Abstract
Patients with inflammatory bowel disease (IBD) are at increased risk for colorectal cancer (CRC). Despite weak supporting evidence, important logistic barriers and high cost, colonoscopy is currently the only recommended approach to CRC surveillance in patients with IBD. As such, there is imperative to explore alternative or complementary strategies with potential to improve the efficiency and effectiveness of surveillance in IBD. Given our increasing understanding of tumorigenesis in IBD and the accompanying cascade of molecular alterations, there is a strong rationale to pursue biomarker assays for this application. Stool-based DNA testing with advanced technology has been shown to be highly discriminatory for detection of sporadic colorectal cancer and advanced precancers. In early observations, stool DNA testing also shows promise for the accurate detection of IBD-associated colorectal neoplasms. These findings raise important clinical and translational questions about how to best evaluate and develop this technology, and devise clinical algorithms that will complement colonoscopy to improve patient outcomes.
Keywords: colorectal neoplasms, DNA methylation, early detection of cancer, inflammatory bowel disease, stool DNA
Introduction
Colorectal cancer (CRC) incidence and overall mortality are higher among those with inflammatory bowel disease (IBD) than in the general population [Herrinton et al. 2012]. Although data supporting its efficacy are limited, colonoscopy remains the only strategy widely used for CRC surveillance in IBD. Colonoscopic surveillance has been particularly advocated for IBD subsets at greatest risk for CRC. Factors known to increase CRC risk in IBD include duration and extent of chronic ulcerative colitis (CUC) or Crohn’s colitis (CD), degree of histological activity, family history of CRC and the presence of primary sclerosing cholangitic (PSC) [Nuako et al. 1998; Itzkowitz and Harpaz, 2004; Cairns et al. 2010; NICE, 2011]. Those with PSC-IBD are at especially high risk of colorectal neoplasia (CRN, CRC + dysplasia) [Jess et al. 2012b] and at >100-fold increased cholangiocarcinoma risk [Bergquist et al. 2002], leading to a >40% mortality rate from hepatic and extrahepatic malignancies [Bergquist et al. 2002]. In the United States, current guidelines recommend that patients undergo every other year colonoscopy after 8–10 years of chronic colitis extending above the rectum and annual colonoscopy for patients with chronic colitis and PSC [Farraye et al. 2010]. British guidelines recommend a colonoscopy 10 years after symptom onset and base subsequent surveillance intervals on inflammation severity and extent, PSC history and family history [NICE, 2011].
Justification for colonoscopic surveillance in IBD is based on soft evidence. While no randomized, controlled trials have been performed, there are several case series that suggest benefit [Nugent et al. 1991; Jonsson et al. 1994]. Case-control studies have demonstrated apparent improvement in overall survival [Lashner et al. 1990; Choi et al. 1993; Karlen et al. 1998] and time to colectomy [Lashner et al. 1990]. However, a Cochrane systematic review pooling these data did not demonstrate a benefit in CRC-related mortality, with 8 of 110 patients under surveillance and 13 of 117 patients not under surveillance meeting that endpoint [relative risk (RR), 0.81; 95% confidence interval (CI), 0.17–3.83] [Collins et al. 2006].
The same study concluded that there may be indirect evidence of cost-effectiveness to surveillance colonoscopy in CUC [Collins et al. 2006] despite the considerable resources consumed [Jonsson et al. 1994]. On the assumption that surveillance increases life expectancy, surveillance colonoscopy was modeled to be effective and cost-competitive compared with other commonly accepted practices such as screening for breast and cervical cancers [Provenzale et al. 1998]. However, such modeling appears to require a very high threshold for cumulative cancer incidence in at-risk IBD patients where the lifetime cancer rate must exceed 25% in order to be cost-effective [Delco and Sonnenburg, 2000]. While estimates vary, that threshold is not supported by current data [Herrinton et al. 2012], especially at the population level [Jess et al. 2012a].
The effectiveness of any screening or surveillance regimen is the product of test sensitivity, access to testing and patient compliance. Surveillance colonoscopy in IBD is no exception [Connell et al. 1994]. However, loss to follow up is assumed to be low or not subjected to sensitivity analysis in several cost-effectiveness models [Provenzale et al. 1998; Sonnenberg et al. 2000; Rubenstein et al. 2009]. This assumption is questioned by recent data showing less than 25% colonoscopy compliance among ulcerative colitis patients within a 2-year window, even among patients with high access [Velayos et al. 2010].
As colonoscopy itself is typically used as a gold standard test, we do not know the absolute sensitivity and specificity for detection of cancer or dysplasia by surveillance colonoscopy with biopsies [Delco and Sonnenburg, 2000]. Because of sobering accounts of endoscopically missed cancers [Lim et al. 2003; Kisiel et al. 2013], and because precancerous dysplasia can be treated endoscopically [Rubin et al. 1999; Odze et al. 2004; Kisiel et al. 2012a], significant effort has been made to improve the diagnostic yield of colonoscopy. Image-enhancing techniques such as chromoendoscopy may identify more dysplastic lesions by targeted rather than random biopsies, but require special training and sometimes extended endoscopy time. In addition, a recent meta-analysis of chromoendoscopy studies showed only a 7% increased diagnostic yield over white light implying that more than 14 of these tests would need to be performed to detect one dysplastic lesion over the use of white light colonoscopy alone [Subramanian et al. 2011].
Understanding the need for improved program sensitivity and compliance, what additional qualities would be found in the ideal screening or surveillance test? Patient-friendly features should include noninvasiveness and avoidance of cathartic preparation; an ideal test should allow off-site sample collection with no lost work time and no diet or medication restrictions. All of these are features of stool DNA (sDNA) testing, a technology with high analytic sensitivity and the potential for enhanced user rates. In screening average risk populations for sporadic CRC, sDNA testing was incorporated into recent practice guidelines [Levin et al. 2008; Rex et al. 2009] and technical advances have since significantly improved overall test performance. Until recently, sDNA had not been studied in the detection of IBD-associated CRN (IBD-CRN). Early feasibility studies are promising [Kisiel et al. 2013], suggesting that sDNA has the potential to overcome current barriers to surveillance for IBD-CRN.
