Abstract
Embryonic patterning displays remarkable consistency from individual to individual despite frequent environmental perturbations and diverse genetic contexts. Stochastic influences on the cellular environment may cause transcription rates to fluctuate, but these fluctuations rarely lead to developmental defects or disease. Here we characterize a set of recessive alleles of the Toll pathway component tube that destabilize embryonic dorsoventral patterning in Drosophila melanogaster. Females bearing these tube alleles generate embryos of an unusually wide range of dorsalized phenotypes, with the distributions across this range being unique for each allele. We determine that the mutant lines have in common a retrotransposon insertion upstream of the tube transcription start site. Genetic and molecular approaches demonstrate that this insertion dramatically reduces maternal expression of tube, thereby uncovering the inherent variability in gene expression. We further find that additional transposable element insertions near the tube gene synergistically enhance the phenotype caused by the sensitizing upstream insertion. These studies document how phenotypic variability can arise from normally occurring fluctuations around reduced mean expression and illustrate the contribution of transposons, individually and combinatorially, to such a state.
Keywords: transcriptional fluctuation, stochasticity, transposable elements, embryonic patterning, phenotypic variability
Gene expression often varies over time within a single cell or among cells of the same tissue. This variation exists, in part, because stochastic forces influence transcription (Chubb and Liverpool 2010; Kaern et al. 2005; Lehner and Kaneko 2011). The source of stochasticity itself varies and includes both the sporadic fluctuations in local transcription factor abundance and the dynamic nature of chromatin. Cells generally buffer this transcriptional noise, avoiding any detrimental effects and thus displaying a property termed phenotypic robustness (Flatt 2005; Waddington 1942). Developmental biologists have long been intrigued by the way wild-type organisms achieve robust patterning by dampening the effects of environmental, genetic, and stochastic perturbations (Arias and Hayward 2006; Houchmandzadeh et al. 2002; Porcher and Dostatni 2010).
Survival requires maintaining transcript levels of essential genes above a threshold value. Gene expression at levels significantly above the threshold is one potential means of lessening the effects of noisy gene expression; expression levels swing back and forth around an average, but the entire range lies above the threshold. In this model, the detrimental effects of stochastic forces on phenotype in a wild-type organism are minimized.
For the vast majority of Drosophila melanogaster genes, changing dosage does not affect survival, as demonstrated by investigations of segmental aneuploids (flies in which particular autosomal regions of the genome are present in only a single copy or in three copies). In their landmark investigation of the D. melanogaster genome, Lindsley et al. (1972) demonstrated the existence of at most 20 loci, and more likely just one, that are haploinsufficient for viability. This finding, together with subsequent studies, revealed that nearly all genes are normally expressed at levels greater than that required for survival, consistent with the idea that surplus gene expression contributes to phenotypic robustness.
On the basis of the aforementioned model, one would predict that fluctuations produced by stochastic forces would be revealed if an additional influence, such as a mutation, reduced the average expression level of a gene to near or below the threshold. The phenotype would then vary with changes in expression, essentially becoming a readout of the probabilistic nature of underlying molecular interactions.
With the exception of temperature-sensitive and other conditional missense alleles, mutations that disrupt developmental patterning typically result in a consistent and relatively narrow phenotypic range. Occasionally, however, phenotypic hypervariability emerges (Raj et al. 2010). What distinguishes these rare cases? It may be that some processes, like transcription, are more sensitive to perturbations than others. To investigate this phenotypic phenomenon, we focused our attention on a set of mutations exhibiting hypervariable disruption of Toll signaling.
In the D. melanogaster embryo, the Toll pathway establishes dorsoventral polarity. Females bearing mutations that block Toll signaling produce dorsalized embryos, with the severity of dorsalization corresponding to the extent of reduction in signaling (Supporting Information, Figure S1 and Anderson and Nüsslein-Volhard 1984; Huang et al. 1997). Generally, isogenic females bearing a mutation in a Toll pathway gene produce embryos of a very narrow phenotypic range. The mutations that we have studied, which affect the Toll pathway adaptor protein Tube, instead cause an unusually wide range of phenotypes. We have used these tube mutants as a model system to study variable gene expression and phenotypic robustness.
Materials and Methods
Fly stocks, site-specific male recombination, precise excision, and cuticle preparation
Alleles tub2, Df(3R)XM3, tubR5.6, tub6, tub7, tub8, and tub9 have been described previously (Hecht and Anderson 1993; Letsou et al. 1991). The tubste allele was identified on a st e marker chromosome obtained in the 1980s from the K. V. Anderson lab. The wild type (tub+/tub+), unless otherwise noted, was P{His2Av-EGFP.C}2/SM6a, obtained from Bloomington Drosophila Stock Center. Df(3R)XM3 served as tubDf and tubR5.6 served as tubnull in all experiments, except where otherwise noted. CG14646CB-0692-3 (CB06923), GS7007, and GS13951 were obtained from the Drosophila Genetic Resource Center at the Kyoto Institute of Technology. Site-specific recombination and precise excision were performed with the use of a transposase source from the stock T(2;3)apXa, apXa/CyO, H[PΔ2-3]HoP2.1; Sb obtained from Bloomington Drosophila Stock Center. Site-specific recombination was conducted as previously described (Chen et al. 1998). Precise excision was conducted by generating males bearing the Δ2−3 source and the tub9 chromosome, collecting their female progeny, mating them with wild-type males at 18°, and assaying for increased fecundity. Genomic DNA from potential excisants was amplified via polymerase chain reaction (PCR) and sequenced to identify precise excisants. For all experiments examining dorsalization, cuticles from embryos (1−2 d after fertilization) raised at 25° were prepared and scored as previously described (Wieschaus and Nüsslein-Volhard 1986), unless otherwise noted.
Survival assays
Survival assays were performed essentially as described previously (Romeo and Lemaitre 2008). Males (2−4 days posteclosion) were stabbed with a needle dipped in a 20% glycerol suspension of purified fungal spores; the fungus used was Fusarium oxysporum f. sp. lycopersici (obtained from the Fungal Genetics Stock Center). Flies were incubated at 29° for the duration of the experiment. Survival was assayed over 4 d.
