Abstract
Nanopore technologies are being developed for fast and direct sequencing of single DNA molecules through detection of ionic current modulations as DNA passes through a pore’s constriction1,2. Here we demonstrate the ability to resolve changes in current that correspond to a known DNA sequence by combining the high sensitivity of a mutated form of the protein pore Mycobacterium smegmatis porin A (MspA)3 with phi29 DNA polymerase (DNAP)4, which controls the rate of DNA translocation through the pore. As phi29 DNAP synthesizes DNA and functions like a motor to pull a single-stranded template through MspA, we observe well-resolved and reproducible ionic current levels with median durations of ~28 ms and ionic current differences of up to 40 pA. Using six different DNA sequences with readable regions 42–53 nucleotides long, we record current traces that map to the known DNA sequences. With single-nucleotide resolution and DNA translocation control, this system integrates solutions to two long-standing hurdles to nanopore sequencing2.
In nanopore DNA sequencing, a pore inserted into a membrane permits the flow of ionic current when a voltage is applied across the membrane. As a strand of DNA passes through the pore, it causes changes in current that can be related to the sequence of the DNA. Such a strategy offers the promise of rapidly sequencing long single molecules of DNA2, amplification-free sample preparation1 and direct detection of epigenetic modifications such as base methylation3,5. Recent progress toward nanopore sequencing has included determining the resolution and characterizing the recognition sites of biological nanopores MspA3,6 and α-hemolysin7-9 as well as developing a method to slow DNA translocation though a nanopore4,6,10. However, to our knowledge no system has yet been reported that can read nucleotide-specific current levels as an unmodified strand of DNA passes through a nanopore. In this work, we read DNA by detecting current levels associated with single-nucleotide movement of the strand through MspA. Base-calling algorithms will need to be developed to translate these ionic current reads into a DNA sequence.
The suitability of a nanopore for DNA sequencing is influenced by the physical dimensions and biophysical properties of different regions of the pore, including the entrance, the vestibule, and the narrowest region, called the constriction. MspA11 is particularly attractive for nanopore sequencing because it has a short and narrow constriction ~1.2 nm wide and ~0.6 nm long12 (Fig. 1a). Thus, the ionic current through MspA is affected by a smaller number of nucleotides compared with other pores3,7,9. Previously, we engineered mutants of MspA by replacing negative charges in the constriction with neutral residues, which enabled DNA to electrophoretically pass through the pore13. We also added 24 positively charged residues in the vestibule and entrance, which enhanced the rate of entry of DNA into the pore13. This mutant, previously called M2-NNN MspA, is used in the present study and is here designated MspA. When DNA was held statically in the constriction of a mutant MspA by a conjugated NeutrAvidin molecule3, different homopolymer strands resulted in conductance differences of as much as ~0.23 nS, nearly ten times more separation than that observed with the widely used α-hemolysin nanopore (~0.028 nS)7,8. However, this was not sufficient for nanopore sequencing because freely translocating DNA acting under the force of an electric field moves through nanopores at an average rate greater than one nucleotide/μs, ~1,000 times too fast to distinguish nucleotide-specific current changes from noise1,2,13,14. Simple techniques for reducing the velocity of translocating DNA, such as lowering the experimental temperature or increasing the viscosity of the solution, also reduce the current signal and therefore do not improve the signal-to-noise ratio.
Figure 1.