Stool DNA: technology in evolution
The most important technical challenge of sDNA testing is the detection of trace amounts of methylated or mutated human DNA within an ocean of nontarget DNA. Of the total DNA in stool, only 0.01% is human and only 0.5% of those copies may be mutant when exfoliated from a target lesion [Zou et al. 2009]. First-generation sDNA tests were hampered by analytical insensitivity, as they could only detect a 1% mutant to wildtype ratio [Ahlquist et al. 2008] and were performed without stabilizing buffer, which has been shown to prevent bacterial degradation of DNA [Olson et al. 2005; Zou et al. 2006]. Analytical sensitivity is also improved by techniques which enrich target gene sequences by selective capture from stool prior to polymerase chain reaction (PCR) [Zou et al. 2008].
Test performance has been greatly enhanced by a better understanding of the genetic heterogeneity of CRN; a representative panel of markers is necessary for adequate lesion detection. However, markers in the first-generation PreGenPlus test, which included 21 DNA mutations, long-fragment DNA and BAT-26, were only found in 67% of the tissues from screen relevant neoplasms [Ahlquist et al. 2008]. In contrast, selected markers of aberrant DNA methylation are more broadly informative with as few as four markers achieving 100% sensitivity and 100% specificity in tissues [Zou H, 2010]. When combining DNA methylation and mutation markers with advances in stool assay technology, sDNA assay is now remarkably accurate for detection of sporadic CRC and large adenomas (Table 1).
Table 1.
Next-generation stool DNA test performance in case-control studies.
| Sensitivity (%) |
Specificity (%) | ||
|---|---|---|---|
| CRC | Adenoma >1 cm | ||
| Prototypes | |||
| Ahlquist et al. [2012b] | 85 | 64 | 90 |
| Ahlquist et al. [2012a] | 91 | 82 | 91 |
| Optimized assay | |||
| Lidgard et al. [2012a] | 98 | 64 | 91 |
| Lidgard et al. [2012b] | 98 | 60 | 90 |
CRC, colorectal cancer
The complex biology of IBD-CRN
With such substantial gains in sporadic CRN detection, is there sufficient biologic plausibility to justify sDNA in the diagnosis IBD-CRN? Despite important phenotypic and genetic differences described between sporadic and IBD-related CRN, the epigenetic markers of tumorigenesis appear to have substantial overlap. These similarities and differences deserve further review.
In sporadic CRN, clinical and histopathological data suggest that malignant carcinoma arises from clonal expansion of benign adenoma precursors [Fearon and Vogelstein, 1987]. Genetic alterations seen in adenomas and carcinomas have been used to develop models of colorectal tumorigenesis [Fearon and Vogelstein, 1990]. The concept of chromosomal instability involves allelic deletion or loss of chromosomal segments (aneuploidy) leading to a loss of heterozygosity (LOH) of key tumor suppressor genes. Deletions of large segments of 17p or 18q have been described in 70–75% of sporadic CRC [Vogelstein et al. 1988]. Further deletion or mutation in additional tumor suppressor gene alleles such as APC [Vogelstein et al. 1988] and p53 [Finlay et al. 1989] leads to loss of cell cycle checkpoints. Lastly, mutant oncogenes directly stimulate cellular processes important for neoplastic progression, including cell growth, proliferation, migration and angiogenesis; for CRC the most important oncogenes are mutant KRAS and to a lesser extent mutant BRAF, which are mutually exclusive in activation of the epidermal growth factor receptor signaling pathway [Berg and Soreide, 2012].
We are also coming to appreciate how epigenetic changes to the genome influence colorectal tumor biology [Goel and Boland, 2012]. Phosphodiester-bound cytosine–guanine dinucleotides (CpGs) occur in clusters, or ‘islands,’ throughout the genome [Gardiner-Garden and Frommer, 1987], but often in gene regulatory elements. When methyl- groups are covalently bound to cytosines in the CpG islands, gene expression can be silenced without mutation [Graff et al. 1997; Herman et al. 1998; Miyakura et al. 2001]. While promotor hypermethylation is strongly linked with aging, there are also cancer-specific methylation events [Toyota et al. 1999]. Promotor methylation is also strongly associated with mutation events in key oncogenes and tumor suppressors [Suehiro et al. 2008], but the exact mechanism of a possible interaction is presently unclear.
While all of these phenomena have been described in IBD-CRN, several clinical observations have led investigators to search for IBD-specific molecular pathways of tumorigenesis. Classically, IBD-CRN is thought to have several key phenotypic differences from sporadic CRC. Studies reviewing pathology specimens concluded that IBD-CRC was more likely to be synchronous and present with mucinous or signet ring histology [Macdermott, 1985]. Because young age of colitis onset may be an independent risk factor for IBD-CRN [Eaden et al. 2001], IBD-CRCs are also thought to occur at an earlier age than sporadic CRC.
