Background: Nitroalkenes are electrophilic anti-inflammatory mediators that signal via Michael addition and are metabolized in vivo.
Results: Prostaglandin reductase-1 is identified as a nitroalkene reductase.
Conclusion: Prostaglandin reductase-1 reduces fatty acid nitroalkenes to nitroalkanes, inactivating electrophilic reactivity.
Significance: A mammalian enzyme is identified that metabolizes fatty acid nitroalkenes in vivo to silence their signaling reactions.
Keywords: Eicosanoid-specific Enzymes, Inflammation, Reactive Nitrogen Species, Redox Signaling, Thiol, Electrophile, Nitro-fatty Acid, Nitroalkene
Abstract
Inflammation, characterized by the activation of both resident and infiltrated immune cells, is accompanied by increased production of oxidizing and nitrating species. Nitrogen dioxide, the proximal nitrating species formed under these conditions, reacts with unsaturated fatty acids to yield nitroalkene derivatives. These electrophilic products modulate protein function via post-translational modification of susceptible nucleophilic amino acids. Nitroalkenes react with Keap1 to instigate Nrf2 signaling, activate heat shock response gene expression, and inhibit NF-κB-mediated signaling, inducing net anti-inflammatory and tissue-protective metabolic responses. We report the purification and characterization of a NADPH-dependent liver enzyme that reduces the nitroalkene moiety of nitro-oleic acid, yielding the inactive product nitro-stearic acid. Prostaglandin reductase-1 (PtGR-1) was identified as a nitroalkene reductase by protein purification and proteomic studies. Kinetic measurements, inhibition studies, immunological and molecular biology approaches as well as clinical analyses confirmed this identification. Overexpression of PtGR-1 in HEK293T cells promoted nitroalkene metabolism to inactive nitroalkanes, an effect that abrogated the Nrf2-dependent induction of heme oxygenase-1 expression by nitro-oleic acid. These results situate PtGR-1 as a critical modulator of both the steady state levels and signaling activities of fatty acid nitroalkenes in vivo.
Introduction
Activation of the inflammatory response promotes the production of both superoxide (O2⨪) and nitric oxide (•NO). These two species react at diffusion-limited rates, giving rise to the formation of the oxidizing and nitrating product peroxynitrite (ONOO−) (1). The reactivity of ONOO− is broad, with physiological levels of carbon dioxide further promoting nitration reactions secondary to the formation of an unstable nitrosoperoxocarbonate intermediate, which immediately decomposes to generate carbonate radical and nitrogen dioxide (•NO2) (1, 2). During inflammatory responses, neutrophil activation releases myeloperoxidase (MPO),3 which catalyzes hydrogen peroxide (H2O2)-dependent oxidation of both chloride and nitrite (NO2−) with the latter reaction yielding •NO2 (3). Finally, gastric acidification of dietary NO2− induces unsaturated fatty acid nitration (4). The reactions of •NO2 and unsaturated fatty acids during digestion and inflammation yields nitroalkene products such as nitro-oleic acid (NO2-OA), nitro-linoleic acid (NO2-LA) and nitro-conjugated linoleic acid (NO2-CLA) as well as nitroalkene derivatives of other polyunsaturated fatty acids (4–6). The nitroalkene moiety renders the alkenyl carbon β to the NO2 substituent electrophilic. Consequently, nitroalkenes reversibly react with both low and high molecular weight nucleophiles by Michael addition to exert redox-regulated signaling and modulate Cys- and Zn-Cys-dependent protein function (7, 8). The levels of free nitroalkenes detected in healthy volunteers are 0.72 nm and 9.9 pmol/mg creatinine NO2-CLA for human plasma and urine, respectively (4, 9, 10). During acute metabolic and inflammatory stress, free nitroalkene tissue levels can increase in murine hearts from undetectable to 9.5 and 17.3 nm for NO2-OA and NO2-CLA respectively, following an episode of focal cardiac ischemia-reperfusion (11). These concentrations are well within the range of those required for physiological signaling (12, 13).
The addition of synthetic fatty acid nitroalkenes to biological systems induces a range of signaling actions that propagate overall anti-inflammatory and beneficial metabolic responses. In this regard nitroalkenes inhibit neutrophil degranulation, superoxide production, and proinflammatory cytokine release and decrease the expression of adhesion molecules in both immune and endothelial cells (14–17). Additionally, treatment of endothelial cells with nitroalkenes induces the expression of heme oxygenase-1 (HO-1) and HIF-1α target genes (18, 19). This effect is accompanied by Nrf2- and heat shock factor 1-dependent gene up-regulation, stimulating antioxidant defenses and the heat shock response (18, 20). As a result, nitroalkenes exert protective effects in animal models of organ infarction, neointimal hyperplasia, hypertension, and atherogenesis via non-cGMP-dependent mechanisms (11, 13, 21–23). Finally, nitroalkenes also function as partial agonists of PPARγ to ameliorate insulin resistance, normalize glycemia, and improve lipid profiles in animal models of metabolic syndrome (12, 24, 25).
While many signaling actions of nitroalkene-containing lipids have been characterized, recent evidence shows that these molecules are also actively metabolized in biological systems. Notably, <5% of NO2-OA is detectable as the native molecule in the circulation within minutes after intravenous injection (22). Michael addition accounts for one component of nitroalkene loss from the circulation, yielding protein and low molecular weight adducts (22). Additionally, fatty acid nitroalkenes are metabolized via a combination of β-oxidation and reduction to non-electrophilic nitroalkane derivatives (22, 26). Because there is a central role for Michael addition in nitroalkene regulation of Nrf2, NFκB, heat shock factor, and PPARγ signaling (7), the relevance of an enzymatic system that inactivates the electrophilic reactivity of fatty acid nitroalkenes is underscored.
Most reports showing the formation of nitrated fatty acids in vivo are limited to vertebrates (7). However, biosynthesis of other nitroalkene-containing molecules has also been described in Gram-negative bacteria, fungi, plants, and insects (27–30). Several enzymes capable of nitroalkene reduction have been described in these organisms, including pentaerythritol tetranitrate (PETN) reductase from Enterobacter cloacae PB2 and members of the old yellow enzyme family such as Escherichia coli N-ethylmaleimide reductase, Pseudomonas putida M10 morphinone reductase, and OYE1 from Saccharomyces carlsbergensis (31–33).
Herein we report that prostaglandin reductase-1 (PtGR-1) mediates NADPH-dependent reduction of fatty acid nitroalkenes. This vertebrate enzyme modulates steady state nitroalkene concentrations and the downstream signaling actions of these reactive lipid electrophiles.
EXPERIMENTAL PROCEDURES
Materials
Chemicals were of analytical grade and purchased from Sigma unless otherwise stated. Oleic, linoleic, and conjugated linoleic acids were obtained from Nu-Check Prep, Inc. (Elysian, MN), 15-oxo-PGE2 and 13,14-dihydro-15-oxo-PGE2 were purchased from Cayman Chemical (Ann Arbor, MI), and nitroalkenes were synthesized as described previously (4, 34). Nitroalkene concentration was measured daily in all synthetic stocks by using the appropriate extinction coefficients: NO2-OA ϵ268 8.22 mm−1cm−1, ϵ268 NO2-LA 8.65 mm−1cm−1 (20 mm sodium phosphate, pH 7.0), and ϵ312 NO2-CLA 11.2 mm−1cm−1 (methanol) (35, 36).
Measurement of Circulating Nitroalkene Levels
Male Sprague-Dawley rats (Harlan Laboratories, Indianapolis, IN) were injected intravenously with 16 mg/kg NO2-OA. Blood was obtained as a function of time, and plasma was promptly isolated and supplemented with NO2-[13C18]OA internal standard. Nitrated fatty acids were extracted from plasma after protein precipitation with four volumes of acetonitrile and analyzed by LC-MS/MS as described in the following section. Quantification of NO2-OA and nitro-stearic acid (NO2-SA) and its corresponding β-oxidation metabolites was performed by stable isotopic dilution analysis using NO2-OA and NO2-SA calibration curves in the presence of the NO2-[13C18]OA internal standard (34). Preliminary experiments indicate that shorter chain nitroalkenes (6-nitrononenoic acid and 6-nitrododecenoic acid) displayed ionization efficiencies and detector responses that were similar to NO2-OA. NO2-SA was synthesized by sodium borohydride reduction of synthetic NO2-OA, purified (>95% per NMR analysis) and determined to be undistinguishable from the product obtained upon total PtGR-1 reduction. Furthermore, comparison of calibration curves prepared with NO2-SA and NO2-OA in the presence of NO2-[13C18]OA indicated a similar MS response toward both molecules, thus allowing the use of NO2-OA calibration curves for quantification of both saturated and unsaturated nitro fatty acids. For detection of endogenous NO2-CLA and its reduced derivative NO2-dihydro-CLA in human circulation, plasma (1 ml) was treated with 1% sulfanilamide (final concentration) to quench any reactive nitrogen species that could be derived from the acidification of contaminating nitrite and extracted with 2 volumes of hexane/isopropyl alcohol/formic acid 1M (30:20:2) followed by the addition of 1 volume of hexane. The organic phase was isolated, dried under nitrogen, and resolvated in acetonitrile for LC-MS/MS determination. Extracts were supplemented with 15NO2-CLA and 15NO2-dihydro-CLA to confirm endogenous NO2-CLA and NO2-dihydro-CLA identification. Animals were housed in accordance with the Guide for the Care and Use of Laboratory Animals published by the United States National Institutes of Health (NIH Publication No. 85-23, revised 1996). All rodent and clinical studies were approved by the University of Pittsburgh Institutional Animal Care and Use Committee (Approval 12070398) and the Institutional Review Board (Approval 09090105).