Event structure. (a) Crystal structure of M2-NNN MspA. Charged vestibule residues are indicated in blue (negative) or red (positive). (b) A schematic depicting a standard experiment. Roman numerals correspond to positions in the current trace in c. (c) The measured blockage current (Ib) as a fraction of the open pore current (Io) is shown for a sample event. (i) A single MspA pore (purple) in a lipid bilayer (gray). The template strand (black) contains the sequence to be read. A primer strand (blue) is hybridized to the template’s 3′ end. A blocking oligomer (red) with a 3′ end of several abasic sites is adjacent to the primer. The phi29 DNAP (green) binds to the DNA to form a complex that is driven into MspA. A positive voltage is applied to the trans side. The single stranded 5′ end of the DNA-motor complex threads through MspA and the ionic current drops. (ii) The electric force on the captured strand draws the DNA through the phi29 DNAP, unzipping the blocking oligomer. Arrows show the direction of motion of the DNA template strand. The ionic current exhibits distinct steps while nucleotides pass through the pore. (iii) The blocking oligomer is removed and DNA reverses direction (marked by blue dashed line). (iv) The phi29 DNAP incorporates nucleotides into the primer strand, pulling the template toward the cis side. The current repeats previously observed levels in reverse time-order. Two abasic sites produce a high current peak (~0.6 Io) indicated by red Xs. This marker is first seen during unzipping and then again during synthesis. When synthesis is complete, the DNA and DNAP escape to the cis volume, marked by the return to Io.
Control of DNA translocation through a nanopore has been attempted using interspersed regions of double-stranded DNA6 and DNAPs4,10,15-18. The DNA polymerases of Escherichia coli (Klenow fragment)15-17, and of the bacteriophages, T7 (ref. 15) and phi29 (refs. 4,10) have been studied as molecular motors to control the movement of DNA in single-nucleotide steps through α-hemolysin. The monomeric phi29 DNAP (~66.5 kD) has 5′-3′ primer strand extension and 3′-5′ exonuclease functions in the presence of Mg2+ as well as 5′-3′ strand displacement activity19,20. Both the extension and exonuclease functions move DNA in single-nucleotide steps, which can be utilized for controlled DNA translocation through a nanopore. In contrast to Klenow fragment and T7 DNAP, phi29 DNAP is able to synthesize very long stretches of DNA (>70 kb) along a single template strand, and it can synthesize under loads of up to ~37 pN21,22, enough to counteract the electrophoretic force needed to drive DNA through the pore.
In a recently developed technique4, phi29 DNAP is bound to a DNA template and a ‘blocking oligomer’ that prevents extension and excision of a primer annealed at the template’s 3′ end. The single-stranded end of this phi29 DNAP-DNA complex is drawn into the pore until the DNAP rests on the pore’s entrance. The force on the template strand, estimated to be ~10 pN, causes the blocking oligomer to ‘unzip’ 3′ to 5′, exposing the extendable 3′ end of the primer, allowing synthesis to start. Previous experiments using this strategy with α-hemolysin showed that the DNA translocation speed was slowed to 2.5–40 nt/sec (ref. 4), but α-hemolysin lacks the ability to resolve nucleotide-specific current.
Here we combine our previously engineered MspA nanopore with the phi29 DNAP blocking oligomer technique to read well-resolved and distinguishable current levels as DNA is drawn through the pore. In our experiments, we established a single MspA pore in a lipid bilayer separating two chambers (cis and trans) containing 0.3 M KCl buffer solution. A patch-clamp amplifier applied +180 mV to the trans side of the bilayer and measured the ionic current through the pore. An illustration of the experiment and a typical current trace are shown in Figure 1. The current through an open MspA pore was Io = 110 ± 6 pA (mean ± s.d., N = 25). In all experiments, 80–91 nucleotide (nt) DNA template strands containing the section to be read were annealed to a 23-nt primer complementary to the template’s 3′end. The primer had a hairpin on its 5′ end to prevent the phi29 DNAP from acting on the double-stranded end of the DNA. Adjacent to the primer we annealed the blocking oligomer, a 15 or 29 nt complementary DNA oligomer with a 3′ end consisting of seven abasic residues and a three-carbon spacer. Most DNA template strands also contained two adjacent abasic sites, which generate a high ionic current level of ~0.6–0.7 Io as they pass through the pore (see Supplementary Table 1 for sequences).