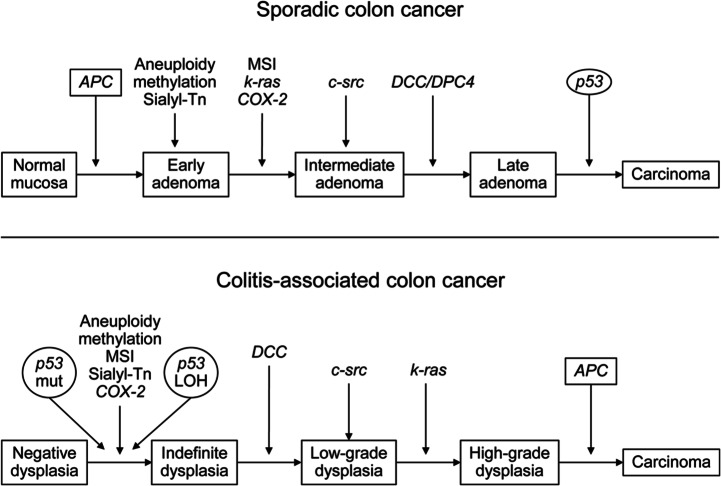
Figure 1 summarizes the literature in support of a separate pathway(s) for IBD-CRN compared with sporadic CRN. While allelic loss of chromosome 17p (which contains p53) has been seen in more than 75% of sporadic CRC, it is infrequently lost in adenomas of any stage [Vogelstein et al. 1988]. In contrast, LOH causing loss of a p53 allele was shown even in nondysplastic epithelium of CUC patients [Burmer et al. 1992], though advanced dysplasia or cancer may have been present elsewhere in the colon.[Brentnall et al. 1994] Abnormal immunochemical tissue staining for p53, a surrogate marker for p53 mutation, was also shown to precede the development of CRC in a historical cohort of IBD patients [Lashner et al. 2003]. In contrast, loss of APC or LOH in 18q (containing the DCC gene), both found in early sporadic adenomas, were infrequent events in epithelium of colitis patients even with high grade dysplasia who had not yet developed cancer [Fogt et al. 1998].
Figure 1.
Comparison of genetic and epigenetic changes in sporadic and colitis-associated CRC. CRC, colorectal cancer; LOH, loss of heterozygosity; Mut, mutation. (Reproduced with permission from Xie and Itzkowitz [2008].)
More recently, aberrant methylation has been shown to be a powerful class of biomarkers in IBD-CRN. Chronic inflammation appears to accelerate age-related methylation of MYOD, ER and p16 in colon tissue of IBD patients but not hMLH1 [Issa et al. 2001]. In a much larger sample, hypermethylation of RUNX3 and MINT1, but not p16 or ER, was seen in nondysplastic tissues of IBD patients known to have cancer in comparison to IBD controls [Garrity-Park et al. 2010]. Methylated EYA4, a marker found in over 80% of sporadic CRN tissues, was also shown to be methylated in IBD-CRC and dysplasia tissues while negative in control IBD tissues [Osborn et al. 2006].
The reality of clinical practice is that CRN in patients with IBD is heterogeneous and multiple tumorigenesis pathways may be occurring in the same individual. Firstly, there are several morphologic categories of IBD-CRN. Endoscopically visible dysplasia may be associated with a mass or lesion [nonadenoma-like dysplasia associated lesion of mass (DALM)]. This may be carpet-like, spreading, ulcerated neoplasm, unresectable by endoscopic techniques [Blackstone et al. 1981]. Lesions may also be discrete and adenomatous in appearance (adenoma-like DALM). Endoscopically unapparent neoplasms found only on random biopsy, termed flat dysplasia are also strongly liked to a subsequent diagnosis of CRC [Thomas et al. 2007]. However, there is no evidence to suggest that sporadic and IBD-CRN are mutually exclusive. Adenoma-like DALMs in IBD patients endoscopically and histologically resemble sporadic adenomas and contain the typical genetic alterations of sporadic adenomas [Walsh et al. 1999]. Adenoma-like DALM (ALD) lesions [Odze et al. 2000] are amenable to conservative endoscopic polypectomy [Rubin et al. 1999; Odze et al. 2004; Kisiel et al. 2012a], which appears to be a safe strategy even in patients with long-standing IBD (Figure 2). Considering the heterogeneity encountered in IBD-CRN surveillance, a panel of diagnostic markers must be able to identify a wide variety of target lesions, including ALD, nonadenoma-like DALM, flat dysplasia and curable stage CRC.
Figure 2.
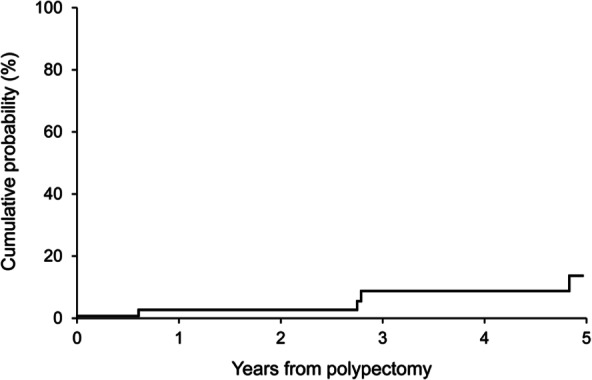
Cumulative probability of subsequent CRC, high-grade dysplasia or flat dysplasia among polypectomy patients from time of polypectomy. CRC, colorectal cancer. (Reproduced with permission from Kisiel et al. [2012a].)
Given these important biological and clinical observations in aggregate, our group hypothesized that identification of ‘early’ events in IBD-CRN tissues, such as LOH, abnormal p53 expression or aberrant methylation, could be identified from exfoliated DNA in stools of patients with IBD-CRN.
Markers representative of IBD-CRC
To determine the best potential candidates for a stool assay, we first studied numerous candidates in tissues. We sequenced a broad panel of candidate exons for mutations in DNA extracted from tissues of 25 case IBD-CRC tumors and 25 control IBD patients matched on sex, age, duration of colitis and comorbid PSC. While mutations were found only in case tissues (100% specificity), only 15 case subjects had a mutation among the aggregate of all markers studied (60% sensitivity) (Figure 3) [Kisiel et al. 2013]. As expected p53 was the most informative marker, although mutations were found on all five p53 exons studied. To replicate these results in the clinical setting would be resource-intensive, requiring DNA sequencing or numerous allele-specific PCR reactions.
