Abstract
BACKGROUND
Both genetic variation at the 17q21 locus and virus-induced respiratory wheezing illnesses are associated with the development of asthma. Our aim was to determine the effects of these two factors on the risk of asthma in the Childhood Origins of Asthma (COAST) and the Copenhagen Prospective Study on Asthma in Childhood (COPSAC) birth cohorts.
METHODS
We tested genotypes at the 17q21 locus for associations with asthma and with human rhinovirus (HRV) and respiratory syncytial virus (RSV) wheezing illnesses and tested for interactions between 17q21 genotypes and HRV and RSV wheezing illnesses with respect to the risk of asthma. Finally, we examined genotype-specific expression of 17q21 genes in unstimulated and HRV-stimulated peripheral-blood mononuclear cells (PBMCs).
RESULTS
The 17q21 variants were associated with HRV wheezing illnesses in early life, but not with RSV wheezing illnesses. The associations of 17q21 variants with asthma were restricted to children who had had HRV wheezing illnesses, resulting in a significant interaction effect with respect to the risk of asthma. Moreover, the expression levels of ORMDL3 and of GSDMB were significantly increased in HRV-stimulated PBMCs, as compared with unstimulated PBMCs. The expression of these genes was associated with 17q21 variants in both conditions, although the increase with exposure to HRV was not genotype-specific.
CONCLUSIONS
Variants at the 17q21 locus were associated with asthma in children who had had HRV wheezing illnesses and with expression of two genes at this locus. The expression levels of both genes increased in response to HRV stimulation, although the relative increase was not associated with the 17q21 genotypes. (Funded by the National Institutes of Health.)
THE FIRST GENOMEWIDE ASSOCIATION study of childhood-onset asthma revealed a susceptibility locus on chromosome 17q21.1 The association of this locus with asthma has since been replicated in both genomewide and candidate-gene association studies,2,3 and the locus represents one of the most consistently associated genetic risk factors for childhood asthma. Variation at the 17q21 locus is associated primarily with childhood-onset asthma,4-6 but not with atopy,4,7,8 and the effects are larger among children who had been exposed to environmental tobacco smoke in early life4,9-11 and in children with reported respiratory infections in infancy.10 The disease-associated variants at this locus are associated with expression levels of two 17q21 genes, GSDMB and ORMDL3, in white cells,5 lymphoblastoid cell lines,12 and CD4+ T cells.13
The onset and progression of asthma result from a complex interplay between genetic background and environmental exposures, particularly in early development. Among the many environmental factors that influence the risk of asthma,14 respiratory infections with viruses15,16 and bacteria17 are the most common triggers of asthma exacerbations in children. Approximately 80% of asthma exacerbations are attributed to respiratory viral infections, with human rhinovirus (HRV) accounting for nearly two thirds of these cases.16 Moreover, infants who have HRV infections with wheezing are at a significantly increased risk for subsequent asthma.18-20 However, exposure to HRV does not lead to wheezing illness in all children, nor does wheezing illness result in asthma in all cases, suggesting that the host genotype also plays a role.
We sought to further elucidate the effect of the genotype at the 17q21 asthma locus and respiratory viral illnesses in early life on the risk of childhood-onset asthma. We hypothesized that these common genetic and environmental risk factors account for a substantial proportion of the overall risk of childhood asthma, in either an additive or an interactive manner. We also used an in vitro model in peripheral-blood mononuclear cells (PBMCs) to identify HRV-responsive genes at the 17q21 locus and to test a potential mechanism underlying the interaction.
METHODS
STUDY PARTICIPANTS
We included data from two cohorts of children — the Childhood Origins of Asthma (COAST) birth cohort and the Copenhagen Prospective Study on Asthma in Childhood (COPSAC) birth cohort — and a group of adult volunteers. Children who participated in the COAST birth cohort provided oral assent when possible, and their parents provided written informed consent; oral and written informed consent were provided by both parents of each child who participated in the COPSAC birth cohort. Adult participants provided written informed consent. The study protocols for the three cohorts were approved by the institutional review board at the University of Wisconsin, the University of Copenhagen, and the University of Chicago, respectively.
For the COAST study, 289 newborns were enrolled in Madison, Wisconsin, between November 1998 and May 2000, as described previously.21 All the children had at least one parent with respiratory allergies, a history of physician-diagnosed asthma, or both. The parents of 214 of the newborns who were of European ancestry gave consent for their child to participate in genetic studies, and data from these children are included in the current analyses. A total of 200 of these children were evaluated for asthma beginning at 6 years of age. The demographic characteristics of the COAST cohort are provided in Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org. The diagnostic criteria for asthma and descriptions of allergic sensitization and phenotypes of viral wheezing illnesses in the COAST cohort are provided in the Methods section of the Supplementary Appendix.
