Abstract
Three new bicyclic depsipeptides, which are related to the previously reported thailandepsins A (1), B (2) and C (3), were discovered from the culture broth of Burkholderia thailandensis E264 when supplemented with amino acid precursors, and were subsequently named as thailandepsins D (4), E (5) and F (6), respectively. Enzyme assays showed that 1–6 are potent histone deacetylase (HDAC) inhibitors, particularly toward HDAC1 which represents class I human HDACs.
Introduction
The study of human diseases has traditionally focused on genetic mutations, but the disruption of the balance of epigenetic networks can also contribute to those diseases, including cancer, syndromes involving chromosomal instabilities, and mental retardation.1,2 Tumorigenesis is a multi-step process that requires several integrated events to allow a cell to grow rapidly without the input of extraneous growth-stimulation signals, and to overcome growth-inhibitory signals and host immune responses.3 Mutations that result in constitutive activation of oncogenes or functional inactivation of tumor-suppressor genes are important tumorigenic events.4 Although these mutations can affect several cellular pathways in the absence of de novo protein synthesis, epigenetic events also play an important role in tumor initiation and progression.
The most common mechanisms of epigenetic regulation are DNA methylation at the C5 position of cytosines, predominantly within the CpG islands,5 and histone modifications at the ε-NH2 group on lysine residues within histone tails.6 Among these, reversible histone acetylation plays a crucial role in chromatin packaging and control of gene expression. Acetyl groups are added to the histone tails by histone acetyltransferases (HATs), which are correlated with nucleosome relaxation and transcriptional activation. Acetyl groups are removed by histone deacetylases (HDACs), which induces transcriptional repression through chromatin condensation. Therefore, the opposing actions of HATs and HDACs allow gene expression to be exquisitely regulated through chromatin remodeling.7 So far, 18 HDACs in the human genome have been identified and they are grouped into four classes, 11 of which are zinc-dependent (classes I, II and IV), while 7 sirtuins are NAD+-dependent (class III).8,9
HDACs have been intensively scrutinized over the past decade for two main reasons. First, they have been linked mechanistically to the pathogenesis of cancer. Second, small molecule HDAC inhibitors have the capacity to reactivate gene expression and to inhibit the growth and survival of tumor cells.10 The remarkable tumor specificity of some of those compounds and their potency in vitro and in vivo underscore the potential of HDAC inhibitors as an exciting new class of agents for the treatment of cancer. SAHA (vorinostat; 7) and FK228 (romidepsin; 8) have been approved by FDA for the treatment of refractory cutaneous T-cell lymphoma,11,12 and many more HDAC inhibitors are in various stages of preclinical or clinical trials.13
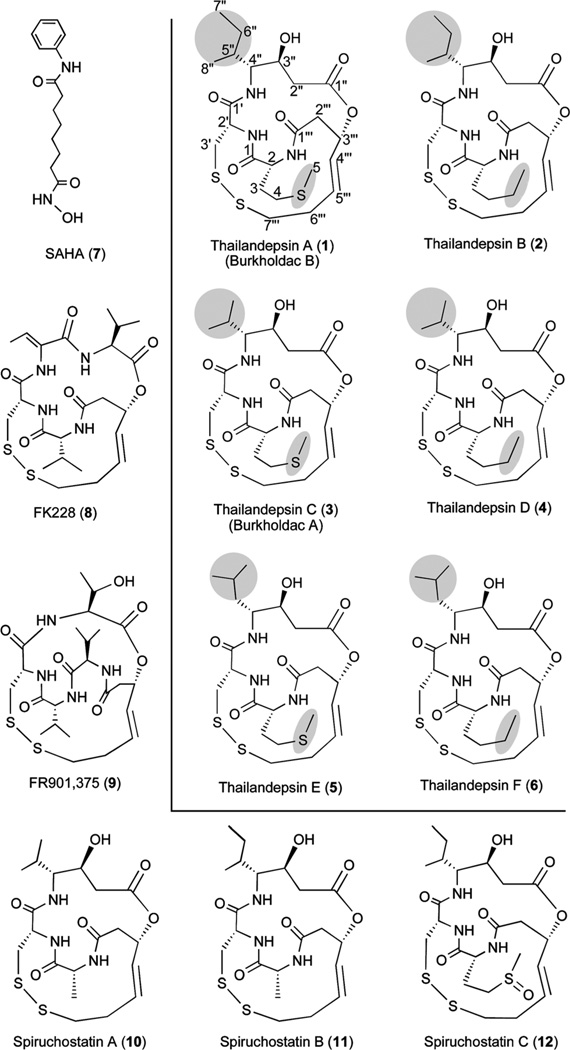
FK228 (8), which is produced by Chromobacterium violaceum no. 968,14 represents a family of natural products that each contains a signature disulfide bond. This family of natural products (Fig. 1) also includes FR901,375 (9) produced by Pseudomonas chlororaphis (no. 2522),15 spiruchostatins A (10) and B (11) by Pseudomonas sp. Q71576,16 spiruchostatin C (12) and its dimer,17 thailandepsins A (1; also known as burkholdac B18) and B (2) reported previously by us,19 and burkholdac A18 (3; independently discovered by us and named sequentially as thailandepsin C), all by Burkholderia thailandensis E264. The biosynthesis of those compounds is proposed to follow an “assembly-line” mechanism in which simple building blocks are assembled stepwise by modular nonribosomal peptide synthetase (NPRS)–polyketide synthase (PKS) systems.19,20 The disulfide bond present in this family of natural products is proven or presumed to mediate the anticancer activity via intracellular reduction to generate a free thiol group (“warhead”) that chelates a Zn2+ ion in the catalytic center of zinc-dependent HDACs, thereby inhibiting the enzyme activities.21
Fig. 1.