Figure 3.
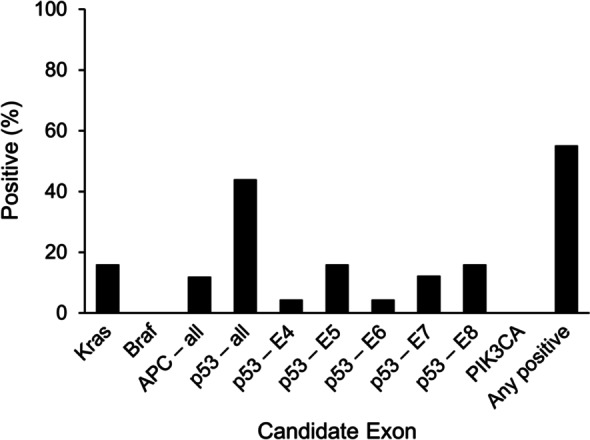
Gene mutations detected in tissue DNA from IBD-associated cancers (n = 25). IBD, inflammatory bowel disease. (Reproduced with permission from Kisiel et al. [2013].)
In contrast, methylated EYA4 alone was found in 22/25 IBD-CRN tissues at 90% specificity; we also demonstrated that other methylated DNA markers present in sporadic CRN were highly discriminant in the same IBD samples [Kisiel et al. 2010]. These methylated DNA marker levels did not appear to be influenced by inflammation severity [Garrity-Park et al. 2010]. In a subsequent prospective stool study, we selected methylated EYA4, Vimentin, BMP3 and NDRG4 (mEYA4, mVIM, mBMP3 and mNDRG4) as candidate markers for detection of IBD-CRN.
sDNA is feasible for detection of IBD-CRN
Stools were collected from patients with biopsy-confirmed IBD-CRN and IBD control patients following negative surveillance colonoscopy. All patients submitted stools in preservative buffer prior to or >1 week after colonoscopy. From stool-extracted DNA, the target genes were enriched by sequence capture, bisulfite treated, and quantitatively assayed by methylation-specific PCR.
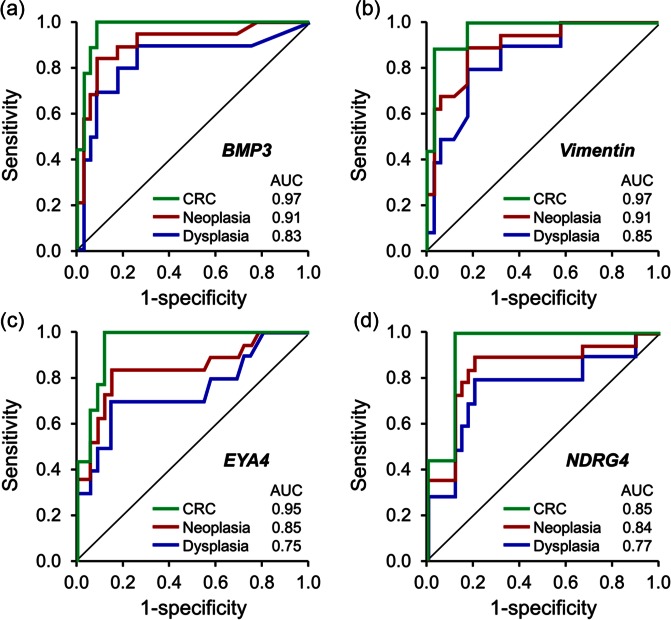
Participants comprised 19 IBD case patients with CRN (9 with cancer and 10 with dysplasia) and 35 IBD controls. All markers individually showed high discrimination for IBD-CRN: areas under the receiver operating characteristics curves [area under curve (AUC)] with mBMP3, mVIM, mNDRG4 and mEYA4 were 0.91, 0.91, 0.84 and 0.85, respectively. For cancer, the AUC with mBMP3, mVIM, mNDRG4 and mEYA4 were 0.97, 0.97, 0.94 and 0.95, respectively (Figure 4) [Kisiel et al. 2013]. At 89% specificity, the combination of mBMP3 and mNDRG4 detected 9/9 (100%) of CRC and 80% of dysplasia (4/4 (100%) of high grade and 4/6 (67%) of low grade). All markers remained highly significant in multivariate analyses which adjusted for inflammation severity, IBD disease extent, duration, severity and comorbid PSC. Importantly, stool marker levels assayed were unaffected by neoplasm site within the colorectum, as we have observed with sporadic colorectal neoplasia [Ahlquist et al. 2012b]. These data establish proof-of-concept for IBD-CRN detection by stool assay of methylated DNA markers [Kisiel et al. 2013].
Figure 4.
Receiver operating characteristics curve for detection of IBD-associated CRC, dysplasia and neoplasia (CRC and dysplasia combined) by stool assay of methylated DNA markers (a) BMP3, (b) Vimentin, (c) EYA4 and (d) NDRG4. AUC, area under curve; CRC, colorectal cancer. (Reproduced with permission from Kisiel et al. [2013].)
sDNA for detection of IBD-CRN: next steps
Feasibility of sDNA for IBD-CRN detection has only just been demonstrated and these results require validation [Kisiel et al. 2013]. Further studies are also needed to evaluate this noninvasive approach as a complement to endoscopic strategies in IBD surveillance cohorts. Those data will help to determine the optimal stool sampling interval and inform algorithms incorporating sDNA as a complement to colonoscopy. Benefits could potentially include a lengthened interval between surveillance colonoscopies in marker-negative patients. This in turn has the potential to reduce the overall high cost of surveillance [Rubenstein et al. 2009]. Further, a noninvasive test that could be performed without bowel cleansing in a patient’s own home might improve compliance with surveillance, which is currently poor [Velayos et al. 2010, Vienne et al. 2011].