For COPSAC, 411 Danish children (all of European ancestry) born to mothers with a history of physician-diagnosed asthma were enrolled between August 1998 and December 2001 in Copenhagen.22 A total of 297 of these children with complete follow-up data from the first 3 years of life and with information on asthma status by 7 years of age were included in this study. The demographic characteristics of the COPSAC cohort are provided in Table S1 in the Supplementary Appendix. The diagnostic criteria for asthma and descriptions of phenotypes of viral wheezing illnesses in the COPSAC cohort are provided in the Methods section of the Supplementary Appendix.
In addition, we recruited 100 unrelated adult volunteers (49 men and 51 women; age range, 19 to 60 years) in Chicago between July and November 2011 in order to study the genotype-specific effects of HRV stimulation on gene-expression patterns in PBMCs. We recruited volunteers by posting flyers (which were approved by the institutional review board) around the University of Chicago campus. Each participant responded to a short medical questionnaire on the day the blood samples were obtained; 23 of the participants reported a previous diagnosis, by a physician, of asthma. According to self-reported ancestry, 84 of the participants were of European descent, 8 were of Asian descent, 3 were Hispanic, and 4 had mixed ancestry; 1 participant did not report ancestry.
GENOTYPING OF 17Q21 VARIANTS
Five asthma-associated 17q21 single-nucleotide polymorphisms (SNPs) — rs9303277, rs11557467, rs12936231, rs2290400, and rs7216389 (Fig. S1 in the Supplementary Appendix) — were genotyped in the COAST cohort. Genotyping was performed at the National Heart, Lung, and Blood Institute Resequencing and Genotyping (RS&G) Service at Johns Hopkins University, with the use of the Custom GoldenGate Genotyping assay (Illumina). Success rates of genotyping ranged from 98.6 to 100%. SNP rs7216389 was selected as a surrogate for the five SNPs that are in near perfect linkage disequilibrium (Fig. S1 in the Supplementary Appendix). This SNP was genotyped in the COPSAC and adult participants with the use of TaqMan assays (Applied Biosystems).
GENETIC ASSOCIATION AND INTERACTION STUDIES
Each SNP was tested for an association with asthma and with phenotypes of viral wheezing illness, with the use of a logistic-regression model if the outcome variable was binary or a linear-regression model if the outcome variable was continuous. The model included the phenotype of interest as the outcome variable and genotype as an explanatory variable. To test for interactions between 17q21 SNP genotypes and viral (HRV or RSV) wheezing illnesses, viral wheezing illness and a term for the interaction of viral wheezing illness with genotype were included as covariates. In addition, the number of wheezing episodes in which another (i.e., non-HRV or non-RSV) virus was present was included as a covariate to ensure that the observed effects were specific to the virus being tested. Details of the statistical methods are provided in the Supplementary Appendix.
HRV16 STIMULATION OF PBMCS AND DNA AND RNA EXTRACTION
A blood sample (20 ml) was obtained from each adult participant. PBMCs were isolated from whole-blood samples with the use of a Ficoll-Paque separation protocol. A total of 4×106 PBMCs from each participant were treated with medium alone for 24 hours, and 4×106 PBMCs were treated with medium containing HRV16 for 24 hours. The multiplicity of infection was 10 plaque-forming units per cell. The remaining cells from each participant were used for DNA extraction. DNA was extracted on the day the blood samples were obtained, with the use of the QIAamp DNA Blood Mini Kit (Qiagen). Total RNA was extracted after a 24-hour incubation period, with the use of the RNeasy Plus Mini Kit (Qiagen). Details of the experimental procedures are provided in the Methods section of the Supplementary Appendix.
COMPLEMENTARY DNA SYNTHESIS AND QUANTITATIVE PCR ASSAY
Complementary DNA (cDNA) was synthesized with the use of the SuperScript III First-Strand Synthesis System and oligo-dT primers (Invitrogen). Primers for five 17q21 genes (IKZF3, ZPBP2, GSDMB, ORMDL3, and GSDMA) and a housekeeping gene (POLR2C) were designed with the use of IDT Sci-Tools (Integrated DNA Technologies) (Table S2 in the Supplementary Appendix). A quantitative polymerase-chain-reaction (qPCR) assay was performed with the use of Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) and the 7900HT Fast Real-Time PCR system (Applied Biosystems).
