Abstract
We previously suggested links between specific XPD mutations in the fetal genome and the risk of placental maldevelopment and preeclampsia, possibly due to impairment of Transcription Factor (TF)IIH-mediated functions in placenta. To identify the underlying mechanisms, we conducted the current integrative analysis of several relevant transcriptome data sources. Our meta-analysis revealed downregulation of TFIIH subunits in preeclamptic placentas. Our overall integrative analysis suggested that, in the presence of hypoxia and oxidative stress, EGFR signaling deficiency, which can be caused by TFIIH impairment as well as by other mechanisms, results in ATF3 upregulation, inducing mediators of clinical symptoms of preeclampsia such as FLT1 and ENG. EGFR- and ATF3-dependent pathways play prominent roles in cancer development. We propose that dysregulation of these canonical cancer molecular pathways occurs in preeclampsia and delineate the relevance of TFIIH, providing etiologic clues which could eventually translate into a therapeutic approach.
We previously reported that specific abnormalities in several DNA repair and transcription genes in the fetal genome were associated with gestational complications such as preeclampsia1. Our initial reports, based on novel clinical observations and systematic genetic epidemiologic investigations in families with trichothiodystrophy (TTD), a rare (affected frequency of 1 in 106) recessive disorder of DNA repair and transcription2, identified significantly increased risk of preeclampsia and other gestational complications1,3. Mutations in XPD (ERCC2), XPB (ERCC3), and TTD-A (GTF2H5), genes involved in the nucleotide excision repair (NER) pathway and in transcription4, can cause TTD. These genes code for subunits of transcription factor (TF)IIH, a component of RNA Polymerase-II (Pol-II) transcription machinery5. In addition to its role in basal and activated transcription, TFIIH also has a role in NER6. Mutations in XPD and XPB can cause other rare recessive DNA repair disorders such as xeroderma pigmentosum (XP)2,7. Our subsequent study of XP and TTD suggested a link between TTD- but not XP-associated XPD mutations in the fetus and the risk of placental maldevelopment and preeclampsia, possibly through impairment of TFIIH-mediated functions in placenta8. The exact mechanism by which these mutations affect placental development remains to be elucidated.
Preeclampsia affects up to 7% of all pregnancies, and is a major cause of severe intrauterine growth restriction (IUGR) and preterm birth9. Preeclampsia has the potential to develop into severe preeclampsia or hemolysis, elevated liver enzymes and low platelets (HELLP) syndrome, potentially fatal conditions for both mother and infant10. Nearly all candidate-gene and genome-wide association studies (GWAS) of preeclampsia to date have focused on markers on the maternal genome. While these studies have identified several potential genetic markers11,12,13,14,15,16,17, major etiologic factor(s) and biologic mechanism(s) leading to preeclampsia and most other gestational complications remain elusive. There have been no integrative transcriptome analyses, including meta-analysis, of preeclampsia reported in the literature. Meta-transcriptome analysis of case-control datasets provides a benefit over individual analysis of each dataset by allowing efficient elimination of false-positive findings pertaining to experimental and design conditions such as those related to platforms and probes. Furthermore, integrative approaches to analysis of microarray data from several data sources relevant to a disease condition are believed to enable extraction of deeper biological insights compared to that achieved through single-dimensional analysis18.
The current study was conducted to decipher the biologic mechanisms which underlie the association between preeclampsia, fetal genotype, and impairment of TFIIH-mediated functions in placenta. We designed an integrative analysis of several relevant data sources containing gene expression arrays of fetal-derived tissues. First, we conducted meta-analysis of gene expression patterns in placenta from case-control studies of preeclampsia. A preeclampsia-specific gene list obtained from this meta-analysis was then interrogated in several relevant data sources, which included expression arrays of normal and time-course placentas (i.e., placenta from first, second and third trimester pregnancies), and XPDTTD fibroblasts (i.e., cells predisposed to preeclampsia). These data sources were specifically chosen in order to filter the differentially-expressed genes and pathways to select those which preceded the development of the clinical symptoms of preeclampsia, as depicted on the Study Design Flow Chart (Figure S1).
Results
Meta-analysis of placental gene expression patterns in preeclampsia case-control studies
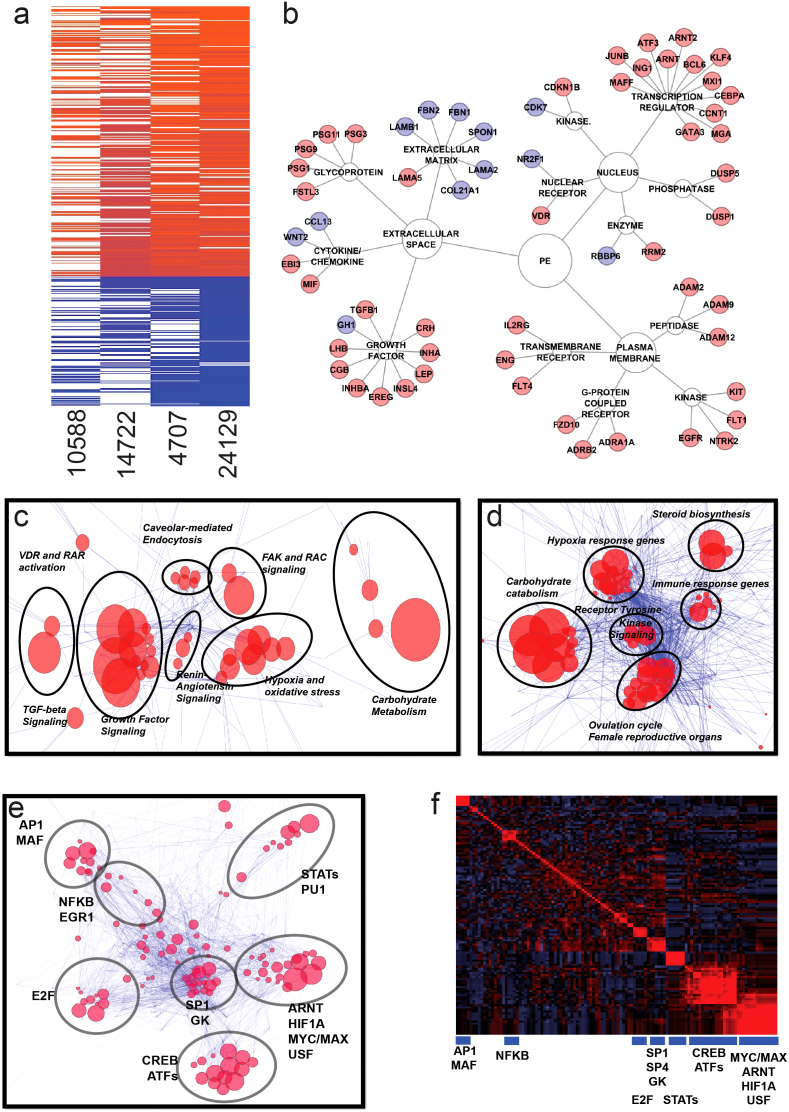
Meta-analysis of datasets GSE1058819, GSE1472220, GSE470721, and GSE2412922, containing gene expression profiles from fetal cell-derived histological subsections of placenta in case-control studies of preeclampsia, revealed 419 differentially-regulated genes (136 downregulated and 283 upregulated) coincident among the four datasets at false discovery rate (FDR) < 0.05 (Figure 1a and Table S1).