Observations are needed to understand the natural history and predictive value of sDNA in IBD-CRN surveillance populations where neoplasia prevalence may vary. Use of a less costly noninvasive alternative to first-line colonoscopy is particularly attractive in IBD populations with low prevalence of CRN. While sDNA is highly specific in the case-control setting, IBD-CRN has relatively low prevalence in the surveillance setting [Toruner et al. 2005]. It needs to be determined if sDNA can maintain high specificity and positive predictive value in surveillance applications.
Beyond the basic ‘false positive’ rate, several more complex scenarios must be carefully studied. It is well established that molecular changes occur in inflamed mucosae prior to the development of histologic dysplasia [Risques et al. 2011]. As we work to better understand the biology of IBD-CRN, a colonoscopy-negative patient with a positive sDNA test may benefit from colonoscopy at shorter endoscopic surveillance intervals. We also know that fields of molecular change can also occur in colonic mucosa at sites distant from a known neoplasm [Garrity-Park et al. 2010; Bista et al. 2011; Kisiel et al. 2011]. Therefore, we will have to determine if sDNA identifies a subset of patients who may benefit from enhanced imaging techniques, including chromoendoscopy.
A third possibility is that sDNA may detect neoplasms above the colon. Some markers of sporadic CRN and IBD-CRN may be aberrantly methylated in other tissues as well [Zou et al. 2005; Yang Wu et al. 2011; Kisiel et al. 2012c; Moinova et al. 2012]. These observations, at first, appear to present a specificity hurdle to sDNA in surveillance. However, IBD patients are at increased risk extra-intestinal cancers [Bergquist et al. 2002; Pedersen et al. 2010]. Therefore, a technology that could be used for surveillance of the entire patient, rather than a single organ, could transform the overall approach to cancer prevention. Identification and use of site-specific markers would be of great value for this application. This concept is bolstered by early data showing that panels of novel methylation markers can classify tumors by their location in the gastrointestinal GI tract (unpublished data).
Observations of sDNA performance in sporadic CRN suggest that important opportunities exist for molecular risk stratification of patients with true positive sDNA results. It is known that mutant DNA [Syngal et al. 2006] and aberrant methylation markers [Kisiel et al. 2012b] clear from stool following resection of sporadic CRC. We must also determine if methylation markers clear from patients after endoscopic ablation or polypectomy of IBD-CRN.
Conclusion
Early results in tissue and stool represent an important first step in the evaluation of sDNA as a noninvasive tool for detection of CRN in IBD patients [Kisiel et al. 2013]. While we corroborate these findings, further studies are indicated to expand our understanding of how to best use this technology. In the short term, prospective cohort studies conducted in the IBD surveillance setting will help determine how this noninvasive tool might improve colonoscopy yield and patient outcomes. Longer range objectives include the study of sDNA in multiorgan surveillance and potential benefits of sDNA on the lowering healthcare costs for IBD.
Footnotes
Funding: J.B.K. was supported by the Maxine and Jack Zarrow Family Foundation of Tulsa, Oklahoma.
Conflict of interest statement: Mayo Clinic is a minor equity investor in, and has licensed intellectual property to, Exact Sciences. J.B.K. and D.A.A. have intellectual property agreements related to sDNA testing and, consistent with Mayo Clinic policy, could share in potential future royalties.
Contributor Information
John B. Kisiel, Division of Gastroenterology and Hepatology, Mayo Clinic, 200 First Street Southwest, Rochester, MN 55905, USA
David A. Ahlquist, Division of Gastroenterology and Hepatology, Mayo Clinic, Rochester, MN, USA
References
- Ahlquist D., Sargent D., Loprinzi C., Levin T., Rex D., Ahnen D., et al. (2008) Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Ann Intern Med 149: 441–450, W481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist D., Taylor W., Mahoney D., Zou H., Domanico M., Thibodeau S., et al. (2012a) The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. Clin Gastroenterol Hepatol 10: 272–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist D., Zou H., Domanico M., Mahoney D., Yab T., Taylor W., et al. (2012b) Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology 142: 248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg M., Soreide K. (2012) EGFR and downstream genetic alterations in Kras/Braf and Pi3k/Akt pathways in colorectal cancer: implications for targeted therapy. Discovery Med 14: 207–214. [PubMed] [Google Scholar]
- Bergquist A., Ekbom A., Olsson R., Kornfeldt D., Loof L., Danielsson A., et al. (2002) Hepatic and extrahepatic malignancies in primary sclerosing cholangitis. J Hepatol 36: 321–327. [DOI] [PubMed] [Google Scholar]
- Bista R., Brentnall T., Bronner M., Langmead C., Brand R., Liu Y. (2011) Using optical markers of nondysplastic rectal epithelial cells to identify patients with ulcerative colitis-associated neoplasia. Inflammatory Bowel Dis 17: 2427–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackstone M., Riddell R., Rogers B., Levin B. (1981) Dysplasia-associated lesion or mass (Dalm) detected by colonoscopy in long-standing ulcerative colitis: an indication for colectomy. Gastroenterology 80: 366–374. [PubMed] [Google Scholar]
