Abstract
Research in obesity and metabolic disorders that involve intestinal microbiota demands reliable methods for the precise measurement of the short-chain fatty acids (SCFAs) and branched-chain amino acids (BCAAs) concentration. Here, we report a rapid method of simultaneously determining SCFAs and BCAAs in biological samples using propyl chloroformate (PCF) derivatization followed by gas chromatography mass spectrometry (GC-MS) analysis. A one-step derivatization using 100 µL of PCF in a reaction system of water, propanol, and pyridine (v/v/v = 8:3:2) at pH 8 provided the optimal derivatization efficiency. The best extraction efficiency of the derivatized products was achieved by a two-step extraction with hexane. The method exhibited good derivatization efficiency and recovery for a wide range of concentrations with a low limit of detection for each compound. The relative standard deviations (RSDs) of all targeted compounds showed good intra- and inter-day (within 7 days) precision (< 10%), and good stability (< 20%) within 4 days at room temperature (23–25 °C), or 7 days when stored at −20 °C. We applied our method to measure SCFA and BCAA levels in fecal samples from rats administrated with different diet. Both univariate and multivariate statistics analysis of the concentrations of these target metabolites could differentiate three groups with ethanol intervention and different oils in diet. This method was also successfully employed to determine SCFA and BCAA in the feces, plasma and urine from normal humans, providing important baseline information of the concentrations of these metabolites. This novel metabolic profile study has great potential for translational research.
Keywords: Propyl chloroformate, Short-chain fatty acids, Branched-chain amino acids, Gas chromatography/mass spectrometry, Biological samples, Targeted metabolomics
1 Introduction
The measurement of short-chain fatty acids (SCFAs) and branched-chain amino acids (BCAAs) (i.e., valine, leucine, and isoleucine) in biological samples receives considerable attention because of the important role SCFAs and BCAAs play in physiological and pathological processes (Hijova et al. 2007; Kawaguchi et al. 2011; Miller 2004; Newgard et al. 2009). SCFAs, also known as “the volatile fatty acids,” contain 2–7 carbon atoms. Straight-chain SCFAs are mainly produced by microbial fermentation of unabsorbed dietary carbohydrates in the gut (Cummings 1981) and branched-chain SCFAs such as isobutyric acid, 2-methylbutyric acid, and isovaleric acid are derived from the catabolism of BCAAs (Macfarlane et al. 2003). SCFAs influence the gut microbiota by stimulating bifidobacteria growth while inhibiting gram-negative facultative and anaerobic bacteria (Andoh et al. 2003), thereby providing important information regarding the bacterial activity in the intestinal tract. SCFAs also act as nutrients for the colonic epithelium, regulate the colonic and intracellular environment (Wong et al. 2006), and modulate cell proliferation and gene expression (Blottiere et al. 2003; Sanderson 2004). Recent studies suggest that SCFAs have a protective effect against gastrointestinal disorders, colon carcinogenesis, and cardiovascular disease (Augenlicht et al. 2002; Kles et al. 2006; Wong et al. 2006).
BCAAs are essential nutrients obtained from food, as they cannot be synthesized de novo by mammals. BCAAs have the unique quality of being poorly metabolized during their first pass through the liver, which allows them to act as a signal to the brain and the periphery organs of the amino acid content of our diet (Herman et al.). BCAAs have been clinically used as nutrition supplements for patients with liver disease or cancer (Holecek ; Muto et al. 2006), because BCAA oxidation provides energy for muscles and other organs. Emerging evidence indicates that SCFAs and BCAAs are associated with metabolic diseases such as obesity and diabetes (Vonk et al. 2011; Wang et al. 2011). In fact, SCFAs stimulate leptin expression and inhibit lipolysis in adipocytes through G-coupled protein receptors, and activate AMPK, which acts as a major cellular fuel gauge and a master regulator of metabolic homeostasis (Samuel et al. 2008). Similarly, BCAAs function as potential regulators of leptin production, glucose homeostasis, cell signaling, and adiposity; abnormal BCAA levels could be a signal for diabetes prediction (Newgard et al. 2009; Wang et al. 2011). Both SCFAs and BCAAs also play important roles in the nervous system as transmitters of neural signals (Fernstrom 2005; Jouvet et al. 2000; Kotani et al. 2009; Soret et al.).
Although a handful of studies investigated the correlation of SCFAs and BCAAs with pathophysiological states, none were able to measure SCFAs and BCAAs simultaneously in biological samples. For example, previous studies employed various techniques, such as direct infusion into capillary electrophoresis (CE) (Arellano et al. 2000; Barbas et al. 1998; Poinsot et al. 2012), high performance liquid chromatography (HPLC) (Agrafiotou et al. 2009; Albert et al. 1997; Kotani et al. 2009; Stein et al. 1992), or gas chromatography (GC) (Bachmann et al. 1979; Zhao et al. 2006). However, these methods have disadvantages of low sensitivity for SCFAs or BCAAs determination in complex biological matrix. Chemical derivatization (Heinrikson et al. 1984; Miwa et al. 1985; Miwa et al. 1987; Schiffels et al. 2011) and solid phase extraction (Horspool et al. 1991; Wang et al. 2010) prior to HPLC analysis were also used, which are time-consuming. Headspace extraction techniques in GC analysis, including purge-and-trap (Mills et al. 1999) or headspace solid-phase microextraction (HS-SPME) (Bianchi et al. 2011; Mills et al. 1999; Perez Olivero et al. 2011) were used for SCFAs analysis, however, these methods are unsuitable for BCAAs determination. To date, no method focusing on simultaneous measurement of SCFAs and BCAAs in complex biological samples has been reported. To develop the ideal method for this purpose, we used the chloroformate derivatization route invented by Dr. Petr Husek (Husek 1998), which has many advantages, such as the short reaction time (< 1 min) at room temperature and the ability to perform the derivatization in aqueous media. Alkyl chloroformates, favorable reagents for treating amino and fatty acids prior to gas chromatography have been applied in metabolomic studies (Qiu et al. 2007; Tao et al. 2008). Dr. Husek has described the use of ethyl chloroformate for plasma amino and organic acids analysis (Husek 1995) and a commercially successful reagent kit has been developed recently (Badawy et al. 2008). BCAAs can be accurately measured by these methods, however, without report the SCFAs were detected. Hence, a comprehensive investigation in chloroformate derivatization of SCFAs and BCAAs for biological samples is required.
In our pilot studies, alkyl chloroformates with different lengths of alkyl groups, including methyl-, ethyl-, propyl-, butyl-, and isobutyl-chloroformate, were compared. Methyl- and ethyl-chloroformates yielded satisfactory derivatization efficiency, but acetic acid derivatives were not separated from the solvent peak and so cannot be measured precisely. Butyl- and isobutyl-chloroformates yielded suboptimal derivatization efficiency. Propyl-chloroformate (PCF) demonstrated a broad coverage of targeted compounds with satisfactory derivatization efficiency, and was, therefore, selected as the derivatization reagent for our study.
We describe here a rapid and quantitative protocol for the simultaneous measurement of SCFA and BCAA levels in various biological matrices using propyl chloroformate derivatiztion procedures and GC-MS analysis.
