Abstract
Periostin (POSTN), a matricellular protein, has been reported to be important in supporting tumor cell dissemination. However, the molecular mechanisms underlying POSTN function within the tumor microenvironment are poorly understood. In this study, we observe that the inducible knockdown of POSTN decreases esophageal squamous cell carcinoma (ESCC) tumor growth in vivo and demonstrate that POSTN cooperates with a conformational missense p53 mutation to enhance invasion. Pathway analyses reveal that invasive esophageal cells expressing POSTN and p53R175H mutation display activation of signal transducer and activator of transcription 1 (STAT1) target genes, suggesting that the induction of STAT1 and STAT1-related genes could foster a permissive microenvironment that facilitates invasion of esophageal epithelial cells into the extracellular matrix. Genetic knockdown of STAT1 in transformed esophageal epithelial cells underscores the importance of STAT1 in promoting invasion. Furthermore, we find that STAT1 is activated in ESCC xenograft tumors, but this activation is attenuated with inducible knockdown of POSTN in ESCC tumors. Overall, these results highlight the novel molecular mechanisms supporting the capacity of POSTN in mediating tumor invasion during ESCC development and have implications of therapeutic strategies targeting the tumor microenvironment.
Keywords: tumor microenvironment, periostin, mutant p53, STAT1, invasion
Introduction
Esophageal cancer comprises two subtypes: esophageal adenocarcinoma and esophageal squamous cell carcinoma (ESCC). ESCC is an aggressive gastrointestinal cancer that is the predominant subtype accounting for the majority of cases in many countries in Asia and Africa.1, 2 Due to a lack of early symptoms, patients with ESCC are often diagnosed at advanced stages of the disease, and clinical outcomes remain dismal. Common risk factors associated with ESCC are smoking tobacco, excessive alcohol use, aromatic hydrocarbons in smoked foods and particular nutritional deficiencies.1 The development of ESCC is a multi-step process, and selective genetic alterations have been identified. For example, aberrant expression of epidermal growth factor receptor (EGFR) and cyclin D1, activation of human telomerase, inactivation of p16Ink4a and p120 catenin and somatic mutations in the DNA-binding domain (DBD) of the p53 tumor-suppressor gene all have been found to be involved in the initiation and progression of ESCC.3 EGFR and cyclin D1 overexpression correlate with squamous dysplasia or neoplastic lesions, which are early events in tumor initiation,4 whereas inactivation of p16Ink4a and p120 catenin and mutations in p53 have been associated with later stages of ESCC progression.2
The majority of human cancers harbor missense mutations in TP53, which not only lead to loss of wild-type p53 transcriptional activity but also an accumulation of mutant p53 protein with gain-of-function activities.5 These missense mutations tend to occur in the DBD of TP53 and result in the loss of wild-type p53 function. Missense mutations in p53 fall into two broad categories known as ‘DNA-contact mutants' or ‘DNA conformational mutants' based on their effect on the thermodynamic stability of p53 protein.6 DNA-contact mutants such as R273H and R248Q have mutations in residues that are involved in DNA binding, whereas DNA-conformational mutants such as R175H, R248W and V143A cause global conformation distortions in the DBD.6 Mutant p53 has been shown to drive a repertoire of target genes that, in turn, regulate a plethora of biological processes such as inhibition of apoptosis, cell migration and invasion.7 Common hotspot mutations such as p53R175H and p53R273H found in human cancers have been genetically engineered into mouse models, respectively, corresponding to p53R172H and p53R270H mice.8 p53R172H and p53R270H heterozygous mice not only develop osteosarcomas and carcinomas but also display a metastatic phenotype similar to p53 heterozygous mice.8, 9 In fact, R175H, R248W and R273H confer a selective growth advantage to increasingly malignant ESCC.10
During tumor progression, acquisition of oncogenic and tumor-suppressor mutations cause cancer cells to activate adjacent stromal components and induce the release of cytokines, growth factors and extracellular matrix (ECM) proteins into the tumor stroma to create a microenvironment permissive for growth and dissemination.11, 12 Recent studies have highlighted the contribution of a subset of ECM proteins known as matricellular proteins to potentiate pro-tumorigenic cell–ECM interactions within the tumor microenvironment.13, 14, 15 This group of proteins is expressed dynamically and is highly elevated during embryonic development but yet shows minimal activity in adult tissues. Matricellular proteins characteristically function as non-structural ECM proteins which modulate cell regulatory pathways mediated by downstream effectors such as integrins or growth factor receptors and promote cell–matrix interactions.13 Wound injury, tissue remodeling, inflammation, cancer and other chronic diseases induce the re-expression of these proteins.16 Important members of this family include tenascin C, osteopontin and periostin (POSTN). In addition, dysregulation of their expression is observed in many solid tumors as well as in sera and is often correlated with poorer prognosis and outcomes in cancer patients, thus implicating the importance of their contributions towards cancer progression.17, 18
Previously, we identified POSTN as a key microenvironmental mediator of ESCC invasion using an organotypic 3D culture system to examine transformed and genetically engineered esophageal cells.19 POSTN is a secreted 90 kDa protein that was identified originally as a cell adhesion molecule responsible for recruitment and attachment of pre-osteoblasts to the periosteum.20 POSTN is a transforming growth factor-beta-inducible protein that has an N-terminal signal peptide sequence, a cysteine-rich Emilin domain, four internal homologous repeats and a hydrophilic C-terminal domain.21 Its four internal repeat domains share structural homology with Fasciclin 1, an insect neuronal cell adhesion protein, and βig-h3, a transforming growth factor-beta-inducible gene.21 The molecular mechanisms underlying POSTN capacity for tumor cell invasion in the microenvironment remain to be elucidated. In this study, using genetic and pharmacological approaches, we find that POSTN cooperates with mutant p53 to support invasion of transformed esophageal cells into the matrix. Bioinformatic network analyses identified the signal transducer and activator of transcription 1 (STAT1) signaling network as a putative pathway induced by POSTN expression in a mutant p53 background, which was validated by expression studies. Moreover, genetic knockdown of STAT1 in invasive and transformed, genetically engineered esophageal cells (EPC-hTERT-EGFR-p53R175H) attenuated invasion into the microenvironment. In addition, and importantly, we noted STAT1 activation in ESCC xenograft tumors that was diminished when genetic knockdown of POSTN was induced, hence highlighting the importance of POSTN in the pathogenesis of ESCC.
Results
Inducible knockdown of POSTN in ESCC tumors lead to decreased tumor growth and invasion
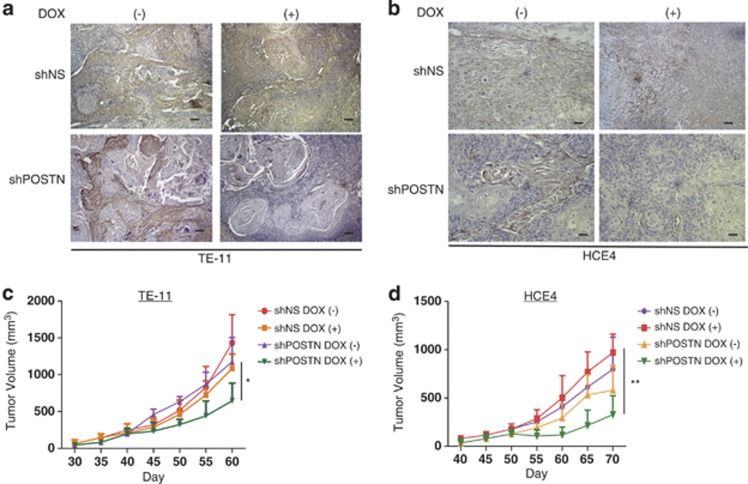
Given that high POSTN expression has been associated with poor patient survival outcomes in ESCC,22 we postulated that POSTN has a key role in promoting ESCC development. Indeed, in immunocompromised mice bearing tumor xenografts of two independent ESCC cell lines (TE11 and HCE4) that were stably transfected with a tetracycline-inducible shRNA targeted to POSTN, we observed that inducible ablation of POSTN expression and deposition in the stroma after initial establishment of these xenograft tumors (Figures 1a and b) led to decreased tumor growth and invasion as well as a decrease in proliferation (Figures 1c and d; Supplementary Figures S1a and S1b), indicating that POSTN contributes functionally in facilitating tumor growth and invasion in ESCC.