- Brentnall T., Crispin D., Rabinovitch P., Haggitt R., Rubin C., Stevens A., et al. (1994) Mutations in the P53 gene: an early marker of neoplastic progression in ulcerative colitis. Gastroenterology 107: 369–378. [DOI] [PubMed] [Google Scholar]
- Burmer G., Rabinovitch P., Haggitt R., Crispin D., Brentnall T., Kolli V., et al. (1992) Neoplastic progression in ulcerative colitis: histology, DNA content, and loss of a P53 Allele. Gastroenterology 103: 1602–1610. [DOI] [PubMed] [Google Scholar]
- Cairns S., Scholefield J., Steele R., Dunlop M., Thomas H., Evans G., et al. (2010) Guidelines for colorectal cancer screening and surveillance in moderate and high risk groups (update from 2002). Gut 59: 666–689. [DOI] [PubMed] [Google Scholar]
- Choi P., Nugent F., Schoetz D., Jr., Silverman M., Haggitt R. (1993) Colonoscopic surveillance reduces mortality from colorectal cancer in ulcerative colitis. Gastroenterology 105: 418–424. [DOI] [PubMed] [Google Scholar]
- Collins P., Mpofu C., Watson A., Rhodes J. (2006) Strategies for detecting colon cancer and/or dysplasia in patients with inflammatory bowel Disease. Cochrane Database Syst Rev: CD000279. [DOI] [PubMed] [Google Scholar]
- Connell W., Lennard-Jones J., Williams C., Talbot I., Price A., Wilkinson K. (1994) Factors affecting the outcome of endoscopic surveillance for cancer in ulcerative colitis. Gastroenterology 107: 934–944. [DOI] [PubMed] [Google Scholar]
- Delco F., Sonnenberg A. (2000) A decision analysis of surveillance for colorectal cancer in ulcerative colitis. Gut 46: 500–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaden J., Abrams K., Mayberry J. (2001) The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut 48: 526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farraye F., Odze R., Eaden J., Itzkowitz S. (2010) Aga technical review on the diagnosis and management of colorectal neoplasia in inflammatory bowel disease. Gastroenterology 138: 746–774, e741–744; quiz e712–743. [DOI] [PubMed] [Google Scholar]
- Fearon E., Hamilton S., Vogelstein B. (1987) Clonal analysis of human colorectal tumors. Science 238: 193–197. [DOI] [PubMed] [Google Scholar]
- Fearon E., Vogelstein B. (1990) A genetic model for colorectal tumorigenesis. Cell 61: 759–767. [DOI] [PubMed] [Google Scholar]
- Finlay C., Hinds P., Levine A. (1989) The P53 Proto-oncogene can act as a suppressor of transformation. Cell 57: 1083–1093. [DOI] [PubMed] [Google Scholar]
- Fogt F., Vortmeyer A., Goldman H., Giordano T., Merino M., Zhuang Z. (1998) Comparison of genetic alterations in colonic adenoma and ulcerative colitis-associated dysplasia and carcinoma. Hum Pathol 29: 131–136. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M. (1987) Cpg islands in vertebrate genomes. J Mol Biol 196: 261–282. [DOI] [PubMed] [Google Scholar]
- Garrity-Park M., Loftus E., Jr., Sandborn W., Bryant S., Smyrk T. (2010) Methylation status of genes in non-neoplastic mucosa from patients with ulcerative colitis-associated colorectal cancer. Am J Gastroenterol 105: 1610–1619. [DOI] [PubMed] [Google Scholar]
- Goel A., Boland C. (2012) Epigenetics of colorectal cancer. Gastroenterology 143: 1442–1460, e1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J., Herman J., Myohanen S., Baylin S., Vertino P. (1997) Mapping patterns of cpg island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J Biol Chem 272: 22322–22329. [DOI] [PubMed] [Google Scholar]
- Herman J., Umar A., Polyak K., Graff J., Ahuja N., Issa J., et al. (1998) Incidence and functional consequences of Hmlh1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A 95: 6870–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrinton L., Liu L., Levin T., Allison J., Lewis J., Velayos F. (2012) Incidence and mortality of colorectal adenocarcinoma in persons with inflammatory bowel disease from 1998 to 2010. Gastroenterology 143: 382–389. [DOI] [PubMed] [Google Scholar]
- Issa J., Ahuja N., Toyota M., Bronner M., Brentnall T. (2001) Accelerated age-related Cpg island methylation in ulcerative colitis. Cancer Res 61: 3573–3577. [PubMed] [Google Scholar]
- Itzkowitz S., Harpaz N. (2004) Diagnosis and management of dysplasia in patients with inflammatory bowel diseases. Gastroenterology 126: 1634–1648. [DOI] [PubMed] [Google Scholar]
- Jess T., Rungoe C., Peyrin-Biroulet L. (2012a) Risk of colorectal cancer in patients with ulcerative colitis: a meta-analysis of population-based cohort studies. Clin Gastroenterol Hepatol 10: 639–645. [DOI] [PubMed] [Google Scholar]
- Jess T., Simonsen J., Jorgensen K., Pedersen B., Nielsen N., Frisch M. (2012b) Decreasing risk of colorectal cancer in patients with inflammatory bowel disease over 30 years. Gastroenterology 143: 375–381, e371. [DOI] [PubMed] [Google Scholar]
- Jonsson B., Ahsgren L., Andersson L., Stenling R., Rutegard J. (1994) Colorectal cancer surveillance in patients with ulcerative colitis. Br J Surg 81: 689–691. [DOI] [PubMed] [Google Scholar]