2 Materials and methods
2.1 Chemicals and reagents
Certified ACS grade sodium hydroxide, sodium carbonate, and anhydrous sodium sulfate were obtained from JT Baker Co. (Phillipsburg, NJ). HPLC grade propanol (PrOH), pyridine (Py), PCF, and hexane, SCFA and BCAA standards, and propyl derivatives (propyl acetate, propyl propanoate, propyl isobutyrate, propyl butyrate, propyl isopentanoate, and propyl hexanoate) were purchased from Sigma-Aldrich (St. Louis, MO). Standard compounds were prepared in ultrapure water (Milli-Q system, Millipore, Billerica, MA) and propyl derivatives that were used to evaluate the derivatization efficiency were prepared in hexane. Stable isotopes (acetic acid-d4, propionic acid-d2, 2-methylbutyric acid-d3, butyric acid-d2, valeric acid-d9, caproic acid-d3, heptanoic acid-d7, valine-d8, and leucine-d10) were obtained from CDN Isotopes (Pointe-Claire, Quebec, Canada) for method recovery evaluation. Caproic acid-d3 was used as an internal standard (IS) for final adjustment on quantity of each compound present in the biological samples.
2.2 Sample preparation
For feces samples, the extraction procedures were performed at 4 °C to protect the volatile SCFAs. A total of 1000 µL of 0.005 M aqueous NaOH containing IS (5 µg mL−1 caproic acid-d3) was added to feces samples (50–150 mg), and the sample was homogenized for 10 min and centrifuged at 13,200 g at 4 °C for 20 min. A 500-µL aliquot of supernatant fecal water was transferred into a 10 mL Corning disposable glass centrifuge tube, and 300 µL of water was added to this aliquot. For urine and plasma samples, 300 µL of each sample and 500 µL of 0.005 M aqueous NaOH containing IS (5 µg mL−1 caproic acid-d3) were mixed in a 10-mL glass centrifuge tube.
An aliquot of 500-µL PrOH/Py mixture solvent (3:2, v/v) and 100 µL of PCF were subsequently added to the glass tube and were vortexed briefly. Then, the derivatization reaction proceeded under ultrasonication for 1 min. After derivatization, the derivatives were extracted by a two-step extraction with hexane. An aliquot of 300-µL hexane was added to the reaction mixture and the sample was vortexed for 1 min followed by centrifugation at 2,000 g for 5 min. An aliquot of 300-µL derivative extraction (upper hexane layer) was transferred to a sampling vial. The extraction procedure was then repeated by adding 200 µL of hexane instead of 300 µL hexane to the reaction mixture. Another 200-µL aliquot of derivative extraction was transferred to the sampling vial with the first extraction. An aliquot of 10-µL n-alkane series was added, serving as the retention index and quality control. Anhydrous sodium sulfate (~10 mg) was added to remove traces of water from hexane. The resultant mixture was briefly vortexed prior to GC-MS analysis.
2.3 GC-MS analysis
Analysis was performed using an Agilent 7890A gas chromatography system coupled to an Agilent 5975C inert XL EI/CI mass spectrometric detector (MSD, Agilent Technologies, Santa Clara, CA). Derivatives were separated using an HP-5ms capillary column coated with 5% phenyl-95% methylpolysiloxane (30 m × 250 µm i.d., 0.25 µm film thickness, Agilent J & W Scientific, Folsom, CA). One microliter of derivatives was injected in split mode with a ratio of 10:1, and the solvent delay time was set to 2.2 min. The initial oven temperature was held at 50°C for 2 min, ramped to 70 °C at a rate of 10 °C min−1, to 85 °C at a rate of 3 °C min−1, to 110 °C at a rate of 5 °C min−1, to 290 °C at a rate of 30 °C min−1, and finally held at 290 °C for 8 min. Helium was used as a carrier gas at a constant flow rate of 1 mL min−1 through the column. The temperatures of the front inlet, transfer line, and electron impact (EI) ion source were set at 260, 290, and 230 °C, respectively. The electron energy was −70 eV, and the mass spectral data was collected in a full scan mode (m/z 30–600).
2.4 Data processing
To align multiple chromatograms, LECO’s ChromaTOF v4.32 software (Leco Corp., St. Joseph, MO) was applied. Raw GC-MS data files were converted to NetCDF files using the Agilent’s MSD Chemstation Data Analysis Application. All of the NetCDF data files were then imported to ChromaTOF and subject to the following processing including baseline correction, smoothing, noise reduction, deconvolution, library searching, and area calculation. Compound identification was performed by comparing both MS spectra and retention times with those of standard compounds. The peak area of each derivatized SCFA or BCAA was calculated using the unique mass selected by ChromaTOF. Subsequently, the “Statistical Compare” function in ChromaTOF software was utilized for the alignment to generate a data table with all the compound information from different chromatograms. Statistical Compare utilized a mass spectral match criterion of 70% when aligning the multidimensional peak data. The data table was exported as a .csv file that included sample names, compounds, retention time (RT), quantification mass, and peak area. The quantification mass for each peak in the data set was selected from the unique mass that was most common in all matching peaks within the retention time window.
3 Results and discussion
3.1 Derivatization optimization
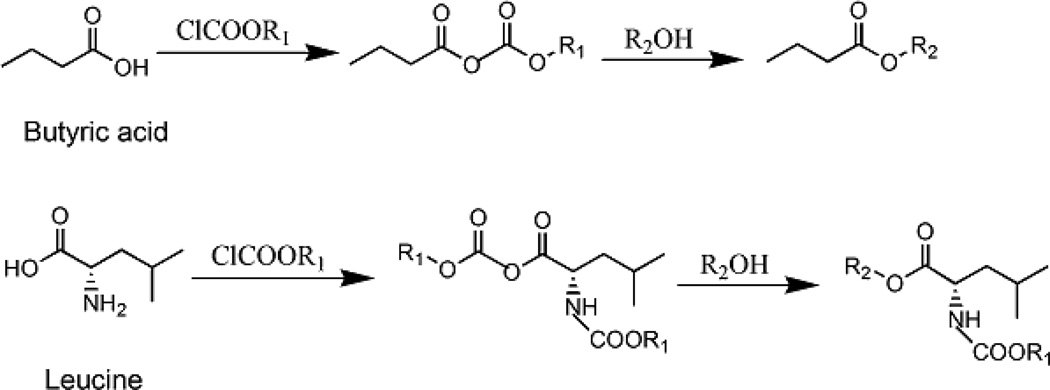
In order to develop an ideal derivatization method based on the reaction of PCF derivatization (Fig. 1), we tested several different parameters: (1) the volume and ratio of water, PrOH, Py, and the pH of the derivatization system, (2) one-step vs. two-step derivatization, and (3) various solvents and procedures for the extraction of propyl ester derivatives. Pooled sample extracts were used to avoid sample variations. Triplicate samples for each method were prepared to ensure reproducibility. Average intensities of each compound were compared across different methods. Feces samples were used for method development and validation. The method was then tailored to plasma and urine samples. Sample weight or volume and sample extraction optimization are provided in Supplementary Information, and supplementary Fig. 1.
Fig 1.
Reaction scheme of two representative compounds, butyric acid and leucine, treated with PCF derivatization.