Figure 1. Identification of preeclampsia-associated genes.
Panel (a). Meta-analysis of four datasets (GSE1058819, GSE1472220, GSE470721, and GSE 2412922) using Fisher's combined probability test. Genes found significant in at least two of four datasets and having the same direction of the fold change were considered for further analysis. Panel (b). Selected representative genes found significant in the meta-analysis grouped based on the type of encoded protein. Upregulated and downregulated genes are marked by red and blue color, respectively. Panel (c). Canonical pathway analysis through the use of IPA (Ingenuity® Systems, www.ingenuity.com) depicting reconstructed pathways containing majority of differentially-regulated genes in preeclampsia. Edge-weighted layout of the network was used to visualize super-clusters of the pathways and genes and ClusterOne Cytoscape plugin52 was used to identify pathways with the highest number of common genes. Those pathways are inside circles outlined by black lines and are named based on the most common types of pathways within the group. Size of the node is inversely proportional to the p-values. Panel (d). Gene Ontology analysis using Biobase International (http://www.biobaseinternational.com) datasets and network construction conducted similar to Panel (c) above. Panel (e). Transcription factor analysis of genes upregulated in preeclmapsia using Biobase software (TRANSFAC database) (http://www.biobaseinternational.com). Genes and their transcription factors were used to construct a network as in Panel (c). Panel (f). Correlogram of transcription factor matrices sharing common genes. Clusters were selected according to the dendrogram. Transcription factors representing groups of transcription factor matrices are shown below blue boxes.
Among genes significantly upregulated in preeclamptic placentas were those coding for growth factors (such as INHBA and TGFB1), growth factor receptors (including EGFR, FLT1 and ENG), and transcription regulators of EGF-dependent pathways (including ATF3) (Figure 1b). Among the genes significantly downregulated in preeclamptic placentas were those coding for extracellular matrix proteins (such as FBN2) (Figure 1b) and proteins involved in lipid metabolism (Table S1). Interestingly, CDK7, which codes for a component of the cdk-activating kinase (CAK) domain of TFIIH, was among significantly downregulated genes in preeclampsia meta-analysis (Figure 1b and Table S1).
Pathway analysis is considered a valuable tool in estimating functions of genes in different systems23. A preeclampsia-specific gene list was used to delineate affected pathways. Canonical pathway analysis revealed several significantly dysregulated pathways in preeclampsia meta-analysis including PTEN, growth hormone, EGF, RAR, VDR/RXR, NRF2-mediated, and HIF1A signaling pathways (Data not shown). Interconnectivity pathway analysis conducted to identify super clusters of affected pathways revealed dominance of growth factor signaling, hypoxia and oxidative stress response, and carbohydrate metabolism (Figure 1c). Similar interconnectivity analysis applied to Gene Ontology (GO) pathway analysis also revealed clusters of overlapping affected pathways as those regulating hypoxia response, tyrosine kinase signaling, immune response, and carbohydrate metabolism, (Figure 1d).
We used the Transcription Factor (TRANSFAC) database followed by network construction of matrices and genes in order to identify transcription factors of the differentially-regulated genes in preeclampsia. The largest group of transcription factor matrices in the preeclampsia-specific gene network was related to oxidative stress response and hypoxia (ARNT, HIF1A and USF) (Figure 1e and 1f). Another large group of predicted transcription factors contained common elements of cAMP response element-binding (CREB) and activating transcription factor (ATF) (Figure 1e and 1f), which is probably indicative of involvement of ATF/CREB transcription complex.
Analysis of gene expression patterns in normal and time-course placenta
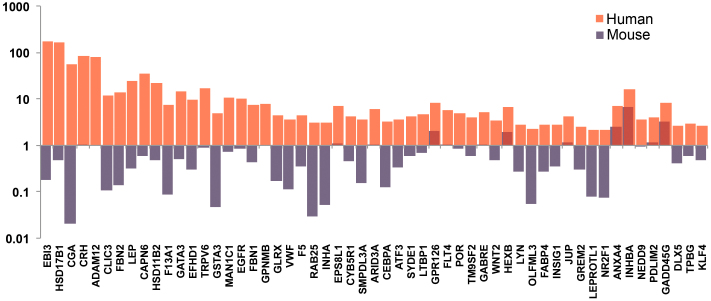
To confirm the relevance of preeclampsia-specific genes identified through the meta-analysis described above, we compared human (GSE 95)24 and mouse (GSE 97)24 datasets containing gene expression patterns of normal tissue with respect to the ratios of expression of preeclampsia-specific genes in placenta versus all other tissues. The rationale for this comparison was that wild-type mice bred for research do not develop preeclampsia spontaneously (i.e., without chemical or surgical manipulation). We found that among 283 genes upregulated in preeclampsia meta-analysis, approximately 50 genes were more than 3-fold higher upregulated in human placentas compared to mouse placentas. We postulated that the genes most relevant to preeclampsia were among these 50 genes, which included EGFR, MAN1C1, ADAM12, ATF3, INHBA, FBN2 and GABRE among others (Figure 2).
Figure 2. Expression of preeclampsia-specific genes in human and mouse placenta versus mean expression in other organs.
Degree of gene overexpression in placenta compared to other normal tissue in human (GSE9524) and mouse (GSE9724) datasets. Figure also depicts comparison of the ratio of expression of each selected gene in placenta versus all other normal tissue between human (GSE9524) and mouse (GSE9724) datasets. Genes used in this comparison were selected from among those identified through meta-analysis of preeclampsia case-control studies (Figure 1, Panel (a)). Y-axis represents the ratio of expression of each gene. Red bars indicate human and blue bars mouse genes.
Preeclampsia is identified clinically as hypertension that first occurs after 20 weeks gestation9; therefore, we speculated that genes differentially-regulated during mid- to term-gestation would be important predictors of molecular pathways impaired in preeclampsia. We conducted time-course analysis of gene expression patterns in normal placenta from first to third trimester in GDS252825. This analysis identified 710 placental genes differentially-expressed during mid- to term-gestation (i.e., 24–40 weeks) compared to first trimester; 303 genes were upregulated and 407 were downregulated (Data not shown). The gene lists obtained from analysis of normal and time-course placentas were used in the integrative analysis described later in this section.