LC-Electrospray Ionization (ESI)-MS/MS Analysis
Fatty acids were resolved by HPLC with a C18 reversed phase column (Gemini 2 × 20 mm, 3 μm; Phenomenex, Torrance, CA) using a water/acetonitrile solvent system containing 0.1% acetic acid and a 0.75 ml/min flow rate. Samples were loaded into the column in either 10 or 30% acetonitrile/acetic acid for determination of 15-oxo-PGE2 and nitro-fatty acid derivatives respectively. After loading, the percentage of organic phase was increased linearly for the next 5.1 min up to 70% for 15-oxo-PGE2 metabolites or 85% for nitro-fatty acids. The column was then washed with 100% organic phase (5.2–6.7min) and re-equilibrated to initial conditions for an additional 2.3 min. MS analysis was performed using either an API 5000 or an API Qtrap 4000 (Applied Biosystems, Framingham, MA) in the negative ion mode with the following settings: source temperature 550 °C, curtain gas 40, ionization spray voltage −4500, ion source gas 1 40, ion source gas 2 40, declustering potential −70V, entrance potential −4 V, collision energy −15V and −35V (PGE2 derivatives and nitrated fatty acids respectively), collision cell exit potential −5 V. Nitrated fatty acids were detected by collision-induced fragmentation of the parent ions to a charged nitro group (m/z 46) following the specific transitions for: NO2-OA (326.2/46), NO2-LA (324.2/46), NO2-CLA (324.2/46), and NO2-[13C18]OA (344.2/46). Transitions corresponding to reduced derivatives were obtained by adding 2 mass units to each parent compound: NO2-SA (328.2/46), NO2-dihydro-LA (326.2/46), and NO2-dihydro-CLA (326.2/46). The molecular mass of the precursor ions for the β-oxidation products was determined by subtracting 28 mass units to the corresponding parent compound as described (22). 15-Oxo-PGE2 and 13,14-dihydro-15-oxo-PGE2 were detected using MRM 349/331 and MRM 351/333, respectively.
Purification of Nitroalkene Reductase from Rat Liver
Rat liver was homogenized using a Potter-Elvehjem tissue grinder and clarified by two successive centrifugations at 15,000 × g and 100,000 × g for 30 min followed by ammonium sulfate fractionation. Nitroalkene reductase activity was maximal in the 60–70% (NH4)2SO4 pellet. This fraction was concentrated, adjusted to 1 m (NH4)2SO4, and subjected to hydrophobic exchange chromatography (HiPrep Phenyl FF, GE Healthcare). The resulting eluate was adjusted to pH 7.0 and subjected to two successive ion exchange chromatographies using DEAE- and sulfopropyl-substituted resins (TOSOH Bioscience). Consistent with previous methods developed for PtGR-1 purification, nitroalkene reductase activity does not bind to cationic resins (37, 38). Finally, the fraction with the highest activity collected from sulfopropyl chromatography was further purified using either an ADP-substituted Sepharose (Sigma) or a Blue-HP (HiTrap BlueHP, GE Healthcare) column leading to comparable results. Similar purification procedures for PtGR-1 have been described previously (37–39). Proteins were resolved by SDS-PAGE, and the three major bands were excised and digested for proteomic analysis following established protocols. Briefly, samples (1–5 μg protein) in 50 mm (NH4)HCO3, pH 8.0, were vacuum-dried, dissolved in 6 m guanidine HCl followed by the addition of 20 mm DTT, and incubated for 1 h at room temperature. Thiols were blocked with 100 mm iodoacetamide for 1 h in the dark, and the sample was digested with sequencing-grade modified trypsin (50:1 protein:trypsin) overnight at 37 °C. Peptides were analyzed on a hybrid linear ion trap-orbitrap mass spectrometer (LTQ Orbitrap, ThermoFisher Scientific) by C18 reversed phase nanoflow HPLC (Ultimate 3000, Dionex) using a 90-min liquid chromatography run and a 60-min MS acquisition time using an integrated electrospray ionization-fused silica capillary column (100 μm inner diameter × 360 μm outer diameter × 200-mm length) packed in-house (Jupiter 5 μm, 300 Å pore size stationary phase; Phenomenex). Full scan MS spectra were collected in the orbitrap; m/z range 350–1800 (r = 60,000 at m/z 400), with the 7 most intense ions selected for collision-induced dissociation (CID) in the ion trap using a normalized CID energy of 35%. Dynamic exclusion of 90 s was used for ions already selected for fragmentation to minimize redundancy. The resulting data set was searched against the UniProt rat (Rattus norvegicus) protein database (10/11 release) using SEQUEST (ThermoFisher Scientific). Peptides with minimum Xc scores of 1.9 for [M+H]1+, 2.2 for [M+2H]2+, and 3.5 for [M+3H]3+ and a minimum ΔCn of 0.08 were considered legitimate.
Nitroalkene Reductase/Prostaglandin Reductase-1 Activity Assay
Nitroalkenes or 15-oxo-PGE2 (10 μm, unless otherwise specified) were incubated with the different fractions from enzyme purification steps in 20 mm sodium phosphate buffer at pH 7.0 supplemented with 100 μm diethylene-triaminepentaacetic acid and 250 μm NADPH at the indicated temperatures and durations. Reactions were stopped by the addition of 4 volumes of methanol, and products were analyzed by LC-MS/MS. Quantification of 15-oxo-PGE2, 13,14-dihydro-15-oxo-PGE2, and nitrated fatty acids was performed using calibration curves prepared from either synthetic or commercially available standards. Electrophilicity of the reaction products was assessed by incubation with 500 mm β-mercaptoethanol in methanol/50 mm sodium phosphate, pH 7.5, (1:1) for 1 h at 37 °C before LC-MS/MS analysis. For enzyme kinetic studies, 36.2 ng of FLAG-hPtGR-1 (11.69 nm, Mr 38,700) were incubated with increasing concentrations of NO2-OA in the presence of 250 μm NADPH. Reactions were performed in 20 mm sodium phosphate buffer, pH 7.0, at 25 °C in the presence of heptadecanoic acid as an internal standard. Rates of NO2-SA formation were determined during the initial 10% of the reaction and non-linearly fitted to Michaelis-Menten kinetics using GraphPad Prism 5.0.
Nitroalkene Reductase/Prostaglandin Reductase-1 Immunodepletion
A solution containing 1 μg of rat liver-purified PtGR-1 in 20 mm sodium phosphate, pH 7.0, supplemented with 100 μm diethylene-triaminepentaacetic acid was preincubated with 0.2 volumes of a 50% Protein A/G slurry (Santa Cruz) to immunoprecipitate nonspecific binding proteins. The resulting solution was divided in half and incubated with or without 2.5 μg of anti-PtGR-1 (ProteinTech) for 1 h at 25 °C. PtGR-1 was immunoprecipitated by the addition of 0.33 volumes of 50% Protein A/G slurry followed by 2-h incubation at 4 °C and centrifugation at 10,000G. Nitroalkene and prostaglandin reductase activities were measured in the resulting supernatants.
Nitroalkene Reductase/Prostaglandin Reductase-1 Expression in HEK293T and HepG2 Cells
HEK293T and HepG2 cells (ATCC, Manassas, VA) were maintained in DMEM and Eagle's minimum essential medium media supplemented with 10% fetal bovine serum (FBS), respectively. Cells were incubated at 37 °C with 95% air and 5% carbon dioxide. To generate the pCMV-3Tag-hPtGR-1, the hPtGR-1 cDNA was inserted into pCMV-3Tag-1 (Stratagene) such that it was tagged in-frame with a 3× FLAG epitope (40). HEK293T cells were plated in 100-mm dishes at 60–70% cell density and allowed to attach overnight. pCMV-3Tag-hPtGR-1 or pCMV-3Tag-1 plasmids were transfected into cells with Lipofectamine 2000 (Invitrogen) overnight. Rat-PtGR-1 overexpression was obtained by transfection with a pCAGGS-EGFP-rat-PtGR-1 plasmid in which rat-PtGR-1 cDNA-tagged with EGFP was ligated into the mammalian expression vector pCAGGS (41, 42). Expression of recombinant as well as endogenous PtGR-1 was assessed by Western blot analysis using FLAG (Santa Cruz), GFP (Sigma), and PtGR-1 (Aviva Systems Biology) antibodies. For experiments in which NO2-OA metabolism by intact cells was followed, growth medium was collected as a function of time, and nitro-fatty acids were measured after protein precipitation by the addition of four volumes of methanol. HO-1 as well as endogenous and recombinant PtGR-1 expression was assessed by immunoblotting after 24 h incubation with 0, 2.5, and 5 μm NO2-OA in the presence of 5% FBS.
FLAG-tagged Human Prostaglandin Reductase-1 Purification for Enzyme Kinetics
HEK293T cells overexpressing FLAG-hPtGR-1 were lysed, and the recombinant enzyme was purified using a FLAG immunoprecipitation kit (Sigma). Purity was assessed by SDS-PAGE developed by silver staining, and protein identity was confirmed by Western blot. Protein concentration in the purified fraction was determined by SDS-PAGE analysis using a BSA calibration curve.
RESULTS
Nitroalkene Reduction Is a Prominent Pathway for Nitro-fatty Acid Metabolism in Vivo
Male rats were injected intravenously with 16 mg/kg NO2-OA. Consistent with previous studies, this nitroalkene fatty acid underwent β-oxidation, yielding dinor- and tetranor-nitroalkenes (Fig. 1, A–C) (22). Notably, a substantial fraction of circulating NO2-OA metabolites were the corresponding saturated derivatives (NO2-SA, dinor-NO2-SA, tetranor-NO2-SA), indicating that nitroalkene reduction is a significant reaction in vivo.