3.1.1 Derivatization solvent system
Water, PrOH, and Py are three essential derivatization solvents, participating in the PCF-triggered derivatization reaction (Husek 1998; Qiu et al. 2007). The optimal volumes of three derivatization solvents were determined to be 300 µL of PrOH, 200 µL of Py, and 800 µL of water, based on the following two experiments:
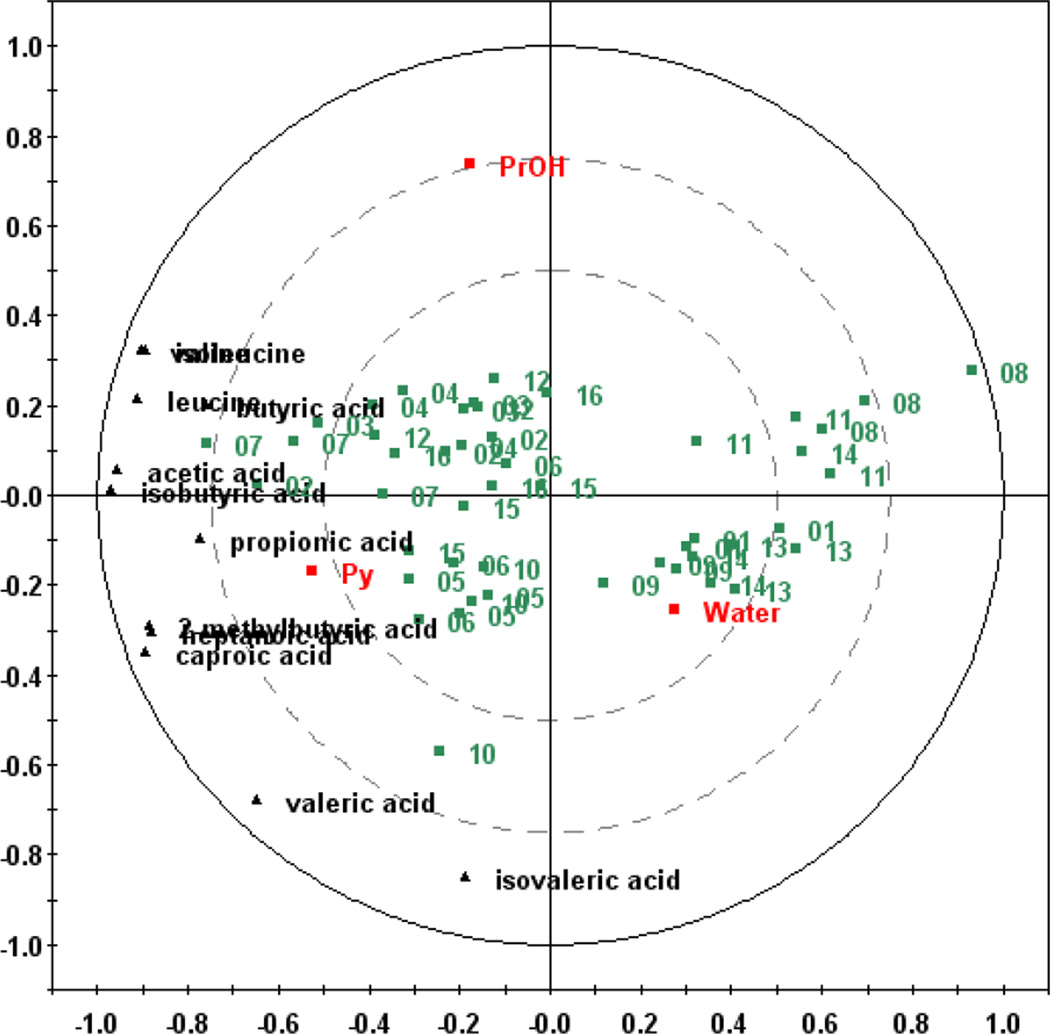
First, we applied an orthogonal design method with three factors and four levels (Table 1a). Nine SCFA and three BCAA peaks were identified and used for method evaluation. The influence of derivatization conditions on the detected signals of SCFAs and BCAAs were first summarized in Fig. 2. A multivariate statistical method, partial least squares (PLS) regression, (Fig. 2) presents the correlation between the volume of derivatization solvents and the intensities of detected compounds (Pan et al. 2010). A two-component PLS model was generated with the 12 resolved peak areas (X matrix, where each peak area is one X-variable) of the 48 analyses (triplicates of 16 methods) and the designed parameters (Y matrix, the amounts of the three derivatization solvents) with R2X, R2Y, and Q2 parameters of 0.839, 0.342, and 0.293, respectively. The PLS bi-plot shown in Fig. 2 is the combination of a two-component PLS model loadings plot and scores plot. The PLS loadings plot shows how the resolved peak areas correlate to the solvent compositions and determines the distribution of experimental analyses shown on the PLS scores plot. Fig. 2 shows that the first PLS component is mainly dominated by Py (positively) and water (negatively), and the second PLS component correlates mainly with PrOH (positively). The samples represented in the first and fourth quadrants appear to have the most negative correlation with the compounds. These samples represent all the methods using 50 µL of Py or with the ratio of water/PrOH larger than 10. These results clearly demonstrate that Py, acting as a catalyst, is beneficial to the derivatization. The results also indicate that the ratio of water to PrOH volume should be restricted. Although PLS result shows different responses of these compounds to different solvent system, the optimal method was selected as a compromise among these responses, and further investigations were carried out.
Table 1.
Experimental design for investigating the derivatization efficiency of water, propanol (PrOH), and pyridine (Py) as derivatization solvents
| a. Experimental Design 1 (Orthogonal experiment design) | ||||
|---|---|---|---|---|
| Expt No. | Water (µL) | PrOH (µL) | Py (µL) | Propyl chloroformate (µL) |
| 01 | 600 | 100 | 50 | 100 |
| 02 | 600 | 200 | 100 | 100 |
| 03 | 600 | 300 | 150 | 100 |
| 04 | 600 | 400 | 200 | 100 |
| 05 | 800 | 100 | 100 | 100 |
| 06 | 800 | 200 | 150 | 100 |
| 07 | 800 | 300 | 200 | 100 |
| 08 | 800 | 400 | 50 | 100 |
| 09 | 1000 | 100 | 150 | 100 |
| 10 | 1000 | 200 | 200 | 100 |
| 11 | 1000 | 300 | 50 | 100 |
| 12 | 1000 | 400 | 100 | 100 |
| 13 | 1200 | 100 | 200 | 100 |
| 14 | 1200 | 200 | 50 | 100 |
| 15 | 1200 | 300 | 100 | 100 |
| 16 | 1200 | 400 | 150 | 100 |
| b. Experimental Design 2 | ||||
|---|---|---|---|---|
| Expt No. | Water (µL) | PrOH (µL) | Py (µL) | Propyl chloroformate (µL) |
| 1 | 800 | 200 | 100 | 100 |
| 2 | 800 | 200 | 200 | 100 |
| 3 | 800 | 200 | 300 | 100 |
| 4 | 800 | 300 | 100 | 100 |
| 5 | 800 | 300 | 200 | 100 |
| 6 | 800 | 300 | 300 | 100 |
Fig. 2.