Quantitative real-time (RT)-PCR, sequencing, and 5′ rapid amplication of cDNA ends (5′ RACE)
RNA was prepared using Trizol (Ambion) or RNeasy kit (QIAGEN) from embryos (0−1.5 hr after fertilization) or adult males (2−5 days after eclosion), and first-strand cDNA was synthesized with the SuperScript II kit (Invitrogen). Quantitative RT-PCR was performed on an iQ5 cycler (BioRad) using iQ SYBR Green Supermix (BioRad). Genomic DNA was prepared from adults as described previously (Huang et al. 2009). Taq Polymerase with ThermoPol buffer (NEB) and Expand HF kit (Roche) were used to amplify the tube transcript region and flanking regions for sequencing. Thermal asymmetric-interlaced (TAIL)-PCR was conducted essentially as described previously (Liu and Whittier 1995), except Phusion (NEB) was used as the polymerase. 5′ RACE was performed using the RLM-RACE kit (Ambion), and Phusion (NEB) was used as the PCR polymerase.
Immunoblotting
Immunoblotting protocols and rabbit α-Tube serum (1:20,000) have been described previously (Letsou et al. 1993; Sun et al. 2004). Rabbit α-Diaphanous (1:5,000) was used as a loading control and was previously described (Afshar et al. 2000). Secondary antibody was goat α-rabbit IgG-peroxidase (1:10,000; Sigma-Aldrich).
Statistics
Quantitative RT-PCR data were analyzed by use of a one-way analysis of variance test followed by a Dunnett post-test (GraphPad PRISM 5).
Results
Phenotypically variable tube alleles
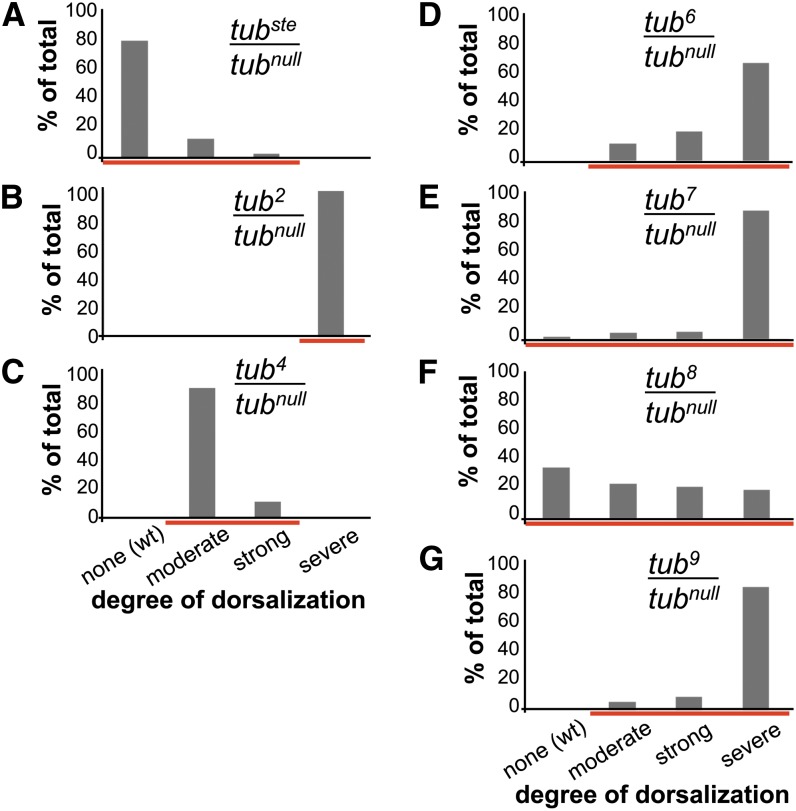
The starting point for these studies was the finding that a particular D. melanogaster chromosome provides variably reduced tube function. Because this tube allele was discovered on a marker chromosome containing visible mutations in the genes scarlet and ebony (st, e), we named it tubste. Females carrying the tubste chromosome in trans to a tube null mutation (tubste/tubnull) produce embryos that span a phenotypic range from strongly dorsalized to wild type (Figure 1A and Figure S1). As stated previously, except in cases of conditional missense alleles, such phenotypic variation is rare. For example, the phenotypes of tub2 and tub4, which disrupt tube function to quite different degrees, are distinct but nevertheless largely invariant (Figure 1, B and C). There is, however, a set of alleles that, like tubste, displays an unusually wide phenotypic range. Hecht and Anderson (1993) have reported isolation of four alleles—tub6, tub7, tub8, and tub9—exhibiting highly variable tube function. To compare the phenotypic variation of tubste with these tube alleles, we analyzed embryonic cuticles. Consistent with the published data, embryos from females carrying any of these alleles in trans to a strong or null tube mutation exhibit a broad range of phenotypes, with a distinct phenotypic distribution for each allele (Figure 1, D−G). Hereafter, we therefore use the term tubvar to refer to the variable alleles tubste, tub6, tub7, tub8, and tub9.
Figure 1.
Embryos from tubvar/tubnull females display a wide range of dorsalized phenotypes compared with conventional alleles, tub2 and tub4. (A−G) Phenotypic distributions of embryonic cuticles from groups of females of the specified genotype. Analysis of tub7 was performed at 18° and in trans to tub2, a strong hypomorphic allele. Red lines highlight phenotypic range. (A) n = 77, (B) n = 56, (C) n = 34, (D) n = 190, (E) n = 160, (F) n = 213, and (G) n = 197.
Phenotypic variability typically arises from heterogeneity in genetic background or the environment. However, when flies are held in a constant environment, a single tubvar/tubnull female, like a population of tubvar/tubnull females, generates embryos with a range of dorsalization (compare Figure 1 and Figure S2; see also Hecht and Anderson 1993). Thus, the phenotypic range does not reflect variation in genetic background or environment, nor does it reflect paternal genotype because tube function in embryonic patterning is strictly maternally contributed (Gerttula et al. 1988). Rather, the tubvar chromosomes must provide variable tube activity, reflecting an alteration in the production, stability, or activity of the tube mRNA or protein.
The observation that single genotypes give rise to widely variable phenotypes suggests a stochastic contribution to tube function. One possible source of stochasticity is the effect of cellular fluctuations on the activity or stability of a protein or RNA transcript. However, sequencing revealed that the Tube proteins encoded by four of the five tubvar alleles are wild-type (the tub6 coding sequence has an asparagine in place of aspartic acid at position 106). Moreover, the noncoding portion of the tube transcription unit is wild-type for all of the tubvar alleles, making a disruption in mRNA processing or stability very unlikely. What then is the source of phenotypic variation for these tubvar alleles?