FIGURE 1.
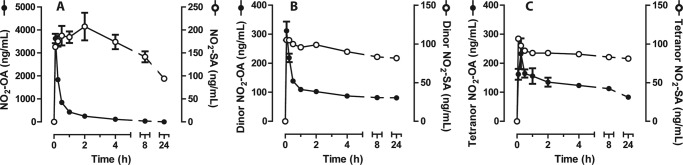
Time-dependent changes in plasma nitroalkene metabolites after a single intravenous injection of NO2-OA in rats. Concentrations of 18-carbon (A), 16-carbon (B), and 14-carbon (C) nitro fatty acid derivatives were calculated by stable isotopic dilution analysis LC-MS/MS using synthetic NO2-[13C18]OA as the internal standard and NO2-OA and NO2-SA calibration curves. Data are the means ± S.E., n = 3 animals per treatment.
Nitroalkene Reductase Purification from Rat Liver
The identity of the enzyme catalyzing nitroalkene reduction is unknown. Given that crude liver homogenates mediated NO2-OA reduction and the importance of the liver in lipid and xenobiotic metabolism (43), liver was chosen as the starting material for nitroalkene reductase purification. Fig. 2A shows a representative reaction analysis where the two-electron reduction of synthetic NO2-OA to NO2-SA is determined by LC-MS/MS. At this stage, spectrophotometric assays based on UV monitoring of NADPH oxidation were complicated by optical interference, sample turbidity, and nonspecific NADPH consumption. The MS-based method sensitively, specifically, and simultaneously reported substrate consumption and product formation regardless of the complexity of the sample, providing analytical rigor within the context of an enzyme purification procedure. As detailed under “Experimental Procedures,” the purification strategy gave fractions displaying increasing enzymatic activity and purity (Fig. 2, B and C). The last purification step, displaying a substantially enriched nitroalkene reductase fraction, consisted of either an ADP-substituted Sepharose (39) or a Blue-HP column (37, 38) that yielded similar results.
FIGURE 2.
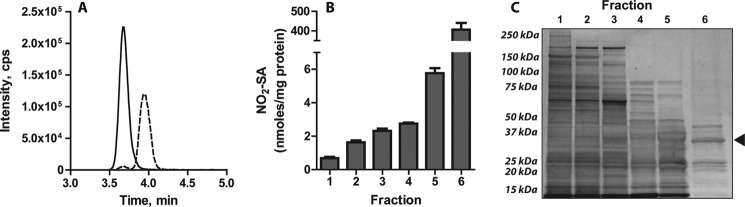
Nitroalkene reductase purification from rat liver. A, shown is a representative LC-MS/MS trace illustrating the conversion of synthetic NO2-OA (MRM 326/46, solid line) into NO2-SA (MRM 328/46, dashed line) by the nitroalkene reductase activity present in the enzyme-purified fraction prepared in B and C. B, NO2-SA production from the reduction of 50 μm NO2-OA in the presence of 250 μm NADPH at 37 °C after 10 min of incubation with different fractions obtained during the purification procedure -. Fractions are: clarified rat liver homogenate (1), 70% (NH4)2SO4 precipitate (2), eluate from hydrophobic interaction chromatography (3), eluate from DEAE chromatography (4), eluate from sulfopropyl chromatography (5), eluate from Blue-HP chromatography (6). Data are the means ± S.D. n = 3. C, silver-stained SDS-PAGE shows increases in protein purity concomitant to the purification procedure. The arrow indicates the migration distance of PtGR-1.
Nitroalkene Reductase Reduces Nitroalkene Moieties to Non-electrophilic Nitroalkanes
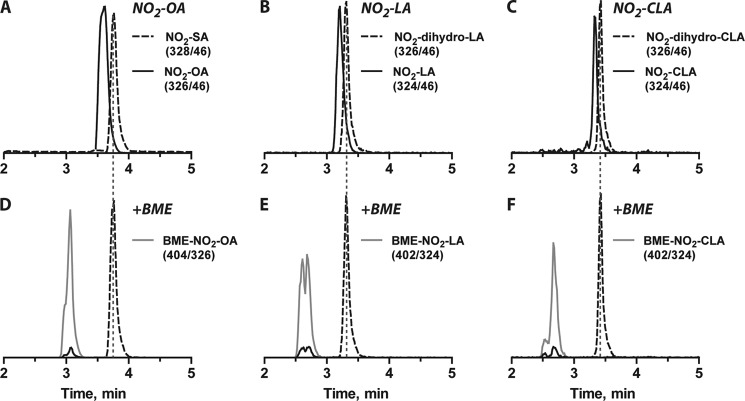
Nitroalkene reductase activity was greater with NADPH than with NADH as the reductant and functioned efficiently within a physiological pH range (not shown). Reaction of NO2-OA, NO2-LA, or NO2-CLA (corresponding to biologically relevant monounsaturated, bis-allylic, and conjugated nitro dienes, respectively) with the purified nitroalkene reductase fraction resulted in stoichiometric nitroalkene reduction to the corresponding nitroalkane or nitroalkane-alkene derivative (Fig. 3, A–C). Notably, even for polyunsaturated fatty acids, the purified nitroalkene reductase fraction was specific for the reduction of the nitroalkene moiety, yielding saturated non-electrophilic products that lost reactivity with β-mercaptoethanol. Only electrophilic nitroalkenes reacted with β-mercaptoethanol to generate the corresponding Michael adducts (Fig. 3, D–F). There was no formation of NO2-SA from either NO2-LA or NO2-CLA reduction, affirming enzyme specificity toward the nitroalkenyl double bond.
FIGURE 3.
Analysis of the electrophilic reactivity of nitro fatty acid derivatives. A–C, shown are representative LC-MS/MS peaks for both native (continuous lines) and reduced forms (dashed lines) of NO2-OA (A), NO2-LA (B), and NO2-CLA (C). D–F, incubation with β-mercaptoethanol (BME) leads to the disappearance of peaks corresponding to the electrophilic NO2-OA (D), NO2-LA (E), and NO2-CLA (F) species, whereas no reaction was observed with the enzymatic reduction products. Early-eluting peaks corresponding to transitions associated with the unsaturated nitroalkenes reflect in-source fragmentation of the β-mercaptoethanol adducts. The gray continuous lines show the appearance of the expected Michael adducts formed upon reaction of β-mercaptoethanol with the corresponding nitroalkenes. Nitroalkenes were incubated with 7.3 μg/ml enzyme-enriched fraction for 1 h at 37 °C, and electrophilicity was assessed as described under “Experimental Procedures.”
Nitroalkene Reductase Identification
Proteomic analysis of the highly enriched fraction identified PtGR-1 as the most abundant protein (top 10 hits summarized in Table 1) with only two additional proteins having reductase/dehydrogenase activity: sorbitol dehydrogenase and biliverdin reductase B. Sorbitol dehydrogenase catalyzes the NAD+-mediated oxidation of an alcohol group in sorbitol to yield fructose, whereas biliverdin reductase B mediates the NAD(P)H-dependent reduction of a double bond between the pyrrolic rings of biliverdin, yielding bilirubin. Nevertheless, both sorbitol dehydrogenase and biliverdin reductase B were identified at much lower yields than PtGR-1 (10-fold less, based on spectral count). Notably, PtGR-1 (MS coverage and Western blot shown in Fig. 4), with ∼80% of the detected collision-induced dissociation spectra located to the 36kDa band (see Fig. 2C), catalyzes the NADPH-mediated reduction of 15-oxo-PGE2 to 13,14-dihydro-15-oxo-PGE2 and reduces electrophilic α,β-unsaturated carbonyls (44, 45). This reinforced the designation of this enzyme as a candidate nitroalkene reductase.
TABLE 1.
Summary of the most abundant proteins detected in the liver nitroalkene reductase-purified fraction
Relative abundance is represented by total spectral count. No. of different peptides: number of unique peptide sequences identified. Unique: number of peptides deemed protein-specific (proteotypic) by Sequest analysis. Total spectral count: total number of CID spectra detected per protein.
| Reference | Protein | No. of different peptides | Unique | Total spectral count |
|---|---|---|---|---|
| P97584 | Prostaglandin reductase-1 | 55 | 55 | 538 |
| P00481 | Ornithine carbamoyltransferase | 33 | 1 | 95 |
| Q63276 | Bile acid-CoA:amino acid N-acyltransferase | 34 | 34 | 82 |
| P27867 | Sorbitol dehydrogenase | 22 | 14 | 44 |
| B5DF65 | Biliverdin reductase B (flavin reductase (NADPH)) | 12 | 12 | 43 |
| P00884 | Fructose-bisphosphate aldolase B | 19 | 0 | 42 |
| B6DYP8 | Glutathione S-transferase | 8 | 1 | 40 |
| P09034 | Argininosuccinate synthase | 21 | 21 | 38 |
| P07824 | Arginase-1 | 18 | 18 | 37 |
| P12346–1 | Serotransferrin | 23 | 0 | 29 |
FIGURE 4.

Identification of PtGR-1 in the nitroalkene reductase fraction. A, the PtGR-1 sequence shows peptide coverage obtained upon digestion and nanoLC-MS/MS analysis of the enriched enzyme fraction. PtGR-1 was found predominantly at the 36-kDa band, consistent with the reported molecular mass for this protein. B, shown is a PtGR-1 immunoblot analysis of the fractions collected during the purification process. Fractions are clarified rat liver homogenate (1), 70% (NH4)2SO4 precipitate (2), eluate from hydrophobic interaction chromatography (3), eluate from DEAE chromatography (4), eluate from sulfopropyl chromatography (5), eluate from Blue-HP chromatography (6).