- Karlen P., Kornfeld D., Brostrom O., Lofberg R., Persson P., Ekbom A. (1998) Is colonoscopic surveillance reducing colorectal cancer mortality in ulcerative colitis? A population based case control study. Gut 42: 711–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisiel J., Garrity-Park M., Taylor W., Smyrk T., Ahlquist D. (2011) Methylated Eya4 gene in non-neoplastic mucosa of ulcerative colitis patients with colorectal cancer: evidence for a field effect. Gastroenterology 140: S348–S349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisiel J., Loftus E., Jr., Harmsen W., Zinsmeister A., Sandborn W. (2012a) Outcome of sporadic adenomas and adenoma-like dysplasia in patients with ulcerative colitis undergoing polypectomy. Inflammatory Bowel Dis 18: 226–235. [DOI] [PubMed] [Google Scholar]
- Kisiel J., Yab T., Nazer Hussain F., Taylor W., Garrity-Park M., Sandborn W., et al. (2013) Stool DNA testing for the detection of colorectal neoplasia in patients with inflammatory bowel disease. Aliment Pharmacol Ther 37: 546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisiel J., Yab T., Simonson J., Devens M., Mahoney D., Ahlquist D. (2012b) Su1903 methylated gene markers in stool before and after colorectal cancer resection: biologic implications for surveillance. Gastroenterology 142: S531–S532. [Google Scholar]
- Kisiel J., Yab T., Taylor W., Chari S., Petersen G., Mahoney D., et al. (2012c) Stool DNA testing for the detection of pancreatic cancer: assessment of methylation marker candidates. Cancer 118: 2623–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashner B., Bauer W., Rybicki L., Goldblum J. (2003) Abnormal P53 immunohistochemistry is associated with an increased colorectal cancer-related mortality in patients with ulcerative colitis. Am J Gastroenterol 98: 1423–1427. [DOI] [PubMed] [Google Scholar]
- Lashner B., Kane S., Hanauer S. (1990) Colon cancer surveillance in chronic ulcerative colitis: historical cohort study. Am J Gastroenterol 85: 1083–1087. [PubMed] [Google Scholar]
- Levin B., Lieberman D., Mcfarland B., Andrews K., Brooks D., Bond J., et al. (2008) Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-society Task Force on Colorectal Ccancer, and the American College of Radiology. Gastroenterology 134: 1570–1595. [DOI] [PubMed] [Google Scholar]
- Lidgard G., Domanico M., Bruinsma J., Gagrat Z., Oldham-Haltom R., Fourrier K., et al. (2012a) Tu1189, an optimized multi-marker stool test for colorectal cancer screening: initial clinical appraisal. Gastroenterology 142: S770. [Google Scholar]
- Lidgard G., Domanico M., Bruinsma J., Light J., Gagrat Z., Oldham-Haltom R., et al. (2012b) An optimized molecular stool test for colorectal cancer screening: evaluation of an automated analytic platform and logistic algorithm. Cancer Research 2012; 72(8, Supplement 1). [Google Scholar]
- Lim C., Dixon M., Vail A., Forman D., Lynch D., Axon A. (2003) Ten year follow up of ulcerative colitis patients with and without low grade dysplasia. Gut 52: 1127–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdermott R. (1985) Review of clinical aspects of cancer of the colon in patients with ulcerative colitis. Dig Dis Sci 30: 114S–118S. [DOI] [PubMed] [Google Scholar]
- Miyakura Y., Sugano K., Konishi F., Ichikawa A., Maekawa M., Shitoh K., et al. (2001) Extensive methylation of Hmlh1 promoter region predominates in proximal colon cancer with microsatellite instability. Gastroenterology 121: 1300–1309. [DOI] [PubMed] [Google Scholar]
- Moinova H., Leidner R., Ravi L., Lutterbaugh J., Barnholtz-Sloan J., Chen Y., et al. (2012) Aberrant vimentin methylation is characteristic of upper gastrointestinal pathologies. Cancer Epidemiol, Biomarkers Prev 21: 594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute for Health and Clinical Excellence (NICE) (2011) Colonoscopic surveillance for prevention of colorectal cancer in people with ulcerative colitis, Crohn’s disease or adenomas. London: National Institute for Health and Clinical Excellence. [PubMed] [Google Scholar]
- Nuako K., Ahlquist D., Mahoney D., Schaid D., Siems D., Lindor N. (1998) Familial predisposition for colorectal cancer in chronic ulcerative colitis: a case-control study. Gastroenterology 115: 1079–1083. [DOI] [PubMed] [Google Scholar]
- Nugent F., Haggitt R., Gilpin P. (1991) Cancer surveillance in ulcerative colitis. Gastroenterology 100: 1241–1248. [PubMed] [Google Scholar]
- Odze R., Brown C., Hartmann C., Noffsinger A., Fogt F. (2000) Genetic alterations in chronic ulcerative colitis-associated adenoma-like dalms are similar to non-colitic sporadic adenomas. Am J Surg Pathol 24: 1209–1216. [DOI] [PubMed] [Google Scholar]
- Odze R., Farraye F., Hecht J., Hornick J. (2004) Long-term follow-up after polypectomy treatment for adenoma-like dysplastic lesions in ulcerative colitis. Clin Gastroenterol Hepatol 2: 534–541. [DOI] [PubMed] [Google Scholar]
- Olson J., Whitney D., Durkee K., Shuber A. (2005) DNA stabilization is critical for maximizing performance of fecal dna-based colorectal cancer tests. Diagn Mol Pathol 14: 183–191. [DOI] [PubMed] [Google Scholar]
- Osborn N., Zou H., Molina J., Lesche R., Lewin J., Lofton-Day C., et al. (2006) Aberrant methylation of the eyes absent 4 gene in ulcerative colitis-associated dysplasia. Clin Gastroenterol Hepatol 4: 212–218. [DOI] [PubMed] [Google Scholar]
- Pedersen N., Duricova D., Elkjaer M., Gamborg M., Munkholm P., Jess T. (2010) Risk of extra-intestinal cancer in inflammatory bowel disease: meta-analysis of population-based cohort studies. Am J Gastroenterol 105: 1480–1487. [DOI] [PubMed] [Google Scholar]