Optimization of the derivatization solvents. The PLS bi-plot shows the correlation between the peak areas for 12 metabolites and the volumes of three derivatization solvents: water, propanol (PrOH), and pyridine (Py). The red squares indicate derivatization solvents; the black triangles indicate detected compounds; and the green squares indicate experimental methods as numbered in Table 1a.
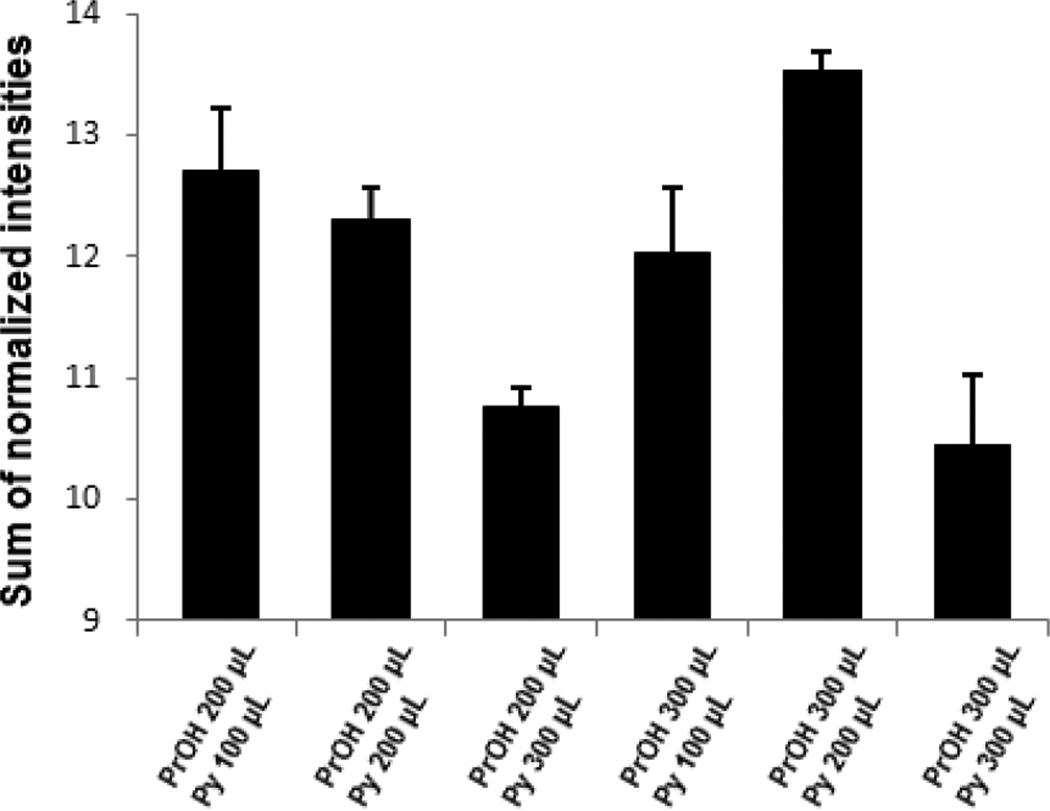
To obtain an optimal derivatization method, a second investigation was performed based on the results above. The second experiment (Table 1b) was conducted to determine the volumes of water, PrOH and Py. Fig. 3 shows the results of total normalized intensities of the metabolites, in which the intensity of each detected metabolite was normalized to the average intensity of that metabolite across all six experiments. These results indicate that Py does not increase derivatization efficiency when the volume of Py equals or is greater than the volume of PrOH. As a result, we concluded that 300 µL of PrOH, 200 µL of Py, and 800 µL of water is the optimal condition for derivatization.
Fig. 3.
Optimization of the derivatization solvents, propanol (PrOH) and pyridine (Py). The bar plots show total intensities (sum) of normalized intensity of each metabolite, which was normalized to the average intensity across the six methods. Error bars indicate the standard error of triplicate samples.
We observed that the derivatization with varying volumes of water and PrOH had a dramatic influence on the derivative extraction procedure. Importantly, increasing the ratio of PrOH to water raised the volume of the resulting hexane layer, resulting in lower final concentrations of derivatives. To eliminate this dilution, water, PrOH, and Py, were supplemented up to a constant volume after the derivatization and before extraction steps.
The optimal pH was determined to be 8.0 with aqueous NaOH solution adjustment. The results of derivatization at different pH levels were compared by evaluating the total normalized intensities of the compounds as described in the section 3.1.1. The reaction system achieved optimal derivatization efficiency in a weak base environment at pH 8.0.
3.1.2 Derivatization procedures
One-step and two-step derivatization procedures were compared, both of which have been applied in previous alkyl chloroformate derivatization methods (Husek 1995; Qiu et al. 2007). Five different procedures were conducted, including two one-step derivatizations (method a and b) and three two-step derivatizations (method c, d, and e). For one-step derivatizations, 50 µL (method a) or 100 µL (method b) of PCF was added to the reaction mixture, followed by a vortex for 10 s and ultrasonication for 1 min. A 500-µL aliquot of hexane was then added for derivative extraction. For the two-step derivatizations, each 50 µL aliquot of PCF was added to the reaction mixture at two consecutive times, followed by vortexing and ultrasonication after each derivatization step. The three different procedures were investigated to determine the pH adjustment and the derivative extraction procedure. In method c, hexane was added between the two derivatization steps. In method d, hexane was also added between the two steps, followed by the addition of 30 µL saturated aqueous NaOH to adjust the pH to 9–10 as previously reported (Qiu et al. 2007). In method e, only 30 µL of saturated aqueous NaOH was added between the two steps, and hexane was added after the two derivatization steps. The results (supplementary Fig. 2) show the total normalized intensities of the metabolites, in which the intensity of each detected metabolite was normalized to the average intensity of that metabolite across all five methods. The intensity from the one-step derivatization using 100 µL of PCF (method b) was higher than two that obtained from methods c and d, and about 5% lower than the one obtained by method e. Since one-step derivatization was markedly simple and its derivatization efficiency was acceptable, we selected method b (100 µL of PCF) for derivatization.
3.1.3 Derivative extraction solvent
Liquid-liquid extraction method was applied to extract derivatives from the reaction system. The aim of this step was not only to switch the sample solvent from aqueous to non-aqueous, but also to reduce the matrix effect and concentrate the analytes. Hexane was selected in our study. Compared with chloroform used in other reports (Husek 1995), hexane in the upper layer facilitates solvent removal and transferring, improving sample-handling efficiency and enabling large-scale automated liquid handling of clinical biological samples (Tao et al. 2008; Zheng et al. 2012). The volume of 500 µL was determined based on the results of volume evaluation. Extraction efficiency was then evaluated by carefully removing the hexane layer completely, and then adding 250 µL of hexane to repeat the extraction procedures, in order to assess the amount of the derivatives that was not extracted in the first extraction. Our results indicate that the derivatization efficiency of most compounds was about 90% (supplementary Table 1). We aimed then to develop a two-step extraction procedure to improve this extraction efficiency. First, 300 µL of hexane was added to reaction system. After 300 µL of the hexane layer was transferred to a sampling vial, another 200 µL of hexane was added to the reaction system. A second 200-µL hexane layer was transferred and combined with the first extraction. The results in supplementary Table 1 show that peak intensities were significantly increased with this method. More than half of the compounds were completely extracted and the extraction efficiency of propyl esters of acetic acid, propionic acid, isobutyric acid, butyric acid, leucine, and isoleucine improved to over 95% efficiency. However, a much lower concentration of SCFAs was observed in biofluids than in fecal samples, including some that were lower than the limit of detection (LOD) in our analytical method. Therefore, the total volume of hexane used in the method should be adjusted to 300 µL to prevent the dilution of analytes for GC-MS analysis.