Figure 1.
Inducible knockdown of POSTN in ESCC tumors lead to decreased tumor growth and invasion. (a) Representative images of knockdown of POSTN expression by immunohistochemistry in tumors formed in vivo by TE-11 cancer cells stably transfected with lentiviral doxycycline-inducible non-specific targeting shRNA (shNS) or shRNA specific to POSTN (shPOSTN) vectors. Left panels represent tumors that were not induced with doxycycline (DOX) and right panels represent confirmation of POSTN knockdown in tumors induced with doxycycline (2 μg/ml). Bars=100 μℳ. (b) Representative images of knockdown of POSTN expression by immunohistochemistry in tumors formed in vivo by HCE4 cancer cells stably transfected with lentiviral doxycycline-inducible non-specific targeting shRNA (shNS) or shRNA specific to POSTN (shPOSTN) vectors. Left panels represent tumors that were not induced with doxycycline and right panels represent confirmation of POSTN knockdown in tumors induced with doxycycline(2 μg/ml). Bars=100 μℳ. (c) Tumor formation of TE-11 cancer cells stably transfected with doxycycline-inducible shNS or shPOSTN (n=10 in each cell line). Cells were subcutaneously injected in lower left flank of NOD-SCID mice, and tumor growth was measured at indicated time points. Doxycycline (2 μg/ml) was administered daily after tumors reached 200 mm3 (n=5 in the treatment group) to induce POSTN knockdown. Error bars represent s.e.m. *P<0.05 (Student's t-test). (d) Tumor formation of HCE4 cancer cells stably transfected with doxycycline-inducible shNS or shPOSTN (n=10 in each cell line). Cells were subcutaneously injected in lower left flank of NOD-SCID mice, and tumor growth was measured at indicated time points. Doxycycline (2 μg/ml) was administered daily after tumors reached 200 mm3 (n=5 in the treatment group) to induce POSTN knockdown. Error bars represent s.e.m. **P<0.01 (Student's t-test).
POSTN cooperates with mutant p53R175H to promote invasion into the mesenchymal ECM
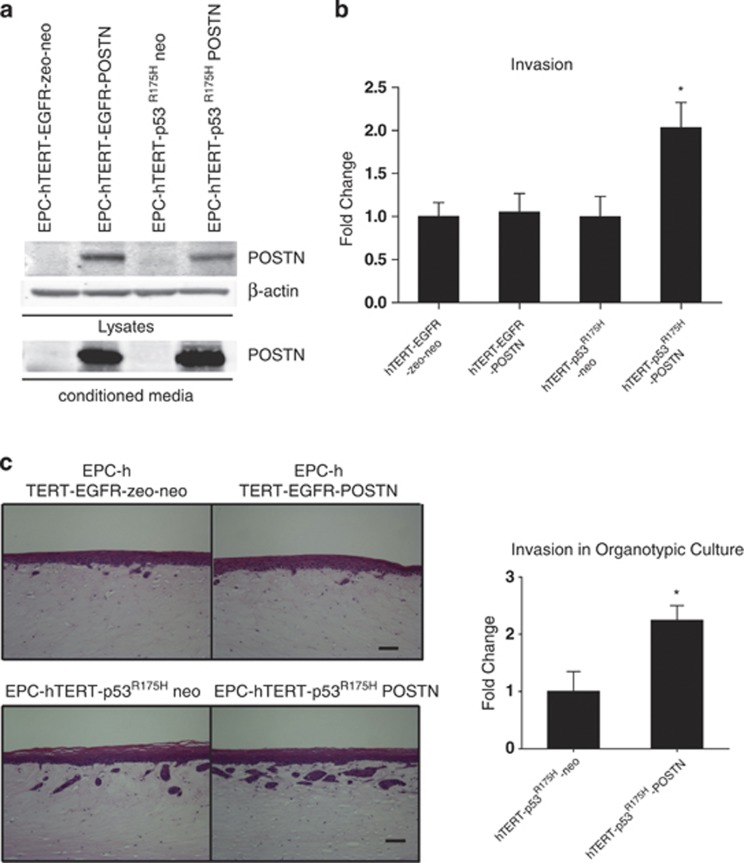
As we have identified POSTN expression to be upregulated in transformed, genetically engineered esophageal cells with p53R175H mutation and overexpressing EGFR (EPC-hTERT-EGFR-p53R175H), both common genetic alterations in ESCC, we hypothesized that the invasive capabilities of POSTN are mediated by either of these genetic alterations. To test this premise, we retrovirally overexpressed POSTN in non-invasive immortalized esophageal cells (EPC-hTERT) singularly expressing each of these genetic alterations (EPC-hTERT-EGFR-zeo and EPC-hTERT-p53R175H) (Figure 2a). Interestingly, while POSTN overexpression in EPC-hTERT-EGFR-zeo cells revealed no increase in invasion in Transwell Boyden invasion assays compared with its empty vector control cell line (EPC-hTERT-EGFR-zeo-neo), a 2-fold increase in invasion was observed when POSTN was overexpressed in EPC-hTERT-p53R175H cells compared with its respective empty vector control cell line (EPC-hTERT-p53R175H-neo) (Figure 2b). We observed the same pattern of invasion when EPC-hTERT-EGFR-POSTN and EPC-hTERT-p53R175H-POSTN cells, together with their respective empty vector control cell lines, when grown in a 3D organotypic culture system (Figure 2c). Invasion of the epithelium into the underlying mesenchymal ECM showed a 2.1 fold increase in EPC-hTERT-p53R175H-POSTN cells compared with its respective empty vector control whereas EPC-hTERT-EGFR-POSTN cells showed minimal differences. Similar findings were observed using an additional set of independently generated cell lines (data not shown). In parallel studies, EPC-hTERT-EGFR-zeo and EPC-hTERT-p53R175H cells were grown in organotypic culture and increasing doses of recombinant POSTN was added to these cultures. We observed no differences in invasion when recombinant POSTN was added to EPC-hTERT-EGFR-zeo cultures but there was a noteworthy increase in invasion when increasing concentrations of recombinant POSTN were added to EPC-hTERT-p53R175H cells (Supplementary Figure S2). Interestingly, mutant p53 alone is seen to be more invasive compared with overexpression of EGFR alone, suggesting that POSTN may act to augment this invasion. Collectively, these data suggest that POSTN cooperates with mutant p53R175H to enhance invasion of esophageal cells into the underlying stromal ECM.
Figure 2.
