Abstract
IFN-ε is a unique type-I interferon (IFN)whose constitutive expression inlung, brain, small intestine, and reproductive tissuesis only partiallyunderstood. Our previous observationthat post-transcriptional events participate in the regulation of IFN-εmRNA expression led us to investigate whether the 5′ and/or 3′un-translated regions (UTR)have regulatory functions. Surprisingly, we found that full-length IFN-ε 5′UTR markedly suppressed mRNA expression under basal conditions. Analysis of the secondary structure of this regionpredicted formation of two stable stem loop-structures, loops 1 and 2. Studies using luciferase constructs harboring various stretches of IFN-ε 5′UTR and mutant constructs in whichthe conformation of loop structures was disrupted showed that loop 1 is essential forregulation of mRNA expression. Incubation of HeLa cell extracts with agarose bound-RNAs harboring IFN-ε loop structures identified importin 9 (IPO9), a molecular transporter and chaperone, as a candidate that associates with these region of the 5′UTR. IPO9 overexpression decreased, and IPO9 silencing increased, basal IFN-ε expression. Our studies uncover a previously undescribedfunction for IPO9 as a specific, and negative, post-transcriptional regulator of IFN-ε expression and they identify key roles for IFN-ε stem loop structure 1 in this process. IPO9-mediated effects on 5′UTRs appear to extend to additional mRNAs, including HIF-1α, that can form specific loop structures.
Introduction
Viral invasion of mammalian cells is followed by un-coating of the viral particles, exposure of nucleic acids, and host cell-mediated replication of the viral genome(1). Recognition of viral nucleic acids by host intracellular receptorsinitiates a cascade of events that culminate in the production of type-I interferons (IFNs) and other cytokines (2, 3). TypeI IFNs are encoded by multiple, usually intron-less, genes, and in humans, most IFNs (e.g., IFN-β, -ω, -κ, -ε, and fourteen IFN-α species) map to chromosome 9 and are expressed in tissue-specific manners (4). IFN-ε, a recently identified member of this family, is expressed in specific tissues, including female reproductive organs such as the ovary and uterus(5). It appears to play a role in immunity and protection against virusesowing to its ability to induce a short-lived and localized mucosal immune response(6). Seminal plasma has been reported to up-regulate expression of IFN-ε in cervical and vaginal tissues(7), a response that may represent an antimicrobial defense mechanismthat evolved to fight infections. Importantly, a recent study demonstrated that the reproductive tracts of female mice deficient in Ifn-ε display increased susceptibility to vaginal infections(8), suggesting that IFN-ε is a cardinal type I IFN that plays key roles as a mediator of innate immunity.
We previously reported that in human cervical cancer cells, IFN-ε mediates STAT1 phosphorylation in response to stimulation with TNF-αand presented evidence indicating that transcriptional and post-transcriptional mechanisms participate in the regulation of IFN-ε expression (9). Gene expression is regulated at the levelsof replication, transcription, mRNA splicing, stabilityand translation, protein post-translational modification and stability, and others(10). Post-transcriptional control of gene expression relies on specific RNA-protein interactions that stabilize mRNAs, promote targeted degradation or prevent access of the ribosome to the translation start codon (10). Such interactions can be mediated by the 5′ or 3′ un-translated regions (UTR)of mRNAs (11, 12). Sequences located in3′UTRsare thought to participate primarily in the regulation of mRNA stability. Anumber of unstable mRNAs, includingthose encoding cytokines, oncogenes, and transcription factors, harbor AU-rich elements (AREs) whose signature sequence (i.e., AUUUA) is located inthe3′UTR (13). 5′UTR-mediatedregulation of expression is associated with events related to the initiation of mRNA translation, although mRNA destabilizing effects also have been noted(11). Mammalian 5′UTRs often form stable secondary structures such as stem-loops positioned between the cap structure and the AUG codon, which can inhibit translation initiation; the extent of this inhibitory effect depends on the thermodynamic stability and position of the structures(14).
In this manuscript, we investigated whether regions in the 5′ and 3′UTRs of IFN–ε mRNA regulate expression. Our motivation to conduct these analyses was based on previousstudiesshowing thatIFN-ε expression is regulated post-transcriptionally(9). We present novel evidence demonstrating that cis 5′UTRsequences negatively regulateconstitutiveIFN-ε expression. This effect is mediated by a 5′UTR stem loop structure that, when disrupted, leads to enhanced mRNA expression. The effect is specific and involves direct or indirect interaction with importin 9 (IPO9), a molecular transporter and chaperonethat belongs to the super-family of karyopherins(15).
Materials and Methods
Cell culture and transfection
The human cervical cancer cell lines HeLa and HeLa S3 were purchased from American Type Culture Collection and were maintained in a 5% CO2 atmosphere at 37°C in Dulbecco’s Modified Eagle Medium (DMEM, Life Technologies Co.) supplemented with 10% FBS (ThermoFisher Scientific). Transient transfection was accomplished by plating HeLa cells at a density of 1.5 × 105 cells per well of 12-well culture plates. After 18–20 h, we transfected 0.5 μg of plasmid DNA using Lipofectamine LTX (Life Technologies Co.), following the manufacturer’s recommendations. We incubated the cells for 24 h, and then determined luciferase activity and/or mRNA levels in cellular extracts. RNA interference (RNAi) was performed by transfection of HeLa cells with non-silencing (control) small interfering RNA (siRNA, Life Technologies Co.) or with siRNAs against IPO9 (Life Technologies Co. #442703) using Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer’s protocol.
IFN-ε5′UTR cloning
We cloned the 5′UTR of human IFN-ε using a 5′ rapid amplification of cDNA ends (RACE) kit (Life Technologies Co.). The template consisted of 1 μg of total RNA from HeLa cells previously transfected with polyinosinic-polycytidylic acid (polyI:C, 100 ng, 6 h), a treatment previously shown to increase IFN-ε mRNA expression (9). We generated cDNA using SuperScript II reverse transcriptase (Life Technologies Co.) combined with gene-specific primer 1 (GSP1: 5′-TTGCTTCATGTCGTTCAAGG-3′). We purified the cDNA and then added a poly (C) tail at the 5′ end using terminal deoxynucleotidyl transferase. Initial amplification was accomplished by PCR (94 °C, 30 s; 65 °C, 30 s; 72 °C, 3 min; 25 cycles followed by 10 min at 72 °C) using gene-specific primer 2 (GSP2: 5′-Tctcccaaccatccagagaaa-3′). A second round of PCR (94 °C, 30 s; 55 °C, 30 s; 72 °C, 3 min; 30 cycles followed by 10 min at 72 °C) was performed using 2.5 μl of a 50X-dilution of the initial PCR product combined with a nested gene-specific primer 3 (GSP3: 5′-gccagcagcaccaacacagt-3′). The forward primer (AUAP: 5′-GGCCACGCGTCGACTAGTAC-3′) was provided by Life Technologies Co. The generated 5′ RACE PCR product was gel-purified, cloned, and sequenced at the University of Utah DNA Sequencing Core Facility.