- Provenzale D., Wong J., Onken J., Lipscomb J. (1998) Performing a cost-effectiveness analysis: surveillance of patients with ulcerative colitis. Am J Gastroenterol 93: 872–880. [DOI] [PubMed] [Google Scholar]
- Rex D., Johnson D., Anderson J., Schoenfeld P., Burke C., Inadomi J. (2009) American College of Gastroenterology guidelines for colorectal cancer screening 2009 [corrected]. Am J Gastroenterol 104: 739–750. [DOI] [PubMed] [Google Scholar]
- Risques R., Lai L., Himmetoglu C., Ebaee A., Li L., Feng Z., et al. (2011) Ulcerative colitis-associated colorectal cancer arises in a field of short telomeres, senescence, and inflammation. Cancer Res 71: 1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein J., Waljee A., Jeter J., Velayos F., Ladabaum U., Higgins P. (2009) Cost effectiveness of ulcerative colitis surveillance in the setting of 5-aminosalicylates. Am J Gastroenterol 104: 2222–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin P., Friedman S., Harpaz N., Goldstein E., Weiser J., Schiller J., et al. (1999) Colonoscopic polypectomy in chronic colitis: conservative management after endoscopic resection of dysplastic polyps. Gastroenterology 117: 1295–1300. [DOI] [PubMed] [Google Scholar]
- Sonnenberg A., Delco F., Inadomi J. (2000) Cost-effectiveness of colonoscopy in screening for colorectal cancer. Ann Intern Med 133: 573–584. [DOI] [PubMed] [Google Scholar]
- Subramanian V., Mannath J., Ragunath K., Hawkey C. (2011) Meta-analysis: the diagnostic yield of chromoendoscopy for detecting dysplasia in patients with colonic inflammatory bowel disease. Aliment Pharmacol Ther 33: 304–312. [DOI] [PubMed] [Google Scholar]
- Suehiro Y., Wong C., Chirieac L., Kondo Y., Shen L., Webb C., et al. (2008) Epigenetic-genetic interactions in the Apc/Wnt, Ras/Raf, and P53 pathways in colorectal carcinoma. Clin Cancer Res 14: 2560–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syngal S., Stoffel E., Chung D., Willett C., Schoetz D., Schroy P., et al. (2006) Detection of stool DNA mutations before and after treatment of colorectal neoplasia. Cancer 106: 277–283. [DOI] [PubMed] [Google Scholar]
- Thomas T., Abrams K., Robinson R., Mayberry J. (2007) Meta-analysis: cancer risk of low-grade dysplasia in chronic ulcerative colitis. Aliment Pharmacol Ther 25: 657–668. [DOI] [PubMed] [Google Scholar]
- Toruner M., Harewood G., Loftus E., Jr., Sandborn W., Tremaine W., Faubion W., et al. (2005) Endoscopic factors in the diagnosis of colorectal dysplasia in chronic inflammatory bowel disease. Inflamm Bowel Dis 11: 428–434. [DOI] [PubMed] [Google Scholar]
- Toyota M., Ahuja N., Ohe-Toyota M., Herman J., Baylin S., Issa J. (1999) Cpg island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A 96: 8681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayos F., Liu L., Lewis J., Allison J., Flowers N., Hutfless S., et al. (2010) Prevalence of colorectal cancer surveillance for ulcerative colitis in an integrated health care delivery system. Gastroenterology 139: 1511–1518. [DOI] [PubMed] [Google Scholar]
- Vienne A., Simon T., Cosnes J., Baudry C., Bouhnik Y., Soule J., et al. (2011) Low prevalence of colonoscopic surveillance of inflammatory bowel disease patients with longstanding extensive colitis: a clinical practice survey nested in the cesame cohort. Aliment Pharmacol Ther 34: 188–195. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Fearon E., Hamilton S., Kern S., Preisinger A., Leppert M., et al. (1988) Genetic alterations during colorectal-tumor development. N Engl J Med 319: 525–532. [DOI] [PubMed] [Google Scholar]
- Walsh S., Loda M., Torres C., Antonioli D., Odze R. (1999) P53 and beta catenin expression in chronic ulcerative colitis–associated polypoid dysplasia and sporadic adenomas: an immunohistochemical study. Am J Surg Pathol 23: 963–969. [DOI] [PubMed] [Google Scholar]
- Yang Wu D., Yab T., Taylor W., Mahoney D., Kisiel J., Devens M., et al. (2011) Aberrant gene methylation in the neoplastic progression of barrett’s esophagus: identification of candidate diagnostic markers. Gastroenterology 140: S222. [Google Scholar]
- Xie J., Itzkowitz S. (2008) Cancer in inflammatory bowel disease. World J Gastroenterol 14: 378–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., Allawi H., Cao X., Domanico M., Harrington J., Taylor W., et al. (2010) Sensitive quantification of vimentin methylation with a novel methylation specific qInvader Technology. Clinical Chemistry 56(6S): A199. [DOI] [PubMed] [Google Scholar]
- Zou H., Harrington J., Klatt K., Ahlquist D. (2006) A Sensitive Method to quantify human long dna in stool: relevance to colorectal cancer screening. Cancer Epidemiol Biomarkers Prev 15: 1115–1119. [DOI] [PubMed] [Google Scholar]
- Zou H., Osborn N., Harrington J., Klatt K., Molina J., Burgart L., et al. (2005) Frequent methylation of eyes absent 4 gene in Barrett’s esophagus and esophageal adenocarcinoma. Cancer Epidemiol Biomarkers Prev 14: 830–834. [DOI] [PubMed] [Google Scholar]
- Zou H., Taylor W., Harrington J., Devens M., Simonson J., Ahlquist D. (2008) A Sensitive method to scan gene mutations in stool: relevance to detection of gastrointestinal neoplasia. Gastroenterology 134: A484. [Google Scholar]
- Zou H., Taylor W., Harrington J., Hussain F., Cao X., Loprinzi C., et al. (2009) High detection rates of colorectal neoplasia by stool DNA testing with a novel digital melt curve assay. Gastroenterology 136: 459–470. [DOI] [PubMed] [Google Scholar]