Abstract
It is generally assumed that sister chromatids are genetically and functionally identical and that segregation to daughter cells is a random process. However, functional differences between sister chromatids regulate daughter cell fate in yeast1 and sister chromatid segregation is not random in Escherichia coli2. Differentiated sister chromatids, coupled with non-random segregation, have been proposed to regulate cell fate during the development of multicellular organisms3. This hypothesis has not been tested because molecular features to reliably distinguish between sister chromatids are not obvious. Here we show that parental “Watson” and “Crick” DNA template strands can be identified in sister chromatids of murine metaphase chromosomes using CO-FISH (Chromosome Orientation Fluorescence in situ Hybridization4) with unidirectional probes specific for centromeric and telomeric repeats. All chromosomes were found to exhibit a uniform orientation with the 5′ end of the short arm on the same strand as T-rich major satellite repeats. The invariable orientation of repetitive DNA was used to differentially label sister chromatids and directly study mitotic segregation patterns in different cell types. Whereas sister chromatids appeared to be randomly distributed between daughter cells in cultured lung fibroblasts and embryonic stem cells, significant (p<0.01) non-random sister chromatid segregation was observed in a subset of colon crypt epithelial cells including cells outside positions reported for colon stem cells5. Our results establish that DNA template sequences can be used to distinguish sister chromatids and follow their mitotic segregation in vivo.
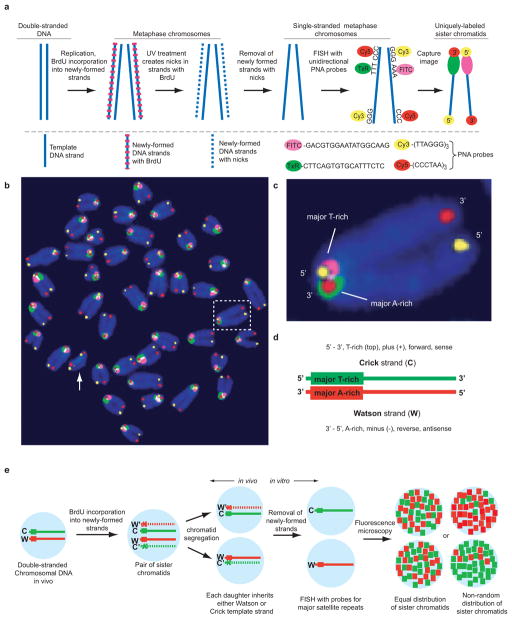
Major satellite repeats have a uniform head-to-tail orientation on mouse chromosomes relative to the centromere6,7. To determine whether this polarity is fixed relative to chromosome ends, we hybridized unidirectional probes specific for major satellite and telomere repeats to single-stranded metaphase chromosomes using CO-FISH (Figure 1a). For the CO-FISH procedure, cells are treated with BrdU for one round of DNA replication resulting in BrdU incorporation exclusively into the newly-formed DNA4,8. Following treatment with Hoechst 33258 (a DNA dye) and UV irradiation, nicks are created exclusively at sites of BrdU incorporation which are then used to remove newly formed DNA by exonuclease treatment and DNA denaturation. The resulting single-stranded chromosomes (containing template DNA only) are hybridized with strand-specific probes (Figure 1a).
Figure 1.
Highly-conserved orientation of telomeric and major satellite DNA in murine chromosomes revealed by four-color CO-FISH. a, Schematic diagram of the CO-FISH procedure. b, Pseudo-color CO-FISH image of murine metaphase chromosomes. Note that major satellite repeats on all chromosomes except Y (arrow, no major satellite DNA) have the same orientation. c, Magnification of the boxed chromosome shown in b. d, Definition of “Watson” and “Crick” DNA template strands based on the uniform orientation of major satellite DNA. e, The relative distribution of Watson and Crick major satellite fluorescence can be used to study sister chromatid segregation patterns in vivo.
Strikingly, all chromosomes except the Y chromosome showed a uniform orientation of major satellite relative to telomeric repeats (Figure 1b). On each chromosome, the 5′ end of the short arm (characterized by C-rich telomere repeats) is adjacent to T-rich major satellite repeat sequences, and the 3′ end of the short arm (characterized by G-rich telomere repeats) is adjacent to A-rich major satellite repeat sequences. All template strands (except those in chromosomes 4 and 18)9 show mutually-exclusive staining with fluorescently-labeled peptide-nucleic acid (PNA) probes specific for either A-rich or T-rich major satellite DNA (Fig 1b, c and Supplementary Figures 1 and 9). Since the orientation of major satellite DNA relative to telomeric DNA is fixed, probes hybridized to major satellite repeats were used to arbitrarily define “Watson” (red fluorescence, Figure 1d) and “Crick” (green fluorescence, Figure 1d) DNA template strands. A similar chromosomal polarity was observed in Mus spretus fibroblasts, with the 5′ end of the short arm adjacent to T-rich minor satellite repeats in most chromosomes (Supplementary Figure 2). Since CO-FISH can differentially label sister chromatids, we adapted the CO-FISH technique to allow us to directly follow chromatid segregation in vivo (Figure 1e).