Plasmid construction
Luciferase reporter constructs were generated in the mammalian expression vector pcDNA3.1/Zeo+ modified to express firefly luciferase cDNA (generated from pGL3-Basic (Promega Co.) by digestion with HindIII and XbaI, as described by Dixon et al (16)). We utilized the 5′RACE PCR product as template to generate constructs harboring stem-loops 1 and 2 (ε1+2), posterior stem-loop 2 (ε2), or no stem-loops (ε0), using Platinum®Pfx DNA Polymerase (Life Technologies Co.). The reverse primer harbored a 5′-NcoI site shown in lower case (5′-TTccatggtgaaggtcaaatatgctac-3′) and forward primers included 5′-XhoI sites: (XhoI-ε1+2: 5′-TGGctcgagTGCCTCAAGGAAAGCATACAA-3′; XhoI-ε2: 5′-TGGctcgagatgtactctgaacaccatga-3′; XhoI-ε0: 5′-TGGctcgagacattagaaaacgaaagcaac-3′). The products were purified, digested with XhoI and NcoI and then cloned into the pcDNA3.1/Zeo+-luciferase(pcDNA-luc) vector described above.
The 3′UTR of IFN-ε mRNA was generated by amplification using primers XbaI-F-3′UTR (5′-AGtctagaGTGGAGGGACTAGAGGACTT-3′) and XbaI-R-3′UTR (5′-CCtctagaGAAGAATCAACCATATTAATG-3′). The product was purified, digested with XbaI, and then cloned into the XbaI site of pcDNA-luc.
A cDNA encoding full-length IPO9 wasobtained by amplification ofHeLa cell cDNA using Platinum® Pfx DNA Polymerase. We used primersNotI-IPO9-F (5′-CTTgcggccgcGATGGCGGCGGCGGCGGCAGCT-3′) and SalI-IPO9-R (5′-CTAGAgtcgacCTAACTAGTGTCCATTGATTC-3′). The amplified products were cloned into the NotI and SalI sites of the mammalian expression vector, p3XFLAG-CMV7.1 (Sigma-Aldrich).
Mutations in the loop and stem structures of IFN-ε 5′UTR were generated using a site-directed mutagenesis kit from Stratagene (La Jolla, CA). Our objective was to generate mutant constructs predicted to have significant destabilization of secondary structures while harboring minimal changes in primary sequence. To achieve this, we made use of CentroidFold, asoftwareprogram that predicts the thermodynamic stability of RNA secondary structures. The primer sequences used to generate mutant IFN-εloops and stem were: 5′-GCATACAATGAATAAcTTATTaTctTACTTCTTCAAAATA-3′ and 5′-TATTTTGAGGAAGTAagAtAATAAgTTATTCATTGTATGC-3′ (loop 1)and 5′-GGGAACCTGAAAATCatAAcTGTAAACTTGGAGAA-3′ and 5′-TTCTCCAAGTTTACAgTTatGATTTTCAGGTTCCC-3′ (loop 2). Mutated nucleotides are shown in lower casefont.
Quantitative RT-PCR
Total RNA was extracted using an RNeasy total RNA isolation kit with on-column DNase I digestion (Qiagen). We used 1 μg of total RNA as template for first-strand cDNA synthesis, combined with oligo(dT) primers and M-MLV reverse transcriptase (Life Technologies Co.). An Opticon 2 Real-Time PCR System (Bio-Rad) was used for quantitative analyses of luciferase and zeocin expression. The sequences of the primers were:
IFN-ε-F: 5′-AGGACACACTCTGGCCATTC-3′;
IFN-ε-R: 5′-TTGCTTCATGTCGTTCAAGG-3′;
Luciferase-F: 5′-ACGGATTACCAGGGATTTCAGTC-3′;
Luciferase-R: 5′-AGGCTCCTCAGAAACAGCTCTTC-3′;
GAPDH-F: 5′-GCACCGTCAAGGCTGAGAAC-3′;
GAPDH-R: 5′-ATGGTGGTGAAGACGCCAG-3′;
CCL5-F: 5′-CTACTCGGGAGGCTAAGGCAGGAA-3′;
CCL5-R: 5′-GAGGGGTTGAGACGGCGGAAGC-3′;
IL6-F: 5′-ATGAACTCCTTCTCCACAAGC-3′;
IL6-R: 5′-AAGAGCCCTCAGGCTGGACTG-3′;
HIF-1α-F: 5′-GAGAAATGCTTACACACAGAAA-3′
HIF-1α-R: 5′-CGGTAATTCTTTCATCACAATA-3′
18S rRNA-F: 5′-ACTCAACACGGGAAACCTCA-3′;
18S rRNA-R: 5′-AACCAGACAAATCGCTCCAC-3′;
Zeocin-F: 5′-GACTTCGTGGAGGACGACTT-3′;
Zeocin-R: 5′-GACACGACCTCCGACCACT-3′.
Amplifications were performed using iQ SYBR Green Supermix (Bio-Rad), according to the manufacturer’s specifications. Cycling conditions were as follows: 50°C, 2 min; 95°C, 3 min; 40 cycles of 95°C (15s) + 58°C (30 s) + 72°C(30 s). A melting curve was generated by acquiring fluorescence measurements while slowly heating to 95°C at a rate of 0.1°C per second. Melting curves and quantitative analysis of the data were performed using an Opticon monitor, version 3.1, as previously reported (17).
Reporter assays
Twenty-four hours after transfection with reporter constructs, we harvested cells using Reporter Lysis Buffer and determined luciferase activity using a commercially available assay system (Promega Co.). When indicated, the cells were co-transfected with a β-galactosidase cDNA (pSV-β-Gal). Fornormalization purposes, we assessed either total protein concentration in the cell lysates or β-galactosidase activity. The latter was determined using a chemiluminescence-based reporter assay (Roche Applied Science). The data presented are representative of at least three independent experiments performed in duplicate, and they are reported as the mean ± SD.
Identification of 5′UTR binding proteins
HeLa S3 cells were grown in suspension in DMEM containing 10% FBS. A cytoplasmic extract was obtained by harvesting 5x108 cells in buffer A [10 mM HEPES-KOH (pH 7.5), 3 mM MgCl2, 14mM KCl, 5% glycerol, 1 mM dithiothreitol, and 0.5% protease inhibitor cocktail (Sigma-Aldrich)]. Followingbrief incubation (15 min on ice), the lysate was homogenized with 20 strokes of a Dounce homogenizer and then subjected to centrifugation (5,000 × g, 15 min). The supernatantwas stored at ′80 °C in small aliquots.