To study an easily resolvable DNA sequence, we used a ‘block homopolymer’ DNA template containing all four bases in short homopolymer sections of adenine (dA3), guanine (dG3) and cytosine (dC5), each separated by thymine (dT3). In control experiments without phi29 DNAP, the DNA translocation was too fast (>1 nt/μs) to read individual nucleotides (Supplementary Fig. 1a). Next, we added phi29 DNAP to the cis volume, but omitted the divalent cations and deoxyribonucleotide triphosphates (dNTPs) necessary for synthesis. We recorded translocation events with current patterns consistent with force-activated unzipping of the blocking oligomer. These events exhibited distinct current levels including a high peak (~0.6 Io) when the two abasic sites passed through the constriction (Supplementary Fig. 1b,c). As expected, after removal of the blocking oligomer, the phi29 DNAP was unable to extend the primer strand owing to the absence of divalent cations and dNTPs. The phi29 DNAP eventually either fell off, allowing cooperative dissociation of the primer in MspA, or continued onwards, unzipping the primer strand (Supplementary Fig. 1b). The duration of these events was generally >20 s.
To allow DNA synthesis to proceed, we added all four standard dNTPs (100 μM each) and 10 mM MgCl2. In five separate experiments using the block homopolymer DNA, we found 33 events with the same distinct current pattern. A typical current trace (Fig. 1c) is characterized by a rapid succession of current steps (levels) with various durations and current values ranging between 0.18 and 0.4 of Io and two peaks with current levels above 0.6 Io. Closer inspection reveals that many levels at the beginning of an event were repeated later in the event, but in the opposite order. These results are consistent with part of each DNA molecule being read twice; once, during stepwise motion of the DNA template toward the cis side, while unzipping the blocking oligomer, and again, during stepwise motion back toward the trans side, while phi29 DNAP was synthesizing. The levels observed during unzipping were symmetrical in time with levels observed during synthesis (Supplementary Figs. 2 and 3). Once phi29 DNAP completed synthesis of the DNA template strand, both the DNA and phi29 DNAP exited to the cis side. The average duration of events containing phi29 DNAP synthesis (~14 s) was shorter than events recorded in the absence of Mg2+ and dNTPs because synthesis is faster than the time for phi29 DNAP to release from the DNA template or to unzip the primer strand. In our experiments with MgCl2 and dNTPs, >10% of the total data acquisition time was spent with a DNA-motor complex threaded through MspA.
The observed duration of each current step was generally shorter during DNA synthesis than during unzipping of the blocking oligomer. Examining levels associated with unzipping and synthesis revealed that both processes appear to be stochastic. Generally, level durations were exponentially distributed (Supplementary Fig. 4) with time constants of τunzip = 100 ± 12 ms and τsyn = 40 ± 10 ms. In many events, a few much longer levels (~1 s) were observed during unzipping or synthesis. In many events, one or more of the expected steps were missing because the duration was too short to be observed or adjacent current levels were indistinguishable.
Occasionally during synthesis, the current returned briefly to its previous current level before continuing on. These easily identifiable ‘toggle’ patterns are consistent with phi29 DNAP back-stepping, owing to its 3′ to 5′ proofreading activity, or failing to incorporate a nucleotide (Supplementary Fig. 5). In many cases, toggles occur numerous times in succession. In rare cases, we observed back-stepping of two sequential nucleotides.
We investigated the effect of dNTP concentration on synthesis by reducing the concentration of dCTP. In these ‘starvation’ experiments we found very long pauses in synthesis as well as toggling between distinct current levels. Furthermore, these experiments allowed us to determine that the distance between the phi29 DNAP’s nucleotide incorporation site and MspA’s constriction is 14–15 nucleotides (Supplementary Fig. 5).
We analyzed the current traces from the MspA-DNA-motor complex in its synthesis phase (Fig. 2a,b) to filter out noise and extract current levels (Fig. 2c). We observed that the temporal ordering of the current levels was preserved across all traces and that the levels could be aligned with the known DNA sequence. The ionic current was lowest (~0.20 Io) when dT3 was centered in MspA’s constriction, highest (~0.40 Io) for dA3 and moderate (~0.37 Io and ~0.30 Io) for dC4 and dG3, respectively. These results are qualitatively consistent with our previous work3 where homopolymer DNA was held statically in MspA by a NeutrAvidin molecule. Those experiments at 1.0 M [KCl] resulted in ~0.15 Io for poly-dT, ~0.25 Io for poly-dC and ~0.28 Io for poly-dA; blockage current for poly-dG could not be reliably determined.