POSTN cooperates with mutant p53R175H to promote invasion into the mesenchymal ECM. (a) Western blot confirming POSTN (90 kDa) overexpression in EPC-hTERT-EGFR and EPC-hTERT-p53R175H cell lines and conditioned media. pFB neo was used as an empty control vector. β-Actin was used as a loading control. (b) Transwell Boyden chamber invasion assay of EPC-hTERT-EGFR and EPC-hTERT-p53R175H cells that overexpress POSTN vs control EPC-hTERT-EGFR-zeo-neo and EPC-hTERT-p53R175H-neo cells. EPC-hTERT-p53R175H-POSTN cells show increased invasion compared with EPC-hTERT-EGFR-POSTN cells and control cell lines. Bar graphs represent fold changes±s.e.m. *P<0.01 (Student's t-test, EPC-hTERT-p53R175H-POSTN cells vs control cells). Note that P<0.05 is statistically significant. Experiments were done in triplicate. (c) Hematoxylin and eosin staining of organotypic cultures comparing EPC-hTERT-EGFR and EPC-hTERT-p53R175H cells that overexpress POSTN vs control EPC-hTERT-EGFR-zeo-neo and EPC2-hTERT-p53R175H-neo cells. EPC-hTERT-p53R175H-POSTN cells show increased invasion into the underlying ECM compared with EPC-hTERT-EGFR-POSTN cells and control cell lines. Bar graphs represent fold changes±s.e.m. *P<0.01 (Student's t-test, EPC-hTERT-p53R175H-POSTN cells vs control cells). Note that P<0.05 is statistically significant. Experiments were done in duplicate. Bar=100 μm.
Restoration of wild-type p53 signaling decreases POSTN expression and invasion into ECM
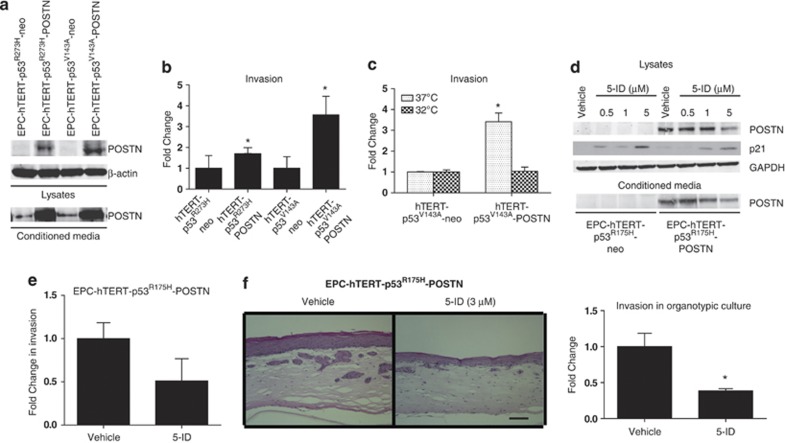
As p53 missense mutations fell into two broad categories of either conformational or DNA-binding mutants that each might lead to the acquisition of differing gain-of-function phenotypes,23 we next wanted to explore whether the ability of POSTN to promote invasion is dependent upon the conformation of mutant p53 as observed with p53R175H or on its DNA-contact-binding abilities. We chose to employ complementary genetic and pharmacological approaches to investigate this function. First, we retrovirally overexpressed POSTN in EPC-hTERT cells stably expressing different p53 point mutations, DNA-contact mutant p53R273H (EPC-hTERT-p53R273H-POSTN) and in a temperature-sensitive conformational mutant, p53V143A (EPC-hTERT-p53V143A-POSTN). The latter conditional mutant expresses p53V143A at 37 °C and induces wild-type p53 tertiary conformation and transcriptional activity at 32 °C. The levels of POSTN expression and secretion along with levels induced by empty vector controls are shown in Figure 3a. Interestingly, although both EPC-hTERT-p53R273H-POSTN and EPC-hTERT-p53V143A-POSTN cells show increased invasion in Boyden Transwell invasion assays compared with their respective empty vector control cells, EPC-hTERT-p53R273H-neo and EPC-hTERT-p53V143A-neo, there was a significant increase in invasion in the EPC-hTERT-p53V143A-POSTN cells compared with EPC-hTERT-p53R273H-POSTN cells (Figure 3b). This increase in invasion is similar to what was observed in EPC-hTERT-p53R175H-POSTN cells. This suggests that the mutation inducing the global conformational change in the p53 DBD may have an important role in regulating the invasive capabilities of POSTN.
Figure 3.
Restoration of wild-type p53 signaling decreases POSTN expression and invasion into ECM. (a) Western blot confirming POSTN expression in EPC-hTERT-p53R273H and EPC-hTERT-p53V143A cell lines and conditioned media. pFB neo was used as an empty control vector. (b) Transwell Boyden Chamber invasion assay showing increase in invasion in EPC-hTERT- p53R273H and mutant p53 temperature-sensitive EPC-hTERT- p53V143A cells overexpressing POSTN compared with control neo cells. Bar graphs represent fold changes±s.e.m. *P<0.003 (Student's t-test, EPC-hTERT-p53V143A-POSTN cells vs control cells). Note that P<0.05 is statistically significant. Experiments were done in triplicate. (c) Transwell Boyden Chamber invasion assay shows decrease in invasion in EPC-hTERT- p53V143A-POSTN cells when wild-type p53 conformation is induced at permissive temperature 32 °C compared with mutant p53 conformation at 37 °C. Bar graphs represent fold changes±s.e.m. *P<0.003 (Student's t-test, EPC-hTERT-p53V143A-POSTN cells vs control cells at 37 °C). Experiments were done in triplicate. (d) Western blot analysis of POSTN expression in EPC-hTERT- p53R175H-POSTN and EPC-hTERT- p53R175H-neo cell lysates and conditioned media after 24 h treatment with 5-ID (Vehicle, 0.5 μℳ, 1 μℳ and 5 μℳ). Immunoblotting for p21 to indicate restoration of wild-type p53 signaling. GAPDH was used as a loading control. (e) Transwell Boyden Chamber invasion assay shows decrease in invasion in EPC-hTERT- p53R175H-POSTN cells after 24 h treatment of 5-ID (3 μℳ). Bar graphs represent fold changes. Experiments were done in triplicate. (f) Hematoxylin and eosin staining of organotypic cultures comparing EPC-hTERT- p53R175H-POSTN cells treated with vehicle and 5-ID (3 μℳ) and show decreased invasion into the ECM after treatment. Bar graphs represent fold changes. Bar=100 μℳ and represent ±s.e.m. *P<0.04 (Student's t-test, EPC-hTERT-p53R175H-POSTN cells, treated with 5-ID vs vehicle-treated cells). Experiments were performed in triplicate.
We decided to interrogate this further by assessing whether the induction of wild-type p53 conformation and signaling can affect the ability of EPC-hTERT-p53V143A-POSTN to invade. As demonstrated in Figure 3c, a similar increase in invasion of EPC-hTERT-p53V143A-POSTN cells as seen in Figure 3b at 37 °C; however, induction of wild-type p53 conformation at 32 °C in EPC-hTERT-p53V143A-POSTN cells showed no increase in invasion compared with its empty vector control cells. To assess whether invasion can be affected pharmacologically by restoring wild-type p53 signaling, we utilized 5-iminodaunorubicin (5-ID), a small molecule compound which has been established previously to restore wild-type 53 signaling such as apoptosis and cell-cycle arrest via induction of p21.24 Treatment of EPC-hTERT-p53R175H-POSTN cells with 5-ID showed a decrease in POSTN expression in a dose-dependent manner (Figure 3d). In addition, treatment of EPC-hTERT-p53R175H-POSTN cells with 5-ID at a concentration with minimal toxicity to the cells, showed a decrease in invasion (Figure 3e) as well as a significant reduction in invasion into the ECM when grown in organotypic culture (Figure 3f). POSTN secretion into the conditioned media harvested from organotypic culture was also diminished with treatment of 5-ID (Supplementary Figure S3). In aggregate, these results indicate that mutant p53 contribute to POSTN-mediated invasion into the underlying ECM.