We immobilized RNA on agarose beads essentially as described by Caputi et al.(18). Briefly, the vectors diagrammatically shown in Figure 5A were linearized using NcoI and then were in vitro-transcribed using T7 RiboMAX™ (Promega Co.), following the manufacturer’s recommendations. RNAs(500 pmol) were placed in 400 μl reaction mixtures containing 100 mM sodium acetate (pH 5.0) and 5 mM sodium m-periodate, and were incubated for 1 h in the dark at room temperature. RNAs were precipitated using ethanol, and thenwere resuspended in 400 μl of 100 mM sodium acetate (pH 5.0). Two-hundredμl aliquots of adipic acid dihydrazide agarose beads (Sigma-Aldrich) were washed four times with 10 ml of 0.1 M sodium acetate (pH 5.0). After the final wash, the beads were resuspended in 400 μl of 0.1 M sodium acetate (pH 5.0), mixed with periodate-treated RNAs, and then rotated for 16 h at 4 °C. Next, we washed the beads with 3 × 1 ml of 2 M NaCl and then 3 × 1 ml of buffer D [20 mM HEPES-KOH (pH 7.6), 5% glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol]. We then mixed agarose-bound RNAswith HeLa S3 cytoplasmic extractsand rotated the mixtures for 2 h at 4°C. Beads were decanted(5,000 rpm, 1 min) and washed four times with 1 ml of buffer D. After the final centrifugation, agarose-bound proteins were eluted with2X SDS sample buffer, subjected to electrophoresis on 10% SDS–polyacrylamide gels, and visualized with Coomassie Blue. We excised several bands of interest from the stained gel, and performed in-gel tryptic digestion, as described (19). The resulting tryptic peptides were dissolved in 20 μl of 0.1 % trifluoroacetic acid and then desalted using C18 ZipTip (Millipore, Bedford, MA), according to the manufacturer’s recommendations. Next, the peptides were dissolved in 0.1% formic acid and subjected to nanoflow-LC-MS/MS analyses and identification, according to Ozaki et al.(20).
Figure 5. Effect of IFN-ε 5′UTR stem-loop structures on post-transcriptional events.
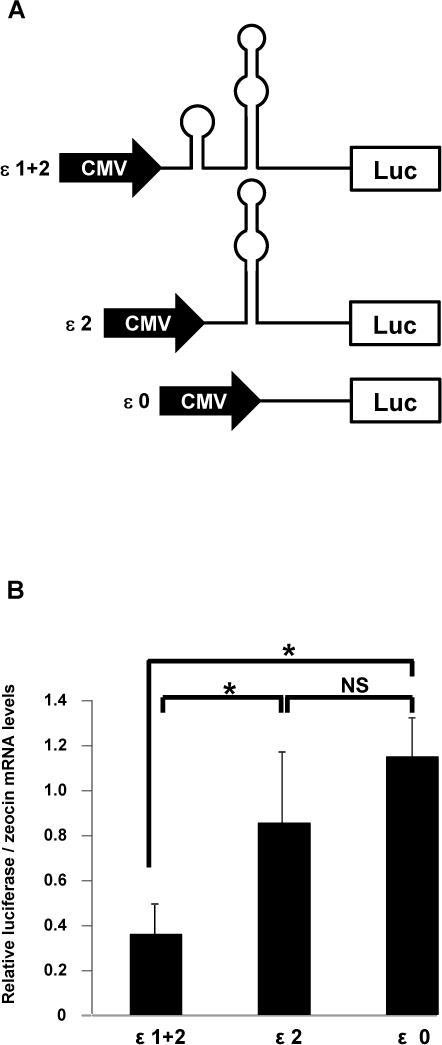
A. We cloned three regions ofIFN-ε 5′UTR upstream of the firefly luciferase cDNA to generate reporter constructs that included loops 1 and 2 (ε1+2), loop 2 (ε2), or no loops (ε0).
B. Reporter constructs diagrammatically represented in A were transfected into HeLa cells as described in Figure 2C. We then determined luciferase mRNA levels using quantitative RT-PCR as described in Materials andMethods. Zeocin mRNA levels served as normalization controls. Data represent the mean ± SD of three separate transfection experiments. *, p < 0.001; NS, not significant.
Results
The 3′-end ofIFN-ε mRNA harborsAUUUApentamers that are not required forconstitutive expression
Our first goal was to investigate whether, in addition to transcriptional regulation(9), post-transcriptional mechanisms regulate expression of the IFN-ε gene. Wefirst hypothesized that one or more AREs might regulate the stability of IFN-ε mRNAunder basal conditions. AREs are usually located in the 3′UTR of mRNAs (21) and sequence analyses revealed the presence of two AUUUA elements in the 3′UTR of IFN-ε mRNA (Figure 1A). We cloned this regionusing RACEand then generated a mammalian expression vectorin whichthe 3′UTR was cloned downstream of the firefly luciferase cDNA (Figure 1B). Transfection ofHeLa cells with this construct resulted in luciferase expression at levelssimilar to those achieved by a construct lacking the 3′UTR (Figure 1C). These results suggest that the 3′UTR is not required for constitutive expression of IFN-ε.
Figure 1. Effect of IFN-ε 3′UTR on post-transcriptional regulatory events.
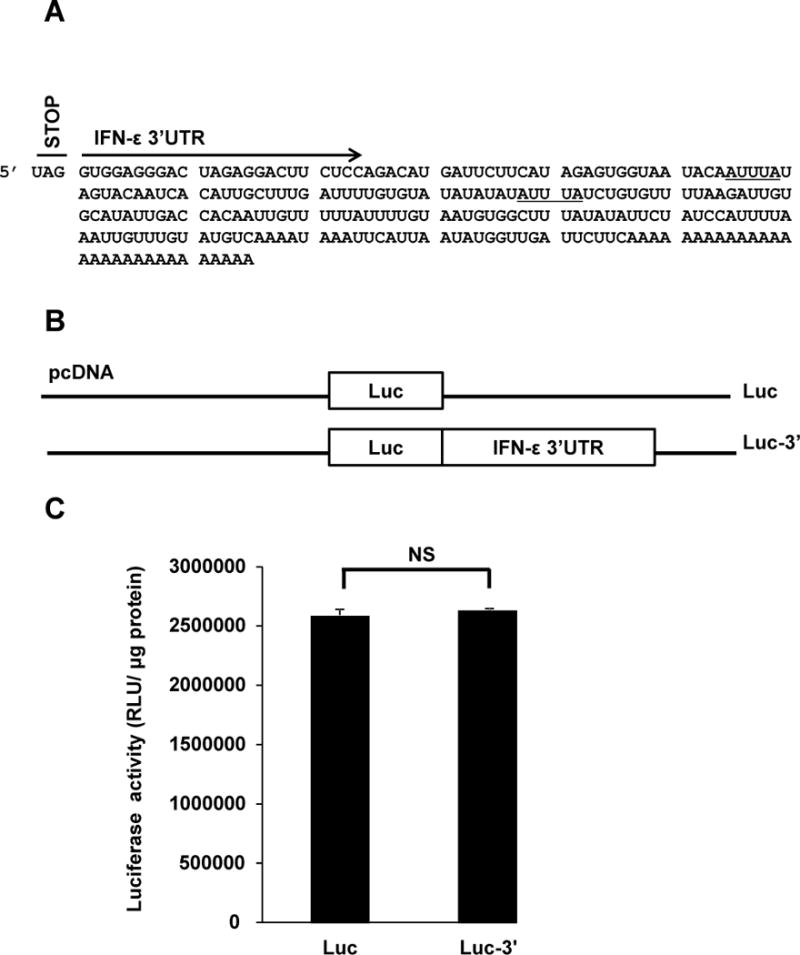
A. DNA sequences immediately downstream of the stop codon UAG represent the 3′UTR of IFN-ε mRNA. TwoAUUUA stretches are underlined.
B. Diagrammatic representation of Luc and Luc-3′ vectors tested in C.
C. We transfected HeLa cells with Luc and Luc-3′, incubated the cells for 24 h, and assessed luciferase activity in cellular extracts. Data represent the mean ± SD from four experiments. NS, not significant.