Stochastic variation in tube expression
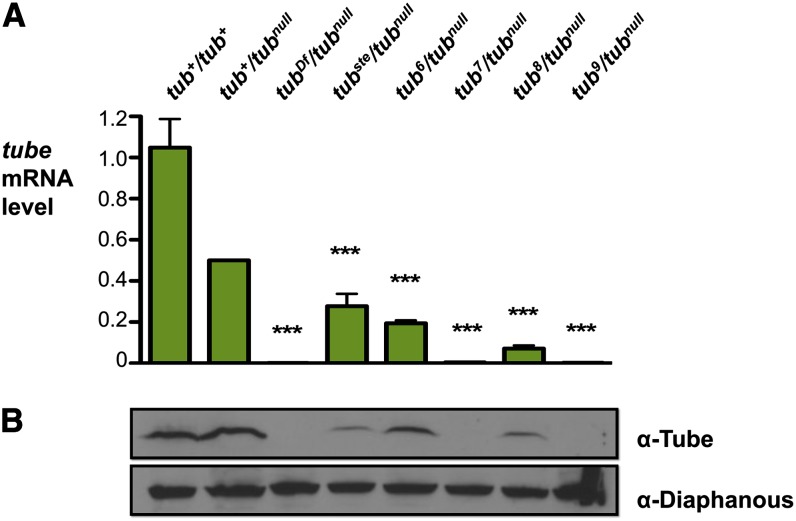
On the basis of these findings, we postulated that the tubvar alleles alter tube transcription. In particular, we envision that these mutations reduce mRNA levels below a threshold amount, revealing phenotypic effects of stochastic gene expression fluctuations. To determine if tubvar alleles on average substantially reduce gene expression, we assayed tube expression in batches of embryos from tubvar/tubnull females by both quantitative RT-PCR (qRT-PCR) of first-strand cDNA and immunoblotting of protein in embryo extract.
All of the tubvar alleles exhibited a marked mean reduction in tube expression. In embryos from tubste/tubnull females, the level of tube mRNA was reduced on average to 28% of the wild-type level (Figure 2A). For tub6/tubnull, tub7/tubnull, tub8/tubnull, and tub9/tubnull, which exhibit a more severe reduction in tube function (see Figure 1), tube mRNA levels were only 0.2–19% of the wild-type level (Figure 2A). Immunoblotting revealed that Tube protein levels also were greatly reduced (Figure 2B). Furthermore, Tube protein levels correlated closely with mRNA levels. On the basis of these findings, we concluded that the mutations responsible for the tubvar phenotypes reduce the production of tube mRNA.
Figure 2.
Maternal tube expression is dramatically reduced in tubvar/tubnull females. (A) Quantitation of tube mRNA in embryos. Quantitative RT-PCR data of tube expression in embryos from females of the specified genotype. Expression data were normalized to rp49 expression and are presented as a fraction of tub+/tubnull expression, with tub+/tubnull set to 0.5. Error bars represent S.E.M., ***P < 0.001. (B) Quantitation of Tube protein in embryos. Immunoblot using α-Tube sera of protein isolated from embryos from females of specified genotype. Loading control is α-Diaphanous.
The finding that tube expression is strongly reduced in tubvar mutants is consistent with a model in which phenotypic variability originates from normally occurring, probabilistic fluctuations in transcription. It remained a possibility, however, that the mutations increase transcriptional noise in addition to reducing average transcription levels. To distinguish between these two models, we measured tube expression in individual embryos from tubste/tubnull, tub8/tubnull, and control females by qRT-PCR.
As shown in Table 1, the tubvar alleles do not enhance transcriptional fluctuations of tube. Rather, the standard deviations (SDs) of tube expression from the tubvar chromosomes were comparable to each other and to the wild type. This finding demonstrates that the mutations responsible for the tubvar phenotypes reduce tube expression without introducing additional variability in gene expression. Said another way, the phenotype of embryos from tubvar females unmasks transcriptional noise.
Table 1. Maternal tube expression is equally variable in tubvar and wild-type females.
| Maternal Genotype | Phenotype | Mean (×10−4) | SD (×10−4) |
|---|---|---|---|
| tub+/tubnull | Wild-type | 25 | 11 |
| tubste/tubnull | Variable | 23 | 9.4 |
| tub8/tubnull | Variable | 8.4 | 8.6 |
| tub4/tubnull | Strongly dorsalized | 3.1 | 1.4 |
Quantitation of tube mRNA in individual embryos from females of the specified genotype by qRT-PCR. Expression data were normalized to rp49 expression. n ≥ 32 for each genotype. qRT-PCR, quantitative real-time polymerase chain reaction.
We also included in our analysis the allele tub4, which produces reduced levels of functioning Tube protein (Letsou et al. 1993) and yet displays an invariant dorsalization phenotype (see Figure 1C). The tub4 chromosome contains a mutation that disrupts splicing, leading to only a very small fraction of tube transcripts being properly spliced and consequently very low amounts of functioning Tube protein. This finding would suggest that low tube gene product levels are required but not sufficient for a variable phenotype. Upon analyzing RNA from individual embryos produced by tub4/tubnull females, we found that the average level of spliced tube mRNA for tub4 was greatly reduced relative to the wild type (Table 1). Furthermore, we found that the SD of tube expression was considerably less than observed from the other genotypes (Table 1). In other words, spliced tube transcript levels varied less from embryo to embryo for tub4 than for other genotypes.
On the basis of our sequence analysis of the transcribed region, it is likely that the tubvar chromosomes are altered for tube transcription initiation or elongation, rather than a cotranscriptional or post-transcriptional process. In contrast, the tub4 chromosome displays defective splicing, a cotranscriptional process, but little variation in mature transcript level. This result can best be explained if the tub4 splicing defect acts as a bottleneck, masking fluctuations in transcription initiation.
We drew two conclusions from the comparison of tubvar and tub4 expression in individual embryos. First, because the tub4 chromosome displayed a reduced average tube expression level with a lower standard deviation than observed from the other genotypes, qRT-PCR of individual embryos introduced little, if any, additional technical variability. Second, low transcript levels do not inherently cause phenotypic variability. Rather, reduced transcript levels combined with naturally occurring transcriptional fluctuations generate the tubvar phenotypic variability.