Figure 2.
Current trace for polymerase synthesis. (a) Illustration of phi29 DNAP during synthesis. (b) Typical current trace for synthesis of ‘block homopolymer’ DNA. At the beginning of the read, multiple thymines (dT4) produce a current level of Ib/Io ~ 0.20. At the end of the read, two abasic residues (XX) produced a high current (Ib/Io ~ 0.61). *, toggles. (c) Mean currents of levels extracted from b are plotted with the associated DNA sequence.
We qualitatively aligned the sequence of the DNA to the levels (Fig. 2c) by assuming equal spacing of nucleotides and matching the position of the abasic peak, the turnaround point and the low currents typically found with multiple thymines. The nucleotides positioned near the center of the constriction dominate the total current of a level, whereas the nucleotides to either side control the current to a lesser extent. This is consistent with previous MspA experiments where each current level was affected by ~four nucleotides in and around the constriction3. For example, when DNA is positioned so that the four nucleotides centered in the constriction are TTAA (Fig. 2a and level 3 of Fig. 2c), the current is intermediate to the current levels for TTTT (level 1) and AAAT (level 5). However, small shifts in the position of nucleotides within the pore constriction may play an important role in the observed current.
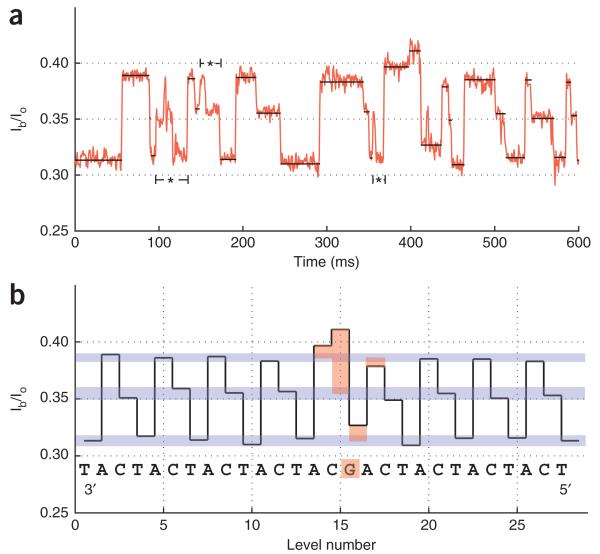
To demonstrate that our system could achieve single-nucleotide resolution, we used an 82-nt DNA template containing repeated instances of the trinucleotide sequence CAT except for one triplet in the center of the sequence that had a single dT substituted with a dG. The majority of events recorded (n = 475 events, N = 5 pores) with this DNA template consisted of a repeated stair-step current motif, except at one location where it changed as a result of the dG substitution (Fig. 3). Based on the results with the block homopolymer DNA template (Fig. 2), the ‘CAT’ stair-step motif during synthesis is consistent with cyclic rotations of TACT→ACTA→CTAC centered about the constriction, where the principal contribution is from the middle two nucleotides (note the DNA is written from 3′ to 5′ from left to right in the traces). The single-nucleotide substitution causes the pattern to be perturbed for about four steps before returning to the CAT motif (Fig. 3). Surprisingly, when ACGA is centered in the constriction, the current in the pore has a relatively high value, even though the adenines are only peripherally involved. This suggests that nucleotide type is not the only determinant of blockage current. Other factors such as secondary structure of the DNA, nucleotide-nucleotide interactions, nucleotide interactions with the pore and exact positioning within the constriction must be considered. However, a single-nucleotide substitution passing through MspA’s constriction alters the current level pattern significantly, indicating that a nucleotide’s identity and position in the strand is encoded in the observed current. This is consistent with our previously reported finding3 using a single-nucleotide polymorphism in NeutrAvidin-anchored DNA.