Compound structures.
Precursor feeding is an effective approach for enhanced production of microbial natural products. In this article we report that, when a combination of cysteine and norleucine was supplemented in the growth medium of B. thailandensis E264, not only the yield of 1–3 increased significantly, but three additional compounds were also produced at elevated levels, which led to the discovery of thailandepsins D (4), E (5) and F (6) (Fig. 1). Compounds 1–6, along with the reference compound 8, were assayed for their inhibitory activities against human recombinant HDAC1, HDAC4 and HDAC6, and it was found that they are potent HDAC inhibitors, particularly toward HDAC1 which represents class I HDACs.
Results and discussion
Improved production of thailandepsins through precursor feeding
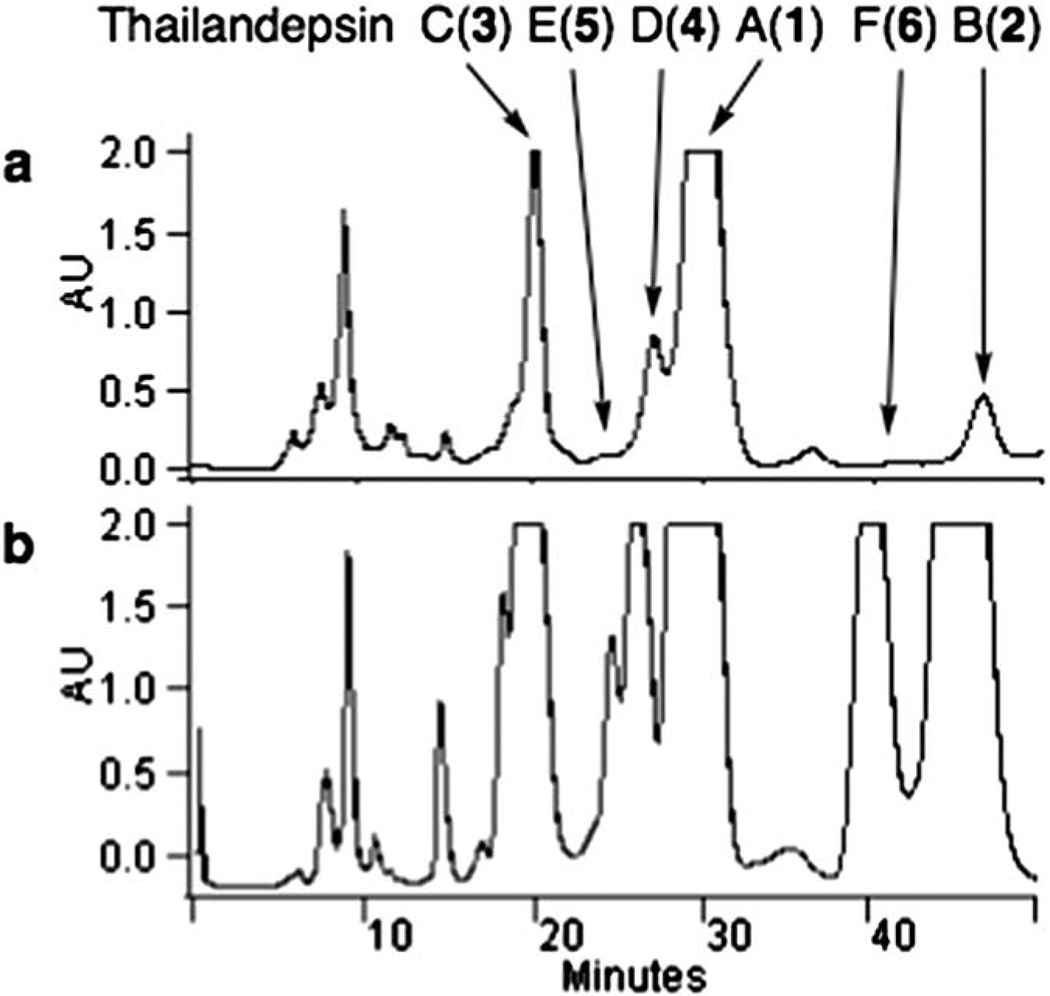
Compounds 1, 2 and 3 all contain one d-cysteine moiety and one complex moiety derived from one cysteine unit and two malonate units, but 2 differs from 1 in having a rare d-norleucine moiety at position 2 (according to annotation in 1) where 1 has a d-methionine moiety, and 3 differs from 1 in having a d-valine moiety at position 4″ where 1 has a d-isoleucine moiety. Although 1 is the predominant compound, it is still produced at less than 2 mg L−1 by our in-flask fermentation not systematically optimized. In an attempt to improve the yields of those bioactive compounds, cysteine in combination with methionine, norleucine or valine was added, each at 0.1% (w/v), to the culture medium of B. thailandensis in the beginning of fermentation. While adding cysteine in combination with methionine or valine slightly improved the yield of 1 and 3 (data not shown), adding cysteine and norleucine resulted in about a 10 fold increase of the yield of 2, and about a 3 fold increase of the yield of 1 and 3. In addition, three additional compounds were also