Effect of the 5′UTR of IFN-ε mRNA on firefly luciferase production under basal conditions
We next focused our attention on the 5′UTR of IFN-ε mRNA. We cloned this region using RACE and found that it harbors 265 ntupstream of the human IFN-εtranslation start site (Figure 2A). We then generated a mammalian expression construct in whichthis region was inserted at the 5′ end of the firefly luciferase cDNA (5′-Luc, Figure 2B). In addition, we created a construct that harbored both the 5′ and 3′UTRs (5′-Luc-3′) to assess potential synergistic effects (Figure 2B). HeLa cells transfected with 5′-Lucexpressed significantly lower levels ofluciferase activity relative to total protein, compared with cells transfected with a construct that lacked UTRs (Figure 2C). These results point at5′UTR-mediated negative regulation of constitutiveIFN-ε expression. Inclusion of IFN-ε3′UTR did not affect 5′UTR-mediated effects (Figure 2C, 5′-Luc-3′) thus confirming lack of participation of the 3′UTR in post-transcriptional regulatory events.
Figure 2. Effect of IFN-ε 5′UTR on post-transcriptional regulatory events.
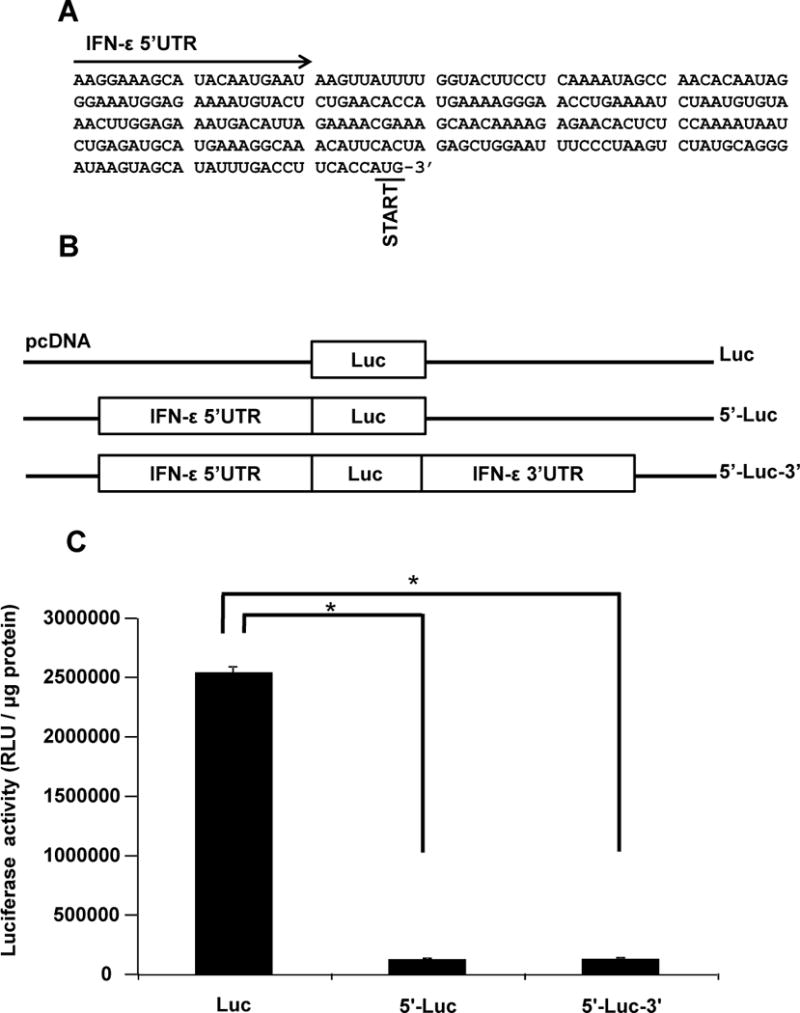
A. DNA sequences immediately upstream of the start codon AUG represent the 5′UTR of IFN-ε mRNA. Two regions showing predicted stem-loop structures are underlined.
B. Diagrammatic representation of Luc, 5′-Luc- and 5′-Luc-3′ vectors tested in C.
C. We transfected HeLa cells with Luc, 5′-Luc- and 5′-Luc-3′, incubated the cells for 24 h, and assessed luciferase activity in cellular extracts. The results obtained with Luc are the same as those presented in Figure 1C and are reproduced in this panel to facilitate direct comparisons. Data represent the mean ± SD from four experiments. *, p < 0.001.
The 5′UTR of IFN-ε mRNA does not affect mRNA stability
Regulation of mRNA stability is a common post-transcriptional mechanism to control protein levels in response to cellular needs. In previous work, we provided evidence suggesting that HeLa cells regulate IFN-ε expression by modulating mRNA stability (9). Thus, we hypothesized that the ability of the 5′UTR of IFN-ε to decrease luciferase mRNA levels (Figure 2C) might involve de-stabilization of the mRNA. To test this, we compared the stability of IFN-ε mRNA in the presence and absence of the 5′UTR, under experimental conditions that precluded transcription, as previously described (9). We observed no 5′UTR-mediated effects on mRNA half-life suggesting that the 5′UTR is not involved in mRNA de-stabilizing effects (not shown).
The 5′UTRof IFN-εmRNA is predicted to form stem-loop structures that affect constitutive mRNAexpression
RNA secondary structures located between the cap structure and the initiation codon can inhibit translation initiation, and the extent of this effect depends on the thermodynamic stability and position of the structure (10). Based on findings presented in Figure 2C, we hypothesized that defined regions in the 5′UTR of IFN-ε mRNA might adopt secondary structures that impactpost-transcriptional events. We conducted computer-aided analyses of the 5′UTR of human IFN-ε mRNA using CentroidFold (www.ncrna.org/centroidfold), a web-based approach frequently employed as a tool to predictRNA secondary structures (22). This analysis identified two sequences potentially capable of forming stem-loop structures: loop 1, spanningbases 23–49 and loop 2 spanning bases 110–142 (Figure 3). The formation of stem-loop structures was consistently predicted by multiple additional algorithms, including KineFold (http://kinefold.curie.fr/, Figure S1A), RNAfold (http://rna.tbi.univie.ac.at, Figure S1B–C), and Mobyle (http://mobyle.pasteur.fr, not shown). Moreover, stem-loop formation at the 5′UTR of IFN-ε is predicted fora variety of primates (Figure 4 (23)), suggesting evolutionarily conserved functions for these regions. We utilized this information to generate firefly luciferase expression constructs in whichIFN-ε mRNA 5′UTRsequences harboring 0-1-2 loopswere cloned between the cytomegalovirus promoter and luciferase cDNA (Figure 5A), and then used these constructs to assess the impact of loop structures on post-transcriptional events. We found that the presence of loop 2 (ε2) did not affect luciferasemRNA expression compared with that resulting from a construct that lacked stem-loop structures (compare ε0 with ε2, Figure 5B), suggesting that this structure does not regulate IFN-ε mRNA expression. In contrast, inclusion of loops 1 and 2 robustly decreased relative luciferasemRNA levels(compare ε0 with ε1+2, Figure 5B). These combined observations suggest that loop 1, or the combined presence of loops 1 and 2, suppresses IFN-ε mRNA expression.