The presence of PtGR-1 activity was confirmed in the purified fraction by following NADPH-dependent 15-oxo-PGE2 reduction by MS analysis (Fig. 5). To probe whether PtGR-1 is responsible for nitroalkene reduction, the effect of increasing concentrations of the substrate 15-oxo-PGE2 on NO2-OA reduction was evaluated (Fig. 5A). There was a dose-dependent inhibition of nitroalkene reduction upon the addition of both equimolar and 10-fold excess concentrations of 15-oxo-PGE2, consistent with substrate competition for the catalytic site. Moreover, 15-oxo-PGE2 reduction by PtGR-1 is inhibited by indomethacin (46); thus, it was reasoned that if PtGR-1 is also responsible for nitroalkene reduction then both activities should be indomethacin-sensitive. Accordingly, indomethacin dose-dependently inhibited both NADPH-dependent 15-oxo-PGE2 and NO2-OA reduction in the purified fraction (Fig. 5B). Nitroalkene reduction was also inhibited by the thiol-oxidizing agent dithionitrobenzoate (data not shown). Treatment of the purified fraction with dithionitrobenzoate induced similar extents of inhibition of both nitroalkene and 15-oxo-PGE2 reduction. In aggregate, Fig. 5, A and B, supports that PtGR-1 was responsible for the reduction of nitroalkene-containing fatty acids. At this juncture, the nitroalkene reductase fraction that was utilized was not purified to homogeneity (see Fig. 2C). Therefore, to confirm the identity of PtGR-1 as a nitroalkene reductase, anti-PtGR-1 was used to immunodeplete PtGR-1 from the nitroalkene reductase-enriched fraction. This strategy completely abolished the ability of the purified fraction to catalyze both NADPH-dependent reduction of 15-oxo-PGE2 and NO2-OA (Fig. 5C), reinforcing the notion that PtGR-1 is responsible for nitroalkene reductase activity.
FIGURE 5.
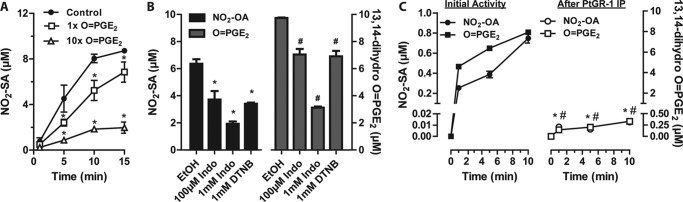
PtGR-1 is responsible for nitroalkene reduction in purified liver fractions. A, shown is dose-dependent inhibition of NO2-OA (10 μm) reduction by 10 μg/ml PtGR-1-purified fraction in the presence of 15-oxo-PGE2. Data are the means ± S.D. n = 3. Dose-dependent inhibition by 15-oxo-PGE2 is statistically significant as determined by two-way ANOVA and the Bonferroni post-test (*, p < 0.01 versus control). B, shown is the effect of indomethacin (Indo) and dithionitrobenzoate (DTNB) on both NO2-OA and 15-oxo-PGE2 reduction by 7.5 μg/ml enzyme-purified fraction after a 10-min incubation. * and #, p < 0.0001 versus respective ethanol vehicle control (EtOH) by one-way ANOVA and Bonferroni post-test. Data are the means ± S.D. n = 3. C, shown is concomitant loss of NO2-OA and 15-oxo-PGE2 reductase activities upon PtGR-1 immunoprecipitation from a solution of enzyme-purified fraction containing ∼1 μg of PtGR-1. Data are the means ±S.D. n = 3. Inhibition is significant for both OA-NO2 and 15-oxo-PGE2 reduction as determined by two way ANOVA and Bonferroni post-test versus non-immunodepleted controls. * and #, p < 0.0001.
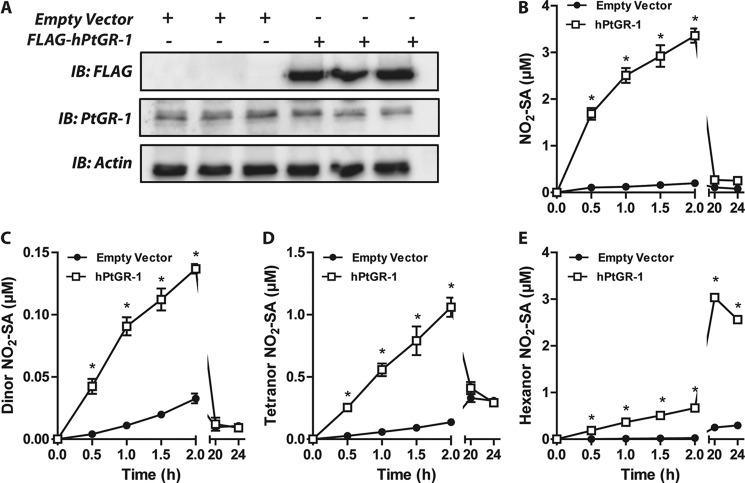
PtGR-1 Overexpression in HEK293T Cells Promotes Nitroalkene Reductase Activity
Both the rat and the human PtGR-1 were transfected and overexpressed in HEK293T cells (Figs. 6A and Fig. 8B). Overexpression of human PtGR-1 significantly increased the formation of NO2-SA and its β-oxidation metabolites (Fig. 6, B–E) in concert with decreased generation of shorter-chain unsaturated derivatives (supplemental Fig. 1). Similar results were observed upon overexpression of the rat isoform of PtGR-1 (not shown). Analysis of the reduction of NO2-OA by affinity-purified recombinant human PtGR-1 (Fig. 7) yielded typical Michaelis-Menten kinetics (Km ∼ 85 nm) and relatively high kcat/Km ratios (4.87 × 106 m−1min−1), consistent with the observation that nitroalkenes are reduced under physiological conditions to non-electrophilic products (Fig. 1, A–C).
FIGURE 6.
Human PtGR-1 overexpression increases cellular nitroalkene reductase activity. A, HEK293T cells were transfected either with an empty or a FLAG-tagged human PtGR-1 expression vector. Recombinant FLAG-hPtGR-1 migrates at ∼40 kDa (shown via anti-FLAG immunoblotting (IB)), and endogenous PtGR-1 was detected at 36kDa. B–E, time-dependent generation of reduced NO2-OA metabolites in HEK293T growth media upon cell transfection with either empty vector or human PtGR-1 gene. The initial NO2-OA was 15 μm. Data are the means ±S.E. n = 3. hPtGR-1 transfection increases the generation of the reduced 18-, 16-, 14-, and 12-carbon metabolites of NO2-OA as determined by two-way ANOVA and Bonferroni post tests. *, p < 0.01.
FIGURE 8.
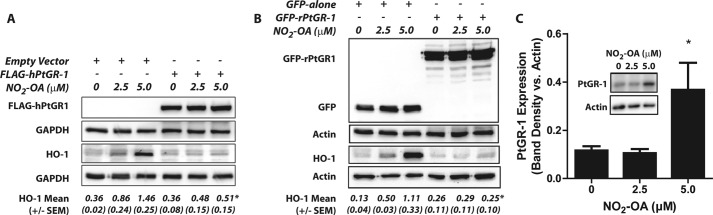
PtGR-1 modulates cellular nitroalkene signaling. A and B, representative Western blots show loss of dose-dependent HO-1 induction by NO2-OA in HEK293T cells transfected with either human (A) or rat (B) PtGR-1 expression vectors. Numbers below each blot represent mean band densitometry values obtained after normalization to the corresponding loading control with S.E. values between brackets. PtGR-1 overexpression inhibits HO-1 induction as determined by two way ANOVA and the Bonferroni post-test. n = 3. *, p < 0.01. C, NO2-OA treatment induces PtGR-1 expression in HepG2 cells (see the inset) as determined by two way ANOVA and the Bonferroni post-test. *, p < 0.01. PtGR-1 band densities are normalized to actin. Data are the means ±S.D. n = 3.
FIGURE 7.
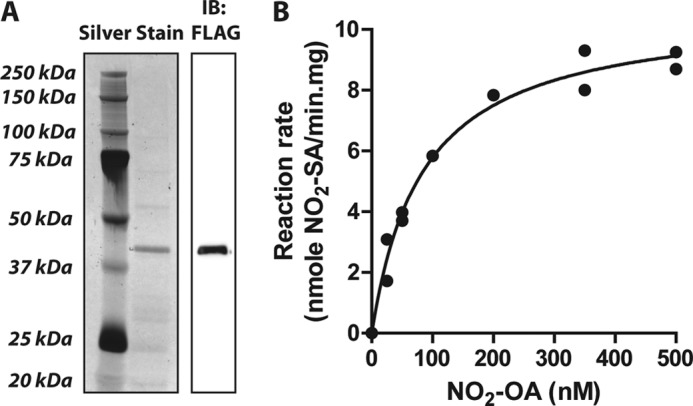
Kinetic analysis of NO2-OA reduction by purified human PtGR-1. A, shown is a representative silver-stained SDS-PAGE and FLAG immunoblot (IB) of the purified FLAG-hPtGR-1 protein utilized for kinetic analysis (purity >90%). B, shown is a Michaelis-Menten plot for the enzymatic reduction of NO2-OA by the recombinant FLAG-hPtGR-1 fraction shown in panel A. Parameters are Km = 84.71 nm, VMAX = 10.67 nmol/min·mg, kcat = 0.413 min−1, R2 = 0.9839.
