Abstract
The HMCM[CG]CBCA experiment (J. Am. Chem. Soc. (2003), 125, 13868–13878) correlates methyl carbon and proton shifts to Cγ, Cβ, and Cα resonances for the purpose of resonance assignments. The relative sensitivity of the HMCM[CG]CBCA sequence experiment is compared to a divide-and-conquer approach to assess whether it is best to collect all of the methyl correlations at once, or to perform separate experiments for each correlation. A straightforward analysis shows that the divide-and-conquer approach is intrinsically more sensitive, and should always be used to obtain methy-Cγ, Cβ, and Cα correlations. The improvement in signal-to-noise associated with separate experiments is illustrated by the detection of methyl-aliphatic correlations in a 65 kDa protein-DNA complex.
Introduction
NMR has made a significant contribution to our understanding of the role of dynamics in the function of proteins, enzymes, and nucleic acids (Mittermaier and Kay, 2006; Markwick et al, 2008; Gobl and Tjandra, 2012; Kleckner and Foster, 2010; Shajani and Varani, 2006). Historically, NMR studies on proteins and nucleic acids have been restricted to relatively small molecules, on the order of 25 kDa in the case of proteins. The reduction in proton-proton dipolar relaxation provided by protein deuteration (Sattler and Fesik, 1996), combined with destructive relaxation interference (TROSY, Tsakos et al, 2006; Fernandez and Wider, 2003; Salzmann et al, 1998), has allowed investigators to obtain near complete assignment of mainchain and Cβ atoms in large proteins (Gardner et al, 1998; McCallum et al, 1999; Salzmann et al, 2000). Deuteration restricts the observation of proton resonances to mainchain HN atoms, limiting NOEs to those protons. This limitation causes challenges in the determination of the tertiary structure of larger proteins because of the reduced number of inter-residue distance constraints. This problem has been circumvented by selective reprotonation of the protein, either randomly at all aliphatic sites (LeMaster and Richards, 1988), or focusing solely on methyl groups from Ile, Leu, and Val (Goto and Kay, 2000), Thr (Sinha et al, 2011;Velyvis et al, 2012), or Ala (Ayala et al, 2009).
There are a number of advantages associated with labeling methyl groups (Ruschak and Kay, 2010). The dense network of dipolar couplings between the methyl groups within the core of the protein provides distance constraints for structure determination of larger proteins (Mueller et al, 2000), and provides convenient sites for the characterization of dynamics by 13C and 2H relaxation (Tugarinov and Kay, 2005), as well as relaxation dispersion (Baldwin et al, 2010). Thr residues are frequently found at protein-nucleic acid interfaces (Biswas et al, 2009), thus can report on the structure and dynamics of critical regions in these complexes.
The assignment of methyl resonances is prerequisite for the utilization of these groups for protein structure determination or for the interpretation of relaxation measurements. Methyl resonances can be assigned by predicting methyl-methyl NOES, either alone (Xu et al, 2009), or in combination with paramagnetic relaxation enhancement (Veditti et al, 2011). If the protein contains a lanthanide ion binding site, pseudocontact shifts can also be used to assign methyl resonances (John et al, 2007). In cases where the N, HN, CO, Cα, and Cβ assignments are known, methyl resonances can be readily assigned by correlation of the methyl resonances to the already assigned atoms. This correlation can be accomplished using COSY type transfers from the methyl to other atoms. A significant advance in the use of COSY type correlation experiments was attained by labeling Leu and Val such that only one methyl was labeled, thus producing a linear arrangement of coupled carbons for these residues (see Tugarinov and Kay, 2003). This advance in labeling permitted the development of a suite of NMR experiments that correlated methyl resonances to assigned N, HN, CO, Cα, and Cβ atoms, leading to near complete methyl assignments in a 723 residue protein. One of the key experiments developed in that work is the HMCM[CG]CBCA pulse sequence, which simultaneously correlates the methyl carbon and proton shifts to the rest of the carbons on the sidechain in a single spectrum. The HMCM[CG]CBCA experiment has been successfully used to assign methyl resonances in a number of smaller proteins (Wang et al, 2012; Krejcirikova and Tugarinov, 2012; Zhuraviena et al, 2012; Chan et al, 2012). In the case of larger systems, this experiment has been replaced by a “divide-and-conquer” strategy, whereby the shifts of the individual carbons, e.g. Cα, are collected in separate experiments (Sprangers and Kay, 2007). To the best of our knowledge there has not been a clear analysis in the literature regarding the relative sensitivities of each approach, which would provide guidance to users regarding which experiments to perform. A straightforward analysis shows that the divide-and-conquer approach should always be used, regardless of the size of the protein.