ANALYSIS OF GENE-EXPRESSION DATA
Among five genes at the 17q21 locus, IKZF3, GSDMB, and ORMDL3 were successfully amplified in unstimulated and HRV-stimulated PBMCs obtained from 100 participants. Each qPCR reaction was run in duplicate, and only samples with a coefficient of variation of less than 0.02 were included in the analyses. After the exclusions, there were 96 paired samples of unstimulated and HRV-stimulated PBMCs for expression studies of IKZF3, 97 paired samples for expression studies of GSDMB, and 97 paired samples for expression studies of ORMDL3. Additional details of the qPCR data analysis are provided in the Methods section of the Supplementary Appendix.
We used a paired t-test to assess differences in the expression of each 17q21 gene (IKZF3, GSDMB, and ORMDL3) between unstimulated and HRV-stimulated cells. Associations between 17q21 SNPs and gene expression were tested under an additive genetic model.
RESULTS
ASSOCIATIONS IN THE COAST COHORT
All the SNPs showed modest associations with childhood-onset asthma in 200 children in the COAST cohort (P≤0.08) (Table 1 and Fig. 1A). The SNPs that were tested were in nearly perfect linkage disequilibrium (Fig. S1 in the Supplementary Appendix) and had nearly identical minor-allele frequencies (0.49 to 0.50) (Table 1), making it impossible to assign the association signal to any particular SNP at the locus. None of the SNPs were associated with allergic sensitization (P≥0.86) (Table 1), a finding that was consistent with the results of previous studies.4,7,8 The 17q21 SNPs also had significant associations with HRV wheezing illness (P≤0.02) (Table 1) and with the number of HRV wheezing illnesses (P<0.001) (Table 1 and Fig. 1B) in the first 3 years of life. None of the SNPs were associated with either the risk of RSV wheezing illnesses (P≥0.17) or the number of RSV wheezing illnesses (P≥0.47) (Table 1), indicating the specificity of the association with HRV wheezing illness.
Table 1.
Associations of 17q21 Single-Nucleotide Polymorphisms (SNPs) with Asthma, Allergic Sensitization, and Viral Wheezing Illness Phenotypes in the Childhood Origins of Asthma (COAST) Cohort.*
| SNP | Gene | Location | Minor Allele |
Minor-Allele Frequency |
P Value | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Asthma | Allergic Sensitization† |
HRV Wheezing Illness |
No. of HRV Wheezing Illnesses |
RSV Wheezing Illness |
No. of RSV Wheezing Illnesses |
|||||
| rs9303277 | IKZF3 | Intron | C‡ | 0.488 | 0.03 | 0.86 | 0.01 | <0.001 | 0.25 | 0.51 |
| rs11557467 | ZPBP2 | Intron | T‡ | 0.498 | 0.05 | 0.98 | 0.02 | <0.001 | 0.30 | 0.63 |
| rs12936231 | ZPBP2 | Exon | C‡ | 0.493 | 0.07 | 0.95 | 0.02 | <0.001 | 0.22 | 0.57 |
| rs2290400 | GSDMB | Intron | A§ | 0.500 | 0.08 | 0.90 | 0.02 | <0.001 | 0.17 | 0.47 |
| rs7216389 | GSDMB | Intron | T§ | 0.500 | 0.04 | 0.90 | 0.01 | <0.001 | 0.22 | 0.54 |
HRV denotes human rhinovirus, and RSV respiratory syncytial virus.
Allergic sensitization was assessed with the use of an in vitro IgE test. The results were similar when allergic sensitization was assessed with the use of a skin-prick test (all P>0.50).
This was the minor allele in the COAST cohort.
The minor allele in Centre d’Etude du Polymorphisme Humain from Utah (CEU) HapMap samples is shown for this SNP.
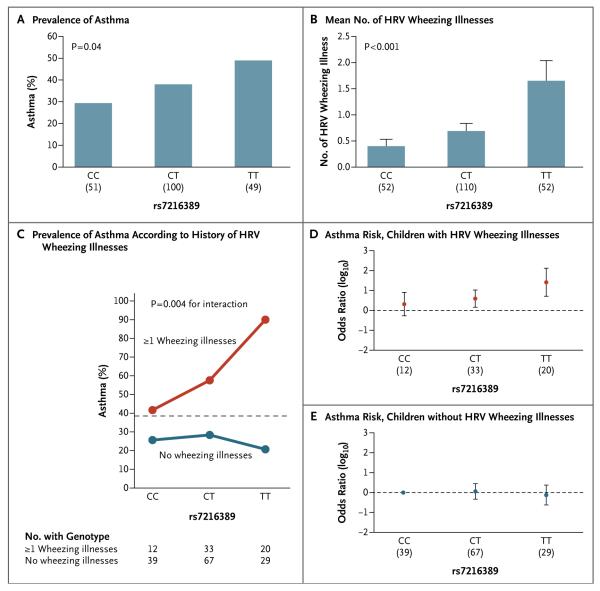
Figure 1. Effects of 17q21 Genotype on Asthma and Human Rhinovirus (HRV) Wheezing Illnesses in the Childhood Origins of Asthma (COAST) Cohort.