Figure 3.
Reading a repetitive DNA template. (a) Example trace for a DNA template composed of repeated ‘CAT’ trinucleotides, with the exception of one ‘CAG’ triplet in the middle of the sequence. (b) Mean currents of levels extracted from a with the associated DNA sequence are shown. The current trace exhibits a repeating pattern of three levels (blue bars) interrupted by the single dG substitution (highlighted in orange). Four levels are affected by the single dG with the largest deviations closest to the substitution. This indicates that the residual current is principally influenced by one or two nucleotides with a lesser influence from neighboring nucleotides.
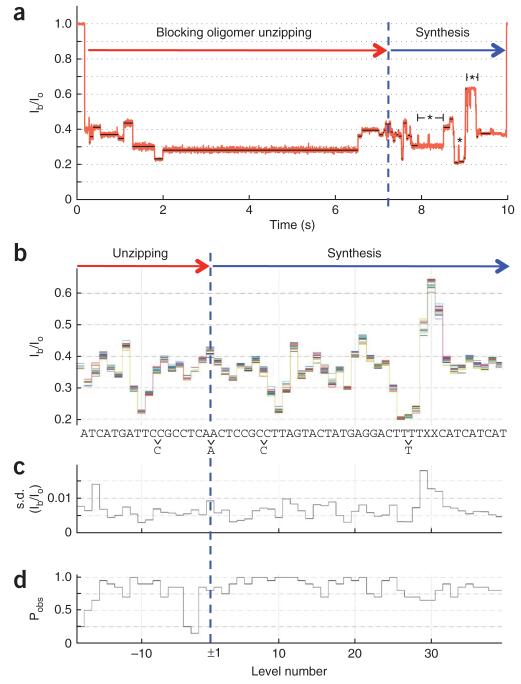
Next, we demonstrated the feasibility of reading heterogeneous DNA with MspA and phi29 DNAP. We tested four DNA heteromeric base sequences (Supplementary Table 1). Analogous to the block homopolymer DNA, these strands also had two abasic residues near the end of the template sequence to produce a clear marker signifying successful completion of synthesis. For each experimental sequence, we obtained consistent current patterns with distinct features corresponding to individual nucleotide steps as DNA passed through MspA’s constriction. Figure 4a shows one sample current trace from several experiments (n = 47, N = 4) recorded with a 91-nt DNA template strand called ‘heteromer DNA 1’. We extracted current levels from 20 events (obtained from two different pores) that exhibited unzipping and synthesis. We formed a consensus current level sequence from multiple events. Next we aligned the events to the consensus using a Needleman-Wunsch algorithm and overlayed the aligned levels (Fig. 4b). Consistent with previous experiments4, the current trace showed a symmetry about a level (dotted blue line in Fig. 4b) that corresponded to the nucleotides in MspA’s constriction when the blocking oligomer is completely removed, demonstrating that current levels observed during unzipping are repeated in opposite time-order during synthesis. As the blocking oligomer used in this experiment was shorter (15 nt) than the length of the template to be read, the number of levels observed during unzipping was fewer than those observed during synthesis, and the abasic peak marking the end of the read was not observed during unzipping.
Figure 4.
Reading heteromic DNA. (a) Example event for heteromer DNA 1. Unzipping and synthesis phases are indicated. (b) Demonstration of the repeatability and similarity of current levels found in multiple events. Time-ordered levels for n = 20 events collected on N = 2 pores are found using a level detection algorithm. A consensus current level sequence was created from multiple events. The time-ordered levels consistent with the consensus sequence are overlaid (one color for each event). The symmetry of the plot about level ± 1 shows that the same levels occur during both unzipping and synthesis (in reverse order). The correlated sequence is shown below. Levels not detected, owing to repeat nucleotides and/or similar currents, are indicated with the unobserved nucleotide beneath the sequence. (c) s.d. derived from the scatter of each overlaid level. The average 1-sigma fluctuation in levels is below 1% of Io. (d) The probability, Pobs, of finding a given level in the proper order. Some levels, such as level 10 or level 21 are found in all events (n = 20). Levels immediately before the transition from unzipping to synthesis are often too fast to be detected, resulting in low Pobs.