produced at elevated levels, which eventually led us to discover thailandepsins D (4), E (5) and F (6) (Fig. 2). Together, all 1–6 were obtained as white, amorphous powder with 90–96% purity and routinely analyzed by HPLC, LC-MS, HR-TOF-MS, CD, IR and NMR. The analytical HPLC traces (indicating purity) of 1–6, and the MS and NMR spectra of 3–6 are available in the ESI‡, and the NMR assignments for 4, 5 and 6 are summarized in Table 1, 2 and 3, respectively.
Fig. 2.
Elevated production of thailandepsins A through F (1–6). HPLC profiling of semi-purified extracts of B. thailandensis E264 culture with (b) or without (a) supplementation of cysteine and norleucine.
Table 1.
NMR spectroscopic data of thailandepsin D (4) in CDCl3
| position | δC, type | δH (J in Hz) | HMBC | |
|---|---|---|---|---|
| Nleu | 1 | 170.4, C═O | 2, 3, NH, NH′ | |
| 2 | 56.8, CH | 4.14, m | 4, NH | |
| 3 | 30.5, CH2 | 1.89, m | 2 | |
| 1.74, m | ||||
| 4 | 28.4, CH2 | 1.39, m | 5, 6 | |
| 5 | 22.3, CH2 | 1.40, m | 3, 6 | |
| 6 | 13.9, CH3 | 0.93, m | ||
| NH | 6.51, d (3.1) | |||
| Cys | 1′ | 169.2, C═O | 2′, 4″, NH″ | |
| 2′ | 55.2, CH | 4.85, m (9.0, 3.5) | ||
| 3′ | 40.1, CH2 | 3.30, m | ||
| 3.14, m | ||||
| NH′ | 6.87, d (9.0) | |||
| Ahhxa | 1″ | 171.7, C═O | ||
| 2″ | 39.6, CH2 | 2.71, m | ||
| 3″ | 69.1, CH | 4.52, m | 2″, 4″ | |
| 4″ | 63.2, CH | 2.85, m | 2″, 5″, 6″, 7″ | |
| 5″ | 29.8, CH | 2.35, m | 3″, 4″, 6″, 7″ | |
| 6″ | 20.6, CH3 | 1.02, d (6.8) | 4″, 5″ | |
| 7″ | 19.8, CH3 | 0.92, s | 4″, 5″ | |
| NH″ | 7.43, d (7.2) | |||
| OH″ | ||||
| acyl | 1‴ | 171.0, C═O | 2, 2‴, NH | |
| 2‴ | 40.8, CH2 | 3.31, m | ||
| 3‴ | 70.9, CH | 5.52, br s (15.7) | 2‴, 4‴ | |
| 4‴ | 129.1, CH | 5.76, d (15.7) | 2‴, 6‴ | |
| 5‴ | 133.1, CH | 6.32, m (12.3) | ||
| 6‴ | 33.1, CH2 | 2.72, m | 4‴ | |
| 2.49, m | ||||
| 7‴ | 40.8, CH2 | 3.30, m | ||
| 2.73, m |
Ahhx: 4-amino-3-hydroxy-5-methylhexanoic acid.