Esophageal cells with mutant p53R175H and POSTN reveal upregulation of STAT1 network and STAT1-dependent target genes
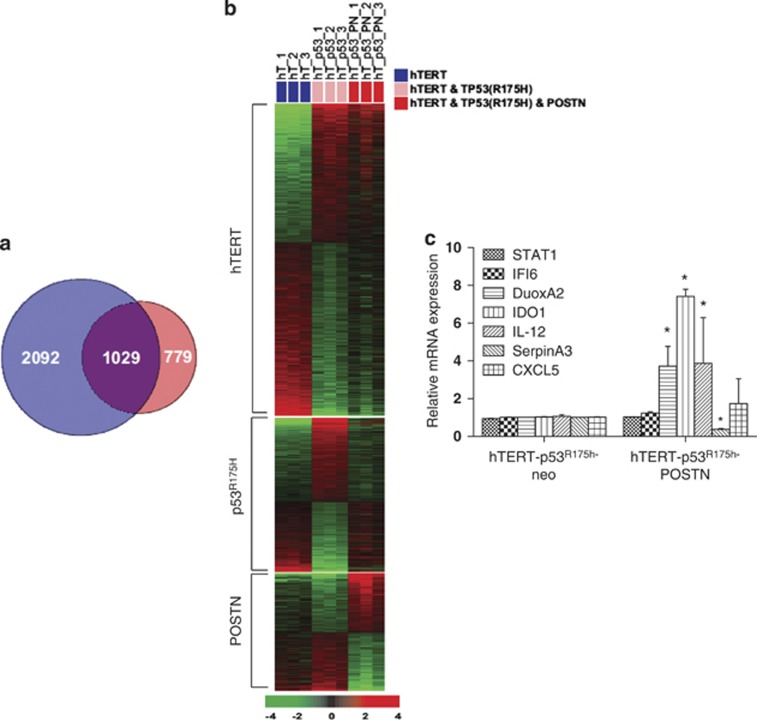
Based on the above findings, we next performed gene expression profiling using mRNA obtained from EPC-hTERT-p53R175H-POSTN, EPC-hTERT-p53R175H-neo and parental EPC-hTERT cells grown in organotypic culture (Figure 4a). Unsupervised hierarchical clustering led us to identify 779 genes, which showed a significant, differential expression in EPC-hTERT-p53R175H-POSTN cells compared with empty vector control and parental cells (Figure 4b and Supplementary Table S1). To aid in our identification of key pathways important in POSTN invasion, we utilized Ingenuity Pathway Analysis software to analyze our gene expression profile data. The STAT1 signaling pathway was found to be the highest represented pathway using Ingenuity Pathway Analysis (Supplementary Figure S4 and Supplementary Table S2). We confirmed the results of the microarray using quantitative reverse transcriptase–PCR validation of STAT1 and downstream STAT1-dependent target genes (IFI6, DUOXA2, IDO1, IL-12, SERPINA3, CXCL5), observing upregulation of STAT1-dependent genes (Figure 4c). Furthermore, western blot analysis shows that STAT1 phosphorylation (Tyr701) is seen only in EPC-hTERT-p53R175H-POSTN cells compared with its empty vector control cells and EPC-hTERT-EGFR-POSTN cells, indicating that STAT1 activation is induced in the context of p53 mutation and POSTN (Supplementary Figure S5).
Figure 4.
Esophageal cells with mutant p53R175H and POSTN reveal activation of the STAT1 signaling pathway. (a) Venn diagram displaying the number of genes with significant differential expression between the compared groups. Gene expression data were generated with RNA isolated from dissected epithelia of EPC-hTERT-p53R175H-POSTN cells grown in organotypic culture (n=3) compared with EPC-hTERT-p53R175H-neo cells (n=3) as well as parental non-invading EPC-hTERT cells (n=3). The blue circle (gene lists hTERT and p53R175H) represents genes differentially expressed between EPC-hTERT and EPC-hTERT-p53R175H-neo (3121). The red circle (gene lists p53R175H and POSTN) represents genes differentially expressed between EPC-hTERT-p53R175H-neo and EPC-hTERT-p53R175H-POSTN (1808). (P<0.001). (b) Heatmap of gene expression data presented in Venn diagram. Expression is based on a log2 scale where red represents upregulation and green represents downregulation. Expression patterns of POSTN not hTERT or p53R175H (779) are specific to expression of POSTN. (c) Quantitative reverse transcriptase–PCR validation of relative mRNA expression of upregulated STAT1-related genes (STAT1, DUOXA2, IDO1, IL-12, CXCL5, IFI6) and downregulated gene (SerpinA3) in microarray in EPC-hTERT-p53R175H-POSTN cells compared with EPC-hTERT-p53R175H-neo cells. Bar graphs represent fold changes±s.e.m. *P<0.05. Experiments performed in triplicate. CXCL, C-X-C motif chemokine ligand; IL, interleukin; IDO, indoleamine 2,3-dioxygenase; IL-12, interleukin-12.
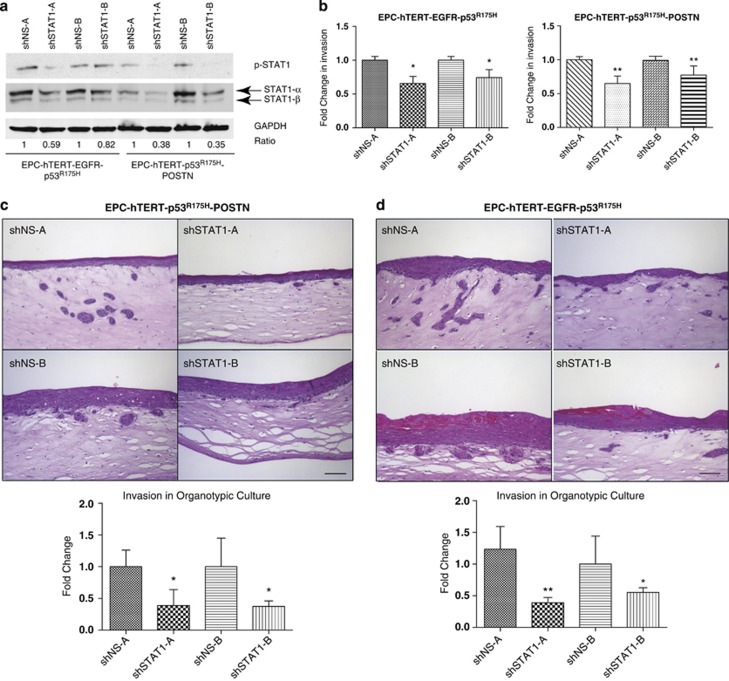
STAT1 knockdown in invasive EPC-hTERT-p53R175H-POSTN and transformed EPC-hTERT-EGFR-p53R175H cells show decrease in invasion
To test whether STAT1 functionally affects invasion of invasive esophageal cells overexpressing POSTN (EPC-hTERT-EGFR-p53R175H and EPC-hTERT-p53R175H-POSTN), an RNA interference approach using two independent shRNAs to transduce stable knockdown of STAT1 in invasive EPC-hTERT-p53R175H-POSTN cells and in transformed, genetically engineered EPC-hTERT-EGFR-p53R175H cells was used (Figure 5a). Knockdown of STAT1 in both cell lines showed a modest, yet significant, decrease in invasion in Transwell Boyden invasion assays compared with their respective empty vector controls (Figure 5b). Moreover, when grown in organotypic culture, both cell lines with knockdown of STAT1 display showed greater reduction in invasion into the stroma as well as a decrease in expression of downstream effectors of STAT1 signaling (Figures 5c and d, Supplementary Figure S6).
Figure 5.