Analysis of gene expression patterns in hypoxic trophoblasts
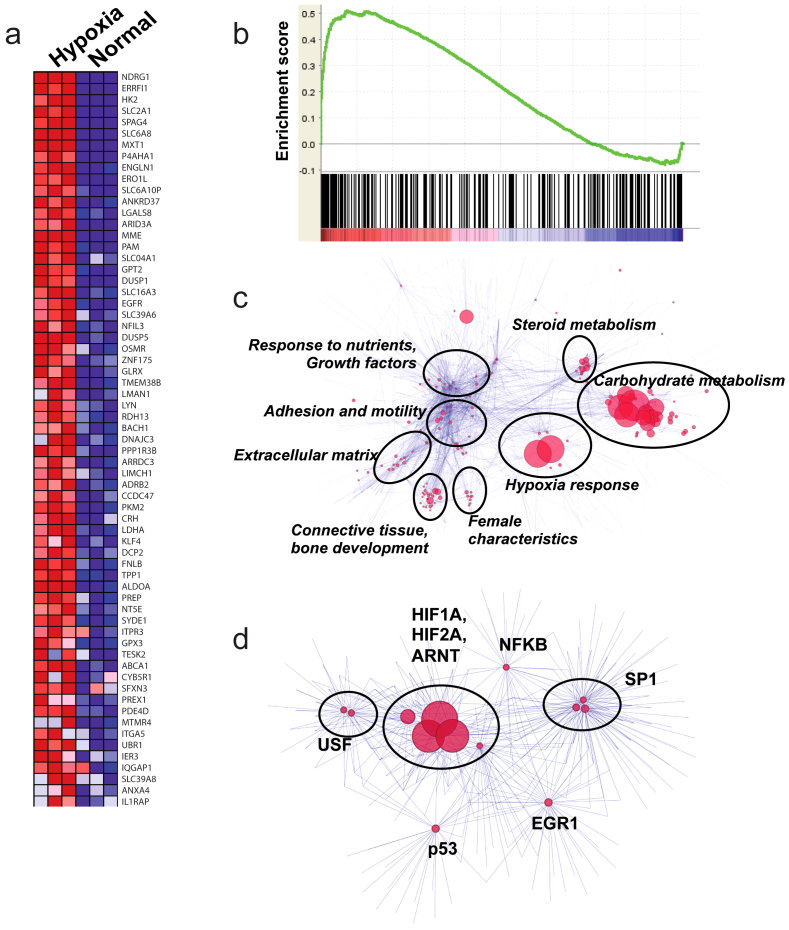
Hypoxia (along with oxidative stress response) was among the dominant affected pathways in the preeclampsia meta-analysis (Figures 1c and 1d) with HIF1A signaling being the most significant regulator of differentially-expressed genes in this pathway (Figures 1e and 1f). In order to test whether hypoxia was the main regulator of a significant number of genes in the preeclampsia gene list, we interrogated preeclampsia-specific genes in hypoxic human trophoblasts (Swan 71 human trophoblast cells under chemical hypoxia by CoCl2-treatment) in an in vitro system (GSE31679)26. We found that among 283 genes upregulated in preeclampsia meta-analysis, 65 were also upregulated in hypoxic trophoblasts (Figure 3a) (total number of upregulated genes in hypoxic trophoblasts was 480). The Gene Set Enrichment Analysis (GSEA) of hypoxic trophoblasts identified significant similarity (enrichment score of 0.51, q < 0.1) between genes upregulated in preeclampsia and those upregulated in hypoxia, confirming hypoxia as a major regulator of preeclampsia gene signature (Figure 3b). Since GSEA of hypoxic trophoblasts using the list of genes downregulated in preeclampsia was not significant, we focused on upregulated genes for pathway (Figure 3c) and transcription factor analysis (Figure 3d). Among upregulated genes coincident in preeclampsia and in hypoxic trophoblasts were EGFR, PI3K and ADRB2. GO analysis of all coincident upregulated genes showed dominance of pathways related to hypoxia and carbohydrate metabolism (Figure 3c). Transcription factor analysis showed dominance of HIF1A, HIF2A, and ARNT in regulating the gene signature of hypoxic trophoblasts (Figure 3d).
Figure 3. Transcriptome analysis of hypoxic trophoblasts.
Panel (a). Heatmap constructed using 283 genes upregulated in preeclampsia meta-analysis demonstrating the 65 genes significantly upregulated in hypoxic trophoblasts. Panel (b). GSEA of hypoxic trophoblasts with respect to genes upregulated in preeclampsia. The analysis demonstrates significant correlation (p < 0.001, Enrichment Score = 0.50) between the majority of upregulated genes in preeclampsia and in hypoxic trophoblasts. Panel (c). GO analysis of the gene signature of hypoxic trophoblasts using network visualization and analysis tools (as described in the Methods). Size of the nodes is inversely proportional to the p-values. Groups identified using clustering algorithm are shown in circles, most common GO terms are listed next to the circles. Panel (d). Transcription factor analysis of the genes upregulated in hypoxic trophoblasts using network visualization and analysis tool. Groups identified using clustering algorithm are shown in circles, most common transcription factors are listed next to the circles. Size of the nodes is inversely proportional to the p-value.
We found no evidence of upregulation of FLT1 or ENG in hypoxic trophoblasts, in contrast to the belief that hypoxia has a direct role in FLT1 and ENG induction. To confirm these findings, we also analyzed transcriptome profiling of umbilical vein endothelial cells during hypoxia and after reoxygenation (GSE 1041)27; our analysis showed that FLT1 level was reduced during hypoxia and restored after reoxygenation, suggesting that hypoxia alone was not sufficient to drive expression of FLT1 (Data not shown).
Analysis of gene expression patterns in XPDTTD fibroblasts
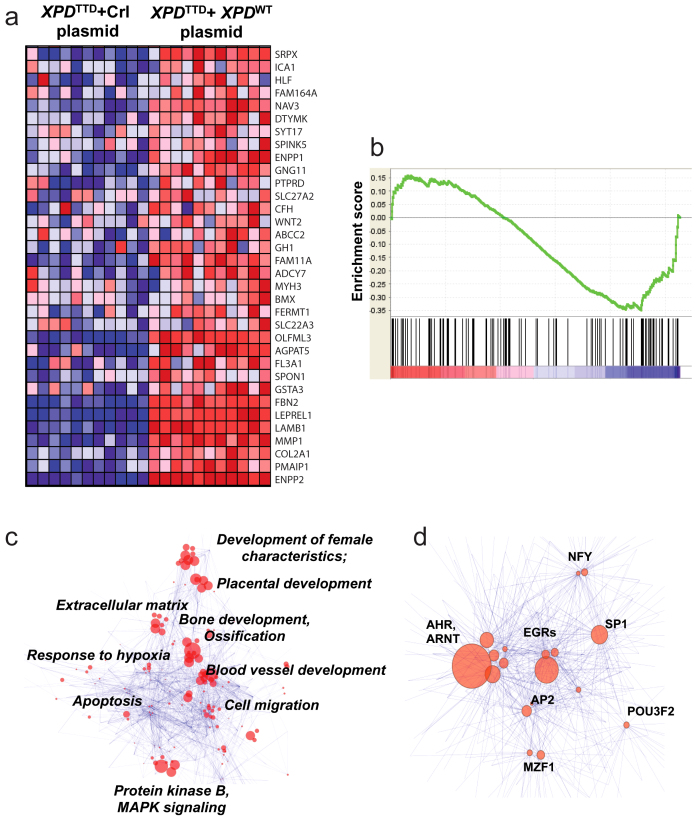
Global gene expression profiling of XPDTTD fibroblasts (i.e., fibroblasts from TTD patients with mutations in XPD) versus XPDTTD fibroblasts transfected with the wild type XPD in a dataset obtained through personal communications28 identified 660 genes differentially-regulated in XPDTTD fibroblasts. The majority of the 314 upregulated and 346 downregulated genes were involved in cell cycle and growth (data not shown). Similar to the previous analysis of hypoxic trophoblasts, we performed GSEA using genes upregulated and downregulated in preeclampsia to detect similarities between gene signatures of XPDTTD mutant cells and those of preeclampsia. We found that genes downregulated in XPDTTD mutant cells had significant similarity (q < 0.1) with those downregulated in preeclampsia (Figures 4a and 4b), suggesting potential similarities between downstream pathways affected by XPD mutations and pathways affected in preeclampsia.
Figure 4. Transcriptome analysis of XPDTTD fibroblasts.