3.2 Method validation
Method validation was performed for the quantitative analysis of the SCFAs and BCAAs. A stock standard mixture was prepared containing 14 SCFAs and 3 BCAAs (Table 2) at a concentration of 1 mg mL−1 for each standard. The stock solution was further diluted to a series of concentrations, ranging from 500 to 0.001 µg mL−1. The calibration curve was constructed by plotting the peak-area ratio of each standard to IS versus concentration. LOD was obtained accordingly (Table 2).
Table 2.
Linearity, LOD, and intra- and inter-day precision of 17 standards
| Compound | Calibration equationa | Linear Range (µg mL−1) |
r2 b | n | LODc | Intra-day precision RSD (%) (n = 6) |
Inter-day precision RSD (%)d (n = 12) |
|
|---|---|---|---|---|---|---|---|---|
| pg on column | Signal /noise |
|||||||
| Acetic acid | y = 0.0081x + 0.1450 | 1–500 | 0.9939 | 9 | - | - | 0.54 | 6.40 |
| Propionic acid | y = 0.0125x + 0.1951 | 0.5–250 | 0.9986 | 9 | - | - | 0.63 | 3.46 |
| Isobutyric acid | y = 0.0124x + 0.0200 | 0.2–100 | 0.9956 | 9 | 100 | 16 | 1.62 | 1.97 |
| Butyric acid | y = 0.0177x + 0.0216 | 0.5–100 | 0.9972 | 8 | 500 | 38 | 0.45 | 3.05 |
| 2-Methylbutyric acid | y = 0.0041x + 0.0045 | 0.2–50 | 0.9929 | 8 | 1000 | 18 | 5.68 | 7.41 |
| Isovaleric acid | y = 0.0079x + 0.0040 | 0.1–50 | 0.9984 | 9 | 400 | 29 | 2.98 | 4.36 |
| Valeric acid | y = 0.0171x + 0.0042 | 0.2–20 | 0.9973 | 7 | 100 | 28 | 1.33 | 3.31 |
| 2-Methylpentanoic acid | y = 0.0134x + 0.0002 | 0.025–2.5 | 0.9998 | 7 | 250 | 41 | 3.12 | 3.20 |
| 3-Methylpentanoic acid | y = 0.0195x + 0.0001 | 0.0125–1.25 | 0.9995 | 7 | 100 | 22 | 1.19 | 1.98 |
| Isocaproic acid | y = 0.0165x + 0.0001 | 0.0125–1.25 | 0.9999 | 7 | 250 | 31 | 1.06 | 1.18 |
| Caproic acid | y = 0.0048x + 0.0002 | 0.01–10 | 0.9981 | 10 | 20 | 95 | 1.34 | 3.46 |
| 2-Methylhexanoic acid | y = 0.0102x − 0.0001 | 0.0125–1.25 | 0.9999 | 7 | 50 | 16 | 5.99 | 6.67 |
| 4-Methylhexanoic acid | y = 0.0134x + 0.0001 | 0.0125–1.25 | 0.9995 | 7 | 500 | 86 | 5.33 | 5.58 |
| Heptanoic acid | y = 0.0213x + 0.0007 | 0.005–5 | 0.9969 | 10 | 50 | 21 | 2.82 | 3.89 |
| Valine | y = 0.0579x + 0.0236 | 0.25–25 | 0.9931 | 7 | 100 | 39 | 0.42 | 7.66 |
| Leucine | y = 0.0430x + 0.0186 | 0.25–25 | 0.9939 | 7 | 100 | 34 | 1.01 | 7.41 |
| Isoleucine | y = 0.0516x + 0.0022 | 0.05–5 | 0.9967 | 7 | 50 | 70 | 1.25 | 8.43 |
x: concentration (µg mL−1); Y: peak area ratio (area of each compound/area of internal standard [IS]).
r2 : Regression coefficient. Regression coefficients were calculated for linearity for the concentration ranges listed here.
LOD (pg on column) is the lowest calibration standard injected with a signal/noise ratio ≥ 3, where the signal/noise ratio calculation was carried out to display the peak-to-peak values by ChromaTOF software. The LODs of acetic acid and propionic acid were not calculated, as these two compounds could be detected in blank water samples introduced by impurity of derivatization solvents.
The RSD of the inter-day precision is calculated by the peak intensities detected in four different days with three replicates each day.
Intra- and inter-day precision studies were performed using a standard mixture of all the standards at a concentration comparable to the measured quantities in the biological samples. Intra-day precision was determined by successive replicate measurements (n = 6) of the analytes. Inter-day precision was determined by analyzing the samples on four different days (days 1, 2, 4, and 7). The results in Table 2 exhibited excellent intra-day and inter-day precision for most of the compounds with the RSDs below 6% and 9%, respectively.
Stability studies for the derivatized analytes were performed using biological sample matrix under three different storage conditions, i.e., room temperature (23–25 °C), −20 °C, and a 12-h room temperature/12 h −20 °C cycle on days 1, 2, 4, and 7. Prepared samples were separated into four aliquots and stored prior to analysis. Caproic acid, heptanoic acid, and BCAAs showed good stability within seven days under all three storage conditions (supplementary Fig. 4). The rest of compounds showed good stability within seven days at −20 °C, and within four days under the other two conditions.
To determine the derivatization efficiency, standard mixtures were prepared in three different concentrations and processed using our method. In parallel, blank pure water instead of standard mixtures was treated with the same procedures, except the derivative extraction step. Instead of pure hexane, hexane dissolved with commercial available propyl esters was added. Moles of propyl esters and standards in the standard mixtures were equal between the parallel samples, so that the peak areas of the added propyl esters could be used as a benchmark, assuming 100% derivatization efficiency. Derivatization efficiency (supplementary Table 3) showed satisfactory results for most SCFAs and BCAAs ranging from 90.06 to 116.34%.
For recovery studies, biological samples were spiked with three levels of analyte isotopes (low, medium, and high). The medium concentration level was consistent with the analyte concentration in biological samples, and the low and high levels were 50% and 200% of the medium level, respectively. The recovery was calculated according to the ratio of the measured concentration of the analyte isotope versus the spiked isotope concentration. All isotopes exhibited high recovery rates, ranging from 84.11 to 118.79% (supplementary Table 3).