Figure 3. Secondary structure ofIFN-ε5′UTR generated using publicly-available prediction tools, and alignment of the region among several primates.
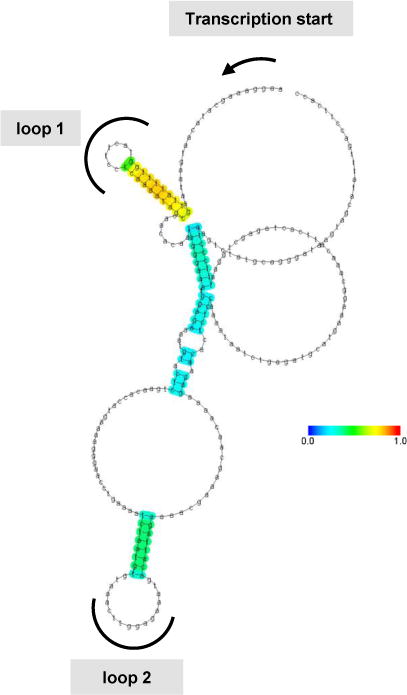
Secondary structure of IFN-ε 5′UTR mRNA as predicted by CentroidFold.
Figure 4. Alignment of IFN-ε 5′UTR among several primates.
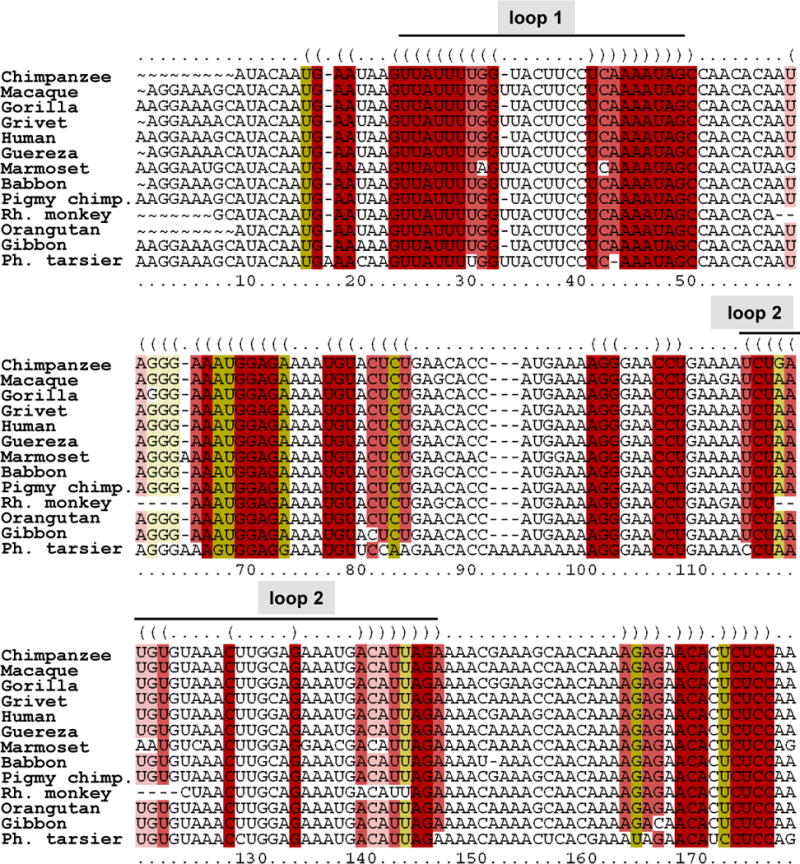
Multiple alignment of IFN-ε 5′UTR from several primates was performed using LocARNA (http://rna.informatik.uni-freiburg.de), a tool that simultaneously folds and aligns RNA sequences, and generates a consensus structure (23). Pigmy chimp: pigmy chimpanzee; Ph. tarsier: Philippine tarsier.
Effect of loop structures within 5′UTR of IFN-ε on constitutive mRNA expression
Our next goal was to investigate whether proper conformation ofstem-loop structures is involved inthe regulation of IFN-ε mRNA expression. To accomplish this, we generated a series of 5′UTR mutant constructs in which each of the predicted secondary structures was individually disrupted (Figure S2A, S2B). We found that disruption of loop 1 rescuedluciferase mRNA expressionto a level comparable to that of ε2, which lacks loop 1 (Figure 6). In contrast, disruption of loop 2 had no effect compared with the control (ε1+2). These data are consistent with studies shown in Figure 5 and they suggest that proper folding of loop 1 is essential for constitutive suppression of IFN-εmRNA. Our results also suggest that loop 1, and not the combined presence of loops 1 and 2, is responsible for negative regulation of IFN-ε expression.
Figure 6. Mutation of IFN-ε 5′UTR stem-loop structure 1increases mRNA expression.
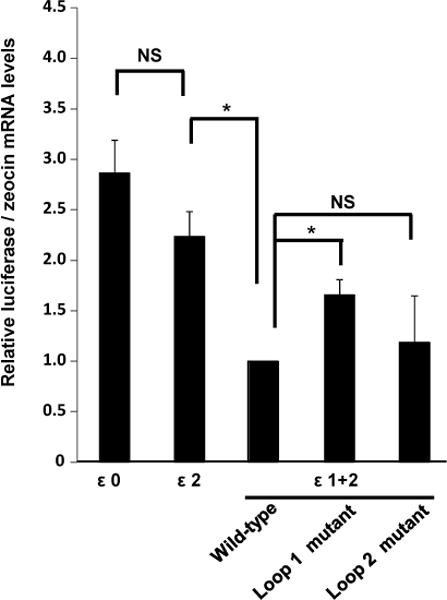
We transfected HeLa cells with ε0, ε2, and ε1+2 and with ε1+2 constructs harboring mutations in loops 1 or 2. We then determined luciferase mRNA levels using quantitative RT-PCR as described in Materials andMethods. Zeocin mRNA levels served as normalization controls. Data represent the mean ± SD of four independent experiments. *, p < 0.01
Identification of IPO9as a protein that binds to IFN-ε 5′UTR stem-loop structures
We next sought toidentify proteins that specifically interact with IFN-ε 5′UTR stem-loops, and determine whether the identified candidates play a role in post-transcriptional regulation of IFN-ε expression. To accomplish this, we generatedRNAs corresponding to IFN-ε 5′UTR harboring both loops (ε1+2) or no loops (ε0). We attached individual RNAs to agarose beads, incubated them withHeLa cell extracts, and then subjected bound proteinsto SDS-PAGE. We found thatremoval ofloops 1 and 2 robustly decreased association with three proteins of apparent m.w. 68, 108, and 118 KDa (Figure 7). We subjected the 100–130 KDa region of the SDS-PAGE gel shown in Figure 7 to mass spectrometric analyses, andidentifiedimportin 9 (IPO9, predicted m.w.=110 KDa), a member of the importin βsuperfamily of nuclear transport receptors, asa candidate involved in post-transcriptional regulation of IFN-ε expression. We were unable to identify the 68 KDa protein owing to technical difficulties.
Figure 7. RNA harboring stem-loop structures 1 and 2 bind 68, 108, and 118 KDa HeLa cell proteins.