Panel (a). Heatmap constructed using 136 genes downregulated in preeclampsia meta-analysis demonstrating the 34 genes significantly-downregulated in XPDTTD Fibroblasts versus XPDTTD Fibroblasts transfected with wild-type XPD. Panel (b). GSEA of XPDTTD Fibroblasts with respect to genes downregulated in preeclampsia. The analysis demonstrates significant correlation (q < 0.1, Enrichment Score = 0.15) between many genes downregulated in preeclampsia and in XPDTTD Fibroblasts. Panel (c). GO analysis of gene signature of XPDTTD Fibroblasts using network visualization and analysis tool (as described in the Methods). Size of the nodes is inversely proportional to the p-values. Groups identified using clustering algorithm; most common GO terms are listed next to the node groups. Panel (d). Transcription factor analysis of the genes downregulated in XPDTTD fibroblasts using network visualization and analysis tool. Groups identified using clustering algorithm; most common transcription factors are listed next to the nodes. Size of the nodes is inversely proportional to the p-values.
Among the most significant GO pathways were those associated with placental development and development of female characteristics (examples included FSTL3, INHBA and ADAMTS1 which were downregulated) as well as response to hypoxia (Figure 4c); these were also among pathways found dysregulated in preeclampsia meta-analysis. Other dowregulated pathways included extracellular matrix and bone and blood vessel development (Figure 4c), which were identified due to the overrepresentation of genes playing roles in extracellular matrix formation, which is consistent with our understanding of TTD as a condition of chronic transcription deficiency. Transcription factor analysis of downregulated genes in XPDTTD showed dominance of AHR and ARNT as well as EGR transcription factor group (Figure 4d).
Integrative analysis of all data sources to delineate mechanisms in preeclampsia
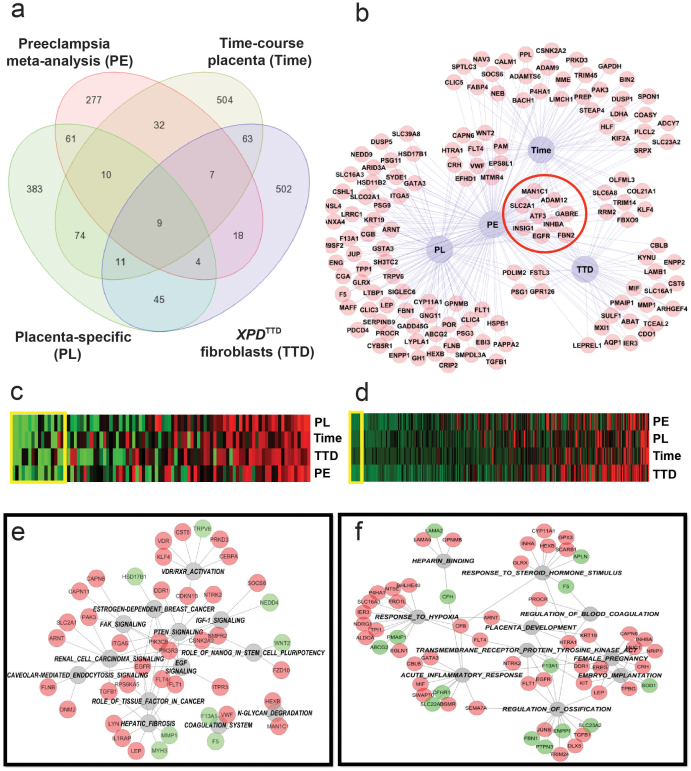
Comparison of the gene lists from the meta-analysis of preeclampsia case-control studies and three of the individual analyses described above (normal human placental gene signature obtained by comparing placenta versus other tissues, placental genes differentially-expressed during mid- to term-gestation compared to first-trimester, and genes differentially-expressed in XPDTTD fibroblasts versus XPDTTD fibroblasts transfected with wild-type XPD) identified nine differentially-regulated genes (EGFR, INHBA, ADAM12, ATF3, FBN2, INSG1, SLC2A1, MAN1C1, GABRE) coincident among all datasets (Figures 5a & 5b). The probability of finding nine common differentially-expressed genes at random among the four datasets was estimated as <10−4. EGFR, ATF3, ADAM12, MAN1C1, SLC2A1, INHBA and INSIG1 were upregulated while GABRE and FBN2 were downregulated in preeclamptic placentas in our meta-analysis (Table S1). The four most significant differentially-regulated genes among all analyzed datasets were EGFR, INHBA, ADAM12 (all belonging to the EGF pathway) and ATF3. EGFR (Figure 3a) and ATF3 (Data not shown) were also upregulated in hypoxic trophoblasts. EGFR was downregulated in XPDTTD fibroblasts suggesting signaling deficiency associated with this factor in cells with XPD-associated TTD mutations.
Figure 5. Integrative analysis of selected datasets related to the development of preeclampsia.
Panel (a). Selected datasets: 1) Gene expression arrays in placenta from four case-control studies of preeclampsia [GSE1058819, GSE1472220, GSE 2412922, and GSE470721], 2) placental gene signature obtained by comparing placenta versus other normal human tissues [GSE9524], 3) genes differentially expressed during mid- to term-gestation compared to first trimester [GDS252825], and 4) genes differentially-expressed in XPDTTD fibroblasts (cells predisposed to preeclampsia) versus XPDTTD fibroblasts transfected with wild-type XPD [obtained through personal communications28]. Individual analyses of each dataset and meta-analysis of preeclampsia case-control studies were done as discussed above. Venn diagrams created to display number of common genes between datasets. Panel (b). Differentially-regulated genes common between preeclampsia and at least one other dataset from panel A shown as a network. Common genes between all four datasets are outlined with a red circle. Panel (c–d). Canonical (Panel (c)) and GO (Panel (d)) pathway analysis using genelists created from analyses depicted in Panel (a). P-values from all analyses were combined using Fisher's test and displayed as a heatmap (green and red depict lowest and highest p-values, respectively). Yellow boxes on heatmaps depict pathways with combined p<0.01. These pathways were used for analyses depicted in Panels e–f. Panels (e–f). Gene-Pathway networks for Canonical (Panel (e)) and GO (Panel (f)) analyses constructed using pathways with combined p < 0.01 (in grey-outlined circles) obtained from analyses in panels (c–d). Red and green circles represent upregulated and downregulated genes, respectively.
We used pathways present in all four datasets to conduct integrative GO and canonical pathway analyses (Figures 5c–5f). Integrative canonical pathway analysis identified EGF-signaling as a key pathway involving the majority of differentially-regulated genes across all datasets based on number of network interactions (Figure 5e). In addition, this analysis revealed dysregulation of PTEN and FAK pathways and upregulation of VDR/RXR, N-glycan degradation and coagulation pathways (Figure 5e). Integrative GO analysis also implicated pathways related to placental development and embryo implantation, transmembrane tyrosine kinase signaling (EGFR, FLT1, FLT4), inflammatory response, and response to hypoxia among the most significant dysregulated pathways (Figure 5f). EGFR-dependent pathways were identified as the most significant and interconnected pathways in both canonical and GO network analyses.
A search of the Human Protein Atlas29 confirmed the presence and localization of some of the significant differentially-regulated gene products identified in our integrative analysis described above (such as EGFR, ATF3, MAN1C1, ADAM12, FBN2, FLT1 and ENG) in either trophoblast or extracellular matrix of human placenta, both of which are mostly of fetal cell origin (Figure S2), further confirming the relevance of these gene products to preeclampsia processes.