3.3 Method application - SCFA and BCAA profiling
To demonstrate the utility of our procedure, this developed method was used to analyze complex biological samples, including feces, plasma, and urine from animal and human subjects. One application aimed to differentiate the concentrations of SCFAs and BCAAs in feces of Sprague-Dawley male rats (16–18 weeks) from three different feeding groups (n = 8 in each group): 1) a “corn oil group” that ingested 35% corn oil in their feed, 2) a “corn oil/ethanol” group that ingested feed containing 35% corn oil and 7% ethanol, and 3) a “medium chain fatty acid (MCFAs)/ethanol” group that ingested feed containing 35% MCFAs and 7% ethanol. Feces were collected after a 15-week dietary intervention. In another experiment, feces, urine and plasma samples were collected from 15 healthy female volunteers, who had maintained control diets for ten days to minimize the dietary-induced metabolism variations. In both rat and human feces samples, 12 metabolites including 9 SCFAs and 3 BCAAs were identified and quantified, while in plasma and urine samples, two of these SCFAs, namely 2-methylbutyric and isovaleric acid, were not detectable (Tables 3). The concentrations of three SCFAs, namely acetic, propanoic, and butyric acid, were predominant in feces samples. In animal experiments, there was an apparent metabolic discrimination between the two groups administered with ethanol and the group fed corn oil only (supplementary Fig. 5). In the groups administrated with ethanol, acetic acid was significantly elevated, consistent with previous reports which indicate that ethanol is oxidized to acetaldehyde and subsequently oxidized by the colonic mucosal or bacterial aldehyde dehydrogenase to acetic acid. This oxidative reaction in vivo showed protective effect against ethanol-induced damage (Israel et al. 1994). The concentration of BCAAs was lower in the ethanol-intake groups, especially in the MCFA/ethanol group. In fact, previously ethanol-fed animals were shown to have more subcutaneous fat and less tissue fat such as perinephric fat compared to animals fed without ethanol. Previous studies reported that BCAAs could be markers for the fat accumulation around organs, even in animals that exhibit overall low body weight (Newgard et al. 2009). Our results with human samples provide important baseline information for the SCFA and BCAA concentrations in human feces, urine and plasma.
Table 3.
Quantitative analysis of SCFAs and BCAAs obtained from rat feces from three groups and different biological samples from humans
| Compounds | Retention time (min) |
Quantitative Mass |
Group Aa (µmol g−1 feces)d (n=8) |
Group Bb (µmol g−1 feces)d (n=8) |
Group Cc (µmol g−1 feces)d (n=8) |
Human Feces (µmol g−1)d (n=15) |
Human Plasma (µM)d (n=15) |
Human Urine (µM)d (n=15) |
|---|---|---|---|---|---|---|---|---|
| Acetic acid | 2.86 | 61 | 30.4 ± 3.46 | 37.1 ± 6.01 | 45.8 ± 13.8 | 35.86 ± 16.8 | 341.1 ± 59.6 | 82.89 ± 60.0 |
| Propionic acid | 4.40 | 75 | 6.80 ± 0.91 | 4.05 ± 0.42 | 5.02 ± 1.5 | 11.40 ± 4.74 | 255.7 ± 60.1 | 108.2 ± 78.1 |
| Isobutyric acid | 5.42 | 89 | 1.30 ± 0.36 | 0.157 ± 0.044 | 0.151 ± 0.11 | 1.71 ± 1.05 | 7.83 ± 0.771 | 0.196 ± 0.103 |
| Butyric acid | 6.45 | 71 | 6.27 ± 3.7 | 3.74 ± 0.58 | 5.90 ± 3.1 | 6.35 ± 3.13 | 1.47 ± 1.13 | 2.98 ± 1.88 |
| 2-Methylbutyric acid | 7.78 | 74 | 2.51 ± 1.5 | 0.623 ± 0.22 | 0.504 ± 0.20 | 1.15 ± 0.88 | nd | nd |
| Isovaleric acid | 7.86 | 61 | 1.13 ± 0.17 | 0.232 ± 0.040 | 0.155 ± 0.086 | 1.28 ± 0.92 | nd | nd |
| Valeric acid | 9.35 | 103 | 0.98 ± 0.20 | 0.31 ± 0.050 | 0.34 ± 0.094 | 1.25 ± 0.52 | 1.29 ± 0.46 | 0.224 ± 0.15 |
| Caproic acid | 12.11 | 99 | 3.89 ± 1.4 | 4.02 ± 1.8 | 3.79 ± 0.89 | 0.0744 ± 0.094 | 4.71 ± 1.1 | 0.969 ± 0.53 |
| Heptanoic acid | 13.87 | 113 | 0.14 ± 0.046 | 0.093 ± 0.082 | 0.070 ± 0.023 | 0.0519 ± 0.082 | 1.94 ± 0.61 | 0.232 ± 0.17 |
| Valine | 16.79 | 158 | 1.64 ± 0.31 | 0.465 ± 0.11 | 0.262 ± 0.11 | 2.023 ± 0.842 | 166.2 ± 17.9 | 26.60 ± 8.01 |
| Leucine | 17.14 | 172 | 4.11 ± 0.67 | 1.41 ± 0.21 | 0.929 ± 0.30 | 2.076 ± 0.837 | 95.08 ± 17.0 | 36.76 ± 6.03 |
| Isoleucine | 17.23 | 172 | 1.34 ± 0.32 | 0.451 ± 0.079 | 0.287 ± 0.10 | 1.47 ± 0.62 | 48.26 ± 7.5 | 9.77 ± 3.8 |
Group A was administrated corn oil.
Group B was administrated corn oil and ethanol.
Group C was administrated MCFAs and ethanol.
Values are reported as means ± standard deviation
nd: not determined
4 Concluding remarks
In this study, we describe a rapid and quantitative protocol for the simultaneous analysis of SCFAs and BCAAs in biological samples using PCF derivatization and GC-MS analysis. Metabolites were derivatized with 100 µL of PCF in a reaction system of water/PrOH/Py (v/v/v = 8:3:2) at pH 8.0 prior to a two-step extraction with hexane. Using our novel method, we analyzed complex biological samples including feces, urine and plasma. Based on its accuracy, simplicity and robustness, our method is an essential tool in clinical applications as well as in translational biomedical research.
Supplementary Material
Acknowledgements
This work was financially supported by the NIH grants 1R01AA020212-01 and 3P30DK056350-10.
Abbreviations
- SCFAs
short-chain fatty acids
- BCAAs
branched-chain amino acids
- PCF
propyl chloroformate
- GC-MS
gas chromatography mass spectrometry
- R.S.D.s
relative standard deviations
- CE
capillary electrophoresis
- HPLC
high performance liquid chromatography
- GC
gas chromatography
- HS-SPME
headspace solid-phase microextraction
- PrOH
propanol
- Py
pyridine
- IS
internal standard
- RT
retention time
- PLS
partial least squares regression
- LOD
limit of detection
- MCFAs
medium chain fatty acids
Contributor Information
Xiaojiao Zheng, Center for Translational Medicine, and Shanghai Key Laboratory of Diabetes Mellitus, Department of Endocrinology and Metabolism, Shanghai Jiao Tong University Affiliated Sixth People’s Hospital, Shanghai 200233, China; School of Pharmacy, Shanghai Jiao Tong University, Shanghai 200240, China.
Yunping Qiu, Center for Translational Biomedical Research, University of North Carolina at Greensboro, North Carolina Research Campus, Kannapolis, North Carolina 28081, USA.
Wei Zhong, Center for Translational Biomedical Research, University of North Carolina at Greensboro, North Carolina Research Campus, Kannapolis, North Carolina 28081, USA.
Sarah Baxter, David H. Murdock Research Institute, North Carolina Research Campus, Kannapolis, North Carolina 28081, USA.
