Abstract
Improved sequencing technologies offer unprecedented opportunities for investigating the role of rare genetic variation in common disease. However, there are considerable challenges with respect to study design, data analysis and replication1. Here, using pooled next-generation sequencing of 507 genes implicated in the repair of DNA in 1,150 samples, an analytical strategy focussed on protein truncating variants (PTVs) and a large-scale sequencing case-control replication experiment in 13,642 individuals, we show that rare PTVs in the p53 inducible protein phosphatase PPM1D are associated with predisposition to breast cancer and to ovarian cancer. PPM1D PTV mutations were present in 25/7781 cases vs 1/5861 controls; P=1.12×10−5, which included 18 mutations in 6,912 individuals with breast cancer; P = 2.42×10−4 and 12 mutations in 1,121 individuals with ovarian cancer; P = 3.10×10−9. Notably, all the identified PPM1D PTVs were mosaic in lymphocyte DNA and clustered within a 370 bp region in the final exon of the gene, C-terminal to the phosphatase catalytic domain. Functional studies demonstrated that the mutations result in enhanced suppression of p53 in response to ionising radiation exposure, suggesting the mutant alleles encode hyperactive PPM1D isoforms. Thus, although the mutations cause premature protein truncation, they do not result in the simple loss-of-function typically associated with this class of variant, but instead likely have a gain-of-function effect. Our results have implications for the detection and management of breast and ovarian cancer risk. More generally, these data provide new insights into the role of rare and of mosaic genetic variants in common conditions, and the utility of sequencing in their identification.
TEXT
There is strong evidence that rare genetic variation is important in breast and ovarian cancer predisposition2,3. In the 1990s, genome-wide linkage analysis and positional cloning led to the identification of the DNA repair genes BRCA1 and BRCA2, rare mutations of which confer substantial risks of both diseases2,3. More recently, through case-control resequencing studies of candidate genes we, and others, have discovered rare variants that confer moderate risks of breast and/or ovarian cancer4-10. These cancers are therefore exemplars of the rare variant-common disease hypothesis.
The successful studies to date have focussed on genes encoding proteins involved in DNA repair such as PALB2, ATM, CHEK2, BRIP1, RAD51C and RAD51D4-10. These genes are characterised by multiple, very rare, loss-of-function mutations, usually protein truncating variants (PTVs), which predispose carriers to breast and/or ovarian cancer4-10. To further investigate the role of DNA repair genes in cancer susceptibility, we sequenced 507 genes (the ‘DNA repair panel’) in 1,150 individuals with breast cancer from the UK, 69 of whom also had ovarian cancer (Supplementary Table 1, Supplementary Fig. 1). To maximise time, sample and cost efficiency we used a pooled approach combining 200 ng of DNA from each of 24 individuals into a single pool which we hybridised to a custom pulldown containing the DNA repair panel (Supplementary Table 2). We performed sequencing using an Illumina HiSeq2000 which generated a minimum coverage per pool of 480× for ≥ 90% of the target region (Supplementary Fig. 2). Sequence variants were called using Syzygy11, the performance of which was evaluated using previously generated data in a subset of the samples. The sensitivity of base substitution calling was 99.6% (439/439 common variants and 24/26 rare variants that were present in 1/24 individuals in a pool). The sensitivity of insertion/deletion calling was 94.4% (51/54 rare insertion/deletions present in 1/24 individuals in a pool, Supplementary Table 3).
We next considered the 34,564 sequence variants called by Syzygy. We first focussed on PTVs because of the strong association of this class of mutation with disease. In total, 1,044 PTVs were called by Syzygy and we used a ’PTV prioritisation method’ to stratify the genes according to the number of different, rare truncating mutations present within the samples12. PPM1D showed the strongest signal in this analysis, and we confirmed by Sanger sequencing that five individuals carried different PPM1D PTVs. Two of these individuals had ovarian cancer in addition to breast cancer.
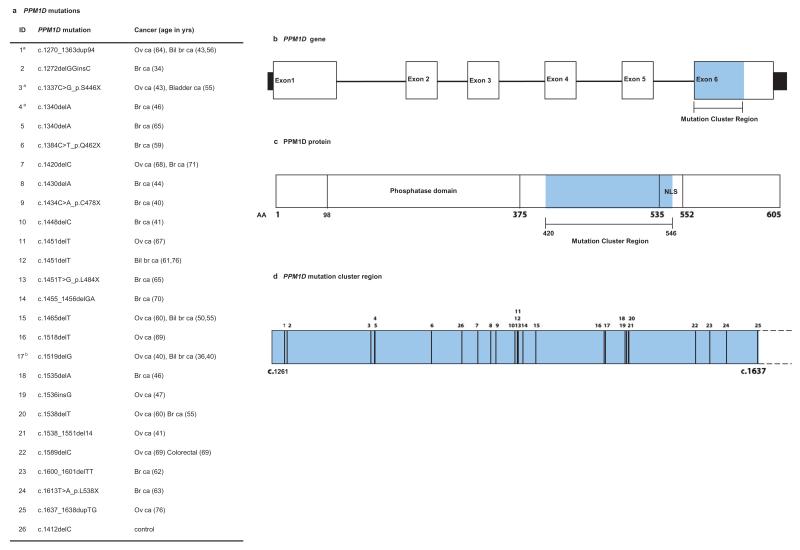
To further explore the role of PPM1D in breast and ovarian cancer susceptibility we next performed a case-control Sanger sequencing analysis of PPM1D in a total of 13,642 individuals; 7,781 unrelated individuals with breast and/or ovarian cancer and 5,861 population controls (Supplementary Table 1). We initially sequenced all PPM1D exons and intron-exon boundaries but after completing this analysis in 3,803 samples we noted that all 10 PTV mutations identified occurred within the last exon of PPM1D, and this clustering was highly significant (P = 8.2×10−6). We thus analysed the remaining 9,839 samples for this mutation cluster region (MCR), identifying a further 16 PTVs (Supplementary Table 1, Fig. 1). In total we identified 25 PPM1D PTVs in individuals with breast and/or ovarian cancer and 1 in controls (P = 1.12×10−5, Fig. 1, Fig. 2a and Supplementary Table 4). This included 18 mutations in 6,912 individuals with breast cancer (P = 2.42×10−4) and 12 mutations in 1,121 individuals with ovarian cancer (P = 3.10×10−9). The histological features of the cancers in PPM1D mutation carriers were diverse, and five individuals had both breast and ovarian cancer (Supplementary Table 5). The case series included 773 individuals with mutations in BRCA1 or BRCA2 (termed ‘BRCA1/2 mutation carriers’), four of whom also carried PTVs in PPM1D (4/773 vs 1/5861 controls, P = 8.30×10−4). We also identified a total of 16 non-synonymous, 14 synonymous and one intronic variant across the cases and controls; there was no evidence for an association with cancer for these variant classes (Supplementary Table 6).