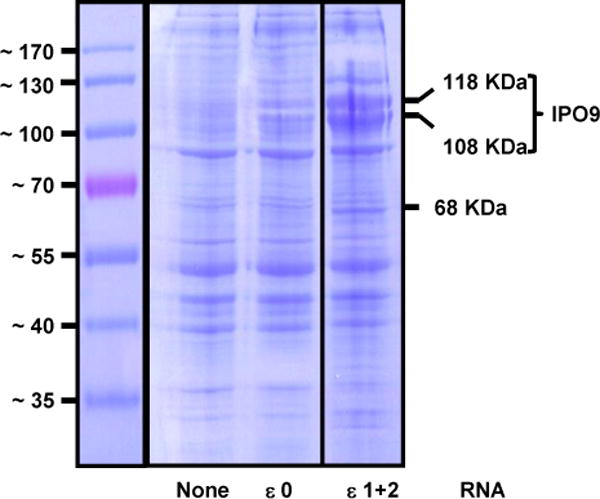
We generated IFN-ε RNAs harboring stem loops 1 and 2 (ε1+2) or no stem-loop structures (ε0) from constructs shown in Figure 5A. The RNAs were covalently attached to agarose beads and the products then were incubated with HeLa cell extracts. Proteins adsorbed to control and RNA-treated beads were subjected to SDS-PAGE and visualized by staining with Coomassie Blue.
IPO9 silencing enhances constitutiveexpression of IFN-ε mRNA
To investigate whether IPO9 functionally affects IFN-ε expression, we silencedits expressionusing siRNA (Figure 8A) and found that decreased IPO9 expression enhancedconstitutive IFN-εmRNA expression (Figure 8B). Stimulation withTNF-αmodestly, but significantly, increased the level of IFN-ε mRNA, in agreement with our previous findings(9). However, IPO9 silencing restoredIFN-ε mRNA to an extent equal to that observed in un-stimulated cells (Figure 8B). These results indicate that IPO9 suppresses constitutive, but not stimulated, IFN-ε mRNA expression. We next investigated whether IPO9 suppressed expression of other mRNAs andfound that constitutive expressionof HIF-1α, predicted to forma large secondary structure within its 5′UTR (Figure S3), was enhanced by IPO9 silencing, in a manner similar to that observed for IFN-ε (Figure 8C). In contrast, the levels of constitutively expressed (housekeeping) gene GAPDH (24)and those of the chemokine CCL5 and the cytokine IL-6 werenot affected by IPO9 silencing (Figure 8D–F). While the 5′UTRs of these mRNAs also are predicted to adopt secondary structures (not shown), the stem loops formed by IFN-ε and HIF-1α appear to be unique.
Figure 8. Knockdown of IPO9 enhancesIFN-εmRNA expression.
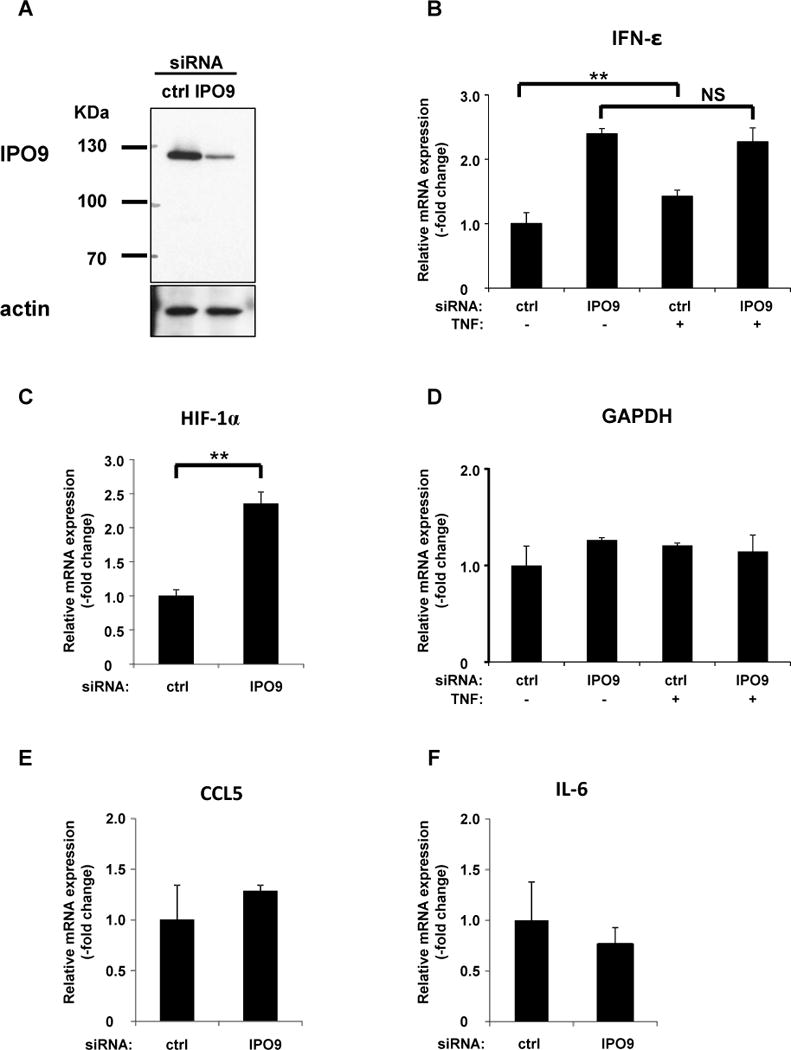
A. We transfected HeLa cells with IPO9siRNAand then incubated the cells for 48 h. Knockdown efficiency of IPO9 was determinedby western blot analyses, as described in Materials andMethods.
B–F. We extracted total RNA from IPO9-silenced and determined mRNA levels of IFN-ε (B), HIF-1α (C), GAPDH (D), CCL5 (E), and IL-6 (F), using quantitative RT-PCR. 18S rRNA levels served as normalization controls. Datarepresent the mean ± SD of three independent experiments. *, p < 0.05; **, p<0.01
Loop 1 is required for IPO9-mediated suppression of IFN-ε expression
To investigate whether the regulatory effects of IPO9 and loop 1 on IFN-ε expression are functionally related, we assessed the impact of IPO9 on inhibition of IFN-ε expression mediated by our 5′UTR constructs. We utilized a two-pronged approach whereby IPO9 was either over-expressed (Figure 9A) or silenced (Figure 9B). These studies showed thatIPO9 regulates INF-ε mRNA levels in a loop-dependent fashion. Increased IPO9 led to marked reductions in the expression of a construct harboring loops 1 and 2 (ε1+2), but had no effect when loops 1 and 2 were deleted (Figure 9A). Conversely, silencing IPO9 enhanced expression of ε1+2, but not that of constructs lacking loop 1 (ε0 and ε2, Figure 9B). These results demonstrate that IPO9 regulates constitutive expression ofIFN-ε mRNA through loop 1 within 5′UTR of IFN-ε. Loop 1 in IFN-ε and a stem loop in HIF-1α (candidate loop structure, Fig. S3) share unique features; the stems are composed of 6-10 bp and the loops harbor 5-7 nucleosides. While this requirement remains to be confirmed experimentally, it appears that the suppressive effect of IPO9 on mRNA expression is dependent on the formation of peculiar secondary structures within 5′UTRs.