Table 2.
NMR spectroscopic data of thailandepsin E (5) in CDCl3
| position | δC, type | δH (J in Hz) | HMBC | |
|---|---|---|---|---|
| Met | 1 | 170.3, C═O | 2, NH, NH′ | |
| 2 | 57.3, CH | 4.34, m | 3, 4, NH | |
| 3 | 38.8, CH2 | 2.66, m | 2 | |
| 4 | 40.5, CH2 | 3.20, d (5.4) | 2′, NH′ | |
| 2.64, m | ||||
| 5 | 15.4, CH3 | 2.17, s | 4 | |
| NH | 7.80, br s | |||
| Cys | 1′ | 168.7, C═O | 2′, NH″ | |
| 2′ | 55.2, CH | 4.81, m | 3′, NH′ | |
| 3′ | 39.4, CH2 | 3.32, m | ||
| 3.20, m | ||||
| NH′ | 6.84, d (9.1) | |||
| Ahhxa | 1″ | 171.4, C═O | 3″ | |
| 2″ | 27.9, CH2 | 2.24, m | 3″, 4″ | |
| 2.04, m | ||||
| 3″ | 70.4, CH | 4.34, m | 2″, 4″, 5″, NH″ | |
| 4″ | 55.9, CH | 3.06, m | 2″, 3″, 5″, NH″ | |
| 5″ | 38.7, CH2 | 2.06, m | 3″, 4″, 6″ | |
| 1.52, m | ||||
| 6″ | 25.1, CH | 1.62, m | 4″, 5″, 7″, 8″ | |
| 7″ | 21.2, CH3 | 0.91, d (1.5) | 5″, 6″ | |
| 8″ | 23.4, CH3 | 0.89, d (1.7) | 5″, 6″ | |
| NH″ | 7.54, d (7.1) | |||
| OH″ | ||||
| acyl | 1‴ | 171.2, C═O | 3.34, m | 2‴, NH |
| 2‴ | 41.4, CH2 | 3.34, m | 4‴ | |
| 2.73, m | ||||
| 3‴ | 70.7, CH | 5.49, br s | 2‴, 4‴ | |
| 4‴ | 129.4, CH | 5.74, d (15.3) | 2‴, 3‴ | |
| 5‴ | 132.9, CH | 6.22, m | 6‴, 7‴ | |
| 6‴ | 32.6, CH2 | 2.72, m | 4‴ | |
| 2.41, m | ||||
| 7‴ | 41.0, CH2 | 3.32, m | ||
| 2.74, m |
Ah(6)hp: 4-amino-3-hydroxy-6-methylheptanoic acid.
Table 3.