Panel A shows the prevalence of asthma according to 17q21 genotype, and Panel B shows the mean number of HRV wheezing illnesses in the first 3 years of life according to 17q21 genotype. Sample sizes for each genotype group are shown in parentheses under the horizontal axis. T bars in Panel B indicate standard errors. Panel C shows the prevalence of asthma among the children according to the whether they had at least one HRV wheezing illness in the first 3 years of life or no HRV wheezing illness. The dashed horizontal line shows the overall prevalence of asthma among children in the COAST cohort. P = 0.006 for the main effect of this single-nucleotide polymorphism (SNP) among children who had at least one HRV wheezing illness in the first 3 years of life, and P = 0.70 among children who did not have an HRV wheezing illness; P = 0.004 for the interaction between the rs7216389 SNP and HRV wheezing illness with respect to the development of asthma. Panel D shows the odds-ratio estimates for asthma risk associated with the three genotypes at rs7216389 among children who had at least one HRV wheezing illness in the first 3 years of life, and Panel E, the odds-ratio estimates among children who did not have any HRV wheezing illnesses in the first 3 years of life. In Panels D and E, the odds ratios are shown relative to the reference group — the children with the CC genotype who did not have any HRV wheezing illnesses. The sample size of each genotype group is shown in parentheses under the horizontal axis in both panels, and I bars indicate 95% confidence intervals.
INTERACTIONS BETWEEN 17Q21 VARIANTS AND VIRAL WHEEZING ILLNESSES
We next examined interactions between the 17q21 genotype and HRV and RSV wheezing illnesses with respect to the risk of asthma in the COAST cohort. We observed significant interactions between 17q21 genotypes and HRV wheezing illness with respect to the subsequent risk of asthma (P≤0.01 for interaction) (Fig. 1C, and Table S3 in the Supplementary Appendix). The association between genotypes and asthma was present in children with HRV wheezing illness (odds ratio for the TT genotype at rs7216389, 26.1; 95% confidence interval [CI], 5.1 to 133.0) (Fig. 1D, and Table S3 in the Supplementary Appendix) but not in children without HRV wheezing illness (odds ratio for the TT genotype at rs7216389, 0.8; 95% CI, 0.2 to 2.4) (Fig. 1E, and Table S3 in the Supplementary Appendix). The odds ratio for childhood asthma among children with the TT genotype by itself was 2.3 (95% CI, 1.0 to 5.2), and the odds ratio for childhood asthma among children with HRV wheezing illness by itself was 5.2 (95% CI, 2.8 to 9.9). In contrast, no significant interaction was observed with RSV wheezing illness (P≥0.30 for interaction) (Table S4 in the Supplementary Appendix). These data suggest an interaction between a common genetic risk factor (17q21 genotype) and a common environmental risk factor (HRV wheezing illness) with respect to childhood-onset asthma.
REPLICATION STUDIES IN THE COPSAC COHORT
We next studied this interaction in the COPSAC birth cohort22 and found that 17q21 genotypes were associated with asthma in this cohort (P = 0.01 for the association with rs7216389)7 and in the subgroup of 297 children included in this study (P = 0.02 for the association with rs7216389). We stratified the children in the COPSAC cohort according to the presence or absence of HRV wheezing illness in the first 3 years of life (Fig. 2A) — an approach similar to the one we used in the COAST cohort.
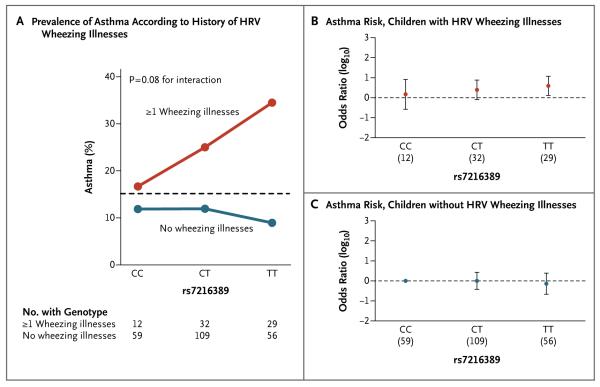
Figure 2. Effects of 17q21 Genotype on Asthma and HRV Wheezing Illnesses in the Copenhagen Prospective Study on Asthma in Childhood (COPSAC) Cohort.