Figure 9. IFN-ε 5′UTR-associated proteins negatively regulate IFN-ε mRNA expression.
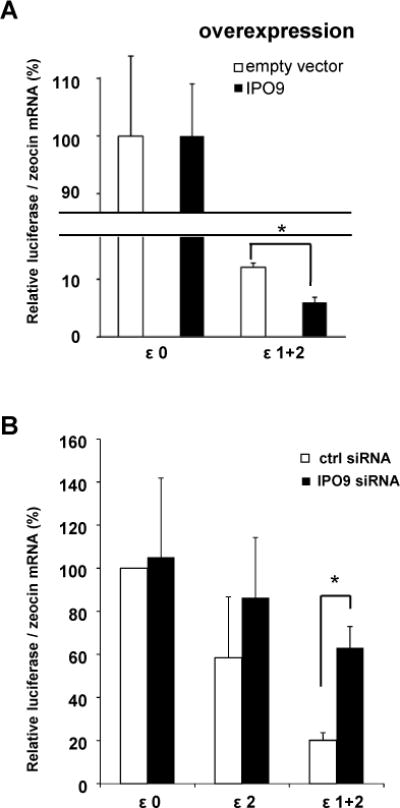
A. We transfected HeLa cells with IPO9 or with empty vector, combined with ε1+2 or ε0 cDNA, and then incubated the cells for 24 h. We determined luciferase mRNAexpression by quantitative RT-PCR; zeocin levels served as normalization controls. Data shown represent the average ± SD of four determinations. *, p < 0.05
B. HeLa cellIPO9 expression was silenced by transfection withsiRNA for 24h. Cells transfected with a non-silencing siRNA served as control. The cells then were transfected with ε1+2, ε2, or ε0 cDNAs and luciferase mRNA levels were determined 24 h later, using quantitative RT-PCR.
IPO9 does not regulateIFN-εexpression at the promoter level
Our final goal was to investigate whether IPO9 transcriptionally regulates IFN-εexpression. To test this, we transfected IPO9-silenced and control HeLa cells with a previously described IFN-ε promoter construct, and then determined promoter activity, as before (9). We found thatIPO9 silencingdid not activate the IFN-ε promoter, in contrast to transfection with polyI:C, which was used as a positive control (Figure 10, (9)). We conclude that IPO9 does not regulateIFN-εexpression at the promoter level.
Figure 10. IPO9 does not regulateIFN-εexpression at the promoter level.
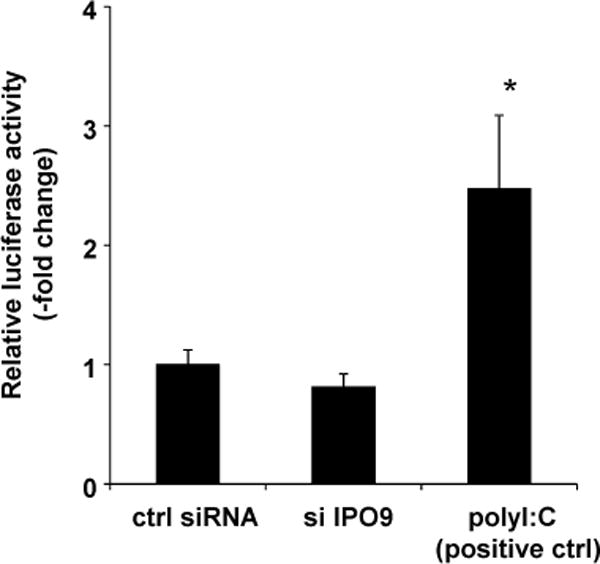
HeLa cell IPO9 expression was silenced by siRNA-mediated transfection for 24h. Non-silencing siRNA-transfected cells served as controls. We thenco-transfected a previously described IFN-ε promoter construct (9) combined witha β-galactosidase vector for 24 h. Our positive control consisted of cells transfected with polyIC (100ng) for 6 h(9). We assessed luciferase and β-galactosidase activities in cellular extracts. The data are expressed in luciferase units normalized for β-galactosidase expression, and they represent the mean ± SD from four experiments. *, p < 0.01
Discussion
Previous studies have shown that IFN-ε expression is induced upon infection with Semliki Forest (5) and vaccinia (14) viruses, and in response to exposure of cervico-vaginal tissues to seminal fluid (7). In addition, RNAi-mediated silencing of IFN-ε significantly inhibitsactivation of STAT1(9), a key transcriptional activator of antiviral, immune, and anti-tumorigenic responses(25). These observations point at the importance of IFN-ε in host immune responses such as those required toestablishantimicrobial, anti-viral, and anti-tumorigenic states in specific organs. The mechanisms that control IFN-ε levels in various tissues, including those of the female reproductive tract, are only partially understood. Previous work rightly focused on transcriptional initiation events shown to participate in the regulation of IFN-εexpression(5, 9). However, this is not the only mechanism whereby the levels of IFN-ε are regulated, as early evidence suggested that post-transcriptionalmechanismsalso are involved(9). In the present study we investigated whether, and how, IFN-ε expression is regulated at the post-transcriptional level since constitutive expression of IFN-εwas recently shown to play essential roles in the protection of female reproductiveorgansfrom sexually-transmitted infections(8). We found evidence supporting a role for the 5′UTRof IFN-ε mRNA in regulation of expression under basal conditions.
5′UTRsoften include cis-elementsthat survey individual mRNAs and affect their stability. Examples includeregions in the 5′UTRs of mRNAs encoding interleukin-2 and growth-related oncogenes(26, 27). Several RNA secondary structure prediction algorithms identified two potential stem-loop structures in IFN-ε 5′UTR, and the studies presented here showevidence supporting a role for loop 1 as a negative regulator of IFN-ε mRNA expression. This inhibitory effect does not appear to involve changes in mRNA stability and, while the precise mechanism involved remains to be elucidated, thermodynamically stablestem-loopshave been shown to stallribosomes and initiate endonucleolytic cleavage events(28).
The inhibitory effect exerted by IFN-ε 5′UTR on mRNA expression may be specific for certain cell types and likely depends on expression of proteins that recognize defined structures or sequences located in the 5′UTR. Our studies identifiedIPO9as protein that binds primarily to stem-loop structure 1 in IFN-ε5′UTR, and negatively impacts mRNA expression. This constitutes the first report identifying IPO9 as a regulator of IFN-ε mRNA expression, a function suggesting roles additional to those previously reported. Importins, including IPO9, are known for their ability to mediate active transport through nuclear pore complexes (29) and effectively suppress the aggregation of their basic import cargoes in polyanionic environments (15). Importins exert their functions through protein-protein interactions; they recognize nuclear localization signalsand prevent protein aggregation by shielding basic patches such as those found in a variety of ribosomal proteins (15). No studies to date have reported direct interactions between importins and nucleic acids, leading us to speculate that in our studies, IPO9-mediated post-transcriptional effects on IFN-ε mRNA expression involves intermediates, such as un-identified RNA-binding protein(s). Our observation that IPO9 inhibitedexpression of IFN-ε and HIF-1α, but not that of other mRNAs, points at specific interactions limited to a sub-set of mRNAs whose 5′UTR fold into defined secondary structures such as IFN-εloop 1.