Materials and Methods
Spectra were acquired on a Bruker AMX spectrometer, operating at 600 MHz (1H) using a standard room temperature probe. The pulse sequences were tested on the 130 residue RNA binding domain of E. coli rho protein (Rho130) and the 245 residue homodimeric EcoRV-DNA complex. Both proteins were perdeuterated and methyl labeled as described by Goto et al (1999). The proteins were expressed from a standard T7 expression system using C3013 cells (New England Biolabs) as the host. Studier’s PG medium (Studier, F. W., 2005) was used with deuterated uniformly 13C labeled glucose, 15N labeled ammonium sulfate, and ~100% D2O. The cells were grown to an A550 of 0.6 at 37°C. At this point, methyl-protonated, 13C-labeled (uniform, except one methyl is 12C) α-ketoisovalerate (100mg/L) and methyl-protonated uniformly deuterated and 13C ketobutyrate (50mg/L), was added to the media, as described by Goto et al (1999). Isotopically labeled compounds were obtained from either Cambridge Isotopes or Sigma Aldrich. The cells were allowed to grow for 60 minutes and isopropylthiogalactoside (IPTG) (1mM) was added to induce protein expression. The cells were harvested 3 hours after induction. Rho130 was purified as described previously (Briercheck et al, 1998) and the purification scheme for EcoRV will be presented elsewhere. The EcoRV-DNA complex was generated by adding the DNA duplex (5′-GCAAAGATATCTTTCG-3′; IDT) to the protein in 1:1 stoichiometric ratio.
Results and Discussion
The HMCM[CG]CBCA sequence was used to acquire methyl correlation spectra on a small 130 residue protein (Rho130) and a 65 kDa EcoRV-DNA complex (see figure 1). Although most (but not all) of the expected correlations were observed for Rho130, the EcoRV-DNA complex gave unexpectedly weak signal in the HMCM[CG]CBCA experiment due to enhanced relaxation of the protein methyls from protons on the non-deuterated DNA. Although it was only possible to obtain a small number of aliphatic-methyl correlations using the HMCM[CG]CBCA experiment, most of the expected correlations were obtained using the divide-and-conquer strategy.
Figure 1.