Fig. 1. Clustering of cancer predisposing mutations in PPM1D.
a, PPM1D mutations and cancer phenotype. b, PPM1D gene with region targeted by mutations (mutation cluster region) in blue; c, PPM1D protein showing position of mutation cluster region downstream of the phosphatase domain and upstream/overlapping the nuclear localisation signal (NLS); d, mutation cluster region showing position of mutations. The numbers above give the position of the mutations and correspond to the IDs in panel (a). Ov ca, ovarian cancer; br ca, breast cancer; bil br ca, bilateral breast cancer.
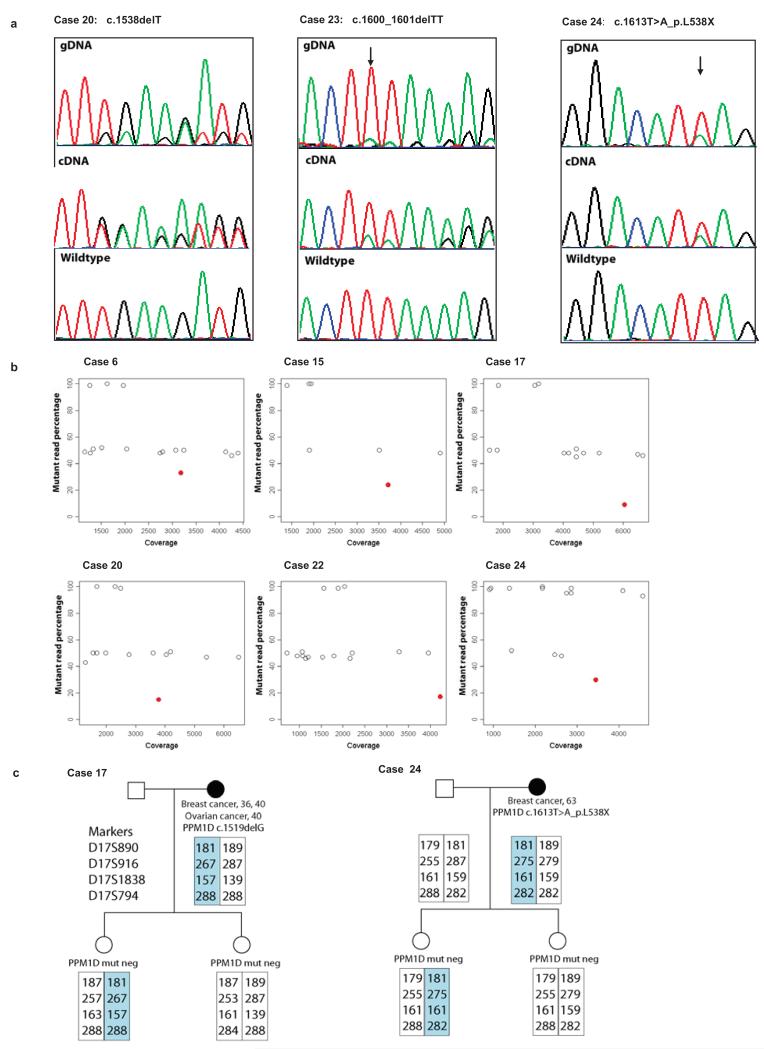
Fig. 2. PPM1D mutations are mosaic in lymphocyte DNA.
a, Sanger sequencing traces showing mutant allele is lower in genomic DNA extracted from peripheral blood lymphocytes (gDNA) than typical for heterozygous mutations. The cDNA analysis demonstrates the mutations lead to truncated products not nmRNA decay. b, deep PCR amplicon sequencing showing heterozygous BRCA1/2 variants at 50% (open dots) whereas the PPM1D mutation is present at a lower percentage (red dots). c, Haplotype analysis in two families. The offspring of PPM1D mutation carriers have different maternal haplotypes spanning PPM1D (highlighted), but neither carry the mutation, indicating that it is either not present, or mosaic in the germline of the proband.
The Sanger sequencing chromatograms for the PPM1D PTVs were unusual for heterozygous mutations as the mutant allele was considerably and consistently lower than the wildtype allele, suggesting the mutations were mosaic in lymphocyte DNA (Fig. 2a and Supplementary Fig. 3). This contrasted with the non-truncating variants which all had normal sequencing profiles. DNA from saliva was available for two individuals and the PTVs were present at similar amplitude to that identified in the corresponding blood derived DNA (Supplementary Fig 3). To further confirm the PTV mutations were bona fide we used two additional mutation detection methods; deep PCR amplicon sequencing13 (Fig. 2b, Supplementary Fig. 4 and Supplementary Table 4) and multiplex ligation-dependent probe amplification (MLPA)14 (Supplementary Fig. 5 and Supplementary Table 7). For the deep PCR amplicon sequencing we generated Nextera libraries of pooled PCR products covering BRCA1, BRCA2 and the PPM1D mutation, which we sequenced using an Illumina MiSeq generating a median coverage of 3387× across the PPM1D mutation (Supplementary Fig. 4 and Supplementary Table 4). This confirmed the PPM1D PTVs were present at a lower proportion than heterozygous polymorphisms in BRCA1 and BRCA2, with a median mutant read percentage of 16% (range 5-34%). Additionally, we sequenced the original DNA repair panel in six cases individually (i.e. unpooled), which again confirmed the mutations were present, but mosaic (Supplementary Fig. 6 and Supplementary Table 4). For three samples we had data from both the deep PCR amplicon sequencing and the DNA repair panel which gave identical mutation percentage results (Supplementary Table 4). Finally, family studies were also consistent with mosaicism; none of 14 relatives carried the PPM1D mutation identified in the proband. Most compellingly, for each of probands 17 and 24, we identified two offspring that had inherited different maternal haplotypes at the PPM1D locus, but neither offspring carried the relevant maternal PPM1D mutation, demonstrating that the mutations were either not present, or mosaic in the germline of the probands (Fig. 2c).