STAT1 knockdown in EPC-hTERT-p53R175H-POSTN and transformed EPC-hTERT-EGFR-p53R175H cells show decrease in invasion. (a) Western blot confirming knockdown total STAT1 and STAT1 phosphorylation in invasive EPC-hTERT-p53R175H-POSTN and in transformed, genetically engineered EPC-hTERT-EGFR-p53R175H cells using two independent shRNAs directed against STAT1 and non-specific shRNAs as controls (A and B represent independently generated cell lines with the same genotype). GAPDH was used as a loading control. (b) Transwell Boyden Chamber invasion assay of EPC-hTERT-p53R175H-POSTN-shSTAT1-A and -B and EPC-hTERT-EGFR-p53R175H-shSTAT1-A and -B cells compared with control EPC-hTERT-p53R175H-POSTN-shNS-A and -B and EPC-hTERT-EGFR-p53R175H-shNS-A and -B cells. Bar graphs represent fold changes±s.e.m. *P<0.04 and 0.02 (Student's t-test, EPC-hTERT-EGFR-p53R175H -shSTAT1-A and -B cells vs control shNS-A and -B cells) and **P<0.001 (Student's t-test, EPC-hTERT-p53R175H-POSTN-shSTAT1-A and -B cells vs control shNS-A and -B cells). Experiments performed in triplicate. (c) Hematoxylin and eosin (H&E) staining of organotypic cultures comparing STAT1 knockdown in EPC-hTERT-p53R175H-POSTN-shSTAT1-A and -B compared with shNS-A and -B controls. Bar graphs represent fold changes±s.e.m. *P<0.01 and 0.02 (Student's t-test, EPC-hTERT-p53R175H-POSTN-shSTAT1-A and -B cells vs control shNS-A and -B cells). Experiments done in triplicate. (d) H&E staining of organotypic cultures comparing STAT1 knockdown in EPC-hTERT-EGFR-p53R175H-shSTAT1-A and -B compared with shNS-A and -B controls. Bar graphs represent fold changes±s.e.m. *P<0.004, **P<0.005 (Student's t-test, EPC-hTERT-EGFR-p53R175H-shSTAT1-A and -B cells vs control shNS-A and -B cells). Experiments done in triplicate.
In line with these results, we next sought to extend these findings to a cohort of matched human primary ESCC tumor gene expression data set25 and analyzed STAT1 expression in this tumor gene expression data set compared with their corresponding adjacent normal tissues. STAT1 expression was found to be significantly elevated in ESCC tumors compared with their adjacent normal tissue (Supplementary Figure S7). Overall, these data demonstrate that STAT1 overexpression is associated with primary ESCC development and that STAT1 has a role in mediating invasion in the ESCC microenvironment.
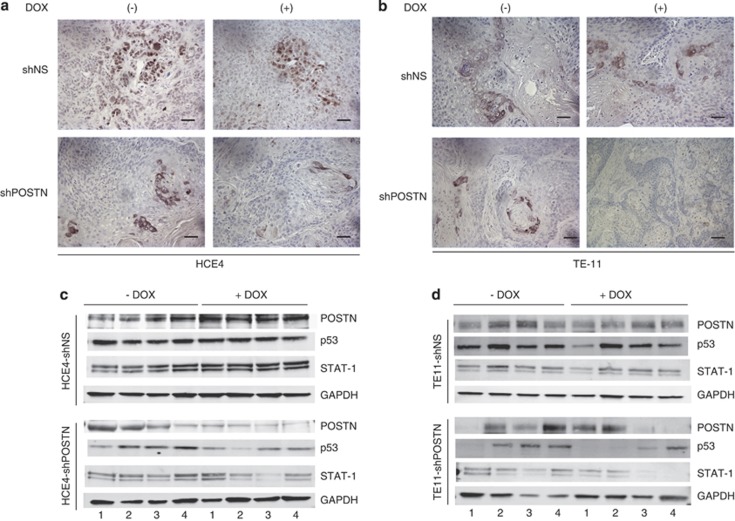
Inducible knockdown of POSTN in ESCC xenograft tumors display decreased p53 expression and STAT1 activation
To investigate the relationship between POSTN and STAT1 activation in vivo, sections from subcutaneous ESCC xenograft tumors (Figures 1a and b) were examined for phospho-STAT1 (Tyr701) by immunohistochemistry. Interestingly, we observed decreased nuclear STAT1 phosphorylation both in ESCC xenograft tumor cells and stroma with induction of POSTN knockdown by doxycycline (Figures 6a and b). Additionally, lysates from these xenograft tumors were analyzed, and we noted that POSTN knockdown in these tumors resulted in decreased STAT1 expression, a concomitant decrease in p53 expression as well as a decrease in downstream STAT1-related genes (Figures 6c and d; Supplementary Figure S8). Collectively, as observed in vitro, these findings imply that POSTN indirectly cooperates with mutant p53 to mediate STAT1 activation in vivo.
Figure 6.
Inducible knockdown of POSTN in ESCC xenograft tumors display decreased p53 expression and STAT1 activation. (a) Phospho-STAT1(Tyr701) expression by immunohistochemistry of tumors formed in vivo by subcutaneous injection of HCE4 cancer cells stably transfected with either lentiviral doxycycline-inducible non-specific targeting shRNA (shNS) or shRNA specific to periostin (shPOSTN) vectors. Left panels represent tumors that were not induced with doxycycline (DOX), and right panels represent tumors induced with doxycycline. Bar=100 μℳ. (b) Phospho-STAT1(Tyr701) expression by immunohistochemistry of tumors formed in vivo by subcutaneous injection of TE-11 cancer cells stably transfected with either lentiviral doxycycline-inducible non-specific targeting shRNA (shNS) or shRNA specific to periostin (shPOSTN) vectors. Left panels represent tumors that were not induced with doxycycline, and right panels represent tumors induced with doxycycline. Bar=100 μℳ. (c) Western blot analysis of STAT1 and p53 expression in four pairs of lysates isolated from HCE4 xenograft tumors transduced with doxycycline-inducible non-specific targeting shRNA (shNS) or shRNA specific to periostin (shPOSTN) with or without doxycycline treatment. Immunoblotting for POSTN expression to confirm doxycycline induced knockdown. GAPDH was used as a loading control. (d) Western blot analysis of STAT1 and p53 expression in four pairs of lysates isolated from TE-11 xenograft tumors transduced with doxycycline-inducible non-specific targeting shRNA (shNS) or shRNA specific to periostin (shPOSTN) with or without doxycycline treatment. Immunoblotting for POSTN expression to confirm doxycycline induced knockdown. GAPDH was used as a loading control.
Discussion
Recent findings have provided mounting evidence for the importance of POSTN in tumor invasion, tumor cell dissemination as well as creating a supportive environment for metastatic colonization.26, 27, 28 However, the molecular mechanisms engaged by POSTN to foster invasion in the tumor microenvironment remain poorly understood. In this study, we demonstrate that POSTN cooperates with mutant p53 in immortalized primary esophageal cells to promote invasion into the underlying ECM. Our finding that the propensity for POSTN to invade is mediated by mutant p53R175H, a p53 DBD conformational mutant found in approximately 6% of human cancers,29 prompted us to test whether this phenotype is recapitulated with other p53 missense mutations. Intriguingly, we observe that POSTN drives invasion to a greater extent when expressed in context of a p53 DBD conformational mutant compared with a p53 DNA-contact mutant, raising the possibility that the dominant-negative ability of p53 conformational mutants to suppress wild-type p53 activities influences the degree of invasion mediated by POSTN.