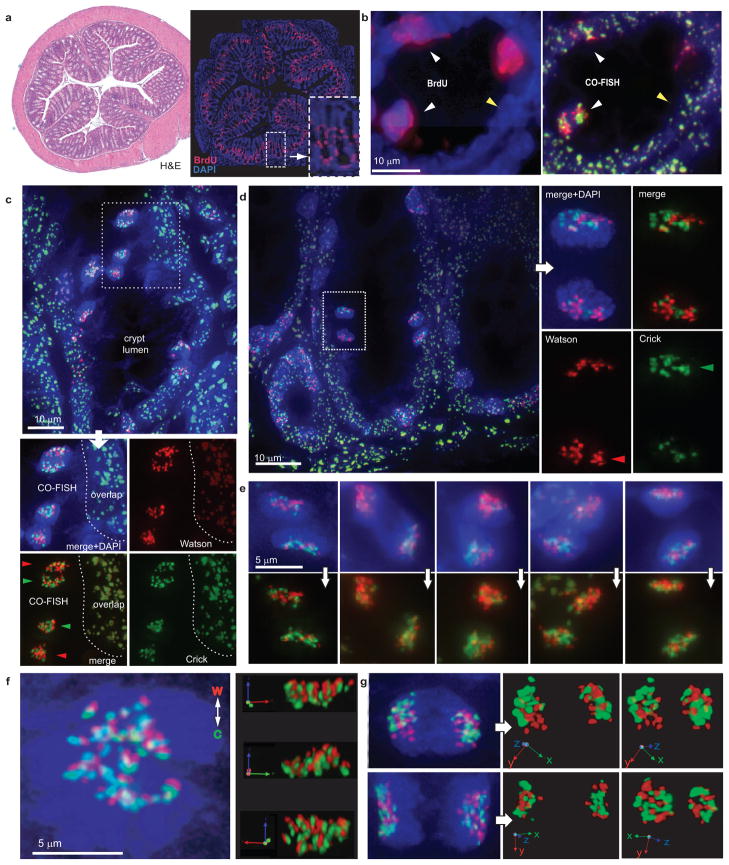
Non-random segregation of DNA strands in mammalian cells was first reported using indirect pulse-chase experiments with nucleotide analogs in dividing murine intestinal crypt epithelial cells10. To directly study the pattern of sister chromatid segregation in such cells, we injected adult mice for 12 hourly intervals with BrdU prior to harvesting of colon tissue which was fixed, sectioned and subjected to CO-FISH with major satellite probes. Only a minority of cells within colon crypts was actively dividing as shown by BrdU incorporation (Figure 2a, right panel and inset). These BrdU-positive cells showed discrete, non-overlapping red and green fluorescent signals (herein referred to as CO-FISH signals) from the strand-specific probes (Figure 2b, white arrowheads) indicating successful generation of single-stranded chromosomes. In contrast, most non-mitotic cells showed overlapping red and green fluorescence from the major satellite probes hybridizing to both strands of double-stranded chromosomes (Figure 2b yellow arrowhead, Figure 2c). Cell pairs exhibiting apparent template strand asymmetry were found at different positions within the colon crypt, including high within the crypt axis (Figure 2c–d, Supplementary Figure 3). Sister nuclei exhibiting reciprocal, asymmetric CO-FISH fluorescence are compatible with non-random distribution of sister chromatids containing either Watson or Crick DNA template strands (Figure 1e, Figure 2e, Supplementary Figure 4 and Supplementary Movie 1). We confirmed that CO-FISH signals in mitotic colon cells from mice subjected to 12 hours of BrdU treatment were exclusively derived from cells after only one round of DNA replication (Supplementary Figure 5) 11. Of note, DNA template strand asymmetry was also observed in colon tissue sections of M. spretus using probes specific for minor satellite repeats (Supplementary Figure 6).
Figure 2.
CO-FISH to study sister chromatid segregation patterns. a, Low magnification of adjacent colon sections stained with H&E (left panel) and DAPI and anti-BrdU antibody (right panel). b, High magnification of a section stained for BrdU (left panel) that was subsequently subjected to CO-FISH (right panel). BrdU-labeled cells show non-overlapping red and green fluorescence (white arrowheads), non-mitotic cells without BrdU show overlapping probe signals (yellow arrowhead). c, Example of CO-FISH (non-overlapping) signals in pairs of post-mitotic cells in colon crypts. d, Post-mitotic cell pairs relatively high in colon crypt with asymmetric CO-FISH fluorescence. e, Examples of asymmetric CO-FISH fluorescence in paired colon cells. f, Non-random alignment of sister chromatids at metaphase (right panels: different projections from Supplementary Movie 2). g, Mirror-image symmetry and clustered CO-FISH fluorescence in paired daughter (see also Supplementary Movies 3 and 4).