At the beginning of synthesis, adenines within MspA’s constriction caused a current level near ~0.4 Io, consistent with the current due to dA3 in experiments with the block homopolymer DNA. Near the end of the sequence, just before the abasic marker, a thymine repeat section resulted in a distinct poly-dT level, ~0.2 Io. The sequence of the heteromer DNA 1 could be aligned to the trace to match expected features (Fig. 4b). For example, the local minimum at level 10 and the peak at level 30 are matched to CTTA and TXXC, respectively. We expect the nucleotides listed to the right and left of each level to contribute most to the current value. The total number of observed current steps was less than the length of the sequence between the turnaround and the abasic peak, indicating that two current steps were unobserved. The suspected locations of these missing levels are indicated (Fig. 4b). These missing levels are thought to be indistinguishable from neighboring current levels. For example, level 27 is associated with the dT5-homopolymer region and likely consists of two steps with indistinguishable currents. The scatter of current values for each level reflects its reproducibility from event to event and from pore to pore. We observed that the average scatter, quantified by the s.d. of current levels, was below 1% of Io (Fig. 4c). This indicates that the positioning of the DNA in the constriction is highly reproducible. The average probability that our level-finding algorithm detected a given level in the proper order between the turn-around point and the abasic peak was 90% (Fig. 4d). The unzipping process was more likely to skip levels, particularly those near the beginning or end of unzipping. The scatter and probability of observing a level are dependent on the level detection and alignment algorithms (Online Methods). Three additional heteromer DNA sequences, ‘heteromer DNA 2’ (n = 121, N = 3), ‘heteromer DNA 3’ (n = 94, N = 3) and ‘heteromer DNA 4’ (n = 142, N = 5) produced similar results (Supplementary Table 1 and Supplementary Figs. 6-16). Differences in nucleotide-specific current levels differed by as much as ~0.36% Io or 40 pA.
In summary, we demonstrated that individual single-stranded DNA molecules traversing through the short and narrow constriction of MspA under the control of phi29 DNAP yielded distinct current levels that were related to the sequence of the nucleotides in MspA’s constriction. We observed current patterns associated with two distinct processes: the 5′ leading–motion of the DNA template, which is consistent with the nearly monotonic unzipping of the blocking oligomer, and the 3′ leading-motion, which is consistent with synthesis by phi29 DNAP. Motion during synthesis is faster than unzipping, and the levels associated with most single-nucleotide advances can be identified. The DNA nucleotides that passed twice through MspA’s constriction—once during unzipping of the blocking oligomer and once during synthesis—yielded redundant current readings.
The patterns of current levels can be related to a known DNA sequence. Automatic sequence extraction algorithms must be developed to sequence unknown DNA. Because a model predicting the current pattern will be complex, deconvolution of a current trace to extract the underlying sequence will likely involve measuring a library of current levels for all possible combinations of ~4-nt sequences. Furthermore, a method to confidently identify extended homopolymer regions must be developed. The path to commercial nanopore sequencing will involve other developments, including a robust platform and parallelization. However, we have shown that the high nucleotide sensitivity of MspA combined with the translocation control of phi29 DNAP enables single-nucleotide discrimination of DNA passing through a nanopore.
ONLINE METHODS
Proteins and DNA oligonucleotides
The M2-NNN-MspA protein was purified from Mycobacterium smegmatis as previously described13. DNA oligonucleotides were synthesized at Stanford University Protein and Nucleic Acid Facility and purified at their facility using column purification methods. DNA templates, primers and blocking oligomers were mixed at relative molar concentrations of 1:1:1.2 and annealed by incubating at 95 °C for 3 min followed by slow-cooling to below 30 °C. Wild-type phi29 DNAP (833,000 U/ml; specific activity 83,000 U/mg) was obtained from Enzymatics. DNA and phi29 DNAP were stored at −20 °C until immediately before use.