Due to the high prevalence of p53 mutations in human cancers, there has been an accelerated interest towards development of therapeutics focused on restoration of wild-type p53 function in tumors.30 Small molecule screens have identified promising small molecule compounds that selectively target and stabilize the core DBD of mutant p53 in tumor cells and restores wild-type p53 activities such as apoptosis and proliferation in vitro.24, 31, 32 Interestingly, a recent study demonstrated the therapeutic efficacy of restoring wild-type p53 in p53R172H mice, which corresponds to human p53R175H, suggesting that the removal of mutant p53 dominant-negative effect on functional wild-type p53 can halt tumor growth and subsequent tumor invasion.33 Using a combination of genetic and pharmacological approaches to restore wild-type p53 activities in invasive cells overexpressing mutant p53, our results of decreased cell motility and invasion are novel. It also establishes for the first time, to our knowledge, that modulation of mutant p53 affects the expression of POSTN as well as its invasive capabilities.
Progression of neoplastic cells in epithelial tissues to advanced malignancy encompasses a variety of biological processes that lead to an acquisition of a pro-invasive, mesenchymal phenotype.34 Initiation of local invasion and dissemination of aggressive carcinomas is often characterized by alterations in cell adhesion molecules that affect cell–cell/cell–matrix interactions and can occur as a result of crosstalk between malignant tumor cells and various components of surrounding neoplastic stroma such as the ECM, inflammatory and endothelial cells and fibroblasts.35 Secreted by tumor cells and stromal components into the stroma, it has been posited that matricellular proteins function to remodel the ECM and initiate downstream intracellular pathways such as integrin and tyrosine kinase receptor signaling that stimulate invasive behavior.36 In general, assorted extracellular matrices and molecules (normal vs tumor associated) have been shown to impart adverse functional effects on cancer cells in vitro.37 POSTN overexpression in clinical samples of several cancers, including oral squamous carcinoma, neuroblastoma, breast and non-small cell lung cancer has been found to be associated with higher malignancy grades and increased propensity for metastastic growth.38, 39, 40 Our finding of increasingly intense POSTN expression correlating with neoplastic tissue22 and invasive ESCC tumors in a genetic mouse model for ESCC strongly suggests that POSTN has a key role with invasion and progression of ESCC.
Moreover, POSTN has been reported to enhance metastatic initiation in the ‘pre-metastatic niche' by regulating the maintenance of Wnt signaling in cancer stem cells.28 In our study, another pathway network activated by POSTN signaling is STAT1. Phosphorylation of STAT1 at Tyr701 is induced by the binding of either Type I or Type II interferons to receptors that lead to the subsequent activation of Janus-activated kinases. Upon activation, phosphorylated STAT1 form homodimers that are translocated into the nucleus to initiate transcription of interferon-stimulated genes. As interferon-stimulated genes are primarily involved in promoting immune anti-pathogenic functions, induction of apoptosis and suppression of cell proliferation;41 STAT1 signaling is generally regarded as a tumor-suppressive pathway. However, recent data have shown that constitutively activated STAT1 signaling is implicated in epithelial cancer invasion and in aggressive tumors, with emerging evidence that increased STAT1 signaling can cause upregulation of genes that promote resistance to genotoxic and cytotoxic stress and subsequent tumor growth during tumor development.41, 42, 43, 44 Thus, these studies suggest that induction of STAT1 and upregulation of STAT1-dependent genes provide tumor cells a selective radioresistant advantage in a cytotoxic tumor microenvironment. In line with these observations, our study showed that knockdown of STAT1 in invasive as well as in transformed esophageal keratinocytes attenuated invasion into the stroma. Therefore, the contribution of POSTN-dependent STAT1 signaling has a key role in mediating invasion into the ECM. Notably, we found that STAT1 is strongly expressed in a cohort of primary human ESCC tumors compared with matched normal tissue, supporting our premise that STAT1 fosters invasiveness of ESCC tumors. Interestingly, the STAT1-dependent target genes that show the highest upregulation (IDO1, DUOX2) in our study are genes that have previously been shown to contribute to a pro-inflammatory microenvironment that promotes cancer progression,45, 46 which suggests that the activation of the STAT1 pathway may be an important mediator in contributing to a microenvironment that is conducive for tumor development.
In summary, our mechanistic findings support the functional role of POSTN in facilitating invasion. We demonstrated the novel finding that POSTN mediates its invasive capabilities through cooperation with mutant p53R175H. Furthermore, we identified that a STAT1 network acts as an effector of POSTN-mediated tumor invasion as underscored by knockdown of STAT1. POSTN appears to be critical in tumor invasion through remodeling of the ECM, and this may be aided, in part, by pro-inflammatory STAT1-dependent resistance against cytotoxic stress (Supplementary Figure S9). This likely creates a niche in the tumor microenvironment that poises tumor cells to metastasize. Indeed, we have observed that knockdown of POSTN in ESCC tumor xenografts leads to a significant decrease in the tumor-initiating cell (CD44hiCD24lo) population (Supplementary Figure S10). The induction of STAT1 and its effectors represents a novel mechanism of action for POSTN to facilitate tumor invasion. These findings represent a platform to explore how POSTN may be exploited as a biomarker for early detection of disease and molecular therapeutics to combat intrinsic tumor radioresistance.
Materials and methods
Cell culture
Stable transduction of transformed EPC-hTERT cells with EGFR and p53R175H retroviral vectors is described previously in Okawa et al.47 All cells were maintained in keratinocyte serum-free medium (SFM) medium (KSFM) (Invitrogen, Carlsbad, CA, USA) supplemented with 40 mg/ml BPE (bovine pituitary extract), 1.0 ng/ml EGF, 100 U/ml penicillin and 100 mg/ml streptomycin (Full KSFM). Cells were grown at 37 °C in a 5% CO2 humidified incubator. For inhibitor studies, 5-ID (3 μℳ) was added to medium.
Genetic knockdown and overexpression studies
Stable transduction of primary esophageal epithelial cells with viral vectors is described previously.19 p53R273H and p53V143A was subcloned into the pBABE-puro retroviral vector. The R273H p53 mutant was prepared using QuikChange site mutagenesis kit (Agilent Technologies, Redwood, CA, USA) according to the manufacturer's instructions. The primers used for R273H p53 mutation is as follows: Sense 5′-GCTTTGAGGTGCATGTTTGTGCCACG-3′ and antisense 5′-CGTGGGCACAAACATGCACCTCAAAGC-3′. All subclones and mutations were verified through DNA sequencing. For POSTN overexpression studies, esophageal epithelial cells were retrovirally infected with pFB-POSTN and pFB-neo. For inducible POSTN knockdown studies, ESCC cells were stably transfected with human tetracycline-inducible lentiviral pTRIPz-shRNAmir against POSTN or control lentiviral pTRIPz-shscramble virus. For STAT1 knockdown studies, esophageal epithelial cells were infected with human lentiviral shRNAmir against STAT1, nonsilencing control shRNAmir lentiviral vector, retroviral pSIREN-DsRed-shRNA against STAT1 or control retroviral non-specific control pSIREN-DsRed virus, all of which were kindly provided by Dr Andy Minn (University of Pennsylvania, Philadelphia, PA, USA). Forty-eight hours after infection, cells were selected in 300 μg/ml G418 (shscramble/shSTAT1), 0.5 μg/ml puromycin (p53 R273H/p53 V143A, shcramble/shPOSTN) for 5 days or by flow cytometry cell sorting for DsRed (shscramble/shSTAT1) FACSVantage SE with FACSDiva Option (BD Biosciences, San Jose, CA, USA). Expression of mutant p53 and POSTN and knockdown of STAT1 was confirmed by western blot.
Organotypic culture
Esophageal keratinocytes were grown in organotypic culture as means of recreating their microenvironment by supplying ECM components such as collagen and laminin, as previously described.47 For inhibitor studies, 5-ID (3 μℳ) was added to organotypic culture media. The amount of invasion was determined as described previously.48 Esophageal epithelium from organotypic cultures was peeled off and snap-frozen in liquid nitrogen before storage at −80 °C.