In conclusion, we have identified a novel mechanism for post-transcriptional regulation of IFN-εexpression. A distinct stem-loop structure predicted to form in the 5′UTR directly or indirectly interacts with IPO9, a member of the importin family, negatively regulatingIFN-ε mRNAexpression under basal conditions. These observations provide a novel mechanism for regulation of IFN-ε, and describe a previously un-identified function for IPO9 in post-translational, gene-specific, regulation of expression. This regulatory mechanism also may be utilized to regulate the expression of other mRNAs, including HIF-1α.
Supplementary Material
Acknowledgments
We are indebted to Janeth C. Tayone for excellent technical assistance.
This work was supported in part by two Grants-in-Aid for Scientific Research (KAKENHI) (20591934 and 23590560 to TM, Japan) and by funds awarded to TM by the Kanzawa Medical Research Foundation. Additional support was provided by the National Institutes of Health (5P01CA073992 and P30 CA042014 to Huntsman Cancer Institute for support of core facilities) and by the Huntsman Cancer Foundation.
References
- 1.Brandenburg B, Zhuang X. Virus trafficking - learning from single-virus tracking. Nat Rev Microbiol. 2007;5:197–208. doi: 10.1038/nrmicro1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matikainen S, Siren J, Tissari J, Veckman V, Pirhonen J, Severa M, Sun Q, Lin R, Meri S, Uze G, Hiscott J, Julkunen I. Tumor necrosis factor alpha enhances influenza A virus-induced expression of antiviral cytokines by activating RIG-I gene expression. J Virol. 2006;80:3515–3522. doi: 10.1128/JVI.80.7.3515-3522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 4.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 5.Hardy MP, Owczarek CM, Jermiin LS, Ejdeback M, Hertzog PJ. Characterization of the type I interferon locus and identification of novel genes. Genomics. 2004;84:331–345. doi: 10.1016/j.ygeno.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Xi Y, Day SL, Jackson RJ, Ranasinghe C. Role of novel type I interferon epsilon in viral infection and mucosal immunity. Mucosal Immunol. 2012 doi: 10.1038/mi.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharkey DJ, Macpherson AM, Tremellen KP, Robertson SA. Seminal plasma differentially regulates inflammatory cytokine gene expression in human cervical and vaginal epithelial cells. Mol Hum Reprod. 2007;13:491–501. doi: 10.1093/molehr/gam028. [DOI] [PubMed] [Google Scholar]
- 8.Fung KY, Mangan NE, Cumming H, Horvat JC, Mayall JR, Stifter SA, De Weerd N, Roisman LC, Rossjohn J, Robertson SA, Schjenken JE, Parker B, Gargett CE, Nguyen HP, Carr DJ, Hansbro PM, Hertzog PJ. Interferon-epsilon protects the female reproductive tract from viral and bacterial infection. Science. 2013;339:1088–1092. doi: 10.1126/science.1233321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumiya T, Prescott SM, Stafforini DM. IFN-epsilon mediates TNF-alpha-induced STAT1 phosphorylation and induction of retinoic acid-inducible gene-I in human cervical cancer cells. J Immunol. 2007;179:4542–4549. doi: 10.4049/jimmunol.179.7.4542. [DOI] [PubMed] [Google Scholar]
- 10.Day DA, Tuite MF. Post-transcriptional gene regulatory mechanisms in eukaryotes: an overview. J Endocrinol. 1998;157:361–371. doi: 10.1677/joe.0.1570361. [DOI] [PubMed] [Google Scholar]
- 11.Pierrat B, Lacroute F, Losson R. The 5′ untranslated region of the PPR1 regulatory gene dictates rapid mRNA decay in yeast. Gene. 1993;131:43–51. doi: 10.1016/0378-1119(93)90667-r. [DOI] [PubMed] [Google Scholar]
- 12.Muhlrad D, Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 13.von Roretz C, Gallouzi IE. Decoding ARE-mediated decay: is microRNA part of the equation? J Cell Biol. 2008;181:189–194. doi: 10.1083/jcb.200712054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Day SL, Ramshaw IA, Ramsay AJ, Ranasinghe C. Differential effects of the type I interferons alpha4, beta, and epsilon on antiviral activity and vaccine efficacy. J Immunol. 2008;180:7158–7166. doi: 10.4049/jimmunol.180.11.7158. [DOI] [PubMed] [Google Scholar]
- 15.Jakel S, Mingot JM, Schwarzmaier P, Hartmann E, Gorlich D. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 2002;21:377–386. doi: 10.1093/emboj/21.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dixon DA, Kaplan CD, McIntyre TM, Zimmerman GA, Prescott SM. Post-transcriptional control of cyclooxygenase-2 gene expression. The role of the 3′-untranslated region. J Biol Chem. 2000;275:11750–11757. doi: 10.1074/jbc.275.16.11750. [DOI] [PubMed] [Google Scholar]
- 17.Matsumiya T, Imaizumi T, Yoshida H, Satoh K, Topham MK, Stafforini DM. The levels of retinoic acid-inducible gene I are regulated by heat shock protein 90-alpha. J Immunol. 2009;182:2717–2725. doi: 10.4049/jimmunol.0802933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caputi M, Mayeda A, Krainer AR, Zahler AM. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozaki T, Yamashita T, Ishiguro S. ERp57-associated mitochondrial micro-calpain truncates apoptosis-inducing factor. Biochim Biophys Acta. 2008;1783:1955–1963. doi: 10.1016/j.bbamcr.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 20.Ozaki T, Yamashita T, Ishiguro S. Mitochondrial m-calpain plays a role in the release of truncated apoptosis-inducing factor from the mitochondria. Biochim Biophys Acta. 2009;1793:1848–1859. doi: 10.1016/j.bbamcr.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato K, Hamada M, Asai K, Mituyama T. CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res. 2009;37:W277–280. doi: 10.1093/nar/gkp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA Tools: a web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res. 2010;38:W373–377. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barber RD, Harmer DW, Coleman RA, Clark BJ. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics. 2005;21:389–395. doi: 10.1152/physiolgenomics.00025.2005. [DOI] [PubMed] [Google Scholar]
- 25.Cheon H, Yang J, Stark GR. The functions of signal transducers and activators of transcriptions 1 and 3 as cytokine-inducible proteins. J Interferon Cytokine Res. 2011;31:33–40. doi: 10.1089/jir.2010.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen CY, Gherzi R, Andersen JS, Gaietta G, Jurchott K, Royer HD, Mann M, Karin M. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000;14:1236–1248. [PMC free article] [PubMed] [Google Scholar]
- 27.Tebo JM, Datta S, Kishore R, Kolosov M, Major JA, Ohmori Y, Hamilton TA. Interleukin-1-mediated stabilization of mouse KC mRNA depends on sequences in both 5′- and 3′-untranslated regions. J Biol Chem. 2000;275:12987–12993. doi: 10.1074/jbc.275.17.12987. [DOI] [PubMed] [Google Scholar]
- 28.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 29.Okada N, Ishigami Y, Suzuki T, Kaneko A, Yasui K, Fukutomi R, Isemura M. Importins and exportins in cellular differentiation. J Cell Mol Med. 2008;12:1863–1871. doi: 10.1111/j.1582-4934.2008.00437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