Panel A shows the prevalence of asthma among children in the COPSAC cohort, according to whether they had at least one HRV wheezing illness in the first 3 years of life or no HRV wheezing illnesses. The dashed horizontal line shows the overall prevalence of asthma among children in the COPSAC cohort. Panel B shows the odds-ratio estimates for asthma risk associated with the three genotypes at rs7216389 among children who had at least one HRV wheezing illness in the first 3 years of life, and Panel C, the odds-ratio estimates for asthma risk associated with the three rs7216389 genotypes among children who did not have any HRV wheezing illnesses in the first 3 years of life. In both panels, the odds ratios were relative to the reference group — children with the CC genotype who did not have any HRV wheezing illnesses. In Panels B and C, the numbers in parentheses under the horizontal axis are the numbers of participants with a given genotype. I bars in Panels B and C indicate 95% confidence intervals.
We observed that the overall prevalence of asthma was lower in the COPSAC cohort than in the COAST cohort, a finding that we attribute to two differences between the cohorts. First, the rate of HRV wheezing illness, an important risk factor for asthma, was lower in the COPSAC cohort than in the COAST cohort (24.6% vs. 31.3%). Second, the criteria for the diagnosis of asthma differed between the two cohorts. In COPSAC, the diagnosis of asthma required a response to a 3-month course of inhaled glucocorticoids followed by a relapse after treatment was stopped (persistent asthma). In the COAST study, the diagnosis of asthma was based either on the documented use of rescue albuterol, intermittent use of agents to control asthma, or short courses of oral glucocorticoids only (intermittent asthma) or on the daily use of medications to control asthma (persistent asthma). Nonetheless, we found that in the COPSAC cohort, the association between 17q21 genotypes and asthma was present in children who had had at least one HRV wheezing illness in early life (odds ratio for the TT genotype at rs7216389, 3.9; 95% CI, 1.3 to 11.7) (Fig. 2B, and Table S5A in the Supplementary Appendix) but not in children who had not had an HRV wheezing illness (odds ratio for the TT genotype at rs7216389, 0.7; 95% CI, 0.2 to 2.4; P = 0.08 for interaction) (Fig. 2C, and Table S5A in the Supplementary Appendix). The difference in the P values for interaction between the COAST and COPSAC birth cohorts is probably due to the smaller number of children with asthma in the COPSAC cohort. There was no interaction between 17q21 genotype and RSV wheezing illness in the COPSAC cohort, a finding consistent with that in the COAST cohort (P = 0.29 for interaction) (Table S5B in the Supplementary Appendix). A combined analysis (meta-analysis) of the results in the COAST and COPSAC samples, with the use of Fisher’s test, yielded a P value of 0.003 for the interaction between 17q21 genotypes and HRV wheezing illness and a P value of 0.30 for the interaction between 17q21 genotypes and RSV wheezing illness with respect to the risk of asthma.
TRANSCRIPTIONAL RESPONSE TO HRV STIMULATION
We hypothesized that exposure to HRV might alter the expression levels of one or more of the 17q21 genes, possibly in a genotype-specific fashion. To test this hypothesis, we examined the expression patterns of the five 17q21 genes in unstimulated PBMCs and in HRV-stimulated PBMCs from 100 adults. Of the five genes at this locus, only IKZF3, GSDMB, and ORMDL3 transcripts were amplified in PBMCs in both stimulated and unstimulated conditions. As compared with the expression of IKZF3, GSDMB, and ORMDL3 in unstimulated cells, the expression of these genes in HRV-stimulated cells was increased by a factor of 1.01, 1.29, and 2.17, respectively (P = 0.95, P<0.001, and P<0.001, respectively) (Fig. 3).
Figure 3. Expression of IKZF3, GSDMB, and ORMDL3 in Peripheral-Blood Mononuclear Cells (PBMCs) from the Adult Participants.
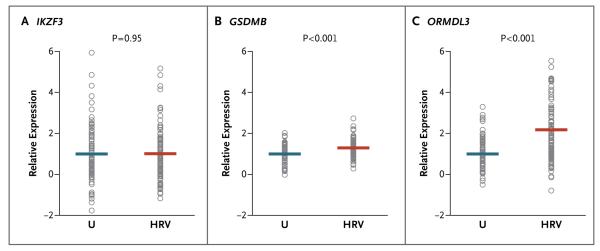
Relative expression levels are shown in unstimulated (U) and HRV-stimulated (HRV) PBMCs. Expression of IKZF3 was measured in 96 paired samples, and expression of GSDMB and ORMDL3 in 97 paired samples. Each circle corresponds to an individual participant. The horizontal lines show the mean expression levels. As compared with the expression of IKZF3, GSDMB, and ORMDL3 in unstimulated cells, the mean expression of these genes in HRV-stimulated cells was increased by a factor of 1.01, 1.29, and 2.17, respectively.