PtGR-1 Modulates Nitroalkene Signaling
Elimination of the electrophilic site upon nitroalkene reduction should be accompanied by a loss in signaling activity (7). Consistent with this notion, overexpression of either the human or rat isoform of PtGR-1 in HEK293T cells inhibited the induction of Nrf2-dependent HO-1 expression by NO2-OA (Fig. 8, A and B). This affirms that PtGR-1 regulates nitroalkene signaling by the reduction of nitroalkenes to non-electrophilic nitroalkane derivatives. Furthermore, Fig. 8C shows that treatment of HepG2 cells with NO2-OA concomitantly leads to induction of significant PtGR-1, indicating the existence of a feedback mechanism capable of modulating nitroalkene signaling via regulation of PtGR-1 expression.
PtGR-1 Reduction Products Are Detected in Human Plasma
NO2-CLA is present in human plasma (4), permitting the evaluation of whether reduced nitroalkane metabolites might also be present in the circulation. To verify this possibility, a reduced 15NO2-dihydro-CLA standard was generated by incubation of synthetic 15NO2-CLA with purified PtGR-1 (Fig. 9, A and B). LC-MS/MS analysis and co-elution with 15N-labeled internal standards reveals that the saturated metabolite of NO2-CLA, NO2-dihydro-CLA, is present in human plasma (Fig. 9, C and D).
FIGURE 9.
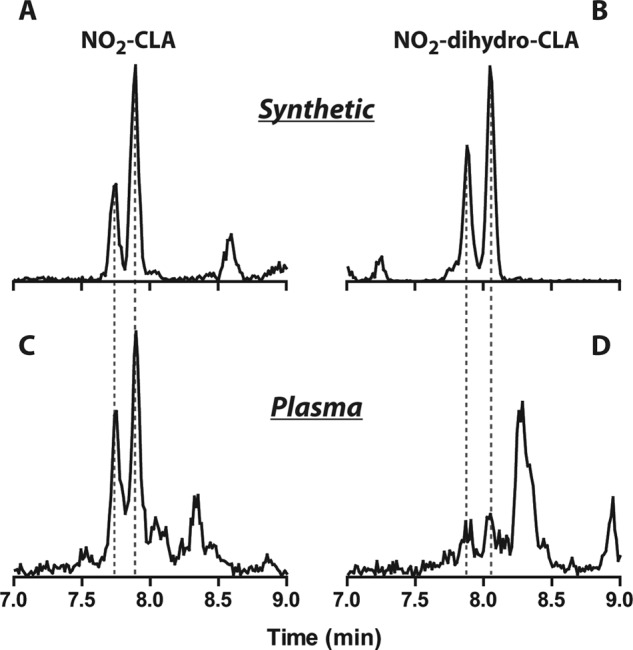
Detection of the reduction product of NO2-CLA (NO2-dihydro-CLA) in human plasma. A and B, representative LC-MS-MS data show the chromatographic profile of exogenous 15NO2-CLA and 15NO2-dihydro-CLA added to human plasma after extraction (MRM: 325/47 and 327/47, respectively). C and D, shown is detection of endogenous NO2-CLA and NO2-dihydro-CLA in healthy human plasma (MRM: 324/46 and 326/46, respectively). The reduced 15NO2-dihydro-CLA derivative was obtained by the reaction of 15NO2-CLA with PtGR-1 in the presence of NADPH for 1 h at 37 °C, pH 7.0. The 15NO2-CLA standard is a mixture of the positional isomers 9-15NO2- and 12-15NO2-CLA, which was resolved into two peaks during LC separations.
DISCUSSION
Partially-reduced oxygen species and oxides of nitrogen such as ONOO− and •NO2 are typically viewed as toxic products of inflammatory or photochemical reactions and indices of the oxidative inactivation of endogenous •NO signaling (47). Although certainly robust mediators of oxidizing and nitrating reactions at high concentrations, it is now apparent that these products also instigate salutary physiological responses. New perspective has come from the study of the electrophilic species that are present in foodstuffs and generated as byproducts of inflammatory reactions. The fact that many electrophiles limit inflammatory responses and promote beneficial metabolic reactions supports that electrophilic species can also regulate physiological homeostasis (7, 48–50).
In particular, electrophilic fatty acid nitroalkenes are generated from the reaction of unsaturated fatty acids with •NO2 derived from (a) NO2− protonation during digestion and subsequent HNO2 disproportionation and (b) •NO and NO2−-dependent oxidative inflammatory reactions (4, 51). The signaling events elicited by nitroalkenes rely on Michael addition between thiols and the electrophilic carbon β to the nitro group (7). Consistent with this notion, nitroalkenes alkylate multiple functionally significant cysteines in the transcriptional regulatory protein Keap1, the p65 subunit of NFκB, and the ligand binding domain of PPARγ, leading to the regulation of >300 genes critical for metabolic, antioxidant defense, and tissue repair responses (12, 15, 52). Although electrophilic reactivity and signaling actions are often similar for NO2-OA, NO2-LA, NO2-CLA, and NO2-arachidonic acid, there are also unique and functionally significant reactions of NO2-OA with xanthine oxidoreductase and NO2-arachidonic acid with cyclooxygenase (53, 54).
Inactivation of the electrophilic nature of nitroalkenes will abrogate any signaling actions that rely on post-translational protein modifications. Whereas nitroalkenes had been observed to be reduced in vivo, a mammalian nitroalkene reductase had not been identified (22, 26). Most current knowledge regarding the metabolism of nitroalkenes comes from bioremediation studies, as bacteria metabolize nitroaromatics by either denitrification or via reduction of the nitro group to a hydroxylamine (27, 33). Additionally, the two-electron reduction of non-aromatic nitroalkenes to nitroalkanes occurs in both bacteria and yeast (55, 56). In particular, OYE (EC 1.6.99.1) the archetypical member of the old yellow enzyme flavoprotein family, reacts with non-native, non-aromatic nitroalkenes such as nitrocyclohexene, nitrostyrene, and nitrovinylthiophene via NADPH-dependent mechanisms to generate the corresponding nitroalkane by the intermediate formation of a nitronate derivative (32). Finally, mammalian enzymes such as thioredoxin reductase, NAD(P)H:quinone oxidoreductases, NADPH:cytochrome P450 reductase, and ferredoxin:NADP+ reductase reduce nitroaromatic compounds either by one- or two-electron transfer to the nitro moiety, yielding the corresponding nitroso, hydroxylamine, or amine derivatives (57–61). The efficiency (kcat/Km) of nitro reduction by these enzymes is typically lower than that reported herein for nitroalkene reduction by PtGR-1 (60, 61), and none of these enzymes was identified in the nitroalkene reductase-purified fraction. Finally, no products suggesting reduction of the nitroalkene to nitroso, hydroxylamine, or amine derivatives were observed either in vivo or in cellular studies, supporting that fatty acid-containing nitroalkenes are not substrates for these reported activities.
PtGR-1 reduced fatty acid nitroalkene derivatives in vivo and in vitro. This is the first report of a mammalian enzyme capable of catalyzing nitroalkene reduction via conjugate hydride addition to the electrophilic π-bond. The similar electron-withdrawing character of nitro and keto groups also suggests that the mechanism of nitroalkene reduction by PtGR-1 is similar to that reported for α,β-unsaturated carbonyls (62). PtGR-1 is the product of the DIG-1 (dithiolethione-inducible gene-1) gene and is identical to the leukotriene-B4 dehydrogenase that catalyzes the oxidation of leukotriene B4 (LTB4) to 12-oxo-LTB4 in humans and rats (37). PtGR-1 is broadly distributed in mammalian tissues and also functions as a NADP(H)-dependent oxidoreductase in the inactivation of 15-oxo-prostaglandin-E2 and other α,β-unsaturated carbon-containing molecules, motivating the more inclusive name alkenal/one oxidoreductase (40, 44, 45, 63). PtGR-1 reduces fatty acid nitroalkenes to non-electrophilic nitroalkanes, as evidenced by MS analysis and a lack of reactivity of products with β-mercaptoethanol. In the case of both the bis-allylic and conjugated isomers of nitro-linoleic acid, only the nitro-containing double bond was reduced. There was no detectable formation of fully double-bond-saturated NO2-SA, demonstrating that reduction by PtGR-1 is specific for the nitroalkene moiety. Comparison of kinetic analyses (Fig. 7) with other reports (40, 45) supports that nitroalkenes exhibit some of the lowest Km values reported for PtGR-1 (Km = 0.001, 0.15, and 0.12 mm for 15-oxo-PGE2, acrolein, and 4-hydroxy-2-nonenal respectively), and kcat/Km ratios reflective of those for α,β-unsaturated carbonyls (kcat/Km = 2.4 × 109, 1.5 × 106, and 3.3 × 107 m−1min−1 for 15-oxo-PGE2, acrolein, and 4-hydroxy-2-nonenal, respectively). This supports that PtGR-1 will efficiently reduce and inactivate nitroalkenes under physiological conditions. Consistent with this, overexpression of PtGR-1 in HEK293T cells shifted nitroalkene metabolism, promoting a decrease in the levels of electrophilic NO2-OA metabolites and a concurrent increase in saturated derivatives. Furthermore, Nrf2-dependent induction of HO-1 by NO2-OA was completely abolished by PtGR-1 overexpression. This reinforces that (a) PtGR-1 is a negative modulator of nitroalkene signaling in vivo and (b) nitroalkene signaling is dependent on the post-translational modification of critical thiols in transcriptional regulatory proteins. Because PtGR-1 expression is also regulated by Nrf2 activation, the linkages between nitroalkene signaling and PtGR-1 induction are strengthened (37, 64). Notably, PtGR-1 was induced in HepG2 cells upon treatment with NO2-OA (Fig. 8), consistent with previous work showing that electrophiles efficiently induce PtGR-1 in these and other cell lines (65). Furthermore, global gene expression studies of human endothelial cells showed that PtGR-1 is among the top four transcripts induced upon treatment with NO2-OA (18). These results support that there is feedback regulation between nitroalkene levels and PtGR-1 expression, which would be expected to modulate the signaling actions of fatty acid nitroalkenes. Finally, the detection of saturated derivatives of NO2-CLA in human circulation suggests that PtGR-1 modulates steady state nitroalkene levels in healthy humans.