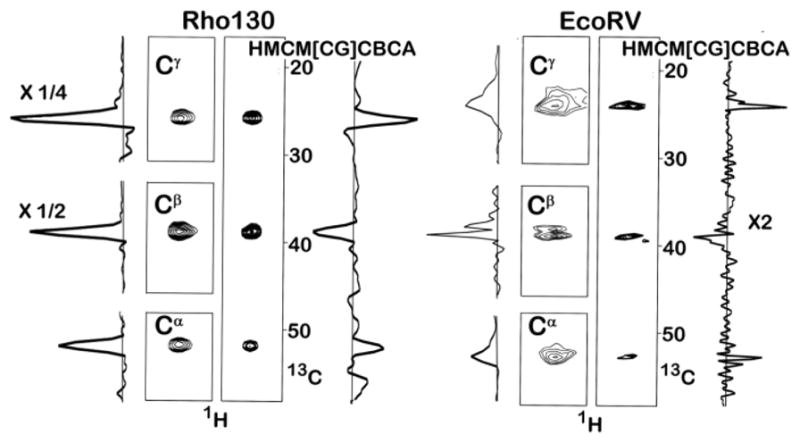
Leucine Methyl Correlation Spectra of Rho130 and the EcoRV-DNA Complex. The methyl carbon region was sampled using 21 complex points in all experiments. The Cα resonances are broadened by unresolved coupling to the 15N amide nitrogen. The concentration of Rho130 was 1.0 mM and spectra were acquired at 30°C. In the case of the Rho130 sample, all four spectra were acquired under identical condition; 8 scans with the same spectral width (50 ppm) and number of points (40) in the non-methyl carbon dimension. The total acquisition time/experiment was 9 hours. In the rho130 spectra the one dimensional trace for the Cβ peaks are divided by two and the trace for the Cγ peak was divided by four. The concentration of the EcoRV-DNA complex was 0.8 mM and the spectra were acquired at 35°C. In the case of EcoRV-DNA sample the spectral widths for the aliphatic carbons in the Cα Cβ, Cγ, and the HMCM[CG]CBCA experiments were 20 ppm, 60 ppm, 20 ppm, and 50 ppm, respectively. These were sampled using 32, 40, 32, and 40 complex points, respectively. A total of 40, 16, 8, and 40 scans were acquired for the Cα, Cβ, Cγ, and HMCM[CG]CBCA experiments, respectively, giving total acquisition times of 35.8, 17.92, 7.17, and 44.8 hours, respectively. The entire one dimensional trace for the HMCM[CG]CBCA experiment is multiplied by two. The increase in signal intensity for one residue from the EcoRV-DNA sample, after scaling for the total experimental time, is 2.0, 8.8, and 6.0 for the Cα, Cβ, Cγ, experiments. These ratios are the lower limit because many correlations could not be observed in the HMCM[CG]CBCA experiment. The Cβ experiment could have been run in ½ the time (8 scans), which would have given a total acquisition time for all three experiments of approximately 52 hours, i.e. not much longer than the entire HMCM[CG]CBCA experiment. Pulse sequences can be found in figure S1, along with additional guidelines for defining the acquisition time for the detection of non-methyl carbons.
The divide-and-conquer experiments are more sensitive than the HMCM[CG]CBCA experiment for a number of reasons. Because the HMCM[CG]CBCA experiment simultaneously correlates the methyl resonances to all of the other aliphatic carbons (e.g. Ile Cγ, Cβ, Cα) it is necessary to produce four different product operator terms (4IzCγxCβz, 2IzCβy, 4IzCβzCαx, 8IzCγxCβyCαx) prior to recording the aliphatic carbon shift. Only the first three lead to single-quantum frequencies, hence one fourth of the signal is lost. Second, since all three carbon peaks are acquired in the same spectrum, the original magnetization from the methyl is divided among the three peaks, thus each peak represents one fourth of the original magnetization. Third, because the Cγ and Cβ frequencies are detected after the magnetization has been transferred to the Cα, all three of the signals suffer the large relaxation loss associated with the long delay required to relay the magnetization from the methyl to the Cα carbon. By acquiring the shift of each carbon in separate experiments only the intensity of the Cα is affected by the long transfer time, effectively increasing the signal of the Cβ and Cγ resonances. Since only one signal is being detected at a time, the full magnetization from the methyl is also detected, increasing the signal by four-fold for the Cγ and Cβ resonances. The increase in signal intensity is smaller for the Cα resonances because it is necessary to lengthen the last polarization transfer step from 7 to 14 ms to allow for complete conversion of the anti-phase term 2CβzCαx to Cαy prior to the evolution period that records the Cα frequency (see supplementary material figure S1 for pulse sequences). Regardless of the increase in polarization transfer time, the sensitivity of the Cα experiment still exceeds that of the HMCM[CG]CBCA experiment, except for very short carbon T2 times, on the order of 10 ms (see figure S3). Although it is necessary to run all three experiments to obtain the same chemical shift information as obtained in the HMCM[CG]CBCA experiment, the additional acquisition time is significantly reduced due to the increase in sensitivity of the Cβ and Cγ experiments. For example, at a carbon T2 of 40 ms, the sensitivities of the Cα, Cβ, and Cγ experiments are approximately 2.8, 5.7, and 11.4 times that of the HMCM[CG]CBCA experiment. In addition to the gains in sensitivity, the Cα and Cγ experiments can be acquired with a much narrower sweepwidth (see figure S4), reducing the acquisition time of the experiment. The increase in sensitivity allows the Cα, Cβ, and Cγ experiments to be acquired in approximately 24/30, 5/30 and 1/30, respectively, of the time required for the HMCM[CG]CBCA experiment and still yield a 2–4 fold increase in intensity.
