Abstract
Upon antigen engagement and proper co-stimulation, naïve lymphocytes exit quiescence and undergo clonal expansion and differentiation into functional effector cells, after which they either die through apoptosis or survive as memory cells. Lymphocytes at different activation stages exhibit distinct metabolic signatures. Emerging evidence highlights a central role for the mechanistic target of rapamycin (mTOR) in bridging immune signals and metabolic cues to direct lymphocyte proliferation, differentiation and survival. Here we review recent advances in understanding the functional significance and signal transduction of mTOR in T cell biology, and the interplay between mTOR signaling and metabolic programs.
INTRODUCTION
Metabolic signatures during lymphocyte activation and differentiation
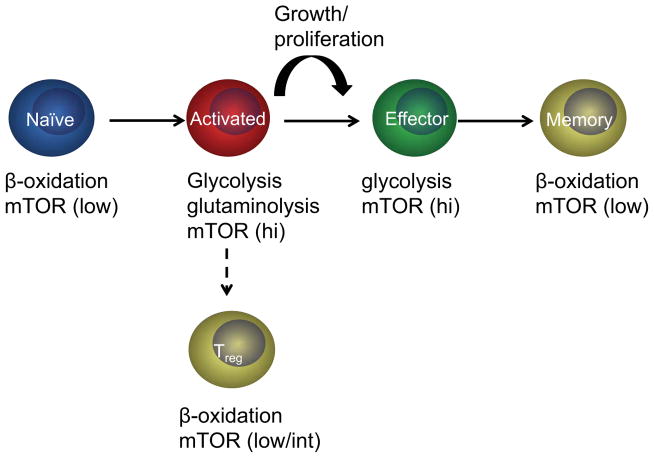
Mammalian cells are usually exposed to a constant supply of nutrients, but they do not normally take up nutrients and proliferate until they are stimulated by extrinsic factors[1]. Naïve lymphocytes, like most cells in normal tissues, have a quiescent status, in which they primarily rely on catabolic metabolism and derive most of their ATP from oxidative phosphorylation, particularly fatty acid β-oxidation[2,3,4**]. Quiescent lymphocytes also break down intracellular components through autophagy to supply molecules for oxidative phosphorylation[5]. Upon antigen recognition and co-stimulation, lymphocytes downregulate fatty acid β-oxidation and rapidly increase glycolytic, glutaminolytic and pentose phosphate pathways to provide biosynthetic materials and energy for cell growth and proliferation[3,4**,6]. Activated and effector T cells preferentially utilize aerobic glycolysis to meet their energy demands, a phenomenon known as the Warburg effect, which is also a metabolic feature of many cancer cells[1]. After clonal expansion and clearance of invading foreign pathogens, most effector T cells undergo apoptosis while some differentiate into long-lived memory cells. Memory T cells, like naïve T cells, are quiescent and have a catabolic metabolism[7,8]. A separate T cell subset, FOXP3+ regulatory T cells (Treg), also exhibits relatively high fatty acid β-oxidation but low glycolysis[9**,10]. Thus, during immune responses, T cells experience two major metabolic switches, from catabolic naïve T cells to anabolic activated/effector T cells and then again transition into catabolic memory T cells (Figure 1). Emerging evidence indicates that metabolism is closely coupled with the differentiation and function of T cells at different stages of their life span[11].
Figure 1.
T cells at different activation stages exhibit distinct metabolic phenotypes and mTOR activities. Naïve T cells rely on catabolism, particularly fatty acid β-oxidation, to maintain homeostasis. They also show low mTOR activity. Antigenic stimulation activates mTOR and markedly upregulates anabolic programs especially glycolysis and glutaminolysis, while downregulating fatty acid β-oxidation. The interplay between mTOR and metabolic programs collectively contributes to antigen-induced T cell growth and proliferation. Effector T cells maintain high glycolytic flux and mTOR activity, but their differentiation to memory T cells is accompanied by a metabolic switch from anabolism to catabolism, particularly upregulation of fatty acid β-oxidation, as well as downregulation of mTOR activity. A separate T cell lineage comprised of FOXP3+ Treg cells also has high fatty acid β-oxidation but low/intermediate mTOR activities.