Partially reduced oxygen species and nitrogen oxides are now appreciated to mediate signaling reactions that are critical for both the maintenance of the homeostasis of an organism and its responses to metabolic and inflammatory stress (66–68). Cell signaling reactions are typically defined by the actuation of a reversible modification that induces specific changes either in protein or gene function, promoting the concerted propagation and amplification of cell and organ responses. Reactive species often display a broad reactivity and impact different targets at diverse reaction rates, thus potentially limiting the specificity of downstream signaling events. The reactivity and downstream responses to different redox mediators will be determined by kinetic parameters, subcellular compartmentalization, and by inactivation reactions mediated by enzymes such as catalase, glutathione peroxidase, and S-nitroso-glutathione reductase, thus modulating both the intensity and anatomic location of productive redox signals in tissue compartments. In this context, inflammatory-derived electrophilic products efficiently mediate cell signaling responses. Importantly, the reaction rates of nitroalkenes and α,β-unsaturated carbonyls with thiols is more than 2 orders of magnitude faster than H2O2 (35). Consequently, nitroalkenes effectively target specific residues in transcriptional regulatory proteins via Michael addition (Cys-285 in PPARγ, Cys-273 and -288 in Keap1, Cys-38 in p65), which can be reversed by thiol exchange with glutathione and further modulated by endogenous hydrogen sulfide generation (12, 15, 49, 52, 69). The formation of nitroalkene-glutathione adducts not only reverses electrophilic modifications but also promotes cellular export via the multidrug resistance protein 1 (MRP-1) and excretion in urine (10, 70).
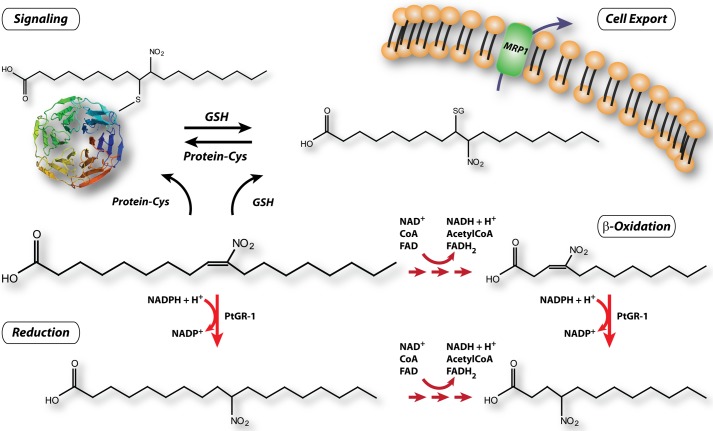
The identification of PtGR-1 as a nitroalkene reductase provides an additional mechanism for inactivation of nitroalkene reactivity, thereby defining the terminal step in the functional cycle of these signaling electrophiles (Scheme 1). Thus, electrophilic fatty acids are signaling mediators that (a) undergo relatively controlled generation during inflammatory conditions and metabolic stress, (b) display specificity in their reactivity, (c) mediate reversible modification of molecular targets, and (d) are inactivated by the efficient enzymatic mechanism reported herein. Although the evidence presented herein implies an important role for PtGR-1 in nitroalkene reduction, no conclusion can be obtained regarding the possible existence of other enzymes also capable of reducing nitroalkenes in extra-hepatic tissues.
SCHEME 1.
Nitroalkene metabolic disposition, reactivity and signaling.
The present results also infer a new component of the multifaceted interactions between eicosanoid, nitric oxide, and nitroalkene metabolism. Comparison of the kinetic parameters that govern NO2-OA reduction by PtGR-1 with those reported for 15-oxo-PGE2 saturation suggest the latter as a preferential substrate for the enzyme. This observation suggests that the signaling actions of nitroalkenes might be influenced by steady state levels of PGE2 and 15-oxo-PGE2. In this scenario, elevated levels of PGE2 and 15-oxo-PGE2 might inhibit nitroalkene reduction by PtGR-1, thus enhancing the protective signaling actions of nitroalkenes during inflammatory conditions where significant increases in cyclooxygenase activity occur (71).
In summary, PtGR-1 mediates the reduction of fatty acid nitroalkenes to non-electrophilic products that lack signaling capabilities. This represents a controlled physiological mechanism for modulating steady state levels as well as downstream metabolic and anti-inflammatory signaling actions of lipid electrophiles.
Acknowledgments
This project used University of Pittsburgh Cancer Institute shared resources that were supported in part by National Institutes of Health P30CA047904.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-HL058115, R01-HL64937, P30-DK072506, and P01-HL103455 (to B. A. F.) and R01 AT006822 (to F. J. S.). B. A. F. and F. J. S. acknowledge financial interest in Complexa, Inc.

This article contains supplemental Fig. 1.
- MPO
- myeloperoxidase
- PtGR-1
- prostaglandin reductase-1
- •NO
- nitric oxide
- •NO2
- nitrogen dioxide
- HNO2
- nitrous acid
- ONOO−
- peroxynitrite/peroxynitrous acid
- NO2-OA
- nitro-oleic acid (equimolar mixture of 9- and 10-nitro-octadec-9-enoic acid)
- NO2-LA
- nitro-linoleic acid (mixture of positional isomers of 9-, 10-, 12-, and 13-nitro-octadeca-9,12-dienoic acid)
- NO2-CLA
- conjugated nitro-linoleic acid (mixture of positional isomers of 9- and 12-nitro-octadeca-9,11-dienoic acid)
- NO2-SA
- nitro-stearic acid (equimolar mixture of 9- and 10-nitro-octadecanoic acid)
- PGE2
- prostaglandin E2
- dinor-NO2-OA (mixture of 7- and 8-nitro-hexadec-7-enoic acid)
- tetranor-NO2-OA (mixture of 5- and 6-nitro-tetradec-5-enoic acid), hexanor-NO2-OA (mixture of 3- and 4-nitro-dodec-3-enoic acid)
- dinor-NO2-SA
- mixture of 7- and 8-nitro-hexadecanoic acid
- tetranor-NO2-SA
- mixture of 5- and 6-nitro-tetradecanoic acid
- hexanor-NO2-SA
- mixture of 3- and 4-nitro-dodecanoic acid
- HO-1
- heme oxygenase-1
- PPAR
- peroxisome proliferator-activated receptor
- ANOVA
- analysis of variance
- MRM
- multiple reaction monitoring.