The predicted gains in sensitivity are essentially realized in practice. In figure 1 we show spectra of Rho130 and the EcoRV-DNA complex that were obtained using the Cα, Cβ, and Cγ and the HMCM[CG]CBCA experiments. Signals from 15 Leu residues in Rho130 gave relative intensities of 1.9+/−0.5, 4.4+/−0.9, and 8.4+/−3.6 for the Cα, Cβ, and Cγ peaks, respectively, compared to the HMCM[CG]CBCA experiment. Note that the sensitivity gain for the Cα experiment is a lower limit since a number of Cα peaks were missing in the HMCM[CG]CBCA experiment, but present in the Cα experiment. In the case of the larger EcoRV-DNA complex, only a few correlations were observed in the HMCM[CG]CBCA experiment hence it is difficult to compare sensitivities. When Cα resonances could be observed, they were approximately two fold less intense in the HMCM[CG]CBCA spectra than in the experiment that only detected the Cα shifts.
In conclusion, because the sensitivity of detecting methyl-aliphatic correlations is significantly increased by recording each carbon shift in a separate experiment, separate experiments should be run, even for smaller proteins. A similar philosophy has also been historically applied when recording correlations between NH resonances and Cβ resonances. In this case, the standard NHCACB experiment, which detects both Cα and Cβ shifts simultaneously, is often run with the delay for magnetization transfer between the Cα and Cβ spins set to 1/2Jcc, causing complete conversion of the Cαx product operator to anti-phase 2CβzCαx, prior to recording of the carbon chemical shift. This approach transfers all of the magnetization from the NH group to the Cβ peak, instead of splitting it between the Cα and Cβ peaks.
Supplementary Material
Acknowledgments
We thank Virgil Simplaceanu for maintenance of the NMR spectrometers and Dr. C. Sanders (Vanderbilt University) for providing the Bruker code for the HMCM[CG]CBCA experiment. This work was supported by the Mellon College of Science at CMU and an NIH Merit grant 5R37-GM029207 to L.J-J.