Chromosomes aligned at the metaphase plate in vivo displayed what appeared to be a polar arrangement of Watson and Crick sister chromatids (Figure 2f and Supplementary Movie 2). In addition, major satellite DNA template strands appeared to be clustered after mitosis (Figure 2g) and often exhibited a marked “mirror-image” asymmetry with territories of red and green fluorescence in one daughter cell mirrored by territories of the opposite color in the other daughter cell (Figure 2g and Supplementary Movies 3 and 4). These observations suggest that pericentric regions of multiple chromosomes cluster in at least some post-mitotic colon cells on the basis of parental DNA template strand sequences. To exclude major rearrangements in nuclear architecture by our CO-FISH procedure, we performed 3-color CO-FISH with both major satellite probes and a telomeric probe (Supplementary Figure 7). Telomeric signals were observed at expected positions adjacent to centric regions (the terminus of the short chromatid arms) and adjacent to the division plane (the terminus of the long chromatid arms) in support of the notion that the CO-FISH procedure does not grossly alter the general morphology and positioning of segregating chromosomes.
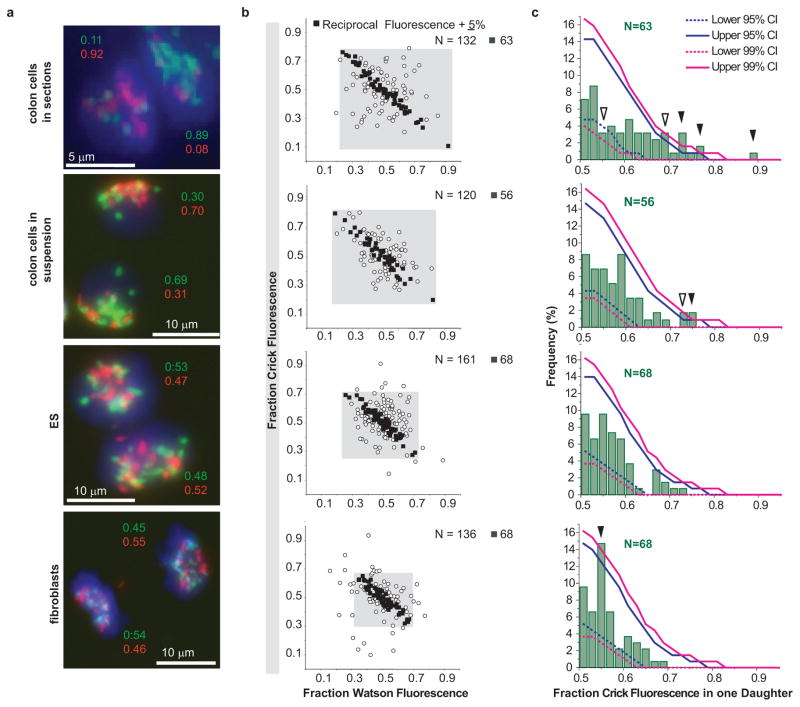
Our qualitative observations suggested that sister chromatids of most chromosomes are segregating non-randomly in a subset of dividing colon epithelial cells. To test if our observations could nevertheless be explained by chance, we quantified the relative Watson and Crick fluorescence in each daughter cell using dedicated software (see Supplementary Figure 8 and Supplementary Methods for details). The measured fluorescence was converted to a relative fluorescence ratio (Figure 3a) based on the reasoning that the total fluorescence from both daughters is the outcome of redistributing a fixed number of DNA template strands from a mother cell to the two daughter cells (Figure 1e). Reciprocal ratios of Watson and Crick fluorescence are in agreement with the expected distribution of chromatids between daughter cells.
Figure 3.
Measurements of Watson and Crick DNA template strand fluorescence in post-mitotic cells. a, Examples of fluorescence measured in the indicated cell types. b, For N cell pairs, the ratio of Watson and Crick fluorescence in one of the daughter cells (arbitrary selection) is plotted. Solid black squares show cells with reciprocal Watson and Crick fluorescence ratios ± 5%, whereas open circles represent cells with Watson/Crick fluorescence ratios outside this arbitrary cutoff. c, The observed Crick fluorescence distributions in selected individual cells (N, black squares in b) was compared to fluorescence distribution values obtained by simulated random segregation. The observed frequency (Y-axis) of Crick fluorescence (X-axis, green histograms) in one daughter cell (with the brightest Crick fluorescence) is plotted. Upper and lower 95% and 99% confidence intervals (CI, solid and dashed blue and magenta lines, respectively) represent the range of fluorescence distributions expected by chance. The values measured in colon tissues sections as well as colon cell suspensions fall outside the range for simulated random segregation (p<0.05: open arrows; p<0.01: solid arrows).