Antibodies and inhibitors
The following antibodies were used for immunoblotting: rabbit polyclonal POSTN (Abcam, Cambridge, UK, ab 14041), p21 (Oncogene Research Products, La Jolla, CA, USA), STAT1 (Cell Signaling, Danvers, MA, USA), N-Cadherin (BD Biosciences), E-Cadherin (BD Biosciences), α-SMA (Sigma, St Louis, MO, USA), ZEB1 (Cell Signaling). β-actin (Sigma) and GAPDH (glyceraldehyde 3-phosphate dehydrogenase; Millipore, Billerica, MA, USA) were used as loading controls. For immunohistochemistry, rabbit polyclonal POSTN (Abcam, ab 14041) and rabbit monoclonal phospho-STAT1 (Tyr701; Cell Signaling) were used. For inhibitor studies, 5-ID (kind gift of Dr El-Deiry) was dissolved in dimethyl sulfoxide at 20 mℳ and diluted before use.
RNA isolation, amplification and microarray studies
Total RNA was isolated using RNeasy Mini Kit (Qiagen, Hilden, Germany) and cDNA was synthesized using Taqman Reverse Transcription Reagents kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. For microarray studies, total RNA isolated from peeled epithelia from organotypic culture was amplified using Illumina Total Prep RNA Amplification Kit (Ambion, Carlsbad, CA, USA); 500 ng total RNA was used for the synthesis of cDNA and followed by amplification and biotin labeling. Each of 1.5 μg biotinylated cRNAs was hybridized to Ilumina Human-6 BeadChip v.4 and signals were developed using Amersham fluorolink streptavidin-Cy3 (GE Healthcare Biosciences, Little chalfont, UK). Gene expression data were collected using an Illumina bead Array Reader confocal scanner (BeadStation 500GXDW; Illumina, San Diego, CA, USA). Gene array data analysis was performed using Illumina BeadStudio software.
Quantitative reverse transcriptase–PCR
Gene-specific primers for SYBR Green real-time PCR was designed by PrimerExpress software (Applied Biosystems) and synthesized by Integrated DNA Technologies, Coralville, IL, USA (rimer sequences in Supplementary Table 3). Real-time PCR was performed and analyzed using ABI PRISM 7000 sequence detection system software (PE Applied Biosystems) and using Power SYBR Green PCR Master Mix (PE Applied Biosystems) according to the manufacturer's instructions. Supplementary Table 3 lists Taqman Expression Assays (Applied Biosystems) used. Relative mRNA expression was determined by normalizing to β-actin expression, which served as an internal control. Assays were performed three times in triplicate.
Western blotting
To confirm protein expression in cell lysates and secreted POSTN expression in collected conditioned media, western blot analyses were performed as described previously.19
Invasion assays
Invasion assays were performed as described previously.19 All experiments were performed at least three times in triplicate.
Immunohistochemistry
Immunohistochemistry was performed using with the Vector Elite kit (Vector Laboratories, Burlingame, CA, USA) using the manufacturer's protocol; its detailed procedures are as previously described.19
Xenograft experiments
Six- to 8-week-old female immunocompromised (NOD/SCID) mice (two groups per cell line, n=10 each) were obtained from National Cancer Institute, (Frederick, MD, USA). The tumors were established by subcutaneous injection of 200 μl (3 × 106 cells) of the cell suspension: Matrigel (1:1 ratio) into the lower left flank of the mice. Tumor dimensions were measured with calipers every 5 days and tumor volume was calculated using volume=(length) × (width)2/2. Doxycycline treatment was initiated 3–4 weeks post cell injection when tumors were approximately 200 mm3. All animal studies were approved by the respective IACUC at the University of Pennsylvania.
Statistical analysis of gene expression data
All statistical analyses were performed using BRB Arraytools Version 3.6 under the R language environment. The microarray data were normalized using the quantile normalization method in the Linear Models for Microarray Data package in the R language environment. The expression level of each gene was log2-transformed before further analysis. The random variance t test with very high stringent cutoff (P<0.001) was used to identify the genes significantly different between the two groups when compared. The first variable indicates parental hTERT cells with P53 mutation only and the second variable with P53 mutation only and P53 mutation and POSTN expression. Canonical pathway analysis was performed by applying Fisher's exact test and using Ingenuity Pathway Analysis database. Primary microarray data are available in the National Center for Biotechnology Information Gene Expression Omnibus public database (microarray platform, GPL10558; microarray data, GSE48999).
Acknowledgments
This work was supported by NIH/NCI P01-CA098101 (GSW, AKS, TJW, JS, YP, MH, HN, PG, AKR), NIH/NCI R01-CA113451 (EC), NIH T32-CA115299 (GSW) and NIH/NIDDK Center for Molecular Studies in Digestive and Liver Diseases (P30-DK050306) and American Cancer Society (RP-10-033-01-CCE). We acknowledge the assistance from the Molecular Pathology and Imaging (D. Budo), Molecular Biology/Gene Expression (G. Wu, S. Keilbaugh) Cell Culture Core ad Transgenic and Chimeric Mouse Core facilities. We are grateful to other members of the Rustgi lab for helpful discussions.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogenesis website (http://www.nature.com/oncsis)
Supplementary Material
References
- Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- Wang LS, Chow KC, Chi KH, Liu CC, Li WY, Chiu JH, et al. Prognosis of esophageal squamous cell carcinoma: analysis of clinicopathological and biological factors. Am J Gastroenterol. 1999;94:1933–1940. doi: 10.1111/j.1572-0241.1999.01233.x. [DOI] [PubMed] [Google Scholar]
- Xu XC. Risk factors and gene expression in esophageal cancer. Methods Mol Biol. 2009;471:335–360. doi: 10.1007/978-1-59745-416-2_17. [DOI] [PubMed] [Google Scholar]
- Sibilia M, Kroismayr R, Lichtenberger BM, Natarajan A, Hecking M, Holcmann M. The epidermal growth factor receptor: from development to tumorigenesis. Differentiation. 2007;75:770–787. doi: 10.1111/j.1432-0436.2007.00238.x. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192:209–218. doi: 10.1083/jcb.201009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Lozano G, Behringer RR. New mouse models of cancer: single-cell knockouts. Proc Natl Acad Sci USA. 2007;104:4245–4246. doi: 10.1073/pnas.0700173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe ML, Dlamini Z. The molecular mechanisms of oesophageal cancer. Int Immunopharmacol. 2005;5:1113–1130. doi: 10.1016/j.intimp.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Christofori G. New signals from the invasive front. Nature. 2006;441:444–450. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- Beacham DA, Cukierman E. Stromagenesis: the changing face of fibroblastic microenvironments during tumor progression. Semin Cancer Biol. 2005;15:329–341. doi: 10.1016/j.semcancer.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Chiodoni C, Colombo MP, Sangaletti S. Matricellular proteins: from homeostasis to inflammation, cancer, and metastasis. Cancer Metastasis Rev. 2010;29:295–307. doi: 10.1007/s10555-010-9221-8. [DOI] [PubMed] [Google Scholar]
- Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister SS, Gifford AM, Greiner AL, Kelleher SP, Saelzler MP, Ince TA, et al. Systemic endocrine instigation of indolent tumor growth requires osteopontin. Cell. 2008;133:994–1005. doi: 10.1016/j.cell.2008.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong GS, Rustgi AK. Matricellular proteins: priming the tumour microenvironment for cancer development and metastasis. Br J Cancer. 2013;108:755–761. doi: 10.1038/bjc.2012.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga-Kalabis M, Santiago-Walker A, Herlyn M. Matricellular proteins produced by melanocytes and melanomas: in search for functions. Cancer Microenviron. 2008;1:93–102. doi: 10.1007/s12307-008-0009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltermann A, Tischler V, Arbogast S, Braun J, Probst-Hensch N, Weder W, et al. Prognostic significance of epithelial-mesenchymal and mesenchymal-epithelial transition protein expression in non-small cell lung cancer. Clin Cancer Res. 2008;14:7430–7437. doi: 10.1158/1078-0432.CCR-08-0935. [DOI] [PubMed] [Google Scholar]
- Michaylira CZ, Wong GS, Miller CG, Gutierrez CM, Nakagawa H, Hammond R, et al. Periostin, a cell adhesion molecule, facilitates invasion in the tumor microenvironment and annotates a novel tumor-invasive signature in esophageal cancer. Cancer Res. 2010;70:5281–5292. doi: 10.1158/0008-5472.CAN-10-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K, Amizuka N, Takeshita S, Takamatsu H, Katsuura M, Ozawa H, et al. Identification and characterization of a novel protein, periostin, with restricted expression to periosteum and periodontal ligament and increased expression by transforming growth factor beta. J Bone Miner Res. 1999;14:1239–1249. doi: 10.1359/jbmr.1999.14.7.1239. [DOI] [PubMed] [Google Scholar]
- Takeshita S, Kikuno R, Tezuka K, Amann E. Osteoblast-specific factor 2: cloning of a putative bone adhesion protein with homology with the insect protein fasciclin I. Biochem J. 1993;294 (Pt 1:271–278. doi: 10.1042/bj2940271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong GS, Habibollahi P, Heidari P, Lee JS, Klein-Szanto AJ, Waldron TJ, et al. Optical imaging of periostin enables early endoscopic detection and characterization of esophageal cancer in mice. Gastroenterology. 2012;S0016-5085:01546–01546. doi: 10.1053/j.gastro.2012.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- Wang W, Kim SH, El-Deiry WS. Small-molecule modulators of p53 family signaling and antitumor effects in p53-deficient human colon tumor xenografts. Proc Natl Acad Sci USA. 2006;103:11003–11008. doi: 10.1073/pnas.0604507103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Hu N, Yang HH, Wang C, Takikita M, Wang QH, et al. Global gene expression profiling and validation in esophageal squamous cell carcinoma and its association with clinical phenotypes. Clin Cancer Res. 2011;17:2955–2966. doi: 10.1158/1078-0432.CCR-10-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Ouyang G, Bai X, Huang Z, Ma C, Liu M, et al. Periostin potently promotes metastatic growth of colon cancer by augmenting cell survival via the Akt/PKB pathway. Cancer Cell. 2004;5:329–339. doi: 10.1016/s1535-6108(04)00081-9. [DOI] [PubMed] [Google Scholar]
- Puglisi F, Puppin C, Pegolo E, Andreetta C, Pascoletti G, D'Aurizio F, et al. Expression of periostin in human breast cancer. J Clin Pathol. 2008;61:494–498. doi: 10.1136/jcp.2007.052506. [DOI] [PubMed] [Google Scholar]
- Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delaloye JF, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2012;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- Wiman KG. Pharmacological reactivation of mutant p53: from protein structure to the cancer patient. Oncogene. 2010;29:4245–4252. doi: 10.1038/onc.2010.188. [DOI] [PubMed] [Google Scholar]
- Bykov VJ, Selivanova G, Wiman KG. Small molecules that reactivate mutant p53. Eur J Cancer. 2003;39:1828–1834. doi: 10.1016/s0959-8049(03)00454-4. [DOI] [PubMed] [Google Scholar]
- Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- Wang Y, Suh YA, Fuller MY, Jackson JG, Xiong S, Terzian T, et al. Restoring expression of wild-type p53 suppresses tumor growth but does not cause tumor regression in mice with a p53 missense mutation. J Clin Invest. 2011;121:893–904. doi: 10.1172/JCI44504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DD. Emerging functions of matricellular proteins. Cell Mol Life Sci. 2011;68:3133–3136. doi: 10.1007/s00018-011-0779-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y, Cukierman E, Godwin AK. Differential expressions of adhesive molecules and proteases define mechanisms of ovarian tumor cell matrix penetration/invasion. PLoS One. 2011;6:e18872. doi: 10.1371/journal.pone.0018872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Yu CY, Dai M, Tam C, Loda M, Auclair D, et al. Elevated serum periostin levels in patients with bone metastases from breast but not lung cancer. Breast Cancer Res Treat. 2003;77:245–252. doi: 10.1023/a:1021899904332. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Lo KM, Chen LB, Auclair D, Nakashima Y, Moriyama S, et al. Expression of periostin, homologous with an insect cell adhesion molecule, as a prognostic marker in non-small cell lung cancers. Jpn J Cancer Res. 2001;92:869–873. doi: 10.1111/j.1349-7006.2001.tb01174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriwardena BS, Kudo Y, Ogawa I, Kitagawa M, Kitajima S, Hatano H, et al. Periostin is frequently overexpressed and enhances invasion and angiogenesis in oral cancer. Br J Cancer. 2006;95:1396–1403. doi: 10.1038/sj.bjc.6603431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodarev NN, Roizman B, Weichselbaum RR. Molecular pathways: interferon/stat1 pathway: role in the tumor resistance to genotoxic stress and aggressive growth. Clin Cancer Res. 2012;18:3015–3021. doi: 10.1158/1078-0432.CCR-11-3225. [DOI] [PubMed] [Google Scholar]
- Khodarev NN, Minn AJ, Efimova EV, Darga TE, Labay E, Beckett M, et al. Signal transducer and activator of transcription 1 regulates both cytotoxic and prosurvival functions in tumor cells. Cancer Res. 2007;67:9214–9220. doi: 10.1158/0008-5472.CAN-07-1019. [DOI] [PubMed] [Google Scholar]
- Schultz J, Koczan D, Schmitz U, Ibrahim SM, Pilch D, Landsberg J, et al. Tumor-promoting role of signal transducer and activator of transcription (Stat)1 in late-stage melanoma growth. Clin Exp Metastasis. 2010;27:133–140. doi: 10.1007/s10585-010-9310-7. [DOI] [PubMed] [Google Scholar]
- Khodarev NN, Roach P, Pitroda SP, Golden DW, Bhayani M, Shao MY, et al. STAT1 pathway mediates amplification of metastatic potential and resistance to therapy. PLoS One. 2009;4:e5821. doi: 10.1371/journal.pone.0005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Antony S, Juhasz A, Lu J, Ge Y, Jiang G, et al. Up-regulation and sustained activation of Stat1 are essential for interferon-gamma (IFN-gamma)-induced dual oxidase 2 (Duox2) and dual oxidase A2 (DuoxA2) expression in human pancreatic cancer cell lines. J Biol Chem. 2011;286:12245–12256. doi: 10.1074/jbc.M110.191031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Chang MY, Parker KH, Beury DW, DuHadaway JB, Flick HE, et al. IDO is a nodal pathogenic driver of lung cancer and metastasis development. Cancer Discov. 2012;2:722–735. doi: 10.1158/2159-8290.CD-12-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okawa T, Michaylira CZ, Kalabis J, Stairs DB, Nakagawa H, Andl CD, et al. The functional interplay between EGFR overexpression, hTERT activation, and p53 mutation in esophageal epithelial cells with activation of stromal fibroblasts induces tumor development, invasion, and differentiation. Genes Dev. 2007;21:2788–2803. doi: 10.1101/gad.1544507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grugan KD, Miller CG, Yao Y, Michaylira CZ, Ohashi S, Klein-Szanto AJ, et al. Fibroblast-secreted hepatocyte growth factor plays a functional role in esophageal squamous cell carcinoma invasion. Proc Natl Acad Sci USA. 2010;107:11026–11031. doi: 10.1073/pnas.0914295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.