PPM1D (protein phosphatase, Mg2+/Mn2+dependent 1D), also known as WIP1 (Wild-type p53-induced phosphatase 1) was first identified in a screen for p53 target genes induced by ionising radiation15. PPM1D encodes a 605 amino acid protein with a N-terminal phosphatase catalytic domain and a C-terminal domain that contains a putative nuclear localisation signal (Fig. 1)16. PPM1D transcription is upregulated in response to various types of DNA damage in a p53-dependent manner. Once upregulated, PPM1D has been shown to dephosphorylate and downregulate several targets, particularly proteins associated with the ATM/ATR-initiated DNA damage response (DDR) and including tumour suppressors with a proven role in cancer susceptibility such as p5317, ATM18 and CHK219. Thus it has been proposed that a primary role of PPM1D is as a homeostatic regulator of the DDR, facilitating return of cells to their normal state after repair of damaged DNA17. There is also accumulating evidence that PPM1D is involved in oncogenesis16. PPM1D amplification and overexpression has been demonstrated in multiple human tumours20, including breast cancers21 and ovarian clear cell carcinoma22, and is a promising therapeutic target22-24.
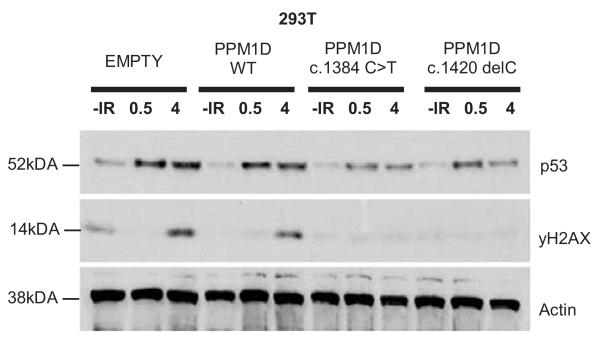
The clustering of PTVs within the 370 bp region corresponding to amino acids 420-546, which is downstream of the phosphatase catalytic domain but precedes or disrupts the nuclear localisation signal25, suggests the PTVs are not acting as simple loss-of-function mutations (Fig. 1). Moreover, all the PTVs were in the last exon and thus predicted to evade nonsense-mediated RNA decay and to result in a truncated protein that retains the phosphatase catalytic domain, rather than in haploinsufficiency25,26. We confirmed this experimentally for three mutations (Fig 2a). To investigate the effect of PPM1D PTVs we generated cDNA expression constructs representing two mutant alleles (PPM1D c.1384C>T; case 6 and PPM1D c.1420delC; case 7) and tested their ability to suppress p53 activation in response to ionising radiation (IR) exposure. As expected, the normal elevation of p53 levels after IR exposure was moderately suppressed in human U2-OS tumour cells transfected with a wildtype PPM1D expression construct, matching previous observations16,17 (Fig. 3). The suppression of p53 was enhanced in cells transfected with the mutant PPM1D expression constructs suggesting that each of these alleles encodes a hyperactive PPM1D isoform, i.e. consistent with a gain-of-function rather than a loss-of-function effect (Fig. 3). Similar effects were also observed in HeLa and 293 cells (Supplementary Fig. 7).
Fig. 3. The effect of mutant PPM1D isoforms on p53 activation.
p53 wildtype U2-0S human osteosarcoma cells were transfected with PPM1D cDNA expression constructs and exposed to ionising irradiation (5 grays). At 30 minute and 4h intervals whole cell lysates were western blotted to estimate the IR-induced activation of p53. Western blots showing p53 and actin (loading control) protein levels at different times (in hours) after IR exposure are shown. ‘Empty’: transfected with empty construct, ‘PPM1D WT’: transfected with wildtype PPM1D construct, ‘PPM1D c.1384C>T’ and ‘PPM1D c.1420delC’: transfected with mutant PPM1D constructs. Suppression of p53 was enhanced in cells transfected with mutant constructs suggesting these alleles encode hyperactive PPM1D isoforms.
To investigate the mechanism of oncogenesis in PPM1D PTV mutation carriers we analysed eight tumours from five individuals. Intriguingly, the PPM1D mutations were not detectable in any of the tumours by Sanger sequencing or MLPA (Supplementary Fig. 8). Through microsatellite analysis we confirmed that the tumours were from the correct individuals and demonstrated loss of heterozygosity at the PPM1D locus in seven of eight tumours, though there was no evidence of PPM1D copy number alteration (Supplementary Fig. 8 and Supplementary Table 8). We microdissected stromal tissue from the ovarian tumour in four cases and undertook deep sequencing across the PPM1D PTV in blood, tumour and stromal DNA. Each mutation was present in the blood, at similar level to that detected previously, absent from the tumour and either absent (two cases) or present at very low level (5/915 reads and 4/5793 reads) in the stroma, consistent with lymphocyte contamination (Supplementary Fig. 8 and Supplementary Table 5).
