] [Google Scholar]
- Piller S. C., Jans P., Gage P. W., Jans D. A. (1998). Extracellular HIV-1 virus protein R causes a large inward current and cell death in cultured hippocampal neurons: implications for AIDS pathology. Proc Natl Acad Sci U S A 95, 4595–4600 10.1073/pnas.95.8.4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping Y. H., Rana T. M. (2001). DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J Biol Chem 276, 12951–12958 10.1074/jbc.M006130200 [DOI] [PubMed] [Google Scholar]
- Qian S., Zhong X., Yu L., Ding B., de Haan P., Boris-Lawrie K. (2009). HIV-1 Tat RNA silencing suppressor activity is conserved across kingdoms and counteracts translational repression of HIV-1. Proc Natl Acad Sci U S A 106, 605–610 10.1073/pnas.0806822106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy V., De Walque S., Van Lint C. (2007). Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies. Subcell Biochem 41, 371–396 [PubMed] [Google Scholar]
- Ranjbar S., Tsytsykova A. V., Lee S. K., Rajsbaum R., Falvo J. V., Lieberman J., Shankar P., Goldfeld A. E. (2006). NFAT5 regulates HIV-1 in primary monocytes via a highly conserved long terminal repeat site. PLoS Pathog 2, e130 10.1371/journal.ppat.0020130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch S., Pulkkinen K., Saksela K., Fackler O. T. (2008). Human immunodeficiency virus type 1 Nef recruits the guanine exchange factor Vav1 via an unexpected interface into plasma membrane microdomains for association with p21-activated kinase 2 activity. J Virol 82, 2918–2929 10.1128/JVI.02185-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravimohan S., Gama L., Barber S. A., Clements J. E. (2010). Regulation of SIV mac 239 basal long terminal repeat activity and viral replication in macrophages: functional roles of two CCAAT/enhancer-binding protein beta sites in activation and interferon beta-mediated suppression. J Biol Chem 285, 2258–2273 10.1074/jbc.M109.075929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refaeli Y., Levy D. N., Weiner D. B. (1995). The glucocorticoid receptor type II complex is a target of the HIV-1 Vpr gene product. Proc Natl Acad Sci U S A 92, 3621–3625 10.1073/pnas.92.8.3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuse S., Calao M., Kabeya K., Guiguen A., Gatot J. S., Quivy V., Vanhulle C., Lamine A., Vaira D. & other authors (2009). Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: implications for treatment of latent infection. PLoS ONE 4, e6093 10.1371/journal.pone.0006093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeth J. F., Collins K. L. (2006). Human immunodeficiency virus type 1 Nef: adapting to intracellular trafficking pathways. Microbiol Mol Biol Rev 70, 548–563 10.1128/MMBR.00042-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr O., Marban C., Aunis D., Schaeffer E. (2003). Regulation of HIV-1 gene transcription: from lymphocytes to microglial cells. J Leukoc Biol 74, 736–749 10.1189/jlb.0403180 [DOI] [PubMed] [Google Scholar]
- Rom S., Rom I., Passiatore G., Pacifici M., Radhakrishnan S., Del Valle L., Piña-Oviedo S., Khalili K., Eletto D., Peruzzi F. (2010). CCL8/MCP-2 is a target for mir-146a in HIV-1-infected human microglial cells. FASEB J 24, 2292–2300 10.1096/fj.09-143503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross H. L., Gartner S., McArthur J. C., Corboy J. R., McAllister J. J., Millhouse S., Wigdahl B. (2001). HIV-1 LTR C/EBP binding site sequence configurations preferentially encountered in brain lead to enhanced C/EBP factor binding and increased LTR-specific activity. J Neurovirol 7, 235–249 10.1080/13550280152403281 [DOI] [PubMed] [Google Scholar]
- Rossi A., Mukerjee R., Ferrante P., Khalili K., Amini S., Sawaya B. E. (2006). Human immunodeficiency virus type 1 Tat prevents dephosphorylation of Sp1 by TCF-4 in astrocytes. J Gen Virol 87, 1613–1623 10.1099/vir.0.81691-0 [DOI] [PubMed] [Google Scholar]
- Sánchez-Del Cojo M., López-Huertas M. R., Mateos E., Alcamí J., Coiras M. (2011). Mechanisms of RNA interference in the HIV-1-host cell interplay. AIDS Rev 13, 149–160 [PubMed] [Google Scholar]