Mingming Su, David H. Murdock Research Institute, North Carolina Research Campus, Kannapolis, North Carolina 28081, USA.
Qiong Li, Center for Translational Biomedical Research, University of North Carolina at Greensboro, North Carolina Research Campus, Kannapolis, North Carolina 28081, USA.
Guoxiang Xie, Center for Translational Biomedical Research, University of North Carolina at Greensboro, North Carolina Research Campus, Kannapolis, North Carolina 28081, USA.
Brandon M. Ore, David H. Murdock Research Institute, North Carolina Research Campus, Kannapolis, North Carolina 28081, USA
Shanlei Qiao, Center for Translational Biomedical Research, University of North Carolina at Greensboro, North Carolina Research Campus, Kannapolis, North Carolina 28081, USA.
Melanie D. Spencer, UNC Nutrition Research Institute, University of North Carolina, Kannapolis, North Carolina 28081, USA
Steven H. Zeisel, UNC Nutrition Research Institute, University of North Carolina, Kannapolis, North Carolina 28081, USA
Zhanxiang Zhou, Center for Translational Biomedical Research, University of North Carolina at Greensboro, North Carolina Research Campus, Kannapolis, North Carolina 28081, USA.
Aihua Zhao, Center for Translational Medicine, and Shanghai Key Laboratory of Diabetes Mellitus, Department of Endocrinology and Metabolism, Shanghai Jiao Tong University Affiliated Sixth People’s Hospital, Shanghai 200233, China.
Wei Jia, Center for Translational Medicine, and Shanghai Key Laboratory of Diabetes Mellitus, Department of Endocrinology and Metabolism, Shanghai Jiao Tong University Affiliated Sixth People’s Hospital, Shanghai 200233, China; Center for Translational Biomedical Research, University of North Carolina at Greensboro, North Carolina Research Campus, Kannapolis, North Carolina 28081, USA.
References
- Agrafiotou P, Sotiropoulos S, Pappa-Louisi A. Direct RP-HPLC determination of underivatized amino acids with online dual UV absorbance, fluorescence, and multiple electrochemical detection. Journal of Separation Science. 2009;32:949–954. doi: 10.1002/jssc.200800636. [DOI] [PubMed] [Google Scholar]
- Albert DB, Martens CS. Determination of low-molecular-weight organic acid concentrations in seawater and pore-water samples via HPLC. Marine Chemistry. 1997;56:27–37. [Google Scholar]
- Andoh A, Tsujikawa T, Fujiyama Y. Role of dietary fiber and short-chain fatty acids in the colon. Current Pharmaceutical Design. 2003;9:347–358. doi: 10.2174/1381612033391973. [DOI] [PubMed] [Google Scholar]
- Arellano M, Jomard P, El Kaddouri S, Roques C, Nepveu F, Couderc F. Routine analysis of short-chain fatty acids for anaerobic bacteria identification using capillary electrophoresis and indirect ultraviolet detection. Journal of Chromatography B. 2000;741:89–100. doi: 10.1016/s0378-4347(00)00066-9. [DOI] [PubMed] [Google Scholar]
- Augenlicht LH, Mariadason JM, Wilson A, Arango D, Yang WC, Heerdt BG, et al. Short chain fatty acids and colon cancer. Journal of Nutrition. 2002;132:3804S–3808S. doi: 10.1093/jn/132.12.3804S. [DOI] [PubMed] [Google Scholar]
- Bachmann C, Colombo JP, Beruter J. Short chian fatty-acids in plasma and brain - quantitative - determination by gas-chromatography. Clinica Chimica Acta. 1979;92:153–159. doi: 10.1016/0009-8981(79)90109-8. [DOI] [PubMed] [Google Scholar]
- Badawy AAB, Morgan CJ, Turner JA. Application of the Phenomenex EZ : faast (TM) amino acid analysis kit for rapid gas-chromatographic determination of concentrations of plasma tryptophan and its brain uptake competitors. Amino Acids. 2008;34:587–596. doi: 10.1007/s00726-007-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas C, Adeva N, Aguilar R, Rosillo M, Rubio T, Castro M. Quantitative determination of short-chain organic acids in urine by capillary electrophoresis. Clinical Chemistry. 1998;44:1340–1342. [PubMed] [Google Scholar]
- Bianchi F, Dall'Asta M, Del Rio D, Mangia A, Musci M, Scazzina F. Development of a headspace solid-phase microextraction gas chromatography-mass spectrometric method for the determination of short-chain fatty acids from intestinal fermentation. Food Chemistry. 2011;129:200–205. [Google Scholar]
- Blottiere HM, Buecher B, Galmiche JP, Cherbut C. Molecular analysis of the effect of short-chain fatty acids on intestinal cell proliferation. Proceedings of the Nutrition Society. 2003;62:101–106. doi: 10.1079/PNS2002215. [DOI] [PubMed] [Google Scholar]
- Cummings JH. Short chain fatty acids in the human colon. Gut. 1981;22:763–779. doi: 10.1136/gut.22.9.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernstrom JD. Branched-chain amino acids and brain function. Journal of Nutrition. 2005;135:1539S–1546S. doi: 10.1093/jn/135.6.1539S. [DOI] [PubMed] [Google Scholar]
- Heinrikson RL, Meredith SC. Amino-acid-analysis by reverse-phase high-performance liquid-chromatography - precolumn derivatization with phenylisothiocyanate. Analytical Biochemistry. 1984;136:65–74. doi: 10.1016/0003-2697(84)90307-5. [DOI] [PubMed] [Google Scholar]
- Herman MA, She PX, Peroni OD, Lynch CJ, Kahn BB. Adipose tissue branched chain amino acid (BCAA) metabolism modulates circulating BCAA levels. Journal of Biological Chemistry. 2010;285:11348–11356. doi: 10.1074/jbc.M109.075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijova E, Chmelarova A. Short chain fatty acids and colonic health. Bratislava Medical Journal-Bratislavske Lekarske Listy. 2007;108:354–358. [PubMed] [Google Scholar]
- Holecek M. Three targets of branched-chain amino acid supplementation in the treatment of liver disease. Nutrition. 2010;26:482–490. doi: 10.1016/j.nut.2009.06.027. [DOI] [PubMed] [Google Scholar]
- Horspool LJI, McKellar QA. Determination of short-chain fatty acids in equine cecal liquor by ion-exchange high-performance liquid-chromatography after solid-phase extraction. Biomedical Chromatography. 1991;5:202–206. doi: 10.1002/bmc.1130050505. [DOI] [PubMed] [Google Scholar]
- Husek P. Simultaneous profile analysis of plasma amino and organic-acids by capillary gas-chromatography. Journal of Chromatography B-Biomedical Applications. 1995;669:352–357. doi: 10.1016/0378-4347(95)00115-y. [DOI] [PubMed] [Google Scholar]
- Husek P. Chloroformates in gas chromatography as general purpose derivatizing agents. Journal of Chromatography B. 1998;717:57–91. [PubMed] [Google Scholar]
- Israel Y, Orrego H, Carmichael FJ. Acetate-mediated effects of ethanol. Alcoholism-Clinical and Experimental Research. 1994;18:144–148. doi: 10.1111/j.1530-0277.1994.tb00894.x. [DOI] [PubMed] [Google Scholar]