We compared the measured CO-FISH fluorescence signals from sectioned colon and preparations of isolated colon cells to two cultured cell types not expected to show non-random segregation patterns: pluripotent embryonic stem (ES) cells and lung fibroblasts (Figure 3a and Supplementary Data Table). In order to avoid selection bias for asymmetry, every cell pair with clear non-overlapping CO-FISH signals was analyzed (Figure 3b). Though this impartial acquisition of data ensures that the measured sister chromatid segregation patterns are not influenced by cell selection, the results will include all recently-divided cells, which may complicate data analysis if chromatid segregation patterns differ between cell types. Nevertheless, cell pairs from colon section and isolated colon cells showed a broader distribution of Watson and Crick fluorescence, compared to cultured ES cells and lung fibroblasts (Figure 3b, gray boxes), reflecting a higher frequency of sister chromatid asymmetry. Up to 50% of cell pairs from all cell types showed an excellent reciprocal ratio of measured fluorescence values between daughter cells, with Watson fluorescence distribution ratios mirrored within 5% by complementary Crick fluorescence distribution ratios (Figure 3b, filled squares, and supplementary Data Table). Cell pairs showing reciprocal fluorescence outside this arbitrary cutoff (Figure 3b, open circles) most likely reflect noise in CO-FISH measurements due to loss of DNA, non-specific fluorescence and other causes.
To test whether the measured asymmetry in colon cells was non-random, we superimposed our observed fluorescence distributions of cell pairs within the arbitrary 5% reciprocal cutoff value to 95% and 99% confidence intervals (CI) calculated from simulated random segregations, representing the range of fluorescence values expected by chance (see Supplementary Figures 9–11 and Supplementary Materials for full discussion of simulated random segregation and statistical analysis). The distribution of Crick template strand fluorescence from sectioned colon tissue and isolated colon cells was outside the 95% or 99% CI calculated for random sister chromatid segregation (Figure 3c; p<0.05 open arrowheads, p<0.01 solid arrowheads). This includes a higher frequency of cell pairs with extreme asymmetry as well as a lower frequency of cell pairs with a symmetrical distribution, than predicted by simulated random segregation. While fewer cell pairs with extreme asymmetry were present or preserved in colon cell suspensions, the results were nevertheless significant (p<0.01). By contrast, in ES and lung fibroblasts the measured fluorescence intensity values were within the 95% and 99% CI calculated for random segregation. The one exception was in lung fibroblasts at the symmetrical 55% fluorescence value, suggesting a skewing of segregation towards a 50:50 distribution of chromatids (Fig 3c, arrowhead, bottom panel). These results support the conclusion that the observed asymmetry of DNA template strand fluorescence in paired colon cells results from non-random segregation of sister chromatids rather than rare random segregation events. We consider it unlikely that this conclusion is flawed by errors in our methods or fluorescence measurements. The inevitable measurement noise from various sources is not expected to affect adjacent daughter cells in opposite ways (skewing for red fluorescence in one daughter and for green fluorescence in the other). On the other hand, we cannot exclude that BrdU incorporation itself somehow affected sister chromatid segregation and further studies are needed to confirm our findings. Of note, we did not observe 100% asymmetric segregation of sister chromatids in any pair of mitotic colon cells. Most likely, a subset of colon cells selectively segregates sister chromatids from a majority but not all chromosomes. Alternatively, a small number of specific chromatids could be selectively captured in a larger proportion of cells. The strand-specific probes in this study are unable to detect minor deviations from random sister chromatid segregation or detect selective segregation of a few or single chromosomes12.
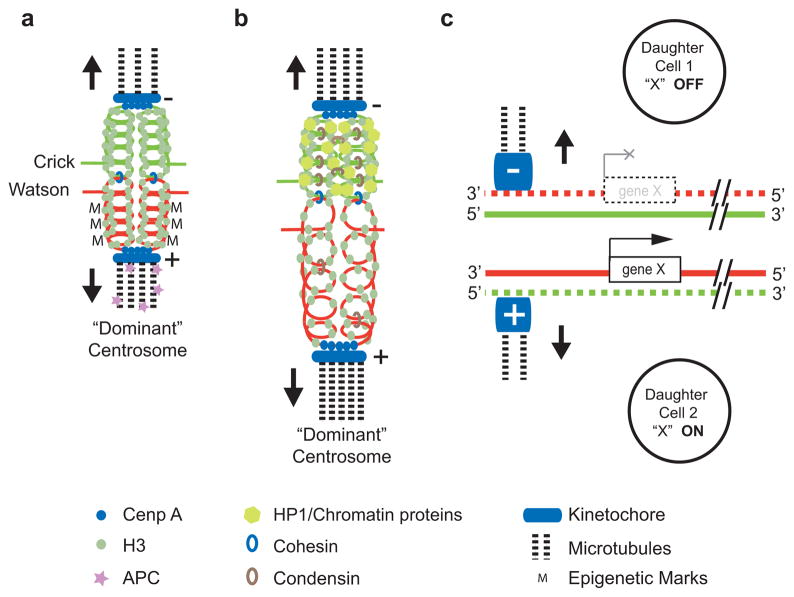