REFERENCES
- 1. Pacher P., Beckman J. S., Liaudet L. (2007) Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 87, 315–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gow A., Duran D., Thom S. R., Ischiropoulos H. (1996) Carbon dioxide enhancement of peroxynitrite-mediated protein tyrosine nitration. Arch Biochem. Biophys. 333, 42–48 [DOI] [PubMed] [Google Scholar]
- 3. Eiserich J. P., Hristova M., Cross C. E., Jones A. D., Freeman B. A., Halliwell B., van der Vliet A. (1998) Formation of nitric oxide-derived inflammatory oxidants by myeloperoxidase in neutrophils. Nature 391, 393–397 [DOI] [PubMed] [Google Scholar]
- 4. Bonacci G., Baker P. R., Salvatore S. R., Shores D., Khoo N. K., Koenitzer J. R., Vitturi D. A., Woodcock S. R., Golin-Bisello F., Cole M. P., Watkins S., St Croix C., Batthyany C. I., Freeman B. A., Schopfer F. J. (2012) Conjugated linoleic acid is a preferential substrate for fatty acid nitration. J. Biol. Chem. 287, 44071–44082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ferreira A. M., Ferrari M. I., Trostchansky A., Batthyany C., Souza J. M., Alvarez M. N., López G. V., Baker P. R., Schopfer F. J., O'Donnell V., Freeman B. A., Rubbo H. (2009) Macrophage activation induces formation of the anti-inflammatory lipid cholesteryl-nitrolinoleate. Biochem. J. 417, 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Manini P., Camera E., Picardo M., Napolitano A., d'Ischia M. (2008) Biomimetic nitration of the linoleic acid metabolite 13-hydroxyoctadecadienoic acid. Isolation and spectral characterization of novel chain-rearranged epoxy nitro derivatives. Chem. Phys. Lipids 151, 51–61 [DOI] [PubMed] [Google Scholar]
- 7. Schopfer F. J., Cipollina C., Freeman B. A. (2011) Formation and signaling actions of electrophilic lipids. Chem. Rev. 111, 5997–6021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bonacci G., Schopfer F. J., Batthyany C. I., Rudolph T. K., Rudolph V., Khoo N. K., Kelley E. E., Freeman B. A. (2011) Electrophilic fatty acids regulate matrix metalloproteinase activity and expression. J. Biol. Chem. 286, 16074–16081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsikas D., Zoerner A. A., Mitschke A., Gutzki F. M. (2009) Nitro-fatty acids occur in human plasma in the picomolar range. A targeted nitro-lipidomics GC-MS/MS study. Lipids 44, 855–865 [DOI] [PubMed] [Google Scholar]
- 10. Salvatore S. R., Vitturi D. A., Baker P. R., Bonacci G., Koenitzer J. R., Woodcock S. R., Freeman B. A., Schopfer F. J. (2013) Characterization and quantification of endogenous fatty acid nitroalkene metabolites in human urine. J. Lipid Res. 54, 1998–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rudolph V., Rudolph T. K., Schopfer F. J., Bonacci G., Woodcock S. R., Cole M. P., Baker P. R., Ramani R., Freeman B. A. (2010) Endogenous generation and protective effects of nitro-fatty acids in a murine model of focal cardiac ischemia and reperfusion. Cardiovasc. Res. 85, 155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schopfer F. J., Cole M. P., Groeger A. L., Chen C. S., Khoo N. K., Woodcock S. R., Golin-Bisello F., Motanya U. N., Li Y., Zhang J., Garcia-Barrio M. T., Rudolph T. K., Rudolph V., Bonacci G., Baker P. R., Xu H. E., Batthyany C. I., Chen Y. E., Hallis T. M., Freeman B. A. (2010) Covalent peroxisome proliferator-activated receptor γ adduction by nitro-fatty acids. Selective ligand activity and anti-diabetic signaling actions. J. Biol. Chem. 285, 12321–12333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J., Villacorta L., Chang L., Fan Z., Hamblin M., Zhu T., Chen C. S., Cole M. P., Schopfer F. J., Deng C. X., Garcia-Barrio M. T., Feng Y. H., Freeman B. A., Chen Y. E. (2010) Nitro-oleic acid inhibits angiotensin II-induced hypertension. Circ. Res. 107, 540–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coles B., Bloodsworth A., Clark S. R., Lewis M. J., Cross A. R., Freeman B. A., O'Donnell V. B. (2002) Nitrolinoleate inhibits superoxide generation, degranulation, and integrin expression by human neutrophils. Novel antiinflammatory properties of nitric oxide-derived reactive species in vascular cells. Circ. Res. 91, 375–381 [DOI] [PubMed] [Google Scholar]
- 15. Cui T., Schopfer F. J., Zhang J., Chen K., Ichikawa T., Baker P. R., Batthyany C., Chacko B. K., Feng X., Patel R. P., Agarwal A., Freeman B. A., Chen Y. E. (2006) Nitrated fatty acids. Endogenous anti-inflammatory signaling mediators. J. Biol. Chem. 281, 35686–35698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ichikawa T., Zhang J., Chen K., Liu Y., Schopfer F. J., Baker P. R., Freeman B. A., Chen Y. E., Cui T. (2008) Nitroalkenes suppress lipopolysaccharide-induced signal transducer and activator of transcription signaling in macrophages. A critical role of mitogen-activated protein kinase phosphatase 1. Endocrinology 149, 4086–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang H., Liu H., Jia Z., Olsen C., Litwin S., Guan G., Yang T. (2010) Nitro-oleic acid protects against endotoxin-induced endotoxemia and multiorgan injury in mice. Am. J. Physiol. Renal Physiol. 298, F754–F762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kansanen E., Jyrkkänen H. K., Volger O. L., Leinonen H., Kivelä A. M., Häkkinen S. K., Woodcock S. R., Schopfer F. J., Horrevoets A. J., Ylä-Herttuala S., Freeman B. A., Levonen A. L. (2009) Nrf2-dependent and -independent responses to nitro-fatty acids in human endothelial cells. Identification of heat shock response as the major pathway activated by nitro-oleic acid. J. Biol. Chem. 284, 33233–33241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rudnicki M., Faine L. A., Dehne N., Namgaladze D., Ferderbar S., Weinlich R., Amarante-Mendes G. P., Yan C. Y., Krieger J. E., Brüne B., Abdalla D. S. (2011) Hypoxia inducible factor-dependent regulation of angiogenesis by nitro-fatty acids. Arterioscler. Thromb. Vasc. Biol. 31, 1360–1367 [DOI] [PubMed] [Google Scholar]
- 20. Bates D. J., Smitherman P. K., Townsend A. J., King S. B., Morrow C. S. (2011) Nitroalkene fatty acids mediate activation of Nrf2/ARE-dependent and PPARγ-dependent transcription by distinct signaling pathways and with significantly different potencies. Biochemistry 50, 7765–7773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cole M. P., Rudolph T. K., Khoo N. K., Motanya U. N., Golin-Bisello F., Wertz J. W., Schopfer F. J., Rudolph V., Woodcock S. R., Bolisetty S., Ali M. S., Zhang J., Chen Y. E., Agarwal A., Freeman B. A., Bauer P. M. (2009) Nitro-fatty acid inhibition of neointima formation after endoluminal vessel injury. Circ. Res. 105, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rudolph V., Schopfer F. J., Khoo N. K., Rudolph T. K., Cole M. P., Woodcock S. R., Bonacci G., Groeger A. L., Golin-Bisello F., Chen C. S., Baker P. R., Freeman B. A. (2009) Nitro-fatty acid metabolome. Saturation, desaturation, β-oxidation, and protein adduction. J. Biol. Chem. 284, 1461–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu H., Jia Z., Soodvilai S., Guan G., Wang M. H., Dong Z., Symons J. D., Yang T. (2008) Nitro-oleic acid protects the mouse kidney from ischemia and reperfusion injury. Am. J. Physiol. Renal Physiol. 295, F942–F949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang H., Liu H., Jia Z., Guan G., Yang T. (2010) Effects of endogenous PPAR agonist nitro-oleic acid on metabolic syndrome in obese zucker rats. PPAR Res. 2010, 601562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gorczynski M. J., Smitherman P. K., Akiyama T. E., Wood H. B., Berger J. P., King S. B., Morrow C. S. (2009) Activation of peroxisome proliferator-activated receptor γ (PPARγ) by nitroalkene fatty acids. Importance of nitration position and degree of unsaturation. J. Med. Chem. 52, 4631–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schopfer F. J., Batthyany C., Baker P. R., Bonacci G., Cole M. P., Rudolph V., Groeger A. L., Rudolph T. K., Nadtochiy S., Brookes P. S., Freeman B. A. (2009) Detection and quantification of protein adduction by electrophilic fatty acids. Mitochondrial generation of fatty acid nitroalkene derivatives. Free Radic. Biol. Med. 46, 1250–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ju K. S., Parales R. E. (2010) Nitroaromatic compounds, from synthesis to biodegradation. Microbiol. Mol. Biol. Rev. 74, 250–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Winkler R., Hertweck C. (2007) Biosynthesis of nitro compounds. Chembiochem. 8, 973–977 [DOI] [PubMed] [Google Scholar]
- 29. Vrkoč J., Ubik K. (1974) 1-Nitro-trans-1-pentadecene as the defensive compound of termites. Tetrahedron Lett. 15, 1463–1464, DOI 10.1016/S0040-4039(01)82519-3 [DOI] [Google Scholar]
- 30. Piskorski R., Hanus R., Vasícková S., Cvacka J., Sobotník J., Svatos A., Valterová I. (2007) Nitroalkenes and sesquiterpene hydrocarbons from the frontal gland of three prorhinotermes termite species. J. Chem. Ecol. 33, 1787–1794 [DOI] [PubMed] [Google Scholar]
- 31. French C. E., Nicklin S., Bruce N. C. (1998) Aerobic degradation of 2,4,6-trinitrotoluene by Enterobacter cloacae PB2 and by pentaerythritol tetranitrate reductase. Appl. Environ. Microbiol. 64, 2864–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meah Y., Massey V. (2000) Old yellow enzyme. Stepwise reduction of nitro-olefins and catalysis of aci-nitro tautomerization. Proc. Natl. Acad. Sci. U.S.A. 97, 10733–10738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Williams R. E., Bruce N. C. (2002) New uses for an old enzyme. The old yellow enzyme family of flavoenzymes. Microbiology 148, 1607–1614 [DOI] [PubMed] [Google Scholar]