These data are intriguing and strongly suggest the mechanism of cancer association in PPM1D mutation carriers differs from that in carriers of mutations in other DNA repair genes associated with predisposition to these cancers. There are several potential explanations. It is possible the mutation was present in the cell of cancer origin but was subsequently lost, perhaps because a PPM1D mutation can act as a driver to initiate oncogenesis, but is not required, or is detrimental to the progression of the resulting cancer. The allele loss we observed at the PPM1D locus could be interpreted as supportive of this hypothesis, but it should be noted that it is not known if the lost allele carried the PPM1D PTV, and loss in this region of 17q is common in these cancers. Alternatively, the absence of the PPM1D mutation in the tumour could be because oncogenesis is being driven by the mutation in circulating blood cells. Another possibility is that the PPM1D mutations are not directly involved in causing breast or ovarian cancer. For example, they could be a separate manifestation of an underlying lesion, perhaps one that causes genomic instability, which can lead to selection and clonal expansion of cells with PPM1D PTVs and also to cancers in other tissues. Clearly, further studies will be required to explain the mechanism of oncogenesis in PPM1D mutation carriers.
Irrespective of the mechanism of the association, our data demonstrate that individuals with mosaic PPM1D PTVs in the mutation cluster region are at increased risk of cancer. The association is not explicable by increasing age, unlike recently reported mosaic chromosomal abnormalities (Supplementary Table 5)27,28. To estimate the cancer risks we undertook a retrospective cohort analysis using information on breast and ovarian cancer occurrence in the 6,577 unrelated individuals negative for BRCA1/2 mutations and controls, by modelling the retrospective likelihood of the observed mutation status conditional on the disease phenotype, as previously described9,29. This approach adjusts for our ascertainment of cases with more extreme phenotypes such as young age of onset or bilateral breast cancer, which we utilise to empower gene discovery4-7,10,30. The relative risk of breast cancer for PPM1D PTV carriers was estimated to be 2.7 (95% CI: 1.3-5.3; P = 5.38×10−3), which translates to approximately 23% cumulative risk by age 80. The relative risk of ovarian cancer was estimated to be 11.5 (95% CI: 4.3-30.4; P = 9.95×10−7), which translates to approximately 18% cumulative risk by age 80. It is noteworthy that we included an unselected hospital-based series of 322 ovarian cancer patients in whom we identified five PPM1D PTVs, suggesting that 1-2% of ovarian cancer patients may harbour mosaic PPM1D mutations.
The frequency of PPM1D PTVs in BRCA1/2 mutation carriers with breast and/or ovarian cancer was also significantly different from population controls (4/773 vs 1/5861; P = 8.30×10−4) and similar to that in cases of breast and/or ovarian cancer without BRCA1/2 mutations (4/773 vs 21/6634; P = 0.56), suggesting that PPM1D PTVs are also associated with increased risks of cancer in BRCA1/2 mutation carriers. Studies of unselected, population-based cancer patients and of larger series of BRCA1/2 mutation carriers would be of value to extend our observations, and to further explore the prevalence and cancer risks associated with PPM1D mutations.
These data provide new insights into ovarian and breast cancer, potentially identifying a novel class of genetic defect that lies somewhere between classic germline genetic predisposition mutations and tumour-specific somatic events. It is also highly plausible that PPM1D mutations are associated with other cancers, and broad evaluation of individuals with other tumour types would be of interest. More generally, the clinical implications of a mosaic cancer predisposition marker that is genetic, but not hereditary, and that is detectable in the blood but not the tumour(s) it is associated with are rather profound, particularly if this phenomenon is observed in other genes/contexts.
Our results also provide insights into genetic variation, particularly in relation to the nature and impact of rare gene mutations associated with disease. Given the truncating mutations we report likely have a gain-of-function effect, the widespread interchangeable use of the terms ‘truncating mutation’ and ‘loss-of-function mutation’ is inappropriate. We believe a more descriptive term such as ‘protein truncating variant’ (PTV), which does not imply the functional consequence of the mutation, is preferable. We also provide evidence that mosaic mutations can have relevance to common disease. Such variants are challenging to detect by Sanger sequencing, but are detectable by next-generation sequencing approaches. It is therefore likely that further examples of mosaic disease-associated mutations will be forthcoming, though studies to define the frequency and characteristics of mosaic mutations in control individuals will be essential, to ensure the implications of such variants in case series are correctly interpreted. Finally, although newer sequencing technologies are making large-scale whole-genome sequencing experiments ever more feasible, it is likely that focussed sequencing experiments with tailored design and analytical prioritisation strategies, such as those employed here, will have utility over the next few years.
ONLINE METHODS
Patients and Samples
Cases
We used lymphocyte DNA from 8,046 individuals affected with breast and/or ovarian cancer that were recruited via two studies. 7,724 cases were recruited through 24 genetics centres in the UK via the Breast and Ovarian Cancer Susceptibility study (BOCS), which recruits women ≥18 years who have had breast cancer and/or ovarian cancer and have a family history of breast cancer and/or ovarian cancer. Each proband was screened for BRCA1 and BRCA2 mutations (by Sanger sequencing and/or heteroduplex analysis) and large rearrangements (by MLPA). The remaining 322 cases are an unselected hospital-based series of women with ovarian cancer who were recruited during treatment for ovarian cancer at the Royal Marsden Hospital. The DNA was extracted from peripheral blood samples except in 11 cases, for whom DNA was extracted from a lymphoblastoid cell line (NB all the PPM1D mutations were identified in peripheral blood-derived DNA). At least 97% of families were of European ancestry, i.e. comparable to the controls. Informed consent was obtained from all participants. The research was approved by the London Multicentre Research Ethics Committee (MREC/01/2/18).
For the Phase 1 pooled DNA repair panel experiment we used lymphocyte DNA from 1,150 women with breast cancer, 69 also had ovarian cancer. 78 of these individuals had one mutation, and one individual had two mutations, in known cancer predisposition genes. These were included as ‘positive controls’ to evaluate variant calling (see below). For the PPM1D case-control sequencing experiment we used 7,781 individuals with breast and/or ovarian cancer. We did not use the case data from the pooled DNA repair panel experiment in the case-control analysis, firstly because the mutation status of individuals cannot be definitively obtained from the pooled experiment as one cannot be certain that every sample is equally represented in a pool, and secondly because the mutation detection method was different to that utilised in the case-control experiment. We used our standard case and control sample trays for the case-control PPM1D sequencing experiment and the sample selection was blind to the pooled DNA repair panel experiment. 885 individuals were part of both experiments.