NMR spectroscopic data of thailandepsin F (6) in CDCl3
| position | δC, type | δH (J in Hz) | HMBC | |
|---|---|---|---|---|
| Nleu | 1 | 170.8, C═O | 2, 2′, NH, NH′ | |
| 2 | 56.7, CH | 4.15, m | 2′″, NH | |
| 3 | 30.4, CH2 | 1.90, m | 2, 4, 5, NH | |
| 1.72, m | ||||
| 4 | 28.3, CH2 | 1.40, m | 2, 3, 5 | |
| 5 | 22.2, CH2 | 1.43, m | 3, 4, 6 | |
| 6 | 13.9, CH3 | 0.93, m | 4, 5 | |
| NH | 6.26, d (3.1) | |||
| Cys | 1′ | 168.8, C═O | 2′, 3′, NH″ | |
| 2′ | 55.0, CH | 4.83, m (8.6, 4.0) | NH′ | |
| 3′ | 40.4, CH2 | 3.29, m | ||
| 3.15, m | ||||
| NH′ | 6.81, d (8.9) | |||
| Ah(6)hpa | 1″ | 171.4, C═O | 2″, 3″ | |
| 2″ | 38.8, CH2 | 2.73, m | 3″, 4″ | |
| 3″ | 70.7, CH | 4.36, m | 2″, 4″, 5″, NH″ | |
| 4″ | 56.0, CH | 3.12, m | 2″, 3″, 5″, NH″ | |
| 5″ | 38.7, CH2 | 2.08, m | 3″, 4″ | |
| 1.46, m | ||||
| 6″ | 25.1, CH | 1.63, m | 5″, 7″, 8″ | |
| 7″ | 21.0, CH3 | 0.93, m | 5″, 6″ | |
| 8″ | 23.4, CH3 | 0.91, m | 5″, 6″ | |
| NH″ | 7.52, d (7.0) | |||
| OH″ | ||||
| acyl | 1′″ | 171.1, C═O | 2′″, NH | |
| 2′″ | 40.9, CH2 | 3.37, m | ||
| 2.70, m | ||||
| 3′″ | 70.6, CH | 5.49, br s | 4′″ | |
| 4′″ | 128.8, CH | 5.73, d (15.4) | 2′″, 3′″, 6′″ | |
| 5′″ | 133.2, CH | 6.40, m (13.5) | ||
| 6′″ | 32.8, CH2 | 2.72, m | 4′″ | |
| 2.45, m | ||||
| 7′″ | 40.9, CH2 | 3.38, m | ||
| 2.73, m |
Ah(6)hp: 4-amino-3-hydroxy-6-methylheptanoic acid.
Structure elucidation of new thailandepsins
Compound 3 has a molecular formula of C22H35N3O6S3, calculated from the molecular cluster ion at m/z 534.1796, [M + H]+ (calculated 534.1758). All NMR spectra (see ESI‡) indicated that 3 is identical to the previously reported burkholdac A.18
The molecular formula of compound 4 was established by HR-TOF-MS as C23H37N3O6S2, calculated from the molecular cluster ion at m/z 516.2277, [M + H]+ (calculated 516.2202). Its CD was determined as [α]D24 − 24.0 (c 0.2, in acetronitrile). Its IR spectrum shows intense absorption bands of amines at νmax 3370 cm−1 (NH), 1670 cm−1 (carbonyl) and 1740 cm−1 (ester band). Its UV spectrum does not have characteristic UV absorbance peaks due to a lack of chromophore. The 1H and 13C NMR spectroscopic data (Table 1) are suggestive of a depsipeptide structure, showing four carbonyl carbon signals at δC 171.7, 171.0, 170.4 and 169.2 and three amide NH protons at δH 7.43, 6.87 and 6.51. The 1H and 13C NMR spectra of 4 (see ESI‡) show great similarity to those of 2,19 except that the corresponding signals (δC 27.6, δH 1.62 and 1.33) of 2 are missing in the spectra of 4. These signals were assigned to the methylene group in 2 and this difference is consistent with the observation of a molecular weight difference of 14 daltons between 2 (530 m/z) and 4 (516 m/z). The COSY spectrum of 4 reveals four spin systems and three of them are the same as that of 2: norleucine, cysteine and the acyl moiety. The sequence of the last spin system is from the methylene protons (2″-H2/δH 2.71) to the two methyl groups (6″-H/δH 1.02 and 7″-H/δH 0.92) through three methine groups (3″-H/δH 4.52, 4″-H/δH 2.85 and 5″-H/δH 2.35), which is also coupled to 3″-OH and 4″-NH. This moiety was determined as Ahhx (4-amino-3-hydroxy-5-methylhexanoic acid). The linkage of these four moieties of 4 was established by HMBC and the final planar structure was further confirmed by 2D NMR analysis. Comparing the planar structure of 4 with 2, the only difference is that the Ahhp (4-amino-3-hydroxy-5-methylheptanoic acid, isoleucine derivative) moiety in 2 is replaced by the Ahhx moiety (valine derivative) in 4.