References
- Ayala I, Sounier R, Use N, Gans P, Boisbouvier J. An efficient protocol for the complete incorporation of methyl-protonated alanine in perdeuterated protein. J Biomol NMR. 2009;43:111–119. doi: 10.1007/s10858-008-9294-7. [DOI] [PubMed] [Google Scholar]
- Biswas S, Guharoy M, Chakrabarti P. Dissection, residue conservation, and structural classification of protein-DNA interfaces. Proteins. 2009;74:643–654. doi: 10.1002/prot.22180. [DOI] [PubMed] [Google Scholar]
- Baldwin AJ, Religa TL, Hansen DF, Bouvignies G, Kay LE. 13CHD2 methyl group probes of millisecond time scale exchange in proteins by 1H relaxation dispersion: an application to proteasome gating residue dynamics. J Am Chem Soc. 2010;132(32):10992–5. doi: 10.1021/ja104578n. [DOI] [PubMed] [Google Scholar]
- Briercheck DM, Wood TC, Allison TJ, Richardson JP, Rule GS. The NMR structure of the RNA binding domain of E. coli rho factor suggests possible RNA-protein interactions. Nat Struct Biol. 1998;5:393–9. doi: 10.1038/nsb0598-393. [DOI] [PubMed] [Google Scholar]
- Chan PHW, Weissbach S, Okon M, Withers SG, McIntosh LP. Nuclear Magnetic Resonance Spectral Assignments of α-1,4-Galactosyltransferase LgtC from Neisseria meningitidis: Substrate Binding and Multiple Conformational States. Biochemistry 2012. 2012;51:8278–8292. doi: 10.1021/bi3010279. [DOI] [PubMed] [Google Scholar]
- Fernandez C, Wider G. TROSY in NMR studies of the structure and function of large biological macromolecules. Current Opinion in Structural Biology. 2003 doi: 10.1016/j.sbi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Gardner KH, Zhang X, Gehring K, Kay LE. Solution NMR Studies of a 42 KDa Escherichia Coli Maltose Binding Protein/β-Cyclodextrin Complex: Chemical Shift Assignments and Analysis. J Am Chem Soc, 1998. 1998;120(45):11738–11748. doi: 10.1021/ja982019w. [DOI] [Google Scholar]
- Göbl C, Tjandra N. Application of Solution NMR Spectroscopy to Study Protein Dynamics Entropy. 2012;14(3):581–598. doi: 10.3390/e14030581. [DOI] [Google Scholar]
- Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE. A robust and cost-effective method for the production of Val, Leu, Ile (delta 1) methyl-protonated 15N-, 13C-, 2H-labeled proteins. J Biomol NMR. 1999;13:369–74. doi: 10.1023/a:1008393201236. [DOI] [PubMed] [Google Scholar]
- Goto NK, Kay LE. New developments in isotope labeling strategies for protein solution NMR spectroscopy. Current Opinion in Structural Biology. 2000;10:585–592. doi: 10.1016/s0959-440x(00)00135-4. [DOI] [PubMed] [Google Scholar]
- John M, Schmitz C, Park AY, Dixon NE, Huber T, Otting G. Sequence-specific and stereospecific assignment of methyl groups using paramagnetic lanthanides. J Am Chem Soc. 2007;7:13749–57. doi: 10.1021/ja0744753. [DOI] [PubMed] [Google Scholar]
- Kleckner IR, Foster MP. An introduction to NMR-based approaches for measuring protein dynamics. Biochimica et Biophysica Acta. 2010 doi: 10.1016/j.bbapap.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejcirikova A, Tugarinov V. 3D-TROSY-based backbone and ILV-methyl resonance assignments of a 319-residue homodimer from a single protein sample. J Biomol NMR. 2012;54:135–143. doi: 10.1007/s10858-012-9667-9. [DOI] [PubMed] [Google Scholar]
- LeMaster DM, Richards FM. NMR sequential assignment of Escherichia coli thioredoxin utilizing random fractional deuteriation. Biochemistry. 1988;27:142–150. doi: 10.1021/bi00401a022. [DOI] [PubMed] [Google Scholar]
- Markwick PRL, Malliavin T, Nilges M. Structural Biology by NMR: Structure, Dynamics, and Interactions. PLoS Comput Biol. 2008;4(9):e1000168. doi: 10.1371/journal.pcbi.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum SA, Hitchens TK, Rule GS. Solution Structure of the Carboxyl Terminus of a Human Class Mu Glutathione S-transferase: NMR Assignment Strategies in Large Proteins. J Mol Biol. 1999;285:2119–2132. doi: 10.1006/jmbi.1998.2428. [DOI] [PubMed] [Google Scholar]