- Jouvet P, Rustin P, Taylor DL, Pocock JM, Felderhoff-Mueser U, Mazarakis ND, et al. Branched chain amino acids induce apoptosis in neural cells without mitochondrial membrane depolarization or cytochrome c release: Implications for neurological impairment associated with maple syrup urine disease. Molecular Biology of the Cell. 2000;11:1919–1932. doi: 10.1091/mbc.11.5.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T, Izumi N, Charlton MR, Sata M. Branched-chain amino acids as pharmacological nutrients in chronic liver disease. Hepatology. 2011;54:1063–1070. doi: 10.1002/hep.24412. [DOI] [PubMed] [Google Scholar]
- Kles KA, Chang EB. Short-chain fatty acids impact on intestinal adaptation inflammation, carcinoma, and failure. Gastroenterology. 2006;130:S100–S105. doi: 10.1053/j.gastro.2005.11.048. [DOI] [PubMed] [Google Scholar]
- Kotani A, Miyaguchi Y, Kohama M, Ohtsuka T, Shiratori T, Kusu F. Determination of short-chain fatty acids in rat and human feces by high-performance liquid chromatography with electrochemical detection. Analytical Sciences. 2009;25:1007–1011. doi: 10.2116/analsci.25.1007. [DOI] [PubMed] [Google Scholar]
- Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proceedings of the Nutrition Society. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- Miller SJ. Cellular and physiological effects of short-chain fatty acids. Mini-Reviews in Medicinal Chemistry. 2004;4:839–845. doi: 10.2174/1389557043403288. [DOI] [PubMed] [Google Scholar]
- Mills GA, Walker V, Mughal H. Headspace solid-phase microextraction with 1-pyrenyldiazomethane in-fibre derivatisation for analysis of faecal short-chain fatty acids. Journal of Chromatography B. 1999;730:113–122. doi: 10.1016/s0378-4347(99)00211-x. [DOI] [PubMed] [Google Scholar]
- Miwa H, Hiyama C, Yamamoto M. High-performance liquid-chromatography of short-chain and long-chain fatty-acids as 2-nitrophenylhydrazides. Journal of Chromatography. 1985;321:165–174. [Google Scholar]
- Miwa H, Yamamoto M. High-performance liquid-chromatographic analysis of serum short-chain fatty-acids by direct derivatization. Journal of Chromatography-Biomedical Applications. 1987;421:33–41. doi: 10.1016/0378-4347(87)80376-6. [DOI] [PubMed] [Google Scholar]
- Muto Y, Sato S, Watanabe A, Moriwaki H, Suzuki K, Kato A, et al. Overweight and obesity increase the risk for liver cancer in patients with liver cirrhosis and long-term oral supplementation with branched-chain amino acid granules inhibits liver carcinogenesis in heavier patients with liver cirrhosis. Hepatology Research. 2006;35:204–214. doi: 10.1016/j.hepres.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metabolism. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Qiu Y, Chen T, Lin J, Chi Y, Su M, et al. An optimized procedure for metabonomic analysis of rat liver tissue using gas chromatography/time-of-flight mass spectrometry. Journal of Pharmaceutical and Biomedical Analysis. 2010;52:589–596. doi: 10.1016/j.jpba.2010.01.046. [DOI] [PubMed] [Google Scholar]
- Perez Olivero SJ, Perez Trujillo JP. A new method for the determination of short-chain fatty acids from the aliphatic series in wines by headspace solid-phase microextraction-gas chromatography-ion trap mass spectrometry. Analytica Chimica Acta. 2011;696:59–66. doi: 10.1016/j.aca.2011.03.063. [DOI] [PubMed] [Google Scholar]
- Poinsot V, Carpene MA, Bouajila J, Gavard P, Feurer B, Couderc F. Recent advances in amino acid analysis by capillary electrophoresis. Electrophoresis. 2012;33:14–35. doi: 10.1002/elps.201100360. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Su M, Liu Y, Chen M, Gu J, Zhang J, et al. Application of ethyl chloroformate derivatization for gas chromatography-mass spectrometry based metabonomic profiling. Analytica Chimica Acta. 2007;583:277–283. doi: 10.1016/j.aca.2006.10.025. [DOI] [PubMed] [Google Scholar]
- Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson IR. Short chain fatty acid regulation of signaling genes expressed by the intestinal epithelium. Journal of Nutrition. 2004;134:2450S–2454S. doi: 10.1093/jn/134.9.2450S. [DOI] [PubMed] [Google Scholar]
- Schiffels J, Baumann MEM, Selmer T. Facile analysis of short-chain fatty acids as 4-nitrophenyl esters in complex anaerobic fermentation samples by high performance liquid chromatography. Journal of Chromatography A. 2011;1218:5848–5851. doi: 10.1016/j.chroma.2011.06.093. [DOI] [PubMed] [Google Scholar]
- Soret R, Chevalier J, De Coppet P, Poupeau G, Derkinderen P, Segain JP, et al. Short-chain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. Gastroenterology. 2010;138 doi: 10.1053/j.gastro.2010.01.053. 1772-U49. [DOI] [PubMed] [Google Scholar]
- Stein J, Kulemeier J, Lembcke B, Caspary WF. Simple and rapid method for determination of short-chain fatty-acids in biological-material is by high-performance liquid-chromatography with ultraviolet detection. Journal of Chromatography-Biomedical Applications. 1992;576:53–61. doi: 10.1016/0378-4347(92)80174-o. [DOI] [PubMed] [Google Scholar]
- Tao X, Liu Y, Wang Y, Qiu Y, Lin J, Zhao A, et al. GC-MS with ethyl chloroformate derivatization for comprehensive analysis of metabolites in serum and its application to human uremia. Analytical and Bioanalytical Chemistry. 2008;391:2881–2889. doi: 10.1007/s00216-008-2220-8. [DOI] [PubMed] [Google Scholar]
- Vonk RJ, Priebe M, Meijer K, Venema K, Rnelofsen H. The interaction of short-chain fatty acids (SCFA) with adipose tissue; relevance for systemic inflammation. Gastroenterology. 2011;140:S860–S860. [Google Scholar]
- Wang L, Xu R, Hu B, Li W, Sun Y, Tu Y, et al. Analysis of free amino acids in Chinese teas and flower of tea plant by high performance liquid chromatography combined with solid-phase extraction. Food Chemistry. 2010;123:1259–1266. [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nature Medicine. 2011;17:U448–U483. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JMW, de Souza R, Kendall CWC, Emam A, Jenkins DJA. Colonic health: Fermentation and short chain fatty acids. Journal of Clinical Gastroenterology. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- Zhao GH, Nyman M, Jonsson JA. Rapid determination of short-chain fatty acids in colonic contents and faeces of humans and rats by acidified water-extraction and direct-injection gas chromatography. Biomedical Chromatography. 2006;20:674–682. doi: 10.1002/bmc.580. [DOI] [PubMed] [Google Scholar]
- Zheng X, Su M, Qiu Y, Jia W. Response to letter to the editor regarding "GC-MS with ethyl chloroformate derivatization for comprehensive analysis of metabolites in serum and its application to human uremia". Analytical and Bioannalytical Chemistry. 2012 doi: 10.1007/s00216-008-2220-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.