Among genes coincident among analyzed datasets (Figure 5b), ATF3 was of particular interest since it was found upregulated in all analyzed datasets in our study including preeclamptic placentas and XPDTTD cells. In addition, transcription factor analysis of preeclampsia-specific genes identified CREB/ATF factors among dominant transcription regulators of preeclampsia (Figures 1e and 1f). These factors were expressed at a higher level in human versus mouse placenta and histologic analysis revealed presence of ATF3 in nuclei of normal placental trophoblasts (Figure S2). ATF3 had never been reported in relation to preeclampsia prior to our current study.
To determine the key role of ATF3 in the mechanism leading to preeclampsia, we analyzed an RNA-sequence dataset containing data on the comparison of untreated versus shATF3-treated K562 cells (i.e., cells with siRNA inhibition of ATF3) (GSE 33816)30. We compared genes identified through this analysis with preeclampsia-specific genes and found 16 genes in common including FLT1, ENG and INHBA (Figures S3a and S3b). FLT1 and ENG (known mediators of clinical symptoms of preeclampsia) as well as INHBA (a known regulator of EGFR signaling) had all been found significantly upregulated in preeclamptic placentas in our meta-analysis. Interestingly, our analysis of shATF3-treated K652 cells found that inhibition of ATF3 results in inhibition of FLT1 and ENG as well as in inhibition of INHBA (Figure S3b). This finding suggests that ATF3 upregulation is an upstream event to the induction of FLT1, ENG and INHBA.
Consistent with the above findings, analysis of a chip-seq dataset (GSM88112631) revealed that in mouse dendritic cells (which are known to express Flt1), ATF3 binds to the promoter region of Flt1 (Figure S3c).
Discussion
Based on our novel clinical observations and subsequent genetic epidemiologic studies of gestational complications associated with TTD1,3,8, we hypothesized that specific mutations in NER and transcription genes such as XPD lead to preeclampsia, possibly through their influence on TFIIH-mediated functions in placenta. Our previous molecular analysis localized the preeclampsia-associated mutations to a c-terminal motif and the helicase surfaces of XPD, most likely affecting XPD's binding to cdk-activating kinase (CAK) and p44 subunits of TFIIH8. Our findings have consistently highlighted the relevance of the fetal genotype to the mechanism leading to preeclampsia1,8. In order to decipher the underlying biologic mechanisms of the proposed associations, we conducted an integrative analysis of gene expression patterns of fetal-derived tissue from several data sources relevant to preeclampsia.
The results of our current integrative analysis are consistent with our previous findings and provide a basis for our prior observed associations between TFIIH impairment and risk of preeclampsia. Our overall analysis implicates lack of EGFR signaling, possibly caused by TFIIH impairment or by other mechanisms, in the presence of hypoxia and oxidative stress resulting in upregulation of ATF3 as one mechanism leading to preeclampsia.
All data sources were carefully selected and all analyses were specifically designed to identify alterations in genes and pathways which preceded the development of clinical symptoms of preeclampsia. For our meta-analysis, we compared gene expression patterns of fetal-derived placental tissues from four case-control studies of preeclampsia. We carefully selected several other relevant data sources (normal human tissue including placenta, time course placentas, hypoxic trophoblasts and XPDTTD fibroblasts) for interrogation of a preeclampsia-specific gene list obtained from the meta-analysis. Three of these gene expression datasets (normal human placenta, time course placenta, and XPDTTD fibroblasts), most relevant to events preceding development of clinical symptoms of preeclampsia, were used for integrative analysis. The rationale for comparison of placenta versus other normal tissue in human was that preeclampsia has been linked to abnormal placental development in various studies including previous genetic and molecular epidemiologic investigations conducted by us1,3,8. Furthermore, since preeclampsia occurs in humans but not in wild-type mice, we interrogated preeclampsia-specific gene list in both human and mouse placenta in order to identify placental genes differentially-expressed between the two species as those would be the most relevant to the development of preeclampsia. The rationale for analysis of time-course placentas was that preeclampsia normally occurs during the second and third trimesters of pregnancy (~24–40 weeks gestation); therefore, genes differentially-expressed during this mid- to term-gestation period compared to the first trimester would be the most relevant to preeclampsia development. The rationale for the use of a dataset containing gene expression patterns of XPDTTD fibroblasts was that these were the only tissues available from TTD patients who are predisposed to preeclampsia and that experimental studies have provided evidence for preservation of disease gene signatures in various tissues used for transcriptome analysis32.
Thus, our integrative analysis used a reductionist approach to systems biology by using specific and relevant knowledge of conditions related to preeclampsia to select high throughput gene expression databases to be used as a filter to identify dysregulated genes and pathways which preceded the development of preeclampsia (Figure S1). Even though the mechanisms which underlie preeclampsia development probably originate during placentation and the initial stages of the clinical symptoms of preeclampsia could start as early as the first trimester, our integrative analysis filtered the dysregulated pathways through datasets of conditions which predispose to preeclampsia, such as gene expression of XPDTTD fibroblasts, thus focusing on preceding mechanisms. The underlying assumption of our integrative study design is that these preceding dysregulated pathways persist until after the development and diagnosis of the major clinical symptoms of preeclampsia (which normally occur after 20 weeks gestation).
Our meta-analysis of placental gene expression patterns in case-control studies of preeclampsia showed significant dysregulation of growth factor receptor signaling, the most prominent being EGFR-related pathways. Dysregulation of EGFR signaling pathway was also noted in XPDTTD cells. Our integrative analysis revealed signatures of hypoxia and oxidative stress as regulators of differentially-expressed genes in preeclampsia and implicated EGFR and ATF3 as the main regulators of preeclampsia development. ATF3 had never been reported in relation to preeclampsia prior to our current study.
Our preeclampsia meta-analysis suggested that a large number of preeclampsia-specific genes were directly induced by hypoxia. Placental hypoxia33 and oxidative stress34 have been implicated in the etiology of preeclampsia in previous studies, although, the exact pathophysiologic mechanisms remain elusive. One speculation is that oxidative stress provokes the release of soluble FLT1 and ENG mediating development of several symptoms of preeclampsia, including high blood pressure and proteinuria34. To identify genes affected by hypoxic conditions, we analyzed gene expression patterns of hypoxic trophoblasts and found that a large proportion of preeclampsia-specific upregulated (but not downregulated) genes were regulated by hypoxia. FLT1 and ENG, however, were not among the genes induced in response to hypoxia. Our analysis of an RNA-seq dataset of ATF3-deficient K562 cells showed that both FLT1 and ENG were significantly downregulated in the absence of ATF3, implicating ATF3 upregulation as an upstream event to induction of these molecules. ATF3 was found significantly upregulated in both preeclamptic palcentas and in XPDTTD fibroblasts in our study. EGFR and ATF3 were also expressed at much higher levels in human than in mouse placenta. Furthermore, ATF3 has been implicated in diabetes mellitus and renal disease, two known conditions associated with increased risk of preeclampsia35.
Our findings also suggest a functional link between EGFR and ATF3. Previous studies suggested an association between EGFR signaling and ATF3 through oxidative stress as a trigger36. ATF3 is known to be part of stress-response genes37 including oxidative stress and hypoxia38. Deficiency in EGFR signaling exacerbates the negative effects of hypoxia and oxidative stress, since EGFR signaling protects from oxidative stress- and hypoxia-induced apoptosis39,40,41. Deficient EGFR signaling is believed to affect not only cell growth, but also cell migration via FAK signaling; consistent with this, cell migration was found among dysregulated pathways in preeclampsia in our analyses.