Compound 5 has the same molecular weight and molecular formula (C23H37N3O6S3) as 1, calculated from the molecular cluster ion at m/z 548.1995, [M + H]+ (calculated 548.1923), but their retention times on LC-MS appeared different, suggesting molecular configuration difference(s). Its CD was determined as [α]D24 − 15.0 (c 0.2, in acetronitrile). Its IR spectrum is identical to that of 4, and its UV spectrum does not have characteristic UV absorbance peaks either. Comparison of the 1H and 13C NMR data of 5 (see ESI‡) with the spectra of 1 (ref. 19) revealed an overall similarity. The COSY spectrum of 5 shows four spin systems and three of them are the same as that of 1: methionine, cysteine and the acyl moiety. However, differences exist in the last spin system where the methyl group 8″-CH3 is connected with 5″-C in 1 and the moiety is Ah(5)hp (4-amino-3-hydroxy-5-methylheptanoic acid, isoleucine derivative), while the same methyl group is connected with 6″-C in 5 and the moiety was determined to be Ah(6)hp (4-amino-3-hydroxy-6-methylheptanoic acid, leucine derivative). The linkage of these four moieties in 5 was established by HMBC. Collectively, 5 was found to be an isomer of 1 with an isoleucine moiety in 1 replaced by a leucine moiety in 5.
Compound 6 has the same molecular weight and molecular formula (C24H39N3O6S2) as 2, calculated from the molecular cluster ion at m/z 530.2424, [M + H]+ (calculated 530.2384), but their retention times on LC-MS are different. Its CD was determined to be [α]D24 − 20.0 (c 0.2, in acetronitrile). Its IR spectrum is identical to that of 4, and its UV spectrum does not have characteristic UV absorbance peaks either. Similarly through comparison of the 1H and 13C NMR data (see ESI‡) and COSY and HMBC spectra of 6 with those of 2,19 it was established that 6 is an isomer of 2 with an isoleucine moiety in 2 replaced by a leucine moiety in 6.
The stereochemistry and absolute configurations of 3, 4, 5 and 6 were assumed by extrapolation from our reported X-ray diffraction structure of 1.22
HDAC inhibitory activities of thailandepsins
Recombinant HDAC1, HDAC4 and HDAC6 were chosen to represent class I, class IIa or class IIb human HDACs, respectively, for HDAC inhibition assays. Compounds 1–6 and reference compound 8 were first reduced by tris(2-carboxyethyl) phosphine hydrochloride (TCEP) prior to assays. The dose–response curves of each compound vs. each HDAC are shown in the ESI‡ Fig. S25, and the calculated IC50 values of each compound vs. each HDAC are listed in Table 4.
Table 4.
The calculated IC50 value of each compound (reduced) vs. each HDAC in µM concentration
| HDAC enzymes | |||
|---|---|---|---|
| Compounds | HDAC1 | HDAC4 | HDAC6 |
| Cpd 8a | 0.00031 | 8.67 | 0.67 |
| Cpd 1a | 0.00028 | 37.22 | 0.83 |
| Cpd 2a | 0.0012 | 57.44 | 1.19 |
| Cpd 3a | 0.00020 | 38.04 | 0.78 |
| Cpd 4a | 0.00048 | 46.97 | 0.93 |
| Cpd 5a | 0.00094 | 58.88 | 1.68 |
| Cpd 6a | 0.0026 | 132.5 | 1.92 |
Indicating compounds in reduced form.
Collectively, all tested compounds in their reduced forms are potent inhibitors of human HDACs with 8 being the most potent one overall. They exhibited outstanding inhibitory activities toward HDAC1 with IC50 values in single- to sub-nM range; their inhibitory activities toward HDAC6 and HDAC4 are notable but are nevertheless 3–5 orders of magnitude weaker than toward HDAC1. It is obvious that all those HDAC inhibitors have a strong degree of selectivity toward class 1 HDACs, and that different molecular constituents with varying sizes or length of side chains have an impact on the HDAC inhibitory activities.
Conclusions
In an effort to improve the yields of three previously reported HDAC inhibitors, 1–3, a series of amino acid precursor combinations were individually added to the fermentation culture of B. thailandensis, which resulted in a significantly increased production of 1–3. Unexpectedly, three additional compounds were also produced at elevated levels, which eventually led to our discovery of 4, 5 and 6, as reported in this communication.
Structurally, all 1–6 belong to the FK228 family of bacterial natural products that share a highly similar bicyclic depsipeptide framework with a signature disulfide bond critical for stability as a prodrug and for bioactivity as a potent HDAC inhibitor when reduced (Fig. 1).21 As it is clear that this class of natural products are produced by a modular NRPS-PKS system,19,20 the nature of chemical building blocks in those molecules is likely dictated by two-fold biochemical mechanisms: substrate specificity as well as substrate promiscuity of individual NRPS modules. Amazingly, the outer ring of their structures, which contains the signature disulfide linkage, is absolutely conserved in all members of this family of natural products. Structural variations exist only at two positions among 1–6: position 2 (according to annotation in 1) is either a methionine or a norleucine moiety, while position 4″ is an isoleucine, a leucine or a valine moiety.