Retrotransposon-mediated disruption of tube expression
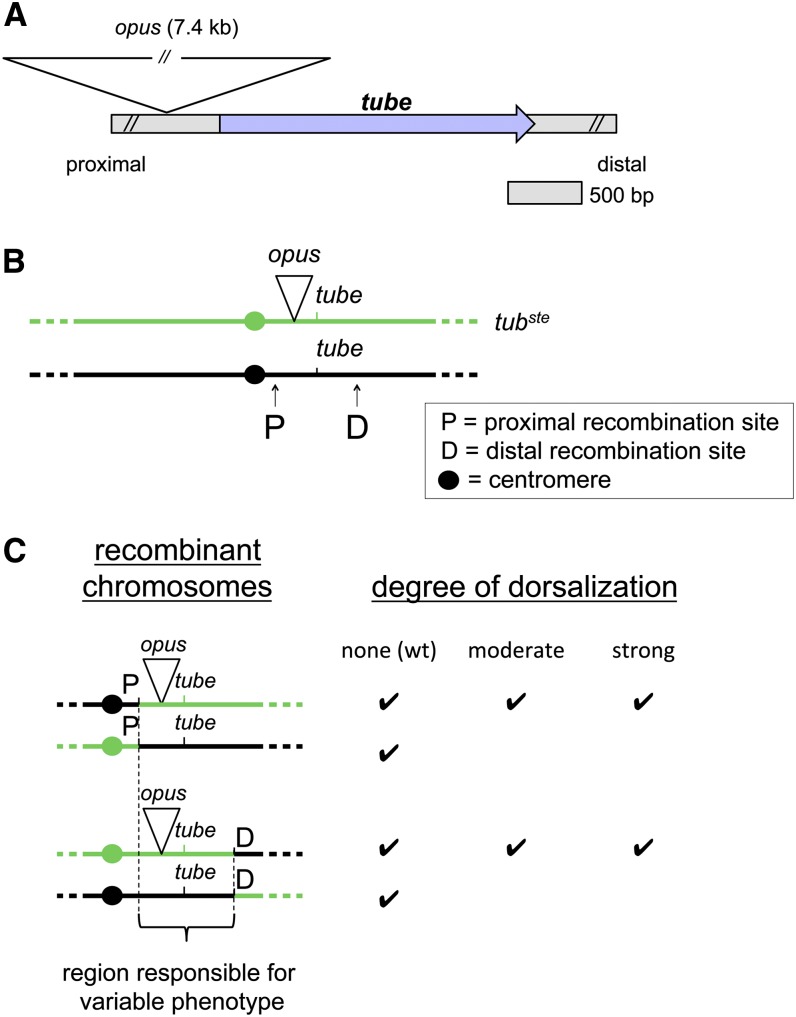
Our investigation revealed that maternal tube expression was diminished in all five tubvar alleles, suggesting that the phenotypic variability is caused by mutations that reduce tube transcription. We therefore set out to find cis-regulatory mutations affecting tube. We have previously demonstrated that a transgene that includes the tube transcription unit and 8 kb of DNA directly upstream rescues Toll signaling in tube deficient embryos (Letsou et al. 1991). We began by amplifying this region of the tubste chromosome with conventional and TAIL PCR (see Material and Methods and Rakyan and Beck 2006), followed by sequencing. In this manner, we discovered a retrotransposon insertion at position −301 relative to the tube transcription start site (Figure 3A). By PCR-based analysis, we further found that each of the tubvar chromosomes, including tub6, contains this −301 insertion but wild-type chromosomes do not (Figure S3). The retrotransposon is a member of a family of insertions called opus elements, which are LTR-containing retrotransposons typically found in 20-30 copies distributed throughout the D. melanogaster genome (Kaminker et al. 2002).
Figure 3.
An opus retrotransposon insertion 301 bp upstream of tube causes variable dorsalization. (A) Schematic of opus insertion site relative to the tube gene in the tubvar chromosomes. (B) Schematic of site-specific recombination technique used to map the mutation responsible for the variable dorsalization in the tubste chromosome. Schematic is not to scale. (C) Phenotypic data from site-specific recombination mapping of the tubste chromosome. Recombination was induced at a P-element insertion site located either 8 kb proximal (P) or 8 kb distal (D) to the tube gene. Phenotypes were assayed in embryos from females bearing the recombinant chromosome in trans to tubnull.
Given the finding that all of the tubvar alleles contain the opus insertion, we made two predictions. First, because the opus insertion is approximately 7.4 kb and is located in the tube promoter region, it is likely required for the variable phenotype observed in all tubvar alleles. In this case, the variable phenotype should map to a region containing the opus insertion. Second, because each allele displays a unique phenotypic profile, the opus insertion is probably the primary event but cannot be the sole source of the variable phenotype. Instead, we hypothesized that the tubvar chromosomes, perhaps with the exception of tubste, are doubly mutant for tube. In this case, we should be able to identify additional, enhancing mutations in some or all of the tubvar chromosomes that are required for their unique phenotypic profiles.
To address the first prediction—whether the opus insertion is required for the tubvar phenotypes—we performed mapping by site-specific male recombination (Chen et al. 1998). We hypothesized that the tubste chromosome might not contain a second mutation because it is the least affected of the tubvar alleles with regard to tube expression and embryonic phenotype. We therefore began by mapping the mutation responsible for the tubste phenotype. Specifically, we induced recombination between the tubste chromosome and a chromosome bearing either of two P element insertions, one approximately 8 kb upstream and one approximately 8 kb downstream of the tube transcription unit (Figure 3B). As shown in Figure 3C, these studies demonstrated that a 25-kb region encompassing tube and the opus insertion was both necessary and sufficient to generate the range of dorsalized phenotypes associated with tubste. Furthermore, sequencing of the entire 25-kb region in the tubste chromosome revealed just seven other changes, each of which were minor sequence variations when compared to wild-type cDNA or genomic sequences (Table S1). These findings are consistent with the hypothesis that the opus retrotransposon is the primary event responsible for the tubvar phenotypic variability and acts by diminishing tube expression.
Using qRT-PCR and 5′ RACE in combination with published modENCODE RNAseq data we determined that the opus insertion disrupts tube expression without affecting the position of the transcription start site (Figure S4). Moreover, the opus insertion site separates the tube transcription start site from the most highly conserved intergenic region upstream of tube (Celniker et al. 2002; Kent et al. 2002). It thus appears that the opus insertion disrupts tube regulation, reducing tube expression without compromising the boundaries of the tube transcript.