Samples and pathology information from mutation-positive families
For families in which a PPM1D mutation was detected, we sought DNA samples from relatives. We also requested tumour material, histopathology information, and immunohistochemical profiles, including hormone receptor receptor and HER2 status for patients with breast cancer, in probands from the hospitals where they had been treated. Representative tumour blocks were retrieved where possible and examined by two histopathologists (DNR & JSR-F) and classified and graded according to the World Health Organisation 2003 classification1,2. Tumours were microdissected under a stereomicroscope and genomic DNA was extracted from tumour and, where possible, stroma using the DNeasy kit (Qiagen) as previously described2.
Controls
We used lymphocyte DNA from 5,861 population-based controls obtained from the 1958 Birth Cohort Collection, an ongoing follow-up of persons born in Great Britain in one week in 1958. Biomedical assessment was undertaken during 2002-2004 at which blood samples and informed consent were obtained for creation of a genetic resource but phenotype data for these individuals is not available. At least 97% of the controls were of European ancestry. (http://www.cls.ioe.ac.uk/studies.asp?section=000100020003).
Sequencing
DNA repair panel sequencing
We identified genes for inclusion on the DNA repair panel from http://www.geneontology.org/ using the search term “DNA repair” (GO:0006281) and from http://string-db.org/ by identifying all genes interacting with ATM, BRCA1, BRCA2, BRIP1, CHEK2 and PALB2 with highest confidence (≥ 0.9). This dataset was manually curated to remove duplicate genes and pseudogenes. CCDS transcripts for the remaining genes were retrieved from UCSC Genome Browser (http://genome.ucsc.edu/ from November 2010) (Supplementary Table 2). Genomic coordinates for all coding exons were identified and targeted in a custom pulldown designed using the Agilent SureSelect Target Enrichment system (Agilent)3. We created 48 pools of DNA that each included 4 μl of 50 ng/μl = 200ng of DNA from 24 individuals. We sheared 80 μl of the pooled DNA using Covaris technology. We prepared libraries without gel size selection or PCR enrichment using the Illumina Genomic PE Sample Prep Kit (Ilumina) and performed target enrichment according to the Agilent SureSelect protocol. Sequencing was performed by the WTCHG High-throughput DNA sequencing and MRC hub in Oxford on an Illumina HiSeq2000 (v2 flow cell, one lane of sequencing per pool) generating 2×100 bp reads. Sequence reads for each pool were mapped to the human reference genome (hg19) using BWA (version 0.5.6)4. Mapped reads were filtered to remove ambiguous alignments with a quality score of 0 and bases with a call quality below 22 were masked. Of the remaining reads for each pool 50-60% fell within the target regions, except for Pool 21 where the on target percentage was significantly lower. Median coverage for each pool achieved for target regions after filtering was between 2849× and 5545×. This corresponded to an average coverage of 119×-231× per sample. All pools had 90% of the target covered at a minimum of 480x. Target regions within the MHC achieved substantially lower coverage and were excluded from further analysis.
We also sequenced the DNA repair panel in six PPM1D PTV positive individuals using Illumina TruSeq kits for library preparation to enable sample indexing. Genomic DNA (1.5 μg) was fragmented and the libraries prepared using the Illumina TruSeq Sample Preparation Kit (index set A). One pool of six libraries (500 ng each) was enriched as before but with the addition of extra blocking primers targeted against the TruSeq index adapter sequences. Sequencing was performed at ICR with an Illumina HiSeq2000 (v3 flowcell, one lane) generating 2×100 bp reads. Mapped reads were filtered to remove ambiguous alignments with a quality score of 0 and bases with a call quality below 22 were masked. Of the remaining reads, 41-43% fell within the target region for each individual. Median coverage of the target for each individual after filtering was between 602x and 690×. All individuals had 90% of the target covered at a minimum of 50×.
PPM1D Sanger sequencing
We designed primers to PCR amplify and Sanger sequence PPM1D using Exon-Primer from UCSC Genome Browser (http://genome.ucsc.edu/ from November 2010). Primers and conditions are available on request. PCR reactions were performed using the QIAGEN Multiplex PCR Kit (Qiagen). Amplicons were unidirectionally sequenced using the BigDye Terminator Cycle sequencing kit and an ABI3730 automated sequencer (ABI PerkinElmer). We analysed the full coding sequence in 2,456 cases and 1,347 controls. As all the mutations identified in these samples were restricted to exon 6 we sequenced the mutation cluster region (c.1261-20-c.1695), but not the rest of the gene, in the remaining 5,325 cases and 4,514 controls. We also sequenced the mutation cluster region in all available samples from relatives of PPM1D PTV positive probands. All sequencing traces were independently analyzed by two individuals who were blind to the others analysis. Each individual analysed the sequencing with both automated software (Mutation Surveyor, SoftGenetics) and manual visual inspection. All putative mutations were confirmed by bidirectional sequencing from a fresh aliquot of the stock DNA. We also undertook Sanger sequencing of the PPM1D cluster region, in triplicate, in DNA from eight tumour samples and four ovarian stromal samples.
For the cDNA sequencing we established lymphoblastoid cell lines from three individuals with PPM1D PTVs (cases 20, 23 and 24). RNA was extracted using RNeasy Minikit (Qiagen) and cDNA synthesised using the ThermoScript RT-PCR system (Invitrogen), employing standard protocols. We amplified the mutation cluster region using a cDNA-specific primer, [Forward_ACCACCAGTCAAGTCACTGG; Reverse_TCTTTCGCTGTGAGGTTGTG] which we sequenced as described above.