EGFR and EGFR-mediated FAK signaling are key regulators of invasion and metastasis in cancer cells. The EGFR signaling pathway is one of the most dysregulated molecular pathways in human cancers. Although the exact mechanism remains to be elucidated, evidence suggests that activated EGFR, acting as a receptor tyrosine kinase, recruits a number of downstream signaling molecules leading to cellular proliferation and promotion of major tumor growth and survival pathways36. ATF3 overexpression has also been shown to play an oncogenic role in vitro and in vivo by protecting against apoptosis42. Thus, our results implicate dysregulation of these cancer promoting pathways in placenta, in the presence of hypoxia and oxidative stress, in preeclampsia development. A recent bioinformatics study reporting similarities between cancer protein-protein interaction networks and dysregulated networks in preeclampsia43 concurs with our findings.
It has been a long-standing observation that placental development histologically and even macroscopically resembles that of a tumor. Survival and growth of the fetus requires normal development of placenta which in humans involves invasion of the maternal decidua by cytotrophoblasts. Placental research in recent years has underlined the striking similarities among the proliferative and invasive properties of trophoblasts and those of cancer cells, which are known to override mechanisms controlling cellular proliferation, invasion and death44. Consistent with the above, EGFR and ATF3 were found expressed in placental trophoblasts in immunohistochemical analyses of normal placenta in our study. Furthermore, EGFR-deficient mice die prenatally from severe blood vessel defects of placental tissue during the second half of the fetal development45 and in vitro ATF3 is critical for reactive oxygen species (ROS)-induced vascular tubule formation35, supporting the potential role of these proteins in placental development.
Another contribution of our meta-analysis of placental gene expression patterns in preeclampsia case-control studies was the identification of 419 preeclampsia-specific genes, which could be the focus of future etiologic studies. A number of the identified genes in our study were not previously reported; these genes included EBI3 (IL27) anti-inflammatory cytokine which was found to be upregulated in preeclampsia.
While the gene signature of hypoxic cells was similar to genes upregulated in preeclamptic placentas, the gene signature of XPDTTD-mutant cells was similar to genes downregulated in preeclampsia, perhaps indicating transcription deficiency in preeclamptic placentas. Interestingly, our preeclampsia meta-analysis also revealed significant downregulation of CDK7, a component of the cdk-activating kinase (CAK) domain of TFIIH, in preeclamptic placentas. CAK complex is required for phosphorylation of RNA Pol-II5. These findings shed light on the association between TTD, TFIIH- and RNA Pol-II-mediated basal transcription and preeclampsia.
RNA Pol-II consists of twelve protein subunits and is found in a complex called the Pol-II holoenzyme, which also includes most of the basal transcription factors (TFIID, TFIIB, TFIIF, TFIIE, TFIIH)5. TFIIH and TFIIE are believed to be part of a set of minimal transcription factors which are necessary for accurate transcription; these molecules are believed to mediate the unwinding of DNA and the early steps in the transcription process46. Therefore, downregulation of components of TFIIH and/or mutations in genes coding for components of TFIIH (as is the case in TTD) leading to dysregulation of RNA Pol-II mediated functions in placenta may be one mechanism leading to preeclampsia. Similarly, mutations in genes coding for components of TFIIH (as is the case in TTD) may also lead to TFIIH impairment and consequently to dysregulation of RNA Pol-II-mediated functions in placenta leading to preeclampsia. Consistent with our findings, levels of basal transcription in placenta have been reported as major modulators of expression of EGFR47 and ATF335; ATF3 is also believed to be responsible for abnormal vascular remodeling in diabetic patients via regulation of p8 (a component of TFIIH complex)35. The above findings point to a potential underlying mechanism for preeclampsia involving impairment of TFIIH and RNA Pol-II mediated functions in placenta, possibly leading to altered expression of EGFR and ATF3.
Thus, drawing from our overall analyses, we propose that hypoxia and oxidative stress combined with lack of EGFR signaling (which in some cases such as TTD can occur due to TFIIH impairment and transcription defect) result in upregulation of ATF3, which induces FLT1 and ENG causing the clinical manifestations of preeclampsia. Our results suggest a link between EGFR signaling deficiency and impaired TFIIH and RNA Pol-II activities. Since EGFR- and ATF3-dependent pathways have been implicated in tumor growth and metastasis, we propose dysregulation of these canonical cancer molecular pathways in placenta leading to preeclampsia. Our findings indicate a potential pathway by which genetic abnormalities in the fetus could initiate the changes that produce the clinical abnormalities seen in preeclampsia. Thus, our findings provide clues into the underlying etiology of preeclampsia which could eventually translate into a therapeutic approach.
Methods
Meta-analysis of placental gene expression patterns in preeclampsia case-control studies
Microarray datasets from case-control studies of preeclampsia containing global gene expression patterns of normal versus preeclamptic placentas downloaded from GEO101 and ArrayExpress48, were used to conduct a meta-analysis. The objective was to compare global gene expression profiles in placentas from normotensive pregnancies (controls) to placentas from preeclamptic pregnancies (cases) across all comparable studies. Per inclusion criteria, only datasets from studies investigating patterns of gene expression on tissue biopsies from fetal cell-derived histological subsections of the placenta (including placental and chorionic villi) were downloaded. Although the biopsied placental tissues may contain a mixture of fetal and maternal cells, we tried to restrict our inclusion criteria to those involving biopsied tissues believed to be predominantly of fetal-cell origin. Other inclusion criteria involved presence of more than five microarrays per group and more than two standard deviations (SD) difference between the groups in Principal Component Analysis (PCA) using first two axes of the PCA. Based on these criteria, we selected four publicly available microarray datasets for our meta-analysis (GSE470721, GSE1058819, GSE1472220, and GSE 2412922). All four datasets ascertained cases of severe preeclampsia as defined by the American College of Obstetricians and Gynecologists49. All four datasets only included cases of early-onset (diagnosis < 34 weeks gestation) preeclampsia and all four studies matched cases and controls based on gestational age19,20,21,22.
Meta-analysis was conducted by calculating the P-values for each gene in each experiment using student's t-test. P-values and fold differences were averaged; if multiples were present, datasets were merged using Gene symbols. We calculated combined p-values using Fisher's combined probability test as follows:
pi: P-value of the test i ; χ2: Chi-square distribution; k: Number of tests; df: degree of freedom; P: P-value. P-values were adjusted using q-value calculation (q < 0.05 was considered significant).
In order to reduce the number of false positive results, we removed all genes with q > 0.1. Furthermore, any gene that was statistically significant (P < 0.05), but had discordant fold change in at least one dataset compared to the other ones, was labeled as non-significant and removed from the analysis. Correction for multiple testing was done using Benjamini and Hochberg False Discovery Rate (FDR)50. Cutoff of q < 0.05 was used for statistically significant result in this analysis of the above datasets.
Individual analysis of other gene expression datasets relevant to preeclampsia
Other microarrays used in the study were downloaded from Gene Expression Omnibus (GEO)10. GSE95 and GSE97 (GDS59624) contained gene expression profiles of normal human and mouse tissues. Individual analyses of datasets GSE9524 and GSE9724 were done through comparison of expression in placenta versus other normal tissue as well as comparison of expression ratios in human versus mouse tissues. A dataset of hypoxic human trophoblasts (Swan 71 human trophoblast cells under chemical hypoxia by CoCl2-treatment in an in vitro system) (GSE31679)26 was downloaded from GEO24. ANOVA models (Method of Moments51) and Partek software version 6.6 (Partek Inc., St. Louis, MO, USA) was used for analysis of the above datasets.