mTOR signaling
The serine/threonine kinase mTOR consists of two distinct complexes: mTOR complex 1 (mTORC1) and 2 (mTORC2). Two scaffold proteins, regulatory associated protein of mTOR (RAPTOR) and rapamycin-insensitive companion of mTOR (RICTOR), are the defining components of mTORC1 and mTORC2, respectively[12]. While mTORC1 is sensitive to rapamycin, mTORC2 can be inhibited by prolonged or high dose of rapamycin treatment in CD4+ T cells[13,14**], but not in effector CD8+ T cells[15*]. Many upstream signals activate mTORC1 pathway through the small GTPase RHEB (RAS homologue enriched in brain). The tuberous sclerosis 1 (TSC1) and TSC2 form a complex that inactivates RHEB through its GAP (GTPase-activating protein) activity, thereby suppressing mTORC1 activity. Further upstream, the PI3K-AKT pathway inactivates TSC1/TSC2 complex while AMP-activated protein kinase (AMPK) enhances its activity. Therefore, TSC1/TSC2 complex functions as a molecular switch that controls mTORC1 activity. S6K1 and 4E-BP1 are two best-characterized downstream targets of mTORC1 that regulates protein translation. Moreover, mTORC1 pathway also promotes glycolysis and lipid biosynthesis while inhibiting autophagy. mTORC2 is activated by PI3K signaling, but detailed mechanism is lacking. mTORC2 controls several AGC family kinases, including AKT, SGK1 and PKC-α and is involved in regulating metabolism, apoptosis and cytoskeletal organization[12]. In particular, phosphorylation of AKT-Ser473 by mTORC2 promotes FOXO1/3a phosphorylation and subsequent cytoplasm translocation and degradation[16,17].
In lymphocytes, diverse environmental signals, including antigens, growth factors, cytokines and nutrients regulate mTOR to direct immune responses and fate decisions[18,19]. Since the roles of mTOR and metabolic pathways have been extensively studied in mature T cells in the periphery, we will mainly focus on these cells. First, we will briefly describe the roles of mTOR in T cell homeostasis under steady state and antigen-triggered activation and differentiation. Second, we will discuss the functional effects and mechanistic basis of mTOR in sensing and propagating diverse immune signals, especially those mediated by TCR, co-stimulation and cytokine receptors. Third, we will present the emerging evidence on mTOR-dependent metabolic reprogramming of T cell responses, by focusing on the interaction between mTOR and transcription factors associated with cell metabolism such as MYC and HIF1, and the potential interplay between mTOR-controlled metabolites and immune signaling. As our discussion focuses on T cells, we refer the readers to an excellent recent review describing the PI3K-AKT-mTOR pathway in B cells[20].
mTOR, A MASTER CONTROLLER IN T CELL BIOLOGY
mTOR in T cell quiescence
The quiescent status of naïve T cells is not a default state determined by the lack of mitogenic stimuli, but is an actively maintained process[7]. Uncontrolled mTORC1 activation by TSC1 deletion in T cells leads to loss of quiescence and predisposes T cells to apoptotic death[21–23]. Consequently, Tsc1−/− mice have markedly reduced peripheral T cell numbers. TSC1-deficient T cells exhibit semi-activated phenotypes, with a larger cell size, increased metabolic gene expression and cell cycle entry. They have enhanced mTORC1 but reduced mTORC2 activity, suggesting a crosstalk between the two complexes[24]. TSC1-deficient mice fail to mount an efficient immune response against bacterial infection[23]. Thus, active control of mTORC1 activity by TSC1 maintains the quiescent status, survival fitness and immune competency of peripheral naïve T cells.
mTOR in CD4+ cell lineage differentiation
Depending on the nature of antigenic stimulation and cytokine milieu, naïve CD4+ T helper cells may differentiate into TH1, TH2 and TH17 effector subsets, or become induced regulatory T cells (iTreg)[25]. The importance of mTOR in T cell differentiation is underscored by the inability of mTOR-deficient CD4+ T cells to differentiate into TH1, TH2 and TH17 effector subsets[26**]. mTORC1 and mTORC2 have distinct functions in directing CD4+ T cell lineage differentiation. RHEB-deficient T cells are defective to differentiate into TH1 or TH17 cells but have normal TH2 differentiation[14]. In contrast, loss of RAPTOR leads to impaired TH17 but normal TH1 differentiation[27]. The reason behind the discrepant phenotypes between Rheb−/− and Raptor−/− T cells is unclear, but this may be related to RHEB-independent activation of mTORC1[28]. Deletion of RICTOR has been shown to impair TH2 differentiation but preserve TH1 and TH17 differentiation[14]. However, a separate study shows that RICTOR-deficient T cells have reduced TH1 and TH2 differentiation due to impaired AKT and PKC-θ activation, respectively[13]. It is worth noting that metabolic activities have not been measured in these genetic models. Hence, how mTORC1 and mTORC2 signaling regulates specific metabolic programs in CD4+ effector T cells remains to be addressed.
In contrast to effector T cell differentiation, iTreg differentiation is negatively regulated by mTOR. PI3K-AKT-mediated mTOR activation inhibits iTreg generation by downregulating FOXP3 and other Treg transcriptional signature genes, whereas rapamycin enhances FOXP3 induction[29*,30*]. TCR ligation induces FOXP3 expression in Mtor−/− T cells even in the absence of exogenous iTreg polarizing cytokines[26]. Both mTORC1 and mTORC2 contribute to this inhibition program[14,26]. Although an increased AKT-mTOR activity has been associated with reduced thymic-derived Treg generation[29*,31*], the underlying mechanisms remain to be fully pinpointed.
mTOR in CD8+ cell effector and memory differentiation and trafficking
In CD8+ T cells, mTORC1 controls glucose uptake and