- Sanghvi V. R., Steel L. F. (2011). A re-examination of global suppression of RNA interference by HIV-1. PLoS ONE 6, e17246 10.1371/journal.pone.0017246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Koyanagi Y. (2011). The mouse is out of the bag: insights and perspectives on HIV-1-infected humanized mouse models. Exp Biol Med (Maywood) 236, 977–985 10.1258/ebm.2011.010294 [DOI] [PubMed] [Google Scholar]
- Savarino A., Mai A., Norelli S., El Daker S., Valente S., Rotili D., Altucci L., Palamara A. T., Garaci E. (2009). “Shock and kill” effects of class I-selective histone deacetylase inhibitors in combination with the glutathione synthesis inhibitor buthionine sulfoximine in cell line models for HIV-1 quiescence. Retrovirology 6, 52 10.1186/1742-4690-6-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnettler E., de Vries W., Hemmes H., Haasnoot J., Kormelink R., Goldbach R., Berkhout B. (2009). The NS3 protein of rice hoja blanca virus complements the RNAi suppressor function of HIV-1 Tat. EMBO Rep 10, 258–263 10.1038/embor.2009.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopman N. C., Willemsen M., Liu Y. P., Bradley T., van Kampen A., Baas F., Berkhout B., Haasnoot J. (2012). Deep sequencing of virus-infected cells reveals HIV-encoded small RNAs. Nucleic Acids Res 40, 414–427 10.1093/nar/gkr719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder A. R., Shinn P., Chen H., Berry C., Ecker J. R., Bushman F. (2002). HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110, 521–529 10.1016/S0092-8674(02)00864-4 [DOI] [PubMed] [Google Scholar]
- Seelamgari A., Maddukuri A., Berro R., de la Fuente C., Kehn K., Deng L., Dadgar S., Bottazzi M. E., Ghedin E. & other authors (2004). Role of viral regulatory and accessory proteins in HIV-1 replication. Front Biosci 9, 2388–2413 10.2741/1403 [DOI] [PubMed] [Google Scholar]
- Sherman M. P., Schubert U., Williams S. A., de Noronha C. M., Kreisberg J. F., Henklein P., Greene W. C. (2002). HIV-1 Vpr displays natural protein-transducing properties: implications for viral pathogenesis. Virology 302, 95–105 10.1006/viro.2002.1576 [DOI] [PubMed] [Google Scholar]
- Simmons A., Gangadharan B., Hodges A., Sharrocks K., Prabhakar S., García A., Dwek R., Zitzmann N., McMichael A. (2005). Nef-mediated lipid raft exclusion of UbcH7 inhibits Cbl activity in T cells to positively regulate signaling. Immunity 23, 621–634 10.1016/j.immuni.2005.11.003 [DOI] [PubMed] [Google Scholar]
- Sloan R. D., Donahue D. A., Kuhl B. D., Bar-Magen T., Wainberg M. A. (2010). Expression of Nef from unintegrated HIV-1 DNA downregulates cell surface CXCR4 and CCR5 on T-lymphocytes. Retrovirology 7, 44 10.1186/1742-4690-7-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solas C., Lafeuillade A., Halfon P., Chadapaud S., Hittinger G., Lacarelle B. (2003). Discrepancies between protease inhibitor concentrations and viral load in reservoirs and sanctuary sites in human immunodeficiency virus-infected patients. Antimicrob Agents Chemother 47, 238–243 10.1128/AAC.47.1.238-243.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strazza M., Pirrone V., Wigdahl B., Nonnemacher M. R. (2011). Breaking down the barrier: the effects of HIV-1 on the blood-brain barrier. Brain Res 1399, 96–115 10.1016/j.brainres.2011.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbramanian R. A., Xu J., Toma E., Morisset R., Cohen E. A., Menezes J., Ahmad A. (2002). Comparison of human immunodeficiency virus (HIV)-specific infection-enhancing and -inhibiting antibodies in AIDS patients. J Clin Microbiol 40, 2141–2146 10.1128/JCM.40.6.2141-2146.