Context-specific expression defects
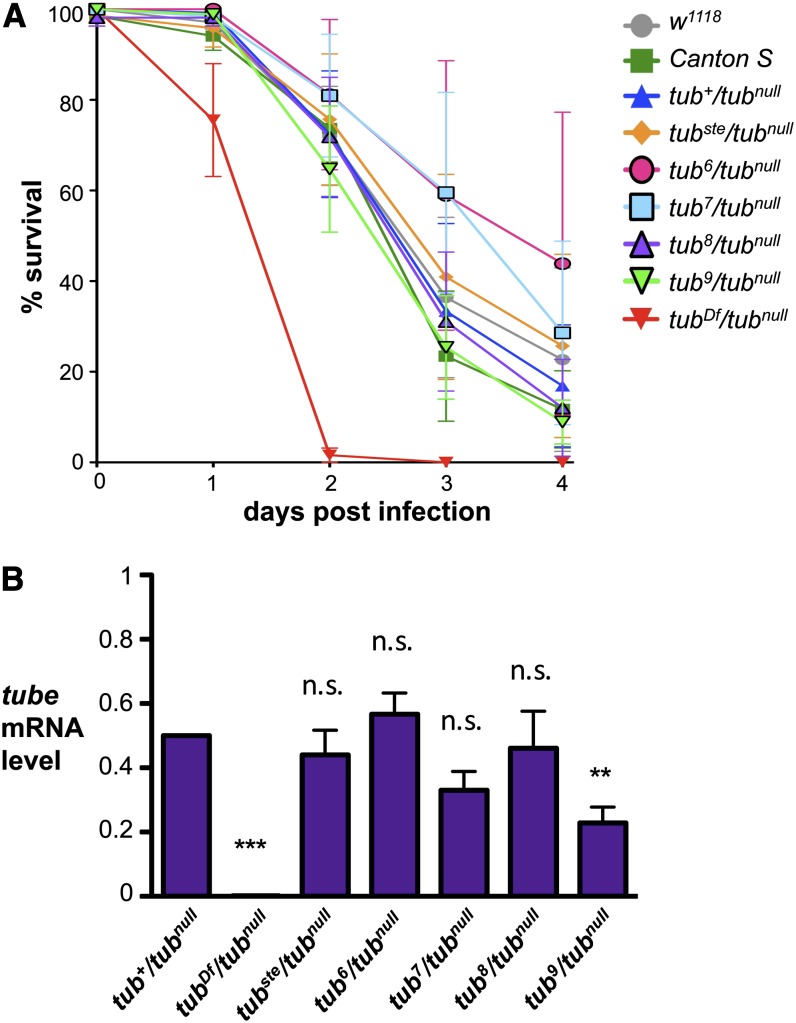
The aforementioned findings indicate that the presence of the opus insertion alters tube expression. Because Tube is required for Toll pathway function in both embryonic patterning and innate immunity, we wondered whether tubvar mutants also display immune defects. To answer this question, we assayed tubvar/tubnull, tubnull/tubnull, and wild-type adult males for survival after septic injury with the fungus Fusarium oxysporum, a specific inducer of Toll signaling. We found that although tubnull/tubnull males died within 2 d after infection, tubvar/tubnull males survived on average for 4 d following infection, indistinguishable from wild-type males (Figure 4A). Thus, the tubvar chromosomes detectably disrupt Toll signaling in embryos but not in adult immune tissues.
Figure 4.
Immune function of tube appears unaffected in tubvar adults. (A) Survival of adult males of specified genotype after septic wounding with F. oxysporum spores. Analysis of tub7 was performed in trans to tub2, a strong hypomorphic allele. Error bars represent SEM. (B) Quantitation of tube mRNA in adults. Quantitative RT-PCR data of tube expression in adult males of the specified genotype. Expression data were normalized to rp49 expression and are presented as a fraction of tub+/tubnull expression, with tub+/tubnull set to 0.5. Error bars represent SEM. **P < 0.01, ***P < 0.001, n.s. = not significant.
We considered two explanations for the distinct effects of the tubvar chromosomes in different in vivo contexts. One possibility is that the threshold level of Tube required for full pathway function during infection is lower than during embryonic development, allowing tubvar/tubnull flies to mount an effective immune response with relatively small amounts of Tube. Alternatively, the effect of the opus insertion on tube expression could differ among tissues. To distinguish between these hypotheses, we measured tube mRNA levels by qRT-PCR in whole adult males. For four of the five variable alleles, tubvar/tubnull adult males exhibited wild-type levels of tube expression (Figure 4B). In the case of tub9, expression in adult males was somewhat reduced compared to tub+/tubnull, but far greater than that observed in embryos. In all cases, it seems that the opus insertion affects the ability of cells to transcribe tube in the ovary but not in the immune tissues. Expression of tube in the different tissues presumably requires distinct regulatory elements, making the effect of the opus element on tube expression context-specific.
Additional mutations and enhancement of the variable phenotype
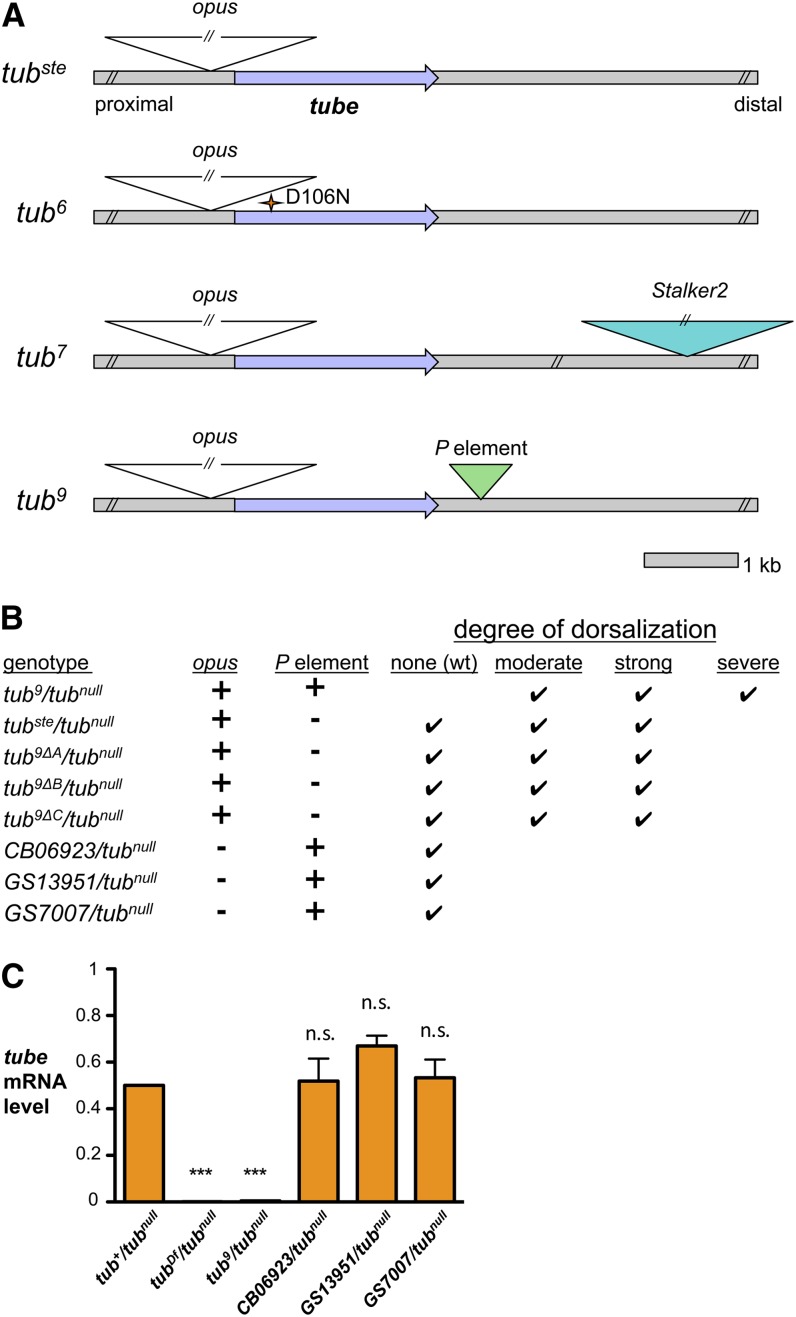
As stated previously, we postulated that the synergistic interactions of the opus insertion with an additional mutation on each of the tubvar chromosomes (except for tubste) cause the distinct phenotypic profile of each allele. As described above, the tub6 chromosome carries a missense mutation in addition to the opus insertion. We were unable to identify a second mutation in the regions flanking tube in the tub8 chromosome (data not shown). However, we were successful in identifying an additional and likely significant mutation in both the tub7 and tub9 chromosomes, both of which had strikingly reduced levels of tube mRNA (see Figure 2).
For the tub7 chromosome, we analyzed the 25-kb region indicated by site-specific recombination experiments to be responsible for the variable phenotype on the tubste chromosome. Using an approach combining conventional PCR, TAIL-PCR, and sequencing, we identified an insertion of Stalker2, another retrotransposon, 6 kb downstream of the tube transcription unit in the tub7 chromosome (see Figure 5A). Stalker2 is an LTR-containing retrotransposon found in approximately 10 copies distributed over the D. melanogaster genome (Kaminker et al. 2002). Genomic sequencing of this 25-kb region revealed no differences between tub7 and tubste other than the Stalker2 insertion. Furthermore, PCR-based analysis demonstrated that the Stalker2 insertion is absent from all other tubvar chromosomes and from wild-type chromosomes (Figure S5). Thus, the Stalker2 insertion appears to be the second mutation in the tub7 chromosome that interacts with the opus insertion to generate its unique phenotype.