Our results provide the first direct data supporting non-random segregation of DNA template strands in mammalian cells in vivo. Non random segregation of sister chromatids has previously been observed in E.coli2 and has been implied from indirect measurements in various eukaryotic cells13,14. Neither the mechanism nor the function of selective sister chromatid segregation is currently known. In order to enable non-random segregation, sister chromatid centromeres as well as the two centrosomes of the mitotic spindle must have distinct marks or properties that enable specific connections (Figure 4). Asymmetry at centrosomes15,16 could result in differences in the timing, the number or the dynamic behavior of microtubules radiating from each pole. Alternatively, such differences could result from proteins enriched at a specific pole (Figure 4a). The Adenomatous Polyposis Coli (APC) tumor suppressor protein could be an example of the latter given its involvement in multiple cellular processes including chromosome segregation and spindle assembly17–21. How sister chromatid centromeres are distinguished is equally enigmatic, but likely depends on differences in (peri-)centric chromatin perhaps via differences in the loading22 or retention23 of (peri-) centric proteins or strand-specific replication24, methylation25 or transcription of centromeric DNA26,27. Centromeric RNA is known to regulate the assembly of centromeres28 and strands of major satellite DNA are differentially transcribed during murine development29. Chromatin differences between sister chromatids could either be directly recognized by factors at asymmetric spindles (Figure 4a) or favor selective attachment to microtubules via changes in elastic properties30 (Figure 4b). We propose that the observed non-random segregation of sister chromatids contributes to cell fate decisions as predicted by the “silent sister” hypothesis (Figure 4c)3. Further studies will test the predictions of this hypothesis that chromatin differences between sister chromatids contribute to differences in gene expression between cells, and thus regulate cell fate in asymmetrically dividing cells.
Figure 4.
Models for the mechanism and function of asymmetric sister chromatid segregation. Only the template strand of double stranded DNA in sister chromatids is shown. a, Uneven distribution of epigenetic marks (M) between sister chromatid centromeres could result in asymmetric nucleation of microtubules or selective capture of microtubules coming from the “dominant” centrosome15,16. b, Differences in higher-order chromatin structure could alter the elastic properties of (peri-)centric chromatin30 and select specific sister chromatids via microtubules originating from the “dominant” centrosome. c, Regulation of cell fate via selective segregation of sister chromatids that differ in epigenetic marks at centromeres and selected genes.
Full Methods
Preparation of cells for CO-FISH analysis
Undifferentiated wild-type murine embryonic stem cells (C2, C57BL/6NTac background) and R1-derived wild-type embryonic stem cells (C1+/+, 129S1 background)31 were obtained from Dr. A. Nagy (Samuel Lunenfeld Research Institute, Toronto) and cultured on gelatin-coated plastic culture dishes in Dulbecco’s Modified Eagle’s Medium containing 20% Fetal Calf Serum (DMEM-FCS) in the presence of 100ng/ml of Leukemia Inhibitory Factor as described32. Murine embryonic fibroblasts were grown in DMEM-FCS. BrdU (Invitrogen, Carlsbad, CA) was added to semi-confluent cultures at a final concentration of 40 μM for 12 hours prior to harvest. Binucleated cells were prepared by adding cytochalasin B (Sigma–Aldrich, 3 μg/ml) to cultures 2 hours prior to collection of cells by trypsinization. Cell were fixed with 3:1 methanol/acetic acid and spun onto microscope slides using a Shandon Cytospin 4 (Shandon Scientific, Runcorn, UK) and. For preparation of metaphase cells colcemid (Sigma-Aldrich, 0.1 μg/ml) was added for one hour prior to harvest. Trypsinized cells were treated with 0.075 M KCl (Stem Cell Technologies, Inc., Vancouver) for 10 minutes prior to fixation with 3:1 methanol/acetic acid using standard cytogenetic procedures. Fixed cells were stored at −20° C. To obtain metaphase spreads for CO-FISH, cells were dropped onto wet microscope slides and dried overnight at room temperature.