Nanopore experiments
Single MspA pores were established in a lipid bilayer with previously described methods13. Briefly, 1,2-diphytanoyl-sn-glycerol-3-phosphocholine (Avanti Polar Lipids) lipid bilayers were formed across a horizontal ~20 μm diameter Teflon aperture. The ~60 μl compartments on both sides of the bilayer contained experimental buffer of 0.3 M KCl, 1 mM EDTA, 1 mM DTT, and 10 mM HEPES/KOH buffered at pH 8.0 ± 0.05. An Axopatch 200B integrating patch clamp amplifier (Axon Instruments) applied a 180 mV voltage across the bilayer (trans side positive) and measured the ionic current through the pore. M2-NNN MspA was added to the grounded cis compartment, yielding a concentration of ~2.5 ng/ml. Once a single pore inserted, the compartment was flushed with experimental buffer to avoid further insertions. The DNA concentration for most experiments was ~1 μM. Phi29 DNAP was added to the cis compartment to a concentration of 0.75 μM. In a standard experiment, 10 mM MgCl2 and 100 μM each of dCTP, dATP, dTTP and dGTP were added to the cis compartment to allow DNA synthesis. All experiments reported here were done at room temperature (23 ± 1 °C). The analog signal was low-pass filtered with a 4-pole Bessel filter at 100 kHz and was digitized at 500 kHz. Data acquisition was controlled with virtual instrument written in LabView (National Instruments).
Data analysis
Current traces were analyzed using custom programs written in Matlab (The MathWorks). Collected data were filtered with a Butterworth filter at 2.5 kHz and then downsampled to 5 kHz. The minimal requirement for an event to be analyzed was a reduction in current <0.9 Io and a duration >2 ms. Events were further filtered with a Savitzky-Golay filter of order 2. To identify levels during DNA synthesis events, we used a custom implemented step detection method using a gradient threshold to detect transitions between unique levels. Adjacent levels were iteratively combined if (i) the difference between the levels’ mean currents was within 1-sigma of the levels’ s.d., (ii) the levels were shorter than 800 μs, or (iii) the difference between the two current level means multiplied by the duration of the subsequent level (a charge-volume) was <20 fC. Toggles were detected as repeated transitions between two levels and were removed to facilitate comparison to other events. For the figures herein, the current traces were filtered further using a 5-point window and downsampled to 1.7 kHz.
Event comparison
To automatically correlate different events’ current levels we used a Needleman-Wunsch sequence alignment algorithm with an affinegap penalty23. Scoring matrix elements for the alignment were generated from the negative square of the normalized current differences multiplied by 50. Affine-gap constants e and d23 were set at −0.25 and −0.21, respectively.
Small voltage offsets, due to electrode potential offsets and differences in ionic conductivity (due to evaporation), reduced comparability between events recorded on different experimental setups. To compensate for this, we allowed for a multiplicative scale factor, ranging between 0.96 and 1.04 and an additive offset ranging from 0.03 and −0.03. These correction factors allowed for proper comparison between levels in two events.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Akeson and G.M. Cherf for getting us started with the blocking oligomer phi29 DNAP technique, sharing their CAT DNA and reading the manuscript. We thank J. Bartlett and G. Nayler for their help running experiments and D. Feldman for writing data acquisition code. This work was supported by the US National Institutes of Health, National Human Genome Research Institute $1000 Genome Program Grants R21HG004145, R01HG005115 and R01HG006321.
Footnotes
Note: Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
E.A.M., I.M.D. and J.H.G. conceptualized the project. E.A.M., I.M.D., A.H.L. and J.H.G. designed the experiments, wrote the paper and contributed equally. E.A.M., I.M.D., A.H.L., K.W.L., M.K.H., N.G. and J.H.G. analyzed the data. K.W.L., M.K.H. and N.G. collected data. M.P. and M.N. produced the MspA mutants. J.H.G. supervised the project.