Figure 5.
Additional mutations in the tubvar chromosomes genetically interact with the opus insertion to enhance the variable phenotype. (A) Schematic of locations of identified mutations in tubvar chromosomes, including opus insertion, tub6 missense mutation, tub7 Stalker2 insertion, and tub9 P-element insertion. (B) Phenotypic ranges of embryos from females of specified genotype in trans to tubnull. Precise excisants (tub9Δ) were generated using a transposase source (Δ2-3). GS7007, GS13951, and CB06923 are chromosomes containing P-element insertions within 4 bp of the tub9 P-element insertion site. (C) Quantitation of tube mRNA in embryos from females bearing P elements downstream of tube. Quantitative RT-PCR expression data were first normalized to rp49 expression and are presented as a fraction of tub+/tubnull expression, with tub+/tubnull set to 0.5. Error bars represent SEM. ***P < 0.001, n.s. = not significant.
In the case of tub9, Hecht and Anderson had found a P-element insertion downstream of the tube gene (Hecht and Anderson 1993). By sequencing we confirmed the presence of a P element, 687 bp long, located 581 bp downstream of the end of the tube transcription unit (see Figure 5A). To test whether this P element synergistically interacts with the opus insertion, we mobilized the P element and assayed for precise excision. We obtained three such excisants and, as reported by Hecht and Anderson, they were fertile (Hecht and Anderson 1993). However, we found that the phenotypic distribution of embryos from the excisant females was not wild-type, as previously reported, but was instead variable and similar to that of embryos from tubste/tubnull females (Figure 5B). Thus, the excision of the P element partially restored tube function in these embryos. This finding demonstrates that the unique tub9 phenotypic profile reflects the combined activities of two mutations, the opus and P element insertions. The opus element is the primary event, and the P element additionally decreases tube expression and correspondingly enhances the dorsalization phenotype of tub9 relative to tubste.
Given the proximity of the P-element insertion in tub9 to the tube gene, we wondered whether such an insertion by itself would perturb tube expression. We could not, however, remove the opus insertion from tub9 because retrotransposons do not excise. Instead, we analyzed P-element insertions in the same location as the P element in tub9 but in a background devoid of the opus insertion. Taking advantage of available collections, we obtained three such P-element insertions, each within four base pairs or less of the tub9 P-element insertion site. All conferred wild-type tube function during embryonic patterning (Figure 5B). Furthermore, tube mRNA levels were wild-type in embryos from females bearing these P elements (Figure 5C). Hence, a P-element insertion at the same location as that in tub9 does not by itself have an effect on tube expression. The simplest explanation for these results is that the P element in tub9 exerts its effect on tube expression exclusively through its synergistic interaction with the upstream opus insertion.
Discussion
Sensitizing mutations and synergistic effects of additional mutations
We have identified a retrotransposon insertion upstream of the tube transcription start site that is specific to the tubvar alleles and that lies within the region responsible for the variable phenotype of tubste. We conclude that the opus insertion at −301 dramatically reduces tube expression, producing a variable phenotype that depends on naturally fluctuating transcriptional levels. The opus insertion is approximately 7.4-kb long and sits between the tube transcription start site and the region of upstream intergenic sequence with the highest conservation among Drosophila species (Celniker et al. 2002; Kent et al. 2002). This conserved region is most likely a regulatory element, suggesting a means by which the opus insertion could interrupt tube expression. The opus insertion could act by simply spatially separating the tube transcription start site from an important regulatory element. Alternatively, the opus insertion could induce epigenetic changes that inhibit the interaction of this conserved region with DNA binding proteins that promote tube transcription.