A microarray dataset containing gene expression profiles of normal fibroblasts as well as XP- and TTD-affected fibroblasts with mutations in the XPD gene was obtained through personal communications28. This dataset contained the following HG-U133 Plus Affymetrix genechips samples: 3 TTD, 3 XP and 3 control fibroblasts untreated or treated with ultraviolet (UV) radiation and collected 4 hours post radiation. In our analyses, UV-treated samples were used to reduce technical variability within the samples. Data were RMA-normalized with adjustments for GC content and probe sequence and were Log2-transformed. Probe intensities of <200 units in less than 10% of samples were excluded from the analysis.
In order to extract gene signatures of XPD-associated TTD, a four-way ANOVA model using the Method of Moments was applied to the dataset51; intra-patient variability was taken into account in these analyses. For the XPDTTD dataset, patients were treated as random variables and included in the ANOVA model. Variables with probe intensity < 100 in more than 75% of samples were excluded from the analyses. An XPDTTD dataset containing fibroblast cell lines treated with either a control plasmid or a plasmid containing the normal XPD gene was analyzed as follows: Affymetrix microarrays for the plasmid transfected dataset were not summarized for the analysis and all 250,000 probes were uploaded separately. The probes for each gene were treated as technical replicates and subjected to paired t-tests (each probe as a pair) and q-values were calculated. Z-score analysis gave similar results to paired t-test (Data not shown).
Integrative analysis involving selected preeclampsia-related datasets
In order to filter the dysregulated genes and pathways in preeclampsia (i.e., genes and pathways involved in both cause and effect) to those genes and pathways which were coincident among all analyzed datasets and therefore more likely to be involved in the etiology, common genes between all preeclampsia-related datasets were identified using a Venn diagram. Common genes and pathways were displayed using Cytoscape 2.8.2 software where edges were labeled in accordance with the gene expression levels.
Pathway analysis
Pathway analysis of different genelists was conducted by using Gene Ontology (GO) analysis using Biobase manually curated datasets (http://www.biobase-international.com/) and Canonical pathway analysis was done using Ingenuity Pathway Analysis, version 8.7103. Significant pathways (q < 0.05) and corresponding genes were exported as networks (based on term-gene relations) into the Cytoscape 2.8.2, where ClusterOne Cytoscape plugin52, was used to identify most interconnected genes and pathways. The most common biological function of the group was then used to label the identified cluster. Network was displayed using Edge-Weighted Spring Embedded layout.
For combined pathway analysis, GO and Canonical pathways were estimated for each of the genelists and combined Fisher's probability test was used to identify relevant pathways for all datasets (q < 0.05). Pathways and their genes were used to construct networks in Cytoscape software, as described earlier.
Transcription factor analyses
Promoter analysis was conducted by using TRANSFAC dataset using BIOBASE (ExPlain 3.0) software (Biological Databases GmbH, Wolfenbuttel, Germany). Promoters were identified in the regions starting from 1.0 kb upstream of the transcription start site (TSS) and ending 500 bp downstream of the genes obtained in Preeclampsia meta-analysis.
Significant matrices (q < 0.05) and corresponding genes were exported as network (based on term-gene relations) into the Cytoscape 2.8.2, where ClusterOne Cytoscape plugin52 was used to identify most interconnected matrices and pathways. Matrices with the largest number of common targets tend to cluster together and form densely-connected sub-networks containing molecules related to the same transcription factor family. Most common matrices of the group were then used to label the identified cluster. As a control in our analyses, we found clustering of transcription factor common structures, such as STAT1-STAT5 clusrter, as well as those with common functions or targets, such as MYC/MAX- HIF1A-ARNT-USF cluster (Figure 1e). Alternatively, transcription factors predicted to regulate same genes were identified using correlogram, calculated based on similarity of the shared genes by the transcription factor matrices (Figure 1f). Overall, we observed good agreement between the two approaches.
Verification of protein expression and localization within the cell
The protein expression database29 (www.proteinatlas.org) was used to verify expression patterns of proteins of interest in placenta.
Analysis of RNA-sequencing data of K562 cells treated with ShRNA inhibitors
RNA-seq data from GSE33816 was analyzed using Partek 6.6 RNA-Seq workflow. In brief, bam files were imported into the Partek and analyzed on gene level via ANOVA method using read number and condition (treatment) variables.
Visualization of chip-seq dataset
BigWig file (GSM88112631) was imported into the UCSC Genome browser and Flt1 promoter region was visualized using the Genome Browser tool.
Author Contributions
R.M. and A.D. conceived the study and analyzed the data. J.L.M., C.S. and X.A. helped with hypothesis development, study design and interpretation of results. A.K. helped with data preparation and analysis. All authors contributed to the completion of the manuscript.
Supplementary Material
Supplementary Figures S1–S3
Supplementary Table
Acknowledgments
The authors thank Dr. AR Lehmann at University of Sussex and Dr. M Hubank at University College, London for providing the XPDTTD gene expression dataset, Dr. A. Morgun at Oregon State University for advice on microarray data analysis, and Dr. V. Nagarajan at National Institute of Allergy and Infectious Diseases (NIAID), NIH for critically reviewing the methods and conclusions of the paper. Technical support provided by Mr. H. Martinez at University at Albany throughout the study has been invaluable. This study was supported in part by a grant to the Center for Social and Demographic Analysis (CSDA) at the University at Albany from NICHD; (R24 HD044943) and a research award to RM from CSDA. This research was also supported by the Intramural Research Program-NICHD, NIH. Opinions, findings and conclusions expressed here are those of the authors and do not necessarily reflect the views of the funding agencies.