Chromosome Orientation Fluorescence in Situ Hybridization (CO-FISH)
Slides were rehydrated in phosphate buffered saline pH 7.4 (PBS, Stem Cell Technologies) for 15 minutes and fixed for 2 minutes in 4% formaldehyde in PBS followed by 3 washes in PBS for 5 minutes each. Slides were treated with freshly-prepared pepsin (P7000, Sigma-Aldrich) at 1mg/ml in acidified water (pH 2.0) at 37°C for 10 minutes followed by 2 washes for 2 minutes each in PBS, a rinse in 2xSSC and treatment with RNaseA (0.1 mg/ml in PBS) for 10 minutes at 37°C. Slides were washed twice with PBS for 5 minutes each, and stained with 100 μl Hoechst 33258 (Sigma-Aldrich) at 1mg/ml under parafilm for 15 minutes at room temperature. The slides were rinsed with 2xSSC, transferred to a tray, covered with a glass cover slip and irradiated with UV light for 30 minutes in a UV Stratalinker 1800 (calculated dose 5.4×103 J/m2). BrdU-substituted DNA strands were digested with 50 μl Exonuclease III (New England Biolabs, Ipswich, MA) at 3000U/ml in buffer supplied by the manufacturer (50 mM Tris-HCl, 5 mM MgCl2, and 5 mM DTT, pH 8.0) at 37°C for 10 minutes under a coverslip. Slides were rinsed 3 times for 5 minutes each in 2xSSC prior to denaturation in 70% formamide in 2xSSC for 1 minute at 70° C and dehydration in ice cold 70%, 90% and 100% ethanol for 2 minutes each. Cells were rehydrated in PBS for 10 minutes, fixed in 4% formaldehyde in PBS for 2 minutes, washed 3 times for 5 minutes each in PBS, dehydrated in ethanol again and air dried. Hybridization mixture (20 μl) was added to each slide, covered with a coverslip and cells were denatured on a hot plate at 80° C for 2 minutes. The hybridization mixture consisted of 10 mM Tris-HCl pH 7.6, 1 mM MgCl2, 70% formamide, 0.25% Blocking Reagent (New England Nuclear, Boston, MA) and 0.5 μg/ml Cy5-labeled (CCCTAA)3 PNA, 0.5 μg/ml Cy3-labeled (TTAGGG)3 PNA (specific for the G- and C-rich telomeres, respectively), 1 μg/ml Fluorescein-labeled GACGTGGAATATGGCAAG PNA specific for the T-rich strand of mouse major satellite DNA33 and 1 μg/ml TexasRed-labeled CTTCAGTGTGCATTTCTC PNA specific for the A-rich strand of mouse major satellite DNA. Fluorescently labeled PNA probes were obtained from Applied Biosystems (Foster City, CA), Panage Inc. (Daejeon, Korea) or Biosynthesis Inc. (Lewisville, TX) without noticeable differences in results. Following hybridization for one hour at room temperature, slides were washed 2 times for 15 minutes each in 70% formamide, 10mM Tris-HCl, 1% BSA and 3 times 5 minutes in TNT (0.1 M Tris-HCl, 0.15 M NaCl, 0.08% Tween-20, pH 7.5). Following dehydration in ethanol, slides were air dried and counterstained with DAPI at 200 ng/ml in DABCO antifade solution34.
CO-FISH on paraffin-embedded tissue sections
C57Bl6/6J mice (2–3 months old) were injected intra-peritoneally with BrdU for 12 hours at 1-hour intervals as described35. For metaphase analysis, BrdU was injected for 0, 8, 12 or 16 hours, with an additional injection of colcemid to arrest cells at metaphase 1 hour prior to tissue harvest. Colon tissue was fixed overnight in 4% formaldehyde in PBS and embedded in paraffin using standard procedures. Tissue sections (6 micron) were baked overnight at 60°C, deparaffinized in xylene 3 times for 15 minutes each at room temperature prior to dehydration in 100% ethanol (2 times for 10 minutes). The previously-described CO-FISH protocol was used for metaphase spreads from cultured cells with the following modifications. Slides were treated in 10 mM citric acid buffer pH 6.0 at 80°C for 45 minutes, washed at room temperature in PBS 2 times for 5 minutes each, and water for 5 minutes followed by pepsin and RNAse treatment as described above. Following incubation with Hoechst and treatment with UV as above, slides were rinsed with 2xSSC for 5 minutes and denatured in 70% formamide 2xSSC for 2 minutes at 72°C, dehydrated, air dried and rehydrated in PBS for 10 minutes prior to treatment with RNAse A and Exonuclease III as above. Slides were washed 2 times for 5 minutes each in 2xSSC, denatured in 70% formamide 2xSSC for 1 minute at 70°C, dehydrated in ethanol and air dried. For hybridization, 40 μl of hybridization mixture containing 1 μg/ml Cy5-labeled GACGTGGAATATGGCAAG or Cy5 labeled GAAGGACCTGGAATATGG PNA specific for the T-rich “Crick” strand of mouse major satellite DNA33 and 1 μg/ml Cy3-labeled CTTGCCATATTCCACGTC specific for the A-rich Watson strand of mouse major satellite DNA was used. Following denaturation for 3 minutes at 80°C and hybridization overnight at room temperature, slides were washed and counterstained with DAPI at 10 ng/ml in PBS for 5 minutes, rinsed 3 times for 5min in PBS, dehydrated in ethanol and covered under DABCO antifade solution for fluorescence microscopy.