Deep PCR amplicon sequencing
In lymphocyte DNA we amplified the PPM1D mutation cluster region and the full coding sequence and intron-exon boundaries of BRCA1 and BRCA2 using the Multiplex PCR Kit (Qiagen). We prepared indexed libraries of the PCR products using Nextera technology (Ilumina)5. We created two pools of 24 indexed libraries which we sequenced using an Illumina MiSeq, generating 2×150 bp reads. Data from 20 individuals passed quality control coverage metrics, generating median coverage greater than 500× across the PPM1D cluster region (average median coverage 3384×).
For the tumour analyses we amplified the mutation cluster region in tumour, stroma and blood DNA using an Illumina Nextera XT library preparation kit and supplied protocol (Illumina). To attain the required 1ng input for tagmentation we also amplified BRCA1 in 24 samples as described above and we then created one pool of 24 indexed libraries which we sequenced using an Illumina MiSeq, generating 2×150bp reads. We visually inspected all sequencing reads present at the mutation site after alignment with Stampy to determine if the PPM1D mutation was present.
NGS data analysis
DNA repair panel data
For the pooled DNA repair panel analysis, variant calling was undertaken with Syzygy (version 1.2.4)6. 402/439 previously validated SNPs with a MAF>5% genotyped through a breast cancer GWAS7 were successfully identified with high confidence and the remaining 37 SNPs were detected at lower confidence. Syzygy also detected 75/80 rare variants (MAF<1%) included in the study as positive controls (24/26 base substitutions, 14/14 insertions, 30/32 deletions and 7/8 complex indels, Supplementary Table 3). Thus sensitivity was 99.6% for base substitutions and 94.4% for rare indels. Frequency estimation for rare variants was assessed by evaluation of 39 BRCA1 and BRCA2 variants at a frequency of one per pool. Syzygy correctly estimated the frequency in 33 of the 35 variants it detected, incorrectly estimating the frequency at two per pool for the remaining two variants.
Deep PCR amplicon sequencing data
For the deep PCR amplicon sequencing and the indexed DNA repair panel sequencing in six individuals, sequence reads were mapped to the human reference genome (hg19) using Stampy version 1.0.148. Duplicate reads were flagged using Picard version 1.60 (http://picard.sourceforge.net). Variant calling was performed with Platypus version 0.1.9 (http://www.well.ox.ac.uk/platypus)9. The mutant read percentage was calculated as the proportion of total reads at the variant location that contained the variant, with a minimum mutant read percentage threshold of 5%.
Variant Annotation
Annotation for all experiments was undertaken with reference to CCDS transcripts from EnsEMBL version 65 identified using a custom Perl script (Supplementary Table 2). Variant calls were annotated for changes with respect to the chosen transcript and assigned a consequence type from the list used by EnsEMBL.
PTV Prioritisation Method
This is a gene-based (rather than the more typical variant-based) strategy that aims to prioritise potential disease-associated genes for follow-up by leveraging two properties of protein truncating variants: (1) the strong association of rare truncating variants with disease, and (2) collapsibility; different PTVs within a gene typically result in the same functional effect and can be combined equally. We implemented the method using the stats package in R. We first outputted all the predicted protein truncating variants: stop gains, coding frameshifts and essential splice site variants (−2, −1, +1, +2, +5). For this experiment we defined ‘rare’ as PTVs that were seen only once in the DNA repair panel data. We next stratified the genes according to the number of different, rare singleton PTVs called. We excluded genes for which samples had been included as positive controls (Supplementary Table 3). PPM1D was the top gene in this analysis. We are undertaking further analyses and follow-up of the DNA repair panel data which we aspire to present in a separate publication.
MLPA
We designed 22 probe pairs targeting PPM1D PTVs (n=18), wildtype PPM1D (n=2), wildtype BRCA1 (n=1) and wildtype CEP112 (n=1) (Supplementary Table 7). We added the synthetic probes to the SALSA MLPA probe mix P200 (MRC Holland). MLPA reactions were performed in triplicate according to the manufacturer’s instructions. MLPA was undertaken in lymphocyte DNA from 17 probands and in eight tumour DNA samples (from five individuals). In brief, probes were hybridised to 150 ng of denatured DNA, amplified by PCR, and separated on an ABI 3130 Genetic Analyzer (Applied Biosystems). Data were analysed using GeneMarker v1.51 software (SoftGenetics).
Microsatellite analysis
We used 5′6-FAM tagged primer pairs and PCR conditions for 17q microsatellite analysis as listed in Supplementary Table 8. 10 μl of a mastermix of 30 μl ROX size standard and 1ml HiDi formamide were added to each reaction post PCR, denatured at 95°C for 5 minutes, and cooled at −20°C for 5 minutes. Reactions were run on a 3730xL genetic analyser (Applied Biosystems) under the fragment analysis protocol. Data were analysed using GeneMarker v1.51 software (SoftGenetics). Microsatellite analysis was undertaken in lymphocyte DNA from 13 individuals from eight families, and in eight tumour DNA samples and four stroma DNA samples from five individuals. Of note, one of these cases (17) harbours both BRCA1 and PPM1D mutations. Both genes are located at chromosome 17q and it is the wild-type BRCA1 allele that is reduced in the tumours and therefore the relevance of the loss of heterozygosity with respect to PPM1D is difficult to deduce.
Cell line and plasmid constructs
The U2OS, HeLa and HEK293 (all p53 wildtype) cell lines were obtained from the American Type Culture Collection (ATCC). Cells were cultured and maintained according to the supplier’s instructions. Cells were transfected with plasmid DNA using Lipofectamine 2000 (Invitrogen). A plasmid containing full-length wildtype PPM1D cDNA (pCMV6 entry-PPM1D) was obtained from Origene, and the PPM1D open reading frame (ORF) subcloned into pCMV6-AN-HA (Origene), generating a construct that could express a PPM1D-N-terminal HA epitope fusion protein. Truncating mutations were introduced into the PPM1D ORF of this construct using the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene). To generate the following mutants, we used the following DNA amplification primers:
PPM1D mutant 1 (c.1384C>T),
forward primer GAGAGAATGTCTAAGGTGTAGTC,
reverse primer GACTACACCTTAGACATTCTCTC,
PPM1D mutant 2 (c.1420delC),
forward primer GATCCAGAACCATTGAAG,
reverse primer CTTCAATGGTTCTGGATC.