References
- Moslehi R. et al. Adverse effects of trichothiodystrophy DNA repair and transcription gene disorder on human fetal development. Clin Genet 77, 365–73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A. R. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie 85, 1101–11 (2003). [DOI] [PubMed] [Google Scholar]
- Tamura D. et al. High-risk pregnancy and neonatal complications in the DNA repair and transcription disorder trichothiodystrophy: report of 27 affected pregnancies. Prenat Diagn 31, 1046–53 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itin P. H., Sarasin A. & Pittelkow M. R. Trichothiodystrophy: update on the sulfur-deficient brittle hair syndromes. J Am Acad Dermatol 44, 891–920; quiz 921–4 (2001). [DOI] [PubMed] [Google Scholar]
- Nikolov D. B. & Burley S. K. RNA polymerase II transcription initiation: a structural view. Proc Natl Acad Sci U S A 94, 15–22 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita M. & Merino C. The transcriptional complexity of the TFIIH complex. Trends Genet 19, 578–84 (2003). [DOI] [PubMed] [Google Scholar]
- Fuss J. O. & Tainer J. A. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair (Amst) 10, 697–713 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moslehi R. et al. Phenotype-specific adverse effects of XPD mutations on human prenatal development implicate impairment of TFIIH-mediated functions in placenta. Eur J Hum Genet 20, 626–31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzan J., Carbonnel M., Piconne O., Asmar R. & Ayoubi J. M. Pre-eclampsia: pathophysiology, diagnosis, and management. Vasc Health Risk Manag 7, 467–74 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciantar E. & Walker J. J. Pre-eclampsia, severe pre-eclampsia and hemolysis, elevated liver enzymes and low platelets syndrome: what is new? Womens Health (Lond Engl) 7, 555–69 (2011). [DOI] [PubMed] [Google Scholar]
- Johnson M. P. et al. The ERAP2 gene is associated with preeclampsia in Australian and Norwegian populations. Hum Genet 126, 655–66 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A. et al. Identification of ACOX2 as a shared genetic risk factor for preeclampsia and cardiovascular disease. Eur J Hum Genet 19, 796–800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses E. K. et al. A genome scan in families from Australia and New Zealand confirms the presence of a maternal susceptibility locus for pre-eclampsia, on chromosome 2. Am J Hum Genet 67, 1581–5 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. P. et al. Genome-Wide Association Scan Identifies a Risk Locus for Preeclampsia on 2q14, Near the Inhibin, Beta B Gene. PLoS One 7, e33666 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roten L. T. et al. A low COMT activity haplotype is associated with recurrent preeclampsia in a Norwegian population cohort (HUNT2). Mol Hum Reprod 17, 439–46 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roten L. T. et al. Association between the candidate susceptibility gene ACVR2A on chromosome 2q22 and pre-eclampsia in a large Norwegian population-based study (the HUNT study). Eur J Hum Genet 17, 250–7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenstad M. H. et al. Genetic and molecular functional characterization of variants within TNFSF13B, a positional candidate preeclampsia susceptibility gene on 13q. PLoS One 5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D. R. & Chinnaiyan A. M. Integrative analysis of the cancer transcriptome. Nat Genet 37 Suppl, S31–7 (2005). [DOI] [PubMed] [Google Scholar]
- Sitras V. et al. Differential placental gene expression in severe preeclampsia. Placenta 30, 424–33 (2009). [DOI] [PubMed] [Google Scholar]
- Winn V. D. et al. Severe preeclampsia-related changes in gene expression at the maternal-fetal interface include sialic acid-binding immunoglobulin-like lectin-6 and pappalysin-2. Endocrinology 150, 452–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa H. et al. Microarray analysis of differentially expressed fetal genes in placental tissue derived from early and late onset severe pre-eclampsia. Placenta 28, 487–97 (2007). [DOI] [PubMed] [Google Scholar]
- Nishizawa H. et al. Comparative gene expression profiling of placentas from patients with severe pre-eclampsia and unexplained fetal growth restriction. Reprod Biol Endocrinol 9, 107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allocco D. J., Kohane I. S. & Butte A. J. Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinformatics 5, 18 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A. I. et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A 101, 6062–7 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn V. D. et al. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology 148, 1059–79 (2007). [DOI] [PubMed] [Google Scholar]
- Hong J. S. et al. A Comparative Analysis of the Transcriptome of Hypoxia and Inflammation in Swan 71 Human Trophoblast Cells: Its Implications in Preeclampsia. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31679 (2012).
- Tang Q. e. a. Endothelial cell response to hypoxia and subsequent reoxygenation. http://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS1968 (2004).
- Offman J. et al. Transcriptional changes in trichothiodystrophy cells. DNA Repair (Amst) 7, 1364–71 (2008). [DOI] [PubMed] [Google Scholar]
- Uhlen M. et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28, 1248–50 (2010). [DOI] [PubMed] [Google Scholar]
- Bansal A. e. a. RNA-seq analysis of K562 cells (K562-shX) with stable integration of a tet-inducible shRNA targeting a candidate (X) transcription factor from ENCODE/HudsonAlpha-MyersLab. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33816 (2012).
- Garber M. et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell 47, 810–22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J. T. & Butte A. J. Identification of discriminating biomarkers for human disease using integrative network biology. Pac Symp Biocomput 27–38 (2009). [PMC free article] [PubMed] [Google Scholar]
- Julian C. G. High altitude during pregnancy. Clin Chest Med 32, 21–31, vii. [DOI] [PubMed] [Google Scholar]
- Hung T. H. & Burton G. J. Hypoxia and reoxygenation: a possible mechanism for placental oxidative stress in preeclampsia. Taiwan J Obstet Gynecol 45, 189–200 (2006). [DOI] [PubMed] [Google Scholar]
- Okamoto A., Iwamoto Y. & Maru Y. Oxidative stress-responsive transcription factor ATF3 potentially mediates diabetic angiopathy. Mol Cell Biol 26, 1087–97 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W. & Lo H. W. Landscape of EGFR signaling network in human cancers: biology and therapeutic response in relation to receptor subcellular locations. Cancer Lett 318, 124–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai T., Wolfgang C. D., Marsee D. K., Allen A. E. & Sivaprasad U. ATF3 and stress responses. Gene Expr 7, 321–35 (1999). [PMC free article] [PubMed] [Google Scholar]
- Hoetzenecker W. et al. ROS-induced ATF3 causes susceptibility to secondary infections during sepsis-associated immunosuppression. Nat Med 18, 128–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy R., Smith S. D., Chandler K., Sadovsky Y. & Nelson D. M. Apoptosis in human cultured trophoblasts is enhanced by hypoxia and diminished by epidermal growth factor. Am J Physiol Cell Physiol 278, C982–8 (2000). [DOI] [PubMed] [Google Scholar]
- Chen S. C., Liu Y. C., Shyu K. G. & Wang D. L. Acute hypoxia to endothelial cells induces activating transcription factor 3 (ATF3) expression that is mediated via nitric oxide. Atherosclerosis 201, 281–8 (2008). [DOI] [PubMed] [Google Scholar]
- Moll S. J., Jones C. J., Crocker I. P., Baker P. N. & Heazell A. E. Epidermal growth factor rescues trophoblast apoptosis induced by reactive oxygen species. Apoptosis 12, 1611–22 (2007). [DOI] [PubMed] [Google Scholar]
- Thompson M. R., Xu D. & Williams B. R. ATF3 transcription factor and its emerging roles in immunity and cancer. J Mol Med (Berl) 87, 1053–60 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejera E., Bernardes J. & Rebelo I. Preeclampsia: a bioinformatics approach through protein-protein interaction networks analysis. BMC Syst Biol 6, 97 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti C., Bruni L., Dangles-Marie V., Pecking A. P. & Bellet D. Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum Reprod Update 13, 121–41 (2007). [DOI] [PubMed] [Google Scholar]
- Dackor J., Caron K. M. & Threadgill D. W. Placental and embryonic growth restriction in mice with reduced function epidermal growth factor receptor alleles. Genetics 183, 207–18 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T. & Kadonaga J. T. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 339, 225–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin de L'hortet A., Gilgenkrantz H. & Guidotti J. E. EGFR: A Master Piece in G1/S Phase Transition of Liver Regeneration. Int J Hepatol 2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca-Serra P. et al. ArrayExpress: a public database of gene expression data at EBI. C R Biol 326, 1075–8 (2003). [DOI] [PubMed] [Google Scholar]
- Schroeder B. M. ACOG practice bulletin on diagnosing and managing preeclampsia and eclampsia. American College of Obstetricians and Gynecologists. Am Fam Physician 66, 330–1 (2002). [PubMed] [Google Scholar]
- Reiner A., Yekutieli D. & Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19, 368–75 (2003). [DOI] [PubMed] [Google Scholar]
- Eisenhart C. The assumptions underlying the analysis of variance. Biometrics 3, 1–21 (1947). [PubMed] [Google Scholar]
- Bader G. D. & Hogue C. W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4, 2 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S3
Supplementary Table