Those subtle structural differences have an impact on their bioactivities. To correlate between the structure and the HDAC inhibitory activities among 1–6, it can be generalized that a methionine moiety at position 2 is somehow better suited for HDAC inhibitory activities than a norleucine moiety, so is a smaller moiety (e.g. valine vs. leucine or isoleucine) at position 4″ than a larger moiety. Perhaps because of a combination of these two factors, 3, which contains both a methionine moiety at position 2 and a valine moiety at position 4″, appeared to be the most potent compound among the six. 3 is even slightly more potent than 8 toward HDAC1, but not so toward HDAC4 or HDAC6. A direct structure–activity relationship between 3 and 8 cannot be readily assessed, because they have structural differences beyond the two discussed points.
To conclude, three new thailandepsins (D, E and F), related to three previously reported thailandepsins (A, B and C), have been discovered from B. thailandensis E264. All those compounds exhibited potent HDAC inhibitory activities, particularly toward class I human HDACs. The chemical nature and size of molecular building blocks affect their biological activities, and additional structure–activity relationship studies may generate synthetic compounds with improved activity and/or selectivity toward specific HDAC isoforms. Finally, the in vitro antiproliferative activities of 1 and 2 against a panel of NCI-60 human cancer cell lines have been reported recently;19 the same in vitro antiproliferative activities of 3–6 will be pursued accordingly.
Experimental section
General natural product chemistry
ProStar 210 HPLC (Varian) was used with a preparative C18 column (10 µm particle size, 21.2 × 250 mm, Agilent) for compound purification, and with an Eclipse XDB-C18 column (5 µm particle size, 2.1 × 50 mm, Agilent) for compound purity analysis. LC-MS was performed on an Agilent 1100 series LC-MSD Trap SL equipped with the same Eclipse XDB-C18 column. CD measurement was achieved on a JASCO DIP-370 digital polarimeter. IR spectra were obtained with a Quantum Microscope with a resolution of 4 cm−1 (ATI Mattson). HR-TOF-MS was performed on an Agilent LC/MSD-TOF mass spectrometer. 1H, 13C and two-dimensional NMR spectra in CDCl3 were acquired with a Bruker DRX 300 MHz NMR spectrometer; chemical shifts are reported in δ (ppm) units. Organic solvents are reagent grade for extraction and HPLC grade for separation. Resins were purchased from Sigma-Aldrich.
Bacterial strains, reagents and fermentation
Burkholderia thailandensis E264 (ATCC 700388), a Gram-negative β-proteobacterium strain originally isolated from a rice paddy in central Thailand, was purchased from the American Type Culture Collection (ATCC). Fermentation of B. thailandensis E264 strain in a total volume of 25 L of modified M9 medium was achieved as previously described,19 except that additional amino acids were supplemented at 0.1% (w/v) each.
Extraction and purification
After fermentation, resins and cells were collected by centrifugation and subsequently lyophilized to dryness. The dry mass was extracted with ethyl acetate and the organic phase extract was concentrated under reduced pressure to an oily residue. Silica gel chromatography was used to fractionate the crude extract. After being washed by hexane–ethyl acetate (1 : 2 ratio, v/v), target compounds were eluted by ethyl acetate and the organic solvent was subsequently removed under reduced pressure to yield a semi-purified mixture. This mixture was resuspended in acetonitrile and subjected to preparative HPLC under isocratic condition with a mobile phase of 40% acetonitrile/water (v/v), a flow rate of 8 ml min−1 and UV detection at 200 nm. Compounds 1–6 were eluted at 28–32 min, 44–48 min, 18–21 min, 25–27 min, 24–25 min, and 39–42 min, respectively (Fig. 2). The overall yields of 1–6 were approximately 1.0 mg L−1, 1.1 mg L−1, 0.6 mg L−1, 0.3 mg L−1, 0.2 mg L−1, and 0.5 mg L−1, respectively.
HDAC inhibition assays
The HDAC-Glo™ I/II Assay and Screening System (Promega) was used to determine the HDAC inhibitory activities of testing compounds 1–6 and the reference compound 8. Recombinant human HDAC1, HDAC4 and HDAC6 were purchased from BPS Bioscience Inc. Assays were performed according to reagent suppliers’ protocols.