Western Blot Analysis of P53 levels
U2OS, HeLa and HEK293 cells were transfected with PPM1D expression constructs and 24 hours after transfection, cells were exposed to gamma irradiation (5 Gy) from an X ray source. Whole cell lysates were generated from transfected cells after irradiation (at 30 minute and four hour time points) and subjected to protein electrophoresis. Immunoblotting of electrophoresed lysates was performed using antibodies specific for p53 (9282S - Cell Signaling Technology) and actin (sc-1616, Santa Cruz Biotech).
Frequency and Risk Estimation
Statistical analyses were performed using the stats package in R. The significance of mutation clustering was modelled under a binomial distribution where the probability of observing a mutation in the last exon, which comprises 31% of the coding sequence, was 0.31. The frequency in BRCA1/BRCA2 carriers and non-carriers was compared using a two-sided test of proportions. Risk estimation was implemented using a competing risks retrospective likelihood model incorporating age at onset according to a proportional hazards model. Since individuals screened for PPM1D mutations were selected on the basis of both personal and family history of breast or ovarian cancer, standard methods of analysis that ignore the sampling frame would yield biased estimates of the risk ratios. To address this, we analysed data within a retrospective cohort approach by modelling the conditional likelihood of the observed genotypes given the disease phenotypes, using information on breast and ovarian cancer occurrence in the set of 6,577 unrelated individuals negative for BRCA1/2 mutations (BRCA1/2 mutation-positive individuals from the BOCS series and all the unselected ovarian case series were excluded) and controls. Male controls were included in the analysis, but were not considered to be at risk of developing breast or ovarian cancer. We assumed a competing risks model, under which, each individual was at risk of developing breast or ovarian cancer. This has been shown to provide unbiased estimates of the risk ratios for breast and ovarian cancer where a genetic variant may be associated with one or both of the diseases10. We estimated the PPM1D mutation carrier frequency in the population and breast and ovarian cancer risk ratios simultaneously. Since mutation screened probands may have been selected on the basis of bilateral breast cancer diagnosis or on the basis of both breast and ovarian cancer diagnosis we allowed for the risks of breast or ovarian cancer diagnosis after the first cancer diagnosis, including the risk of contralateral breast cancer. This model assumes that the increased breast cancer (including contralateral) or ovarian cancer risk after the first cancer diagnosis is entirely due to the susceptibility as defined by the model, with no additional variation in risk. Site-specific cancer risks were assumed to be independent conditional on genotype. Therefore the incidence of cancer at the second site was assumed to be the same as if the preceding cancer had not occurred, with the exception of contralateral breast cancer incidence after the first breast cancer, which was assumed to be half the overall breast cancer incidence, since only one breast was at risk. In all models females were censored at age 80 years. We assumed that the breast and ovarian cancer incidences depend on the underlying PPM1D genotype through models of the form: λ(t) = λ0(t)exp(βx)where λ0(t) is the baseline incidence at age t in non-mutation carriers, β is the log risk ratio associated with the mutation and x takes value 0 for non-mutation carriers and 1 for mutation carriers. The overall breast and ovarian cancer incidences, over all genotypes, were constrained to agree with the population incidences for England and Wales in the period of 1993-199711, as described previously12.13. The models were parameterised in terms of the mutation frequencies and log-risk ratios for breast and ovarian cancer. Parameters were estimated using maximum likelihood estimation and were implemented in the pedigree analysis software MENDEL14. The variances of the parameters were obtained by inverting the observed information matrix. To obtain confidence intervals for the risk ratios and perform hypothesis testing, log risk ratios were assumed to be normally distributed. A Wald test-statistic was used to test the null hypothesis that β=0 for both breast and ovarian cancer. Since PPM1D mutations were not found to segregate within families, we did not take into account precise family histories or pedigree information and therefore did not incorporate the effects of other susceptibility genes.
Acknowledgements
We thank all the subjects and families that participated in the research and D. Dudakia, J. Bull, R. Linger for their assistance in recruitment. We are indebted to Mike Stratton for discussions of the data and to Ann Strydom for extensive editorial assistance. We thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics, Oxford (funded by Wellcome Trust grant reference 090532/Z/09/Z and MRC Hub grant G0900747 91070) for the generation of the Phase 1 Sequencing data. This work was funded by the Institute of Cancer Research, The Wellcome Trust, Cancer Research UK and Breakthrough Breast Cancer. We acknowledge support by the RMH-ICR National Institute for Health Research (NIHR) Specialist Biomedical Research Centre for Cancer. We acknowledge the use of DNA from the British 1958 Birth Cohort collection funded by the MRC grant G0000934 and the Wellcome Trust grant 068545/Z/02. A.C.A. is a Cancer Research UK Senior Cancer Research Fellow (C12292/A11174). P.D. is supported by a Wolfson-Royal Society Merit Award. K.S. is supported by the Michael and Betty Kadoorie Cancer Genetics Research Programme.