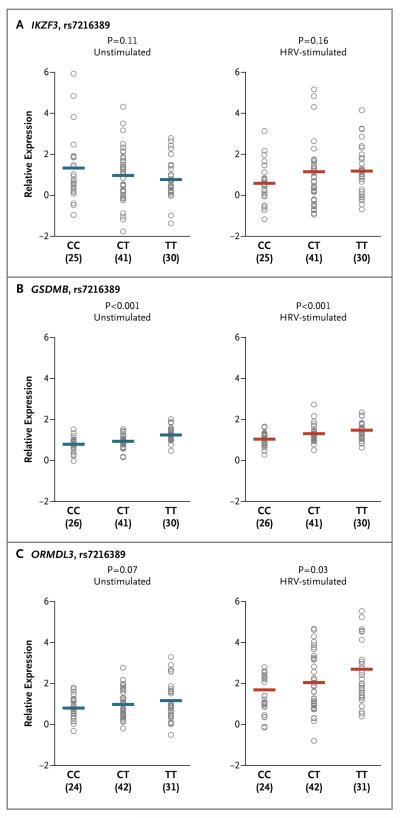
To examine genotype-specific effects on transcript levels of the three expressed genes in unstimulated cells and in HRV-stimulated cells, we stratified participants according to genotype at rs7216389. The rs7216389 genotype was associated with transcript levels of GSDMB and ORMDL3, but not IKZF3, in both unstimulated and HRV-stimulated cells (Fig. 4), a finding that was consistent with the results of previous studies.5,12,13 The relative increase in the expression of GSDMB and ORMDL3 after exposure to HRV was not, however, associated with 17q21 genotype (Fig. S2 in the Supplementary Appendix). These same trends were observed when we limited our studies to participants of European ancestry. In addition, in genomewide gene-expression data from the same 100 participants, the 17q21 genotypes were not significantly associated with expression levels of any non-17q21 genes in either unstimulated or HRV-stimulated cells (data not shown).
Figure 4. Association between rs7216389 Genotype and Expression of 17q21 Genes in Unstimulated and HRV-Stimulated PBMCs.
Relative expression levels of IKZF3, GSDMB, and ORMDL3 are shown according to 17q21 genotype in unstimulated and HRV-stimulated PBMCs. The sample size of each genotype group is shown in parentheses under the horizontal axis. Each circle corresponds to an individual participant. The horizontal lines show the mean expression levels. The P values indicate the significance of the association between the number of T alleles and gene expression.
DISCUSSION
We have found a significant interaction, in both the COAST and COPSAC birth cohorts, between 17q21 genotypes and HRV wheezing illness in early life with respect to the development of childhood-onset asthma, such that associations between 17q21 genotypes and asthma are restricted to children with HRV wheezing illness in early childhood. These findings are consistent with those of a previous study that showed interactions between SNPs at the 17q21 locus and retrospective reports of respiratory illnesses in early life (P = 0.02 for interaction)10 and with reports of more significant associations between 17q21 variants and asthma among children exposed to environmental tobacco smoke in early life than among those without such exposure.4,9-11 Exposure to environmental tobacco smoke is associated with increased rates of early viral illnesses,23 and a large body of literature links both exposure to environmental tobacco smoke and early viral illnesses to an increased risk of asthma.18-20,23 Our results implicate an interaction between HRV wheezing illness in early childhood and 17q21 genotype. The odds ratio for childhood asthma among children in the COAST cohort who were TT homozygotes at rs7216389 and had had a wheezing HRV illness was 26.1 (95% CI, 5.1 to 133.0), as compared with odds ratios for childhood asthma of 2.3 (95% CI, 1.0 to 5.2) with the TT genotype by itself and 5.2 (95% CI, 2.8 to 9.9) with HRV wheezing illness by itself; the effects of each are not merely additive but are due to an interaction.