Given that all of the tubvar alleles contain the opus insertion and several other small polymorphisms absent from the reference genome, it seems likely that a common progenitor chromosome was used to generate each of the tubvar alleles (Hecht and Anderson 1993). We speculate that the opus insertion provided a sensitized background for mutagenesis, leading to the recovery of the tub6, tub7, tub8, and tub9 chromosomes. Our data support a model in which each of the tubvar chromosomes, except tubste, contain a second, enhancing mutation, which acts synergistically with the opus insertion to substantially disrupt tube gene function. Despite the fact that tub8 also displays a phenotype distinct from that observed in tubste, we have not identified a second mutation in the tub8 chromosome. It may be, therefore, that the tub8 chromosome contains multiple or complex changes that cannot be as easily dissected by site-specific recombination mapping.
We identified two different types of mutations that interact with the opus insertion to increase the severity of the variable dorsalization. The second mutation on the tub6 chromosome, a missense mutation, appears to mildly decrease Tube protein function without affecting tube expression. Embryos from tub6/tubnull females are more severely dorsalized, on average, than those from tubste/tubnull females despite similar mRNA levels (see Figure 2A). Embryos from tub6/tubnull females are not completely dorsalized, however, demonstrating that the Tube protein encoded by tub6 retains at least some activity. The wild-type survival of tub6/tubnull males following fungal infection further demonstrates this functionality. We speculate that their wild-type survival is due to the elevated levels of tube gene product in males relative to embryos. An excess of Tube protein in these males would mitigate the shortcoming of reduced Tube activity. In the embryos, however, low Tube protein levels in combination with reduced functionality generate the unique phenotypic profile of tub6.
In the cases of tub7 and tub9, we identified a second transposable element insertion in each chromosome, which we believe work synergistically with the opus insertion to further reduce tube mRNA levels. The insertions that we found in the course of our studies of the tubvar chromosomes represent three distinct families of transposable elements, giving us examples of the complexity of genetic interactions that are possible among transposable elements. Each element contains distinct sets of cis regulatory sequences, which on a genome-wide scale, allows for seemingly endless combinations of potential genetic perturbations of a gene locus via alterations to the local chromatin landscape.
Our studies of the tub9 chromosome provide evidence that transposable element insertions that are innocuous on their own can induce profound alterations in local gene expression when located near other transposable element insertions. If a single insertion is benign on its own, such as the tub9 P element, it will not be selected against. This safety from selection allows transposable element insertions to accumulate around the genome, sensitizing many loci to additional insertions. One might expect more published examples of similar hypervariable phenotypes given the abundance of transposable elements in eukaryotic genomes. We speculate, however, that in-depth studies of such mutations are underrepresented in the literature because they lack the robust phenotypes required by most traditional genetic approaches.
Gene-proximal transposable elements and gene regulation
The fact that gene-proximal transposable element insertions can cause dramatic and complex regulatory changes on neighboring genes is relevant to our understanding of intergenic DNA. Transposable elements comprise approximately 10–20% of the D. melanogaster genome and more than 45% of the human genome (Burns and Boeke 2012; Ganko et al. 2006). The D. melanogaster genome contains at least 96 families of transposable elements, each identified based on their unique sequence composition and each ranging in euchromatic copy number from 1 to approximately 150 per genome (Kaminker et al. 2002). In wild populations, the frequency with which transposable element insertion sites are shared is low (Biemont et al. 1994; Charlesworth and Lapid 1989; Charlesworth et al. 1992) or, in other words, subpopulations of flies harbor unique collections of transposable element insertion sites. The diversity of insertion sites produces many opportunities for adaptive modulations in gene expression.
In the case of the tubvar alleles, the transposable element insertions produce a detrimental phenotype. However, a similar effect on a non-essential gene could lead to a tempered modulation of gene activity, a change that could potentially improve organismal fitness. Such effects have been reported in both D. melanogaster and mammalian models involving particular subsets of neurons that show elevated transposition rates (Perrat et al. 2013; Thomas and Muotri 2012). In at least one case, the resulting de novo insertions altered local gene expression and cell fate (Muotri et al. 2005). One possibility is that derepression of transposon mobility is an evolutionary adaptation to generate genomic diversity, and subsequently gene expression diversity, among genetically identical cells.
Surplus gene expression as a source of phenotypic robustness
We find that tubste/tubnull females on average express only 28% of the wild-type level of maternal tube transcripts and yet approximately 82% of their embryos develop wild-type dorsoventral axes. This finding suggests that the threshold of tube expression needed for a wild-type phenotype is considerably less than 50% of the wild-type level. It seems energetically unfavorable for an organism to produce so much more mRNA and protein than necessary. However, surplus gene expression may be a molecular mechanism to buffer the effects of stochastic influences. This model is simpler than the alternative of attempting to minimize or eliminate fluctuations in transcription factor abundance or activity.
In the case of early embryonic development, the guaranteed abundance of a gene product above a threshold level is particularly important because there is no opportunity for feedback regulation—all germline gene expression is completed before fertilization. Thus, a mutation that significantly perturbs maternal gene expression would have an irreversible effect on gene product levels in the oocyte and, ultimately, the phenotype of the progeny.
It would be interesting to look for the extent of surplus expression in other maternally expressed genes, especially those essential to embryonic survival, like tube, and compare them to the expression profiles of zygotically expressed genes. One possibility is that surplus expression is less common among zygotically expressed genes because there is the opportunity for positive feedback regulation. In this case, the female germline would have unique epigenetic or transcriptional mechanisms to ensure surplus expression of gene products that are to be transferred into the oocyte. A more complete understanding of this problem will require the determination of threshold levels of gene expression for additional genes.
Supplementary Material
Acknowledgments
We thank Sherry Alexander, Tina Chung, Yan Hu, Chris Lee, Brooke White Jenkins, and Jasmine Manubay for assistance in this project and Gary Karpen and Scott Rifkin for helpful discussions. We also thank the Troemel and McGinnis labs at UC San Diego for sharing equipment and technical advice. This work was supported by NIH grant RO1 GM05054516 (to S.A.W.) and by the NIH predoctoral training grant T32 GM008666 (support for A.W.C.).