References
- 1.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 2.Turnbull C, Rahman N. Genetic predisposition to breast cancer: past, present, and future. Annu Rev Genomics Hum Genet. 2008;9:321–345. doi: 10.1146/annurev.genom.9.081307.164339. [DOI] [PubMed] [Google Scholar]
- 3.Gayther SA, Pharoah PD. The inherited genetics of ovarian and endometrial cancer. Curr Opin Genet Dev. 2010;20:231–238. doi: 10.1016/j.gde.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rahman N, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renwick A, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38:873–875. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- 6.Seal S, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 7.Meijers-Heijboer H, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 8.Meindl A, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42:410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 9.Loveday C, et al. Germline RAD51C mutations confer susceptibility to ovarian cancer. Nat Genet. 2012;44:475–476. doi: 10.1038/ng.2224. [DOI] [PubMed] [Google Scholar]
- 10.Loveday C, et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011;43:879–882. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivas MA, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snape K, et al. Predisposition gene identification in common cancers by exome sequencing: insights from familial breast cancer. Breast Cancer Res Treat. 2012;134:429–433. doi: 10.1007/s10549-012-2057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caruccio N. Preparation of next-generation sequencing libraries using Nextera technology: simultaneous DNA fragmentation and adaptor tagging by in vitro transposition. Methods Mol Biol. 2011;733:241–255. doi: 10.1007/978-1-61779-089-8_17. [DOI] [PubMed] [Google Scholar]
- 14.Schouten JP, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiscella M, et al. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci U S A. 1997;94:6048–6053. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu X, et al. The type 2C phosphatase Wip1: an oncogenic regulator of tumor suppressor and DNA damage response pathways. Cancer Metastasis Rev. 2008;27:123–135. doi: 10.1007/s10555-008-9127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu X, Nguyen TA, Donehower LA. Reversal of the ATM/ATR-mediated DNA damage response by the oncogenic phosphatase PPM1D. Cell Cycle. 2005;4:1060–1064. [PubMed] [Google Scholar]
- 18.Shreeram S, et al. Regulation of ATM/p53-dependent suppression of myc-induced lymphomas by Wip1 phosphatase. J Exp Med. 2006;203:2793–2799. doi: 10.1084/jem.20061563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujimoto H, et al. Regulation of the antioncogenic Chk2 kinase by the oncogenic Wip1 phosphatase. Cell Death Differ. 2006;13:1170–1180. doi: 10.1038/sj.cdd.4401801. [DOI] [PubMed] [Google Scholar]
- 20.Bulavin DV, et al. Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nat Genet. 2002;31:210–215. doi: 10.1038/ng894. [DOI] [PubMed] [Google Scholar]
- 21.Natrajan R, et al. Tiling path genomic profiling of grade 3 invasive ductal breast cancers. Clin Cancer Res. 2009;15:2711–2722. doi: 10.1158/1078-0432.CCR-08-1878. [DOI] [PubMed] [Google Scholar]
- 22.Tan DS, et al. PPM1D is a potential therapeutic target in ovarian clear cell carcinomas. Clin Cancer Res. 2009;15:2269–2280. doi: 10.1158/1078-0432.CCR-08-2403. [DOI] [PubMed] [Google Scholar]
- 23.Rayter S, et al. A chemical inhibitor of PPM1D that selectively kills cells overexpressing PPM1D. Oncogene. 2008;27:1036–1044. doi: 10.1038/sj.onc.1210729. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi R, et al. Optimization of a cyclic peptide inhibitor of Ser/Thr phosphatase PPM1D (Wip1) Biochemistry. 2011;50:4537–4549. doi: 10.1021/bi101949t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chuman Y, et al. PPM1D430, a novel alternative splicing variant of the human PPM1D, can dephosphorylate p53 and exhibits specific tissue expression. J Biochem. 2009;145:1–12. doi: 10.1093/jb/mvn135. [DOI] [PubMed] [Google Scholar]
- 26.Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L. Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA. 2008;14:563–576. doi: 10.1261/rna.815108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs KB, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44:651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurie CC, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44:642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnes D, et al. Evaluation of assosciation methods for analysing modifiers of disease risk in carriers of high risk mutations. Genet. Epidemiol. 2012;36:274–291. doi: 10.1002/gepi.21620. [DOI] [PubMed] [Google Scholar]
- 30.Antoniou AC, Easton DF. Polygenic inheritance of breast cancer: Implications for design of association studies. Genet Epidemiol. 2003;25:190–202. doi: 10.1002/gepi.10261. [DOI] [PubMed] [Google Scholar]
References for Online Methods
- 1.Tavassoli FA, Devilee P. Pathology and Genetics of Tumours of the Breast and Female Genital Organs. IARC Press; Lyon, France: 2003. [Google Scholar]
- 2.Hernandez L, et al. Genomic and mutational profiling of ductal carcinomas in situ and matched adjacent invasive breast cancers reveals intra-tumour genetic heterogeneity and clonal selection. J. Pathol. 2012;227:42–52. doi: 10.1002/path.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caruccio N. Preparation of next-generation sequencing libraries using Nextera technology: simultaneous DNA fragmentation and adaptor tagging by in vitro transposition. Methods Mol. Biol. 2011;733:241–255. doi: 10.1007/978-1-61779-089-8_17. [DOI] [PubMed] [Google Scholar]
- 6.Rivas MA, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbull C, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lunter G, Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rimmer A, Mathieson I, Lunter G, McVean G. Platypus: An Integrated Variant Caller. 2012 www.well.ox.ac.uk/platypus. [Google Scholar]
- 10.Barnes D, et al. Evaluation of association methods for analysing modifiers of disease risk in carriers of high risk mutations. Genet. Epidemiol. 2012;36:274–291. doi: 10.1002/gepi.21620. [DOI] [PubMed] [Google Scholar]
- 11.Cancer incidence in five continents. VIII. IARC Sci. Publ.; 2002. pp. 1–781. [PubMed] [Google Scholar]
- 12.Antoniou AC, et al. Evidence for further breast cancer susceptibility genes in addition to BRCA1 and BRCA2 in a population-based study. Genet. Epidemiol. 2001;21:1–18. doi: 10.1002/gepi.1014. [DOI] [PubMed] [Google Scholar]
- 13.Antoniou AC, Easton DF. Polygenic inheritance of breast cancer: Implications for design of association studies. Genet. Epidemiol. 2003;25:190–202. doi: 10.1002/gepi.10261. [DOI] [PubMed] [Google Scholar]
- 14.Lange K, Weeks D, Boehnke M. Programs for Pedigree Analysis: MENDEL, FISHER, and dGENE. Genet. Epidemiol. 1988;5:471–472. doi: 10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]