Isolation of paired cells from colon
C57Bl6/6J mice (2–3 months old) were injected intra-peritoneally with BrdU for 12 hours at 1-hour intervals as described35. Paired cells from colon were isolated by modification of a published method35. Briefly, colon tissues were dissected from BrdU-treated mice and placed in ice-cold PBS. Faeces was cleared by flushing the colon with a syringe filled with ice-cold PBS. Colons were subsequently cut longitudinally and minced into 1cm pieces then incubated in 15 ml of predigestion solution (5 mM EDTA, 1 mM DTT, 1x PBS) for 30 minutes at 37° C. The resulting cell suspension was centrifuged for 5 minutes at 1200 rpm at 20° C. Supernatant was aspirated and the cell pellet was resuspended in 15 ml of digestion solution (prepared by mixing 50 mg of collagenase type XI (Sigma–Aldrich) and 100 mg of dispase II (Sigma–Aldrich) into 100 ml of PBS) for 90 minutes at 37° C. After incubation, the cell suspension was centrifuged for 5 minutes at 1200 rpm at 20° C. Supernatant was aspirated and discarded. Cell pellet was resuspended in PBS, vortexed for 20 s and passed through a 100 μm cell strainer (BD Falcon). Cells were treated with 0.075M KCl for 10 minutes at 37° C prior to fixation with 3:1 methanol/acetic acid. Fixed cells were stored at −20° C, then spun onto microscope slides and subject to CO-FISH analysis as above.
Fluorescence microscopy, image acquisition and selection
Fluorescence signals were captured on an Axioplan microscope (Zeiss, Germany) equipped with filters for DAPI, FITC, Cy3, Cy5, and Texas Red (Chroma Technology, Rockingham, and Semrock, Rockchester) using an Axiocam MRm digital camera controlled by Metasystems ISIS software (Altlussheim – Germany). Alternatively, images were acquired on a Coolsnap HQ digital camera attached to an inverted microscope (IX70 Olympus) fitted to an imaging system (DeltaVision RT, Applied Precision, Seattle) equipped with similar filter sets. Gray-scale (12 bit) images at the wavelengths of interest were acquired through a high-numerical-aperture 63x/1.4 or 60x/1.4 oil immersion lens. For tissue sections, a stack of images at 0.15–0.5 micron intervals was acquired to cover the entire thickness of the section. Fluorescence signals in individual image planes were projected onto a single image plane using ISIS software (Metasystems) or SoftWoRx (Applied Precision) software before or after deconvolution.
Acquisition of image stacks was limited to informative cell pairs defined as cell pairs where both nuclei appeared to be intact and did not overlap with neighboring nuclei. To avoid ascertainment bias, image stacks from every informative cell pair were acquired. Since only a few informative cell pairs were present on individual slides, images were acquired from multiple slides to generate sufficient data for statistical analysis. Details of quantitative image analysis and statistical analysis are provided as Supplementary Information.
Supplementary Material
Acknowledgments
Kathleen Lisaingo helped acquire images for this study, Jiepeng Tan helped with image analysis and Cam Smith assisted with in vivo studies. We thank Sam Aparicio and members of the Lansdorp lab for discussions and Louis Lefebvre for critically reading the manuscript. This study was funded in part by a grant from the Canadian Institutes of Health Research (RMF-92093) the Canadian Cancer Society and the Terry Fox Foundation. We are thankful to the Canada Foundation for Innovation, the British Columbia Knowledge Development Fund, the British Columbia Cancer Foundation, the Blossom Fund of the University of British Columbia and the Mahon family for funding of equipment used in this work.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions E.F. helped with the design of the experiments, image acquisition, data analysis, interpretation of results and writing of the paper. E.C. performed most of the CO-FISH experiments. A.H. performed most of the mouse work. L.B. acquired some images for this study. S.P. performed analysis of digital data and helped with statistical analysis which was performed by S.M. D.H. helped with the design of the study and interpretation of results. P.M.L. conceived the study, helped with image acquisition, interpretation of results and writing of the paper.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Klar AJ. Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature. 1987;326 (6112):466–470. doi: 10.1038/326466a0. [DOI] [PubMed] [Google Scholar]
- 2.White MA, Eykelenboom JK, Lopez-Vernaza MA, Wilson E, Leach DR. Non-random segregation of sister chromosomes in Escherichia coli. Nature. 2008;455 (7217):1248–1250. doi: 10.1038/nature07282. [DOI] [PubMed] [Google Scholar]
- 3.Lansdorp PM. Immortal strands? Give me a break. Cell. 2007;129 (7):1244–1247. doi: 10.1016/j.cell.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 4.Meyne J, Goodwin EH. Strand-specific fluorescence in situ hybridization for determining orientation and direction of DNA sequences. Methods Mol Biol. 1994;33:141–145. doi: 10.1385/0-89603-280-9:141. [DOI] [PubMed] [Google Scholar]