There are some limitations of our study. First, because HRV is the most common virus encountered, we cannot exclude the possibility that the absence of an observed interaction with RSV is due to insufficient power. Second, we analyzed viral exposure in children with wheezing illness, and we cannot establish causality; it may be that HRV wheezing illness is merely a marker of the underlying predisposition for asthma and is not causal. Finally, we were unable in this study to evaluate the role of concomitant bacteria, which also triggers episodes of wheezing.17
Although the functions of the genes at the 17q21 locus are not well understood and the mechanisms leading to asthma are unknown, it has previously been suggested that ORMDL3 may play a role in viral respiratory infections.24 The ORMDL3 protein localizes to the endoplasmic reticulum membrane, where it regulates endoplasmic reticulum–mediated calcium signaling and the unfolded-protein response,25 probably through the ATF6 pathway.26 The induction and manipulation of unfolded-protein–response signaling are mechanisms through which viruses protect host cells from death mediated by endoplasmic reticulum stress.27 The significant up-regulation of ORMDL3 transcript in HRV-stimulated cells and the association between 17q21 variants and the number of HRV wheezing illnesses are consistent with this role and suggest that overexpression of ORMDL3 may increase the efficiency of the infection or viral replication in respiratory epithelial cells, which are the site of HRV infection and replication, and possibly reduce the ability of these cells to repair themselves after HRV infection. Little is known about GSDMB, other than that it belongs to the cancer-associated gasdermin domain–containing protein family28,29 and might be involved in secretory pathways28 and stem-cell proliferation in normal epithelia.29 Further studies of the effects of the 17q21 genotype in response to HRV infection in respiratory epithelial cells and in the presence of concomitant bacteria could shed additional light on the mechanism for the observed interaction. Moreover, studies of the effects of RSV and of other respiratory viruses on 17q21 gene-expression levels would allow us to determine whether the increased expression levels of ORMDL3 and GSDMB are specific to HRV stimulation.
In summary, this study establishes a role of 17q21 variants in the development of HRV wheezing illnesses during early childhood and shows that the effects of the 17q21 genotype on the susceptibility to asthma are seen primarily in the subgroup of children who have had an HRV wheezing illness in early childhood. Our results underscore the importance of studying the joint effects of genetic and environmental risk factors to gain a fuller understanding of the mechanisms underlying complex diseases.
Supplementary Material
Acknowledgments
Supported by grants (HL070831 and AI070503) from the National Institutes of Health. Dr. Jackson was supported by KL2 grant UL1TR000427 from the National Institutes of Health. The Lundbeck Foundation, Danish Council for Strategic Research, and Danish Pediatric Asthma Center provided the core funding for the COPSAC research unit.
We thank Nick Levinsky for his help with the HRV stimulation protocols; Maitane Arruabarrena Orbegozo for technical assistance; Dagan Loisel, Darren Cusanovich, and Yoav Gilad for helpful discussions and comments on an earlier draft of the manuscript; all the study participants; and the COAST and COPSAC research teams.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Moffatt MF, Kabesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 2.Akhabir L, Sandford AJ. Genomewide association studies for discovery of genes involved in asthma. Respirology. 2011;16:396–406. doi: 10.1111/j.1440-1843.2011.01939.x. [DOI] [PubMed] [Google Scholar]
- 3.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242:10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouzigon E, Corda E, Aschard H, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008;359:1985–94. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 5.Halapi E, Gudbjartsson DF, Jonsdottir GM, et al. A sequence variant on 17q21 is associated with age at onset and severity of asthma. Eur J Hum Genet. 2010;18:902–8. doi: 10.1038/ejhg.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bisgaard H, Bønnelykke K, Sleiman PM, et al. Chromosome 17q21 gene variants are associated with asthma and exacerbations but not atopy in early childhood. Am J Respir Crit Care Med. 2009;179:179–85. doi: 10.1164/rccm.200809-1436OC. [DOI] [PubMed] [Google Scholar]
- 8.Wu H, Romieu I, Sienra-Monge JJ, Li H, del Rio-Navarro BE, London SJ. Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood asthma. Allergy. 2009;64:629–35. doi: 10.1111/j.1398-9995.2008.01912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flory JH, Sleiman PM, Christie JD, et al. 17q12-21 Variants interact with smoke exposure as a risk factor for pediatric asthma but are equally associated with early-onset versus late-onset asthma in North Americans of European ancestry. J Allergy Clin Immunol. 2009;124:605–7. doi: 10.1016/j.jaci.2009.05.047. [DOI] [PubMed] [Google Scholar]