- Mittermaier A, Kay LE. New Tools Provide New Insights in NMR Studies of Protein Dynamics. Science. 2006;312:224–228. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- Mueller GA, Choy WY, Yang D, Forman-Kay JD, Venters RA, Kay LE. Global Folds of Proteins with Low Densities of NOEs Using Residual Dipolar Couplings: Application to the 370-Residue Maltodextrin-binding Protein. J Mol Biol. 2000;300:197–212. doi: 10.1006/jmbi.2000.3842. [DOI] [PubMed] [Google Scholar]
- Ruschak AM, Kay LE. Methyl groups as probes of supra-molecular structure, dynamics and function. J Biomol NMR. 2010;46:75–87. doi: 10.1007/s10858-009-9376-1. [DOI] [PubMed] [Google Scholar]
- Salzmann M, Pervushin K, Wider G, Senn H, Wüthrich K. TROSY in triple-resonance experiments: New perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci U S A. 1998;95(23):13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann M, Pervushin K, Wider G, Senn H, Wüthrich K. NMR Assignment and Secondary Structure Determination of an Octameric 110 kDa Protein Using TROSY in Triple Resonance Experiments. J Am Chem Soc. 2000;122:7543–7548. doi: 10.1021/ja0003268. [DOI] [Google Scholar]
- Sattler M, Fesik SW. Use of deuterium labeling in NMR: overcoming a sizeable problem. Structure. 1996 Nov 15;4:1245–1249. doi: 10.1016/s0969-2126(96)00133-5. [DOI] [PubMed] [Google Scholar]
- Shajani Z, Varani G. NMR studies of dynamics in RNA and DNA by 13C relaxation. Biopolymers. 2007;86:348–359. doi: 10.1002/bip.20650. [DOI] [PubMed] [Google Scholar]
- Sinha K, Jen-Jacobson L, Rule GS. Specific labeling of threonine methyl groups for NMR studies of protein-nucleic acid complexes. Biochemistry. 2011;50:10189–91. doi: 10.1021/bi201496d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprangers R, Kay LE. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007;445:618–22. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- Studier FW. Protein expression and purification. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Tugarinov V, Kay LE. Ile, Leu, and Val Methyl Assignments of the 723-Residue Malate Synthase G Using a New Labeling Strategy and Novel NMR Methods. J Am Chem Soc. 2003;125:13868–13878. doi: 10.1021/ja030345s. [DOI] [PubMed] [Google Scholar]
- Tugarinov V, Kay LE. Quantitative 13C and 2H NMR relaxation studies of the 723-residue enzyme malate synthase G reveal a dynamic binding interface. Biochemistry. 2005;44:15970–7. doi: 10.1021/bi0519809. [DOI] [PubMed] [Google Scholar]
- Tzakos AG, Grace CRR, Lukavsky PJ, Riek R. NMR Techniques for Very Large Proteins and RNAs in Solution. Annu Rev Biophys Biomol Struct. 2006;35:319–42. doi: 10.1146/annurev.biophys.35.040405.102034. [DOI] [PubMed] [Google Scholar]
- Velyvis A, Ruschak AM, Kay LE. An economical method for production of (2)H, (13)CH3-threonine for solution NMR studies of large protein complexes: application to the 670 kDa proteasome. PLoS One. 2012;7(9):e43725. doi: 10.1371/journal.pone.0043725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venditti V, Fawzi NL, Clore GM. Automated sequence- and stereo-specific assignment of methyl-labeled proteins by paramagnetic relaxation and methyl-methyl nuclear Overhauser enhancement spectroscopy. J Biomol NMR. 2011;51:319–328. doi: 10.1007/s10858-011-9559-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Vu A, Lee K, Dahlquist FW. CheA–Receptor Interaction Sites in Bacterial Chemotaxis. J Mol Biol. 2012;422:282–290. doi: 10.1016/j.jmb.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Matthews S. MAP-XSII: an improved program for the automatic assignment of methyl resonances in large proteins. J Biomol NMR. 2013;55:179–87. doi: 10.1007/s10858-012-9700-z. [DOI] [PubMed] [Google Scholar]
- Zhuravleva A, Clerico EM, Gierasch LM. An Interdomain Energetic Tug-of-War Creates the Allosterically Active State in Hsp70 Molecular Chaperones. Cell. 2012;151:1296–1307. doi: 10.1016/j.cell.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