- 34. Woodcock S. R., Bonacci G., Gelhaus S. L., Schopfer F. J. (2013) Nitrated fatty acids. Synthesis and measurement. Free Radic. Biol. Med. 59, 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baker L. M., Baker P. R., Golin-Bisello F., Schopfer F. J., Fink M., Woodcock S. R., Branchaud B. P., Radi R., Freeman B. A. (2007) Nitro-fatty acid reaction with glutathione and cysteine. Kinetic analysis of thiol alkylation by a Michael addition reaction. J. Biol. Chem. 282, 31085–31093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baker P. R., Lin Y., Schopfer F. J., Woodcock S. R., Groeger A. L., Batthyany C., Sweeney S., Long M. H., Iles K. E., Baker L. M., Branchaud B. P., Chen Y. E., Freeman B. A. (2005) Fatty acid transduction of nitric oxide signaling. Multiple nitrated unsaturated fatty acid derivatives exist in human blood and urine and serve as endogenous peroxisome proliferator-activated receptor ligands. J. Biol. Chem. 280, 42464–42475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Primiano T., Li Y., Kensler T. W., Trush M. A., Sutter T. R. (1998) Identification of dithiolethione-inducible gene-1 as a leukotriene B4 12-hydroxydehydrogenase. Implications for chemoprevention. Carcinogenesis 19, 999–1005 [DOI] [PubMed] [Google Scholar]
- 38. Yokomizo T., Izumi T., Takahashi T., Kasama T., Kobayashi Y., Sato F., Taketani Y., Shimizu T. (1993) Enzymatic inactivation of leukotriene B4 by a novel enzyme found in the porcine kidney. Purification and properties of leukotriene B4 12-hydroxydehydrogenase. J. Biol. Chem. 268, 18128–18135 [PubMed] [Google Scholar]
- 39. Hansen H. S. (1979) Purification and characterization of a 15-ketoprostaglandin δ 13-reductase from bovine lung. Biochim. Biophys. Acta 574, 136–145 [DOI] [PubMed] [Google Scholar]
- 40. Yu X., Egner P. A., Wakabayashi J., Wakabayashi N., Yamamoto M., Kensler T. W. (2006) Nrf2-mediated induction of cytoprotective enzymes by 15-deoxy-δ12,14-prostaglandin J2 is attenuated by alkenal/one oxidoreductase. J. Biol. Chem. 281, 26245–26252 [DOI] [PubMed] [Google Scholar]
- 41. Niwa H., Yamamura K., Miyazaki J. (1991) Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 [DOI] [PubMed] [Google Scholar]
- 42. Primiano T., Gastel J. A., Kensler T. W., Sutter T. R. (1996) Isolation of cDNAs representing dithiolethione-responsive genes. Carcinogenesis 17, 2297–2303 [DOI] [PubMed] [Google Scholar]
- 43. Sevior D. K., Pelkonen O., Ahokas J. T. (2012) Hepatocytes. The powerhouse of biotransformation. Int. J. Biochem. Cell Biol. 44, 257–261 [DOI] [PubMed] [Google Scholar]
- 44. Anggård E., Larsson C., Samuelsson B. (1971) The distribution of 15-hydroxy prostaglandin dehydrogenase and prostaglandin-δ13-reductase in tissues of the swine. Acta Physiol. Scand. 81, 396–404 [DOI] [PubMed] [Google Scholar]
- 45. Dick R. A., Kwak M. K., Sutter T. R., Kensler T. W. (2001) Antioxidative function and substrate specificity of NAD(P)H-dependent alkenal/one oxidoreductase. A new role for leukotriene B4 12-hydroxydehydrogenase/15-oxoprostaglandin 13-reductase. J. Biol. Chem. 276, 40803–40810 [DOI] [PubMed] [Google Scholar]
- 46. Clish C. B., Sun Y. P., Serhan C. N. (2001) Identification of dual cyclooxygenase-eicosanoid oxidoreductase inhibitors. NSAIDs that inhibit PG-LX reductase/LTB(4) dehydrogenase. Biochem. Biophys. Res. Commun. 288, 868–874 [DOI] [PubMed] [Google Scholar]
- 47. Beckman J. S., Koppenol W. H. (1996) Nitric oxide, superoxide, and peroxynitrite. The good, the bad, and ugly. Am. J. Physiol. 271, C1424–C1437 [DOI] [PubMed] [Google Scholar]
- 48. Higdon A., Diers A. R., Oh J. Y., Landar A., Darley-Usmar V. M. (2012) Cell signalling by reactive lipid species. New concepts and molecular mechanisms. Biochem. J. 442, 453–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. West J. D., Marnett L. J. (2006) Endogenous reactive intermediates as modulators of cell signaling and cell death. Chem. Res. Toxicol. 19, 173–194 [DOI] [PubMed] [Google Scholar]
- 50. Kansanen E., Jyrkkänen H. K., Levonen A. L. (2012) Activation of stress signaling pathways by electrophilic oxidized and nitrated lipids. Free Radic. Biol. Med. 52, 973–982 [DOI] [PubMed] [Google Scholar]
- 51. Rocha B. S., Gago B., Barbosa R. M., Lundberg J. O., Radi R., Laranjinha J. (2012) Intragastric nitration by dietary nitrite. Implications for modulation of protein and lipid signaling. Free Radic. Biol. Med. 52, 693–698 [DOI] [PubMed] [Google Scholar]
- 52. Kansanen E., Bonacci G., Schopfer F. J., Kuosmanen S. M., Tong K. I., Leinonen H., Woodcock S. R., Yamamoto M., Carlberg C., Ylä-Herttuala S., Freeman B. A., Levonen A. L. (2011) Electrophilic nitro-fatty acids activate NRF2 by a KEAP1 cysteine 151-independent mechanism. J. Biol. Chem. 286, 14019–14027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kelley E. E., Batthyany C. I., Hundley N. J., Woodcock S. R., Bonacci G., Del Rio J. M., Schopfer F. J., Lancaster J. R., Jr., Freeman B. A., Tarpey M. M. (2008) Nitro-oleic acid, a novel and irreversible inhibitor of xanthine oxidoreductase. J. Biol. Chem. 283, 36176–36184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Trostchansky A., Bonilla L., Thomas C. P., O'Donnell V. B., Marnett L. J., Radi R., Rubbo H. (2011) Nitroarachidonic acid, a novel peroxidase inhibitor of prostaglandin endoperoxide H synthases 1 and 2. J. Biol. Chem. 286, 12891–12900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stuermer R., Hauer B., Hall M., Faber K. (2007) Asymmetric bioreduction of activated C=C bonds using enoate reductases from the old yellow enzyme family. Curr. Opin. Chem. Biol. 11, 203–213 [DOI] [PubMed] [Google Scholar]
- 56. Messiha H. L., Munro A. W., Bruce N. C., Barsukov I., Scrutton N. S. (2005) Reaction of morphinone reductase with 2-cyclohexen-1-one and 1-nitrocyclohexene. Proton donation, ligand binding, and the role of residues histidine 186 and asparagine 189. J. Biol. Chem. 280, 10695–10709 [DOI] [PubMed] [Google Scholar]
- 57. Boland M. P., Knox R. J., Roberts J. J. (1991) The differences in kinetics of rat and human DT diaphorase result in a differential sensitivity of derived cell lines to CB 1954 (5-(aziridin-1-yl)-2,4-dinitrobenzamide). Biochem. Pharmacol. 41, 867–875 [DOI] [PubMed] [Google Scholar]
- 58. Cenas N., Prast S., Nivinskas H., Sarlauskas J., Arnér E. S. (2006) Interactions of nitroaromatic compounds with the mammalian selenoprotein thioredoxin reductase and the relation to induction of apoptosis in human cancer cells. J. Biol. Chem. 281, 5593–5603 [DOI] [PubMed] [Google Scholar]
- 59. Knox R. J., Boland M. P., Friedlos F., Coles B., Southan C., Roberts J. J. (1988) The nitroreductase enzyme in walker cells that activates 5-(aziridin-1-yl)-2,4-dinitrobenzamide (Cb-1954) to 5-(aziridin-1-yl)-4-hydroxylamino-2-nitrobenzamide is a form of Nad(P)H dehydrogenase (quinone) (Ec-1.6.99.2). Biochem. Pharmacol. 37, 4671–4677 [DOI] [PubMed] [Google Scholar]
- 60. Knox R. J., Chen S. (2004) Quinone reductase-mediated nitro reduction. Clinical Applications. Methods Enzymol. 382, 194–221 [DOI] [PubMed] [Google Scholar]
- 61. Nemeikaite-Ceniene A., Sarlauskas J., Miseviciene L., Anusevicius Z., Maroziene A., Cenas N. (2004) Enzymatic redox reactions of the explosive 4,6-dinitrobenzofuroxan (DNBF). Implications for its toxic action. Acta Biochim. Pol. 51, 1081–1086 [PubMed] [Google Scholar]
- 62. Dick R. A., Kensler T. W. (2004) The catalytic and kinetic mechanisms of NADPH-dependent alkenal/one oxidoreductase. J. Biol. Chem. 279, 17269–17277 [DOI] [PubMed] [Google Scholar]
- 63. Dick R. A., Yu X., Kensler T. W. (2004) NADPH alkenal/one oxidoreductase activity determines sensitivity of cancer cells to the chemotherapeutic alkylating agent irofulven. Clin. Cancer Res. 10, 1492–1499 [DOI] [PubMed] [Google Scholar]
- 64. Zhang Y., Munday R. (2008) Dithiolethiones for cancer chemoprevention. Where do we stand? Mol. Cancer Ther. 7, 3470–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yu X., Erzinger M. M., Pietsch K. E., Cervoni-Curet F. N., Whang J., Niederhuber J., Sturla S. J. (2012) Up-regulation of human prostaglandin reductase 1 improves the efficacy of hydroxymethylacylfulvene, an antitumor chemotherapeutic agent. J. Pharmacol. Exp. Ther. 343, 426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Winterbourn C. C. (2008) Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 4, 278–286 [DOI] [PubMed] [Google Scholar]
- 67. Dickinson B. C., Chang C. J. (2011) Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat. Chem. Biol. 7, 504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sena L. A., Chandel N. S. (2012) Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 48, 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nishida M., Sawa T., Kitajima N., Ono K., Inoue H., Ihara H., Motohashi H., Yamamoto M., Suematsu M., Kurose H., van der Vliet A., Freeman B. A., Shibata T., Uchida K., Kumagai Y., Akaike T. (2012) Hydrogen sulfide anion regulates redox signaling via electrophile sulfhydration. Nat. Chem. Biol. 8, 714–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Alexander R. L., Bates D. J., Wright M. W., King S. B., Morrow C. S. (2006) Modulation of nitrated lipid signaling by multidrug resistance protein 1 (MRP1). Glutathione conjugation and MRP1-mediated efflux inhibit nitrolinoleic acid-induced, PPARγ-dependent transcription activation. Biochemistry 45, 7889–7896 [DOI] [PubMed] [Google Scholar]
- 71. Giroux M., Descoteaux A. (2000) Cyclooxygenase-2 expression in macrophages. Modulation by protein kinase Cα. J. Immunol. 165, 3985–3991 [DOI] [PubMed] [Google Scholar]