- 10.Smit LA, Bouzigon E, Pin I, et al. 17q21 Variants modify the association between early respiratory infections and asthma. Eur Respir J. 2010;36:57–64. doi: 10.1183/09031936.00154509. [DOI] [PubMed] [Google Scholar]
- 11.van der Valk RJ, Duijts L, Kerkhof M, et al. Interaction of a 17q12 variant with both fetal and infant smoke exposure in the development of childhood asthma-like symptoms. Allergy. 2012;67:767–74. doi: 10.1111/j.1398-9995.2012.02819.x. [DOI] [PubMed] [Google Scholar]
- 12.Verlaan DJ, Ge B, Grundberg E, et al. Targeted screening of cis-regulatory variation in human haplotypes. Genome Res. 2009;19:118–27. doi: 10.1101/gr.084798.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy A, Chu JH, Xu M, et al. Mapping of numerous disease-associated expression polymorphisms in primary peripheral blood CD4+ lymphocytes. Hum Mol Genet. 2010;19:4745–57. doi: 10.1093/hmg/ddq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ober C, Vercelli D. Gene-environment interactions in human disease: nuisance or opportunity? Trends Genet. 2011;27:107–15. doi: 10.1016/j.tig.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sykes A, Johnston SL. Etiology of asthma exacerbations. J Allergy Clin Immunol. 2008;122:685–8. doi: 10.1016/j.jaci.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 16.Johnston SL, Pattemore PK, Sanderson G, et al. Community study of role of viral infections in exacerbations of asthma in 9-11 year old children. BMJ. 1995;310:1225–9. doi: 10.1136/bmj.310.6989.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bisgaard H, Hermansen MN, Bønnelykke K, et al. Association of bacteria and viruses with wheezy episodes in young children: prospective birth cohort study. BMJ. 2010;341:c4978. doi: 10.1136/bmj.c4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotaniemi-Syrjänen A, Vainionpää R, Reijonen TM, Waris M, Korhonen K, Korppi M. Rhinovirus-induced wheezing in infancy — the first sign of childhood asthma? J Allergy Clin Immunol. 2003;111:66–71. doi: 10.1067/mai.2003.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyvärinen MK, Kotaniemi-Syrjänen A, Reijonen TM, Korhonen K, Korppi MO. Teenage asthma after severe early childhood wheezing: an 11-year prospective follow-up. Pediatr Pulmonol. 2005;40:316–23. doi: 10.1002/ppul.20273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson DJ, Gangnon RE, Evans MD, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med. 2008;178:667–72. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemanske RF., Jr The Childhood Origins of Asthma (COAST) study. Pediatr Allergy Immunol. 2002;13(Suppl 15):38–43. doi: 10.1034/j.1399-3038.13.s.15.8.x. [DOI] [PubMed] [Google Scholar]
- 22.Bisgaard H, Pipper CB, Bønnelykke K. Endotyping early childhood asthma by quantitative symptom assessment. J Allergy Clin Immunol. 2011;127(5):1155.e2–1164.e2. doi: 10.1016/j.jaci.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 23.DiFranza JR, Aligne CA, Weitzman M. Prenatal and postnatal environmental tobacco smoke exposure and children’s health. Pediatrics. 2004;113:1007–15. [PubMed] [Google Scholar]
- 24.Hirota T, Harada M, Sakashita M, et al. Genetic polymorphism regulating ORM1-like 3 (Saccharomyces cerevisiae) expression is associated with childhood atopic asthma in a Japanese population. J Allergy Clin Immunol. 2008;121:769–70. doi: 10.1016/j.jaci.2007.09.038. [DOI] [PubMed] [Google Scholar]
- 25.Cantero-Recasens G, Fandos C, Rubio-Moscardo F, Valverde MA, Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 2010;19:111–21. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
- 26.Miller M, Tam AB, Cho JY, et al. ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc Natl Acad Sci U S A. 2012;109:16648–53. doi: 10.1073/pnas.1204151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trujillo-Alonso V, Maruri-Avidal L, Arias CF, López S. Rotavirus infection induces the unfolded protein response of the cell and controls it through the non-structural protein NSP3. J Virol. 2011;85:12594–604. doi: 10.1128/JVI.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carl-McGrath S, Schneider-Stock R, Ebert M, Rocken C. Differential expression and localisation of gasdermin-like (GSDML), a novel member of the cancer-associated GSDMDC protein family, in neoplastic and non-neoplastic gastric, hepatic, and colon tissues. Pathology. 2008;40:13–24. doi: 10.1080/00313020701716250. [DOI] [PubMed] [Google Scholar]
- 29.Saeki N, Usui T, Aoyagi K, et al. Distinctive expression and function of four GSDM family genes (GSDMA-D) in normal and malignant upper gastrointestinal epithelium. Genes Chromosomes Cancer. 2009;48:261–71. doi: 10.1002/gcc.20636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.