Footnotes
Communicating editor: H. D. Lipshitz
Literature Cited
- Afshar K., Stuart B., Wasserman S. A., 2000. Functional analysis of the drosophila diaphanous fh protein in early embryonic development. Development 127: 1887–1897 [DOI] [PubMed] [Google Scholar]
- Anderson K. V., Nüsslein-Volhard C., 1984. Genetic analysis of dorsal-ventral embryonic patterning in drosophila, pp. 269–289 in Pattern Formation, edited by Malacinski G. M., Bryant S. MacMillan, New York [Google Scholar]
- Arias A. M., Hayward P., 2006. Filtering transcriptional noise during development: concepts and mechanisms. Nat. Rev. Genet. 7: 34–44 [DOI] [PubMed] [Google Scholar]
- Biemont C., Lemeunier F., Garcia Guerreiro M. P., Brookfield J. F., Gautier C., et al. , 1994. Population dynamics of the copia, mdg1, mdg3, gypsy, and p transposable elements in a natural population of drosophila melanogaster. Genet. Res. 63: 197–212 [DOI] [PubMed] [Google Scholar]
- Burns K. H., Boeke J. D., 2012. Human transposon tectonics. Cell 149: 740–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S. E., Wheeler D. A., Kronmiller B., Carlson J. W., Halpern A., et al. , 2002. Finishing a whole-genome shotgun: Release 3 of the drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Lapid A., 1989. A study of ten families of transposable elements on x chromosomes from a population of Drosophila melanogaster. Genet. Res. 54: 113–125 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Lapid A., Canada D., 1992. The distribution of transposable elements within and between chromosomes in a population of Drosophila melanogaster. I. Element frequencies and distribution. Genet. Res. 60: 103–114 [DOI] [PubMed] [Google Scholar]
- Chen B., Chu T., Harms E., Gergen J. P., Strickland S., 1998. Mapping of drosophila mutations using site-specific male recombination. Genetics 149: 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb J. R., Liverpool T. B., 2010. Bursts and pulses: insights from single cell studies into transcriptional mechanisms. Curr. Opin. Genet. Dev. 20: 478–484 [DOI] [PubMed] [Google Scholar]
- Flatt T., 2005. The evolutionary genetics of canalization. Q. Rev. Biol. 80: 287–316 [DOI] [PubMed] [Google Scholar]
- Ganko E. W., Greene C. S., Lewis J. A., Bhattacharjee V., Mcdonald J. F., 2006. Ltr retrotransposon-gene associations in Drosophila melanogaster. J. Mol. Evol. 62: 111–120 [DOI] [PubMed] [Google Scholar]
- Gerttula S., Jin Y. S., Anderson K. V., 1988. Zygotic expression and activity of the drosophila toll gene, a gene required maternally for embryonic dorsal-ventral pattern formation. Genetics 119: 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht P. M., Anderson K. V., 1993. Genetic characterization of tube and pelle, genes required for signaling between toll and dorsal in the specification of the dorsal-ventral pattern of the drosophila embryo. Genetics 135: 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchmandzadeh B., Wieschaus E., Leibler S., 2002. Establishment of developmental precision and proportions in the early drosophila embryo. Nature 415: 798–802 [DOI] [PubMed] [Google Scholar]
- Huang A. M., Rusch J., Levine M., 1997. An anteroposterior dorsal gradient in the drosophila embryo. Genes Dev. 11: 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, A. M., E. J. Rehm, and G. M. Rubin, 2009 Quick preparation of genomic DNA from drosophila. Cold Spring Harb Protoc 2009: pdb prot5198. [DOI] [PubMed]
- Kaern M., Elston T. C., Blake W. J., Collins J. J., 2005. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 6: 451–464 [DOI] [PubMed] [Google Scholar]
- Kaminker J. S., Bergman C. M., Kronmiller B., Carlson J., Svirskas R., et al. , 2002. The transposable elements of the drosophila melanogaster euchromatin: A genomics perspective. Genome Biol. 3: RESEARCH0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., et al. , 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B., Kaneko K., 2011. Fluctuation and response in biology. Cell. Mol. Life Sci. 68: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letsou A., Alexander S., Orth K., Wasserman S. A., 1991. Genetic and molecular characterization of tube, a drosophila gene maternally required for embryonic dorsoventral polarity. Proc. Natl. Acad. Sci. USA 88: 810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letsou A., Alexander S., Wasserman S. A., 1993. Domain mapping of tube, a protein essential for dorsoventral patterning of the drosophila embryo. EMBO J. 12: 3449–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D. L., Sandler L., Baker B. S., Carpenter A. T., Denell R. E., et al. , 1972. Segmental aneuploidy and the genetic gross structure of the drosophila genome. Genetics 71: 157–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. G., Whittier R. F., 1995. Thermal asymmetric interlaced pcr: Automatable amplification and sequencing of insert end fragments from p1 and yac clones for chromosome walking. Genomics 25: 674–681 [DOI] [PubMed] [Google Scholar]
- Muotri A. R., Chu V. T., Marchetto M. C., Deng W., Moran J. V., et al. , 2005. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435: 903–910 [DOI] [PubMed] [Google Scholar]
- Perrat P. N., Dasgupta S., Wang J., Theurkauf W., Weng Z., et al. , 2013. Transposition-driven genomic heterogeneity in the drosophila brain. Science 340: 91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher A., Dostatni N., 2010. The Bicoid morphogen system. Curr. Biol. 20: R249–R254 [DOI] [PubMed] [Google Scholar]
- Raj A., Rifkin S. A., Andersen E., Van Oudenaarden A., 2010. Variability in gene expression underlies incomplete penetrance. Nature 463: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan V. K., Beck S., 2006. Epigenetic variation and inheritance in mammals. Curr. Opin. Genet. Dev. 16: 573–577 [DOI] [PubMed] [Google Scholar]
- Romeo Y., Lemaitre B., 2008. Drosophila immunity: methods for monitoring the activity of Toll and Imd signaling pathways. Methods Mol. Biol. 415: 379–394 [DOI] [PubMed] [Google Scholar]
- Sun H., Towb P., Chiem D. N., Foster B. A., Wasserman S. A., 2004. Regulated assembly of the toll signaling complex drives drosophila dorsoventral patterning. EMBO J. 23: 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. A., Muotri A. R., 2012. Line-1: creators of neuronal diversity. Front. Biosci. 4: 1663–1668 (Elite Ed) [DOI] [PubMed] [Google Scholar]
- Waddington C. H., 1942. Canalization of development and the inheritance of acquired characters. Nature 150: 563–565 [DOI] [PubMed] [Google Scholar]
- Wieschaus E., Nüsslein-Volhard C., 1986. Looking at embryos, pp. 199–227 in Drosophila: A Practical Approach, edited by Roberts D. B. IRL Press, Oxford [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.