- 5.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449 (7165):1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 6.Garagna S, et al. Pericentromeric organization at the fusion point of mouse Robertsonian translocation chromosomes. Proc Natl Acad Sci U S A. 2001;98 (1):171–175. doi: 10.1073/pnas.98.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin MS, Davidson RL. Centric fusion, satellite DNA, and DNA polarity in mouse chromosomes. Science. 1974;185 (4157):1179–1181. doi: 10.1126/science.185.4157.1179. [DOI] [PubMed] [Google Scholar]
- 8.Bailey SM, Goodwin EH, Cornforth MN. Strand-specific fluorescence in situ hybridization: the CO-FISH family. Cytogenet Genome Res. 2004;107 (1–2):14–17. doi: 10.1159/000079565. [DOI] [PubMed] [Google Scholar]
- 9.Alves P, Jonasson J. New staining method for the detection of sister-chromatid exchanges in BrdU-labelled chromosomes. J Cell Sci. 1978;32:185–195. doi: 10.1242/jcs.32.1.185. [DOI] [PubMed] [Google Scholar]
- 10.Potten CS, Hume WJ, Reid P, Cairns J. The segregation of DNA in epithelial stem cells. Cell. 1978;15 (3):899–906. doi: 10.1016/0092-8674(78)90274-x. [DOI] [PubMed] [Google Scholar]
- 11.Schneider EL, Sternberg H, Tice RR. In vivo analysis of cellular replication. Proc Natl Acad Sci U S A. 1977;74 (5):2041–2044. doi: 10.1073/pnas.74.5.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armakolas A, Klar AJ. Cell type regulates selective segregation of mouse chromosome 7 DNA strands in mitosis. Science. 2006;311 (5764):1146–1149. doi: 10.1126/science.1120519. [DOI] [PubMed] [Google Scholar]
- 13.Bell CD. Is mitotic chromatid segregation random? Histol Histopathol. 2005;20 (4):1313–1320. doi: 10.14670/HH-20.1313. [DOI] [PubMed] [Google Scholar]
- 14.Karpowicz P, et al. The germline stem cells of Drosophila melanogaster partition DNA non-randomly. Eur J Cell Biol. 2009;88 (7):397–408. doi: 10.1016/j.ejcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, et al. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461 (7266):947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315 (5811):518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etienne-Manneville S, Hall A. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature. 2003;421 (6924):753–756. doi: 10.1038/nature01423. [DOI] [PubMed] [Google Scholar]
- 18.Hanson CA, Miller JR. Non-traditional roles for the Adenomatous Polyposis Coli (APC) tumor suppressor protein. Gene. 2005;361:1–12. doi: 10.1016/j.gene.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan KB, et al. A role for the Adenomatous Polyposis Coli protein in chromosome segregation. Nat Cell Biol. 2001;3 (4):429–432. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]
- 20.Kita K, Wittmann T, Nathke IS, Waterman-Storer CM. Adenomatous polyposis coli on microtubule plus ends in cell extensions can promote microtubule net growth with or without EB1. Mol Biol Cell. 2006;17 (5):2331–2345. doi: 10.1091/mbc.E05-06-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamashita YM, Jones DL, Fuller MT. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301 (5639):1547–1550. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- 22.Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176 (6):795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorpe PH, Bruno J, Rothstein R. Kinetochore asymmetry defines a single yeast lineage. Proc Natl Acad Sci U S A. 2009;106 (16):6673–6678. doi: 10.1073/pnas.0811248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lew DJ, Burke DJ, Dutta A. The immortal strand hypothesis: how could it work? Cell. 2008;133 (1):21–23. doi: 10.1016/j.cell.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 25.Luo S, Preuss D. Strand-biased DNA methylation associated with centromeric regions in Arabidopsis. Proc Natl Acad Sci U S A. 2003;100 (19):11133–11138. doi: 10.1073/pnas.1831011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanellopoulou C, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19 (4):489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102 (34):12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouzinba-Segard H, Guais A, Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci U S A. 2006;103 (23):8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudert F, Bronner S, Garnier JM, Dolle P. Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm Genome. 1995;6 (2):76–83. doi: 10.1007/BF00303248. [DOI] [PubMed] [Google Scholar]
- 30.Bouck DC, Bloom K. Pericentric chromatin is an elastic component of the mitotic spindle. Curr Biol. 2007;17 (9):741–748. doi: 10.1016/j.cub.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding H, et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117 (7):873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 32.Gertsenstein M, Lobe C, Nagy A. ES cell-mediated conditional transgenesis. Methods Mol Biol. 2002;185:285–307. doi: 10.1385/1-59259-241-4:285. [DOI] [PubMed] [Google Scholar]
- 33.Horz W, Altenburger W. Nucleotide sequence of mouse satellite DNA. Nucleic Acids Res. 1981;9 (3):683–696. doi: 10.1093/nar/9.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson GD, et al. Fading of immunofluorescence during microscopy: a study of the phenomenon and its remedy. J Immunol Methods. 1982;55 (2):231–242. doi: 10.1016/0022-1759(82)90035-7. [DOI] [PubMed] [Google Scholar]
- 35.Allen JW, Latt SA. Analysis of sister chromatid exchange formation in vivo in mouse spermatogonia as a new test system for environmental mutagens. Nature. 1976;260 (5550):449–451. doi: 10.1038/260449a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.