COMPETING FINANcIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Branton D, et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manrao EA, Derrington IM, Pavlenok M, Niederweis M, Gundlach JH. Nucleotide discrimination with DNA immobilized in the MspA nanopore. PLoS ONE. 2011;6:e25723. doi: 10.1371/journal.pone.0025723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherf GM, et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-Å precision. Nat. Biotechnol. 2012 Feb 14; doi: 10.1038/nbt.2147. advance online publication, doi:10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallace EVB, et al. Identification of epigenetic DNA modifications with a protein nanopore. Chem. Commun. (Camb.) 2010;46:8195–8197. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derrington IM, et al. Nanopore DNA sequencing with MspA. Proc. Natl. Acad. Sci. USA. 2010;107:16060–16065. doi: 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc. Natl. Acad. Sci. USA. 2009;106:7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purnell RF, Mehta KK, Schmidt JJ. Nucleotide identification and orientation discrimination of DNA homopolymers immobilized in a protein nanopore. Nano Lett. 2008;8:3029–3034. doi: 10.1021/nl802312f. [DOI] [PubMed] [Google Scholar]
- 9.Purnell RF, Schmidt JJ. Discrimination of single base substitutions in a DNA strand immobilized in a biological nanopore. ACS Nano. 2009;3:2533–2538. doi: 10.1021/nn900441x. [DOI] [PubMed] [Google Scholar]
- 10.Lieberman KR, et al. Processive replication of single DNA molecules in a nanopore catalyzed by phi29 DNA polymerase. J. Am. Chem. Soc. 2010;132:17961–17972. doi: 10.1021/ja1087612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niederweis M, et al. Cloning of the MspA gene encoding a porin from Mycobacterium smegmatis. Mol. Microbiol. 1999;33:933–945. doi: 10.1046/j.1365-2958.1999.01472.x. [DOI] [PubMed] [Google Scholar]
- 12.Faller M, Niederweis M, Schulz GE. The structure of a mycobacterial outer-membrane channel. Science. 2004;303:1189–1192. doi: 10.1126/science.1094114. [DOI] [PubMed] [Google Scholar]
- 13.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc. Natl. Acad. Sci. USA. 2008;105:20647–20652. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proc. Natl. Acad. Sci. USA. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gyarfas B, et al. Mapping the position of DNA polymerase-bound DNA templates in a nanopore at 5 angstrom resolution. ACS Nano. 2009;3:1457–1466. doi: 10.1021/nn900303g. [DOI] [PubMed] [Google Scholar]
- 16.Wilson NA, et al. Electronic control of DNA polymerase binding and unbinding to single DNA molecules. ACS Nano. 2009;3:995–1003. doi: 10.1021/nn9000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurt N, Wang HY, Akeson M, Lieberman KR. Specific nucleotide binding and rebinding to individual DNA polymerase complexes captured on a nanopore. J. Am. Chem. Soc. 2009;131:3772–3778. doi: 10.1021/ja809663f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benner S, et al. Sequence-specific detection of individual DNA polymerase complexes in real time using a nanopore. Nat. Nanotechnol. 2007;2:718–724. doi: 10.1038/nnano.2007.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanco L, Salas M. Relating structure to function in phi29 DNA polymerase. J. Biol. Chem. 1996;271:8509–8512. doi: 10.1074/jbc.271.15.8509. [DOI] [PubMed] [Google Scholar]
- 20.Salas M, Blanco L, Lazaro JM, de Vega M. The bacteriophage phi29 DNA polymerase. IUBMB Life. 2008;60:82–85. doi: 10.1002/iub.19. [DOI] [PubMed] [Google Scholar]
- 21.Ibarra B, et al. Proofreading dynamics of a processive DNA polymerase. EMBO J. 2009;28:2794–2802. doi: 10.1038/emboj.2009.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanco L, et al. Highly efficient DNA Synthesis by phage phi29 DNA polymerase. J. Biol. Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 23.Durbin R, Eddy S, Krogh A, Mitchison G. Biological Sequence Analysis. ed. 11 Cambridge University Press; Cambridge, UK: 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.