Compounds 1–6 and 8 were first dissolved in DMSO and reduced by tris(2-carboxyethyl)phosphine hydrochloride (TCEP) in a molar ratio of 1 : 1.5 for 20 min at ambient temperature prior to being assayed. A series of fivefold dilutions of each reduced compound were prepared with HDAC-Glo buffer and 25 µl of each dilution was added into a well on a 96-well plate. HDAC enzymes were diluted to the desired concentrations with HDAC-Glo buffer and 25 µl of each diluted enzyme was dispensed into a well to mix with 25 µl of the testing compound at room temperature for 30–60 seconds. After the reaction mixture had been incubated at room temperature for 1 hour, 50 µl of HDAC-Glo reagent was added to each reaction well and mixed for 30–60 seconds. Finally, the plate was incubated at room temperature for an additional 30 minutes before the luminescence was measured on a Synergy HT plate reader (Bio-Tek). The luminescence intensity data were analyzed using GraphPad Prism 5 (GraphPad Software). In the absence of any compound, the luminescence intensity (I100) in each data set was defined as 100% activity. In the absence of HDAC, the luminescence intensity (I0) in each data set was defined as 0% activity. The relative activity (%) in the presence of each compound was calculated according to the following equation: %activity = (I −I0)/(I100 − I0), where I = the luminescence intensity if a compound is present in the reaction. Experiments were performed in triplicate and the calculated mean values were used for plotting.
Supplementary Material
Acknowledgements
This work was supported by a Research Growth Initiative Award from the University of Wisconsin-Milwaukee and an NIH/NCI grant R01 CA152212 (both to Y.-Q.C.).
Footnotes
This article is part of the MedChemComm natural products themed issue.
Electronic supplementary information (ESI) available: HPLC traces of thailandepsins A through F (1–6), HR-TOF-MS and NMR spectra of 3–6. See DOI: 10.1039/c2md20024d
Notes and references
- 1.Egger G, Liang G, Aparicio A, Jones PA. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 2.Yoo CB, Jones PA. Nat. Rev. Drug Discovery. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 3.Johnstone RW. Nat. Rev. Drug Discovery. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 5.Takai D, Jones PA. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taby R, Issa JP. Ca-Cancer J. Clin. 2010;60:376–392. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- 7.Jenuwein T, Allis CD. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Yang XJ, Seto E. Nat. Rev. Mol. Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakagawa T, Guarente L. J. Cell Sci. 2011;124:833–838. doi: 10.1242/jcs.081067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minucci S, Pelicci PG. Nat. Rev. Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 11.FDA. J. Natl. Cancer Inst. 2010;102:219. doi: 10.1093/jnci/djq030. [DOI] [PubMed] [Google Scholar]
- 12.Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. Oncologist. 2007;12:1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- 13.Jones P. Med. Chem. Commun. 2012;3:135–161. [Google Scholar]
- 14.Ueda H, Nakajima H, Hori Y, Fujita T, Nishimura M, Goto T, Okuhara M. J. Antibiot. 1994;47:301–310. doi: 10.7164/antibiotics.47.301. [DOI] [PubMed] [Google Scholar]
- 15.Masakuni O, Toshio G, Takashi F, Yasuhiro H, Hirotsugu U. 3141296. Japan Patent. 1991 (A)
- 16.Masuoka Y, Nagai A, Shin-ya K, Furihata K, Nagai K, Suzuki K, Hayakawa Y, Seto H. Tetrahedron Lett. 2001;42:41–44. [Google Scholar]
- 17.Klausmeyer P, Shipley SM, Zuck KM, McCloud TG. J. Nat. Prod. 2011;74:2039–2044. doi: 10.1021/np200532d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biggins JB, Gleber CD, Brady SF. Org. Lett. 2011;13:1536–1539. doi: 10.1021/ol200225v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Henkes LM, Doughty LB, He M, Wang D, Meyer-Almes FJ, Cheng Y-Q. J. Nat. Prod. 2011;74:2031–2038. doi: 10.1021/np200324x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng Y-Q, Yang M, Matter AM. Appl. Environ. Microbiol. 2007;73:3460–3469. doi: 10.1128/AEM.01751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furumai R, Matsuyama A, Kobashi N, Lee KH, Nishiyama M, Nakajima H, Tanaka A, Komatsu Y, Nishino N, Yoshida M, Horinouchi S. Cancer Res. 2002;62:4916–4921. [PubMed] [Google Scholar]
- 22.Wang C, Cheng Y-Q. Acta Crystallogr., Sect. E: Struct. Rep. Online. 2011;67:o2948–o2949. doi: 10.1107/S1600536811041390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.