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Li H., Wu X., Covarrubias M., Scherer L., Meinking K., Luk B., Chomchan P., Alluin J. & other authors (2011). Interplay between HIV-1 infection and host microRNAs. Nucleic Acids Res 10.1093/nar/gkr961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung T. L., Rice A. P. (2009). miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1. PLoS Pathog 5, e1000263 10.1371/journal.ppat.1000263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swingler S., Brichacek B., Jacque J. M., Ulich C., Zhou J., Stevenson M. (2003). HIV-1 Nef intersects the macrophage CD40L signalling pathway to promote resting-cell infection. Nature 424, 213–219 10.1038/nature01749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirov T. H., Babayeva N. D., Varzavand K., Cooper J. J., Sedore S. C., Price D. H. (2010). Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature 465, 747–751 10.1038/nature09131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N., Hoshino Y., Gold J., Hoshino S., Martiniuk F., Kurata T., Pine R., Levy D., Rom W. N., Weiden M. (2005). Interleukin-10 induces inhibitory C/EBPβ through STAT-3 and represses HIV-1 transcription in macrophages. Am J Respir Cell Mol Biol 33, 406–411 10.1165/rcmb.2005-0140OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trible R. P., Emert-Sedlak L., Smithgall T. E. (2006). HIV-1 Nef selectively activates Src family kinases Hck, Lyn, and c-Src through direct SH3 domain interaction. J Biol Chem 281, 27029–27038 10.1074/jbc.M601128200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet R., Mari B., Lin Y. L., Chable-Bessia C., Bennasser Y., Lebrigand K., Cardinaud B., Maurin T., Barbry P. & other authors (2007). Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315, 1579–1582 10.1126/science.1136319 [DOI] [PubMed] [Google Scholar]
- Tyagi M., Rusnati M., Presta M., Giacca M. (2001). Internalization of HIV-1 Tat requires cell surface heparan sulfate proteoglycans. J Biol Chem 276, 3254–3261 10.1074/jbc.M006701200 [DOI] [PubMed] [Google Scholar]
- Tyagi M., Pearson R. J., Karn J. (2010). Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J Virol 84, 6425–6437 10.1128/JVI.01519-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne R., Easley R., Wu W., Berro R., Pedati C., Klase Z., Kehn-Hall K., Flynn E. K., Symer D. E., Kashanchi F. (2008). Lysine methylation of HIV-1 Tat regulates transcriptional activity of the viral LTR. Retrovirology 5, 40 10.1186/1742-4690-5-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint C. (2000). Role of chromatin in HIV-1 transcriptional regulation. Adv Pharmacol 48, 121–160 10.1016/S1054-3589(00)48005-1 [DOI] [PubMed] [Google Scholar]
- Vardabasso C., Manganaro L., Lusic M., Marcello A., Giacca M. (2008). The histone chaperone protein nucleosome assembly protein-1 (hNAP-1) binds HIV-1 Tat and promotes viral transcription. Retrovirology 5, 8 10.1186/1742-4690-5-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varin A., Decrion A. Z., Sabbah E., Quivy V., Sire J., Van Lint C., Roques B. P., Aggarwal B. B., Herbein G. (2005). Synthetic Vpr protein activates activator protein-1, c-Jun N-terminal kinase, and NF-κB and stimulates HIV-1 transcription in promonocytic cells and primary macrophages. J Biol Chem 280, 42557–42567 10.1074/jbc.M502211200 [DOI] [PubMed] [Google Scholar]
- Verschure P. J., Visser A. E., Rots M. G. (2006). Step out of the groove: epigenetic gene control systems and engineered transcription factors. Adv Genet 56, 163–204 10.1016/S0065-2660(06)56005-5 [DOI] [PubMed] [Google Scholar]
- Vlach J., Garcia A., Jacqué J. M., Rodriguez M. S., Michelson S., Virelizier J. L. (1995). Induction of Sp1 phosphorylation and NF-κB-independent HIV promoter domain activity in T lymphocytes stimulated by okadaic acid. Virology 208, 753–761 10.1006/viro.1995.1207 [DOI] [PubMed] [Google Scholar]
- Wang X., Ye L., Hou W., Zhou Y., Wang Y. J., Metzger D. S., Ho W. Z. (2009). Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood 113, 671–674 10.1182/blood-2008-09-175000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg J. B., Matthews T. J., Cullen B. R., Malim M. H. (1991). Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J Exp Med 174, 1477–1482 10.1084/jem.174.6.1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger L. S., Burnett J. C., Toettcher J. E., Arkin A. P., Schaffer D. V. (2005). Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122, 169–182 10.1016/j.cell.2005.06.006 [DOI] [PubMed] [Google Scholar]
- Weinberger L. S., Dar R. D., Simpson M. L. (2008). Transient-mediated fate determination in a transcriptional circuit of HIV. Nat Genet 40, 466–470 10.1038/ng.116 [DOI] [PubMed] [Google Scholar]
- Williams S. A., Greene W. C. (2007). Regulation of HIV-1 latency by T-cell activation. Cytokine 39, 63–74 10.1016/j.cyto.2007.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer K. W., Watson A. K., Blankson J. N., Clements J. E. (2012). Relationships of PBMC microRNA expression, plasma viral load, and CD4+ T-cell count in HIV-1-infected elite suppressors and viremic patients. Retrovirology 9, 5 10.1186/1742-4690-9-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K., Sharma A., Awasthi S., Matlock E. F., Rogers L., Van Lint C., Skiest D. J., Burns D. K., Harrod R. (2005). HIV-1 Tat interactions with p300 and PCAF transcriptional coactivators inhibit histone acetylation and neurotrophin signaling through CREB. J Biol Chem 280, 9390–9399 10.1074/jbc.M408643200 [DOI] [PubMed] [Google Scholar]
- Wu L. P., Wang X., Li L., Zhao Y., Lu S., Yu Y., Zhou W., Liu X., Yang J. & other authors (2008). Histone deacetylase inhibitor depsipeptide activates silenced genes through decreasing both CpG and H3K9 methylation on the promoter. Mol Cell Biol 28, 3219–3235 10.1128/MCB.01516-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N., Cherepanov P., Daigle J. E., Engelman A., Lieberman J. (2009). The SET complex acts as a barrier to autointegration of HIV-1. PLoS Pathog 5, e1000327 10.1371/journal.ppat.1000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. C., Shen L., Siliciano R. F., Pomerantz J. L. (2009a). Isolation of a cellular factor that can reactivate latent HIV-1 without T cell activation. Proc Natl Acad Sci U S A 106, 6321–6326 10.1073/pnas.0809536106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. C., Xing S., Shan L., O’Connell K., Dinoso J., Shen A., Zhou Y., Shrum C. K., Han Y. & other authors (2009b). Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J Clin Invest 119, 3473–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung M. L., Bennasser Y., Myers T. G., Jiang G., Benkirane M., Jeang K. T. (2005). Changes in microRNA expression profiles in HIV-1-transfected human cells. Retrovirology 2, 81 10.1186/1742-4690-2-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung M. L., Houzet L., Yedavalli V. S., Jeang K. T. (2009a). A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem 284, 19463–19473 10.1074/jbc.M109.010033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung M. L., Bennasser Y., Watashi K., Le S. Y., Houzet L., Jeang K. T. (2009b). Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res 37, 6575–6586 10.1093/nar/gkp707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylisastigui L., Archin N. M., Lehrman G., Bosch R. J., Margolis D. M. (2004). Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS 18, 1101–1108 10.1097/00002030-200405210-00003 [DOI] [PubMed] [Google Scholar]
- Yukl S., Pillai S., Li P., Chang K., Pasutti W., Ahlgren C., Havlir D., Strain M., Günthard H. & other authors (2009). Latently-infected CD4+ T cells are enriched for HIV-1 Tat variants with impaired transactivation activity. Virology 387, 98–108 10.1016/j.virol.2009.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P. D., Haley B. (2005). Ribo-gnome: the big world of small RNAs. Science 309, 1519–1524 10.1126/science.1111444 [DOI] [PubMed] [Google Scholar]
- Zhao R. Y., Li G., Bukrinsky M. I. (2011). Vpr-host interactions during HIV-1 viral life cycle. J Neuroimmune Pharmacol 6, 216–229 10.1007/s11481-011-9261-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman E. S., Sherman M. P., Blackett J. L., Neidleman J. A., Kreis C., Mundt P., Williams S. A., Warmerdam M., Kahn J. & other authors (2006). Human immunodeficiency virus type 1 Vpr induces DNA replication stress in vitro and in vivo. J Virol 80, 10407–10418 10.1128/JVI.01212-06 [DOI] [PMC free article] [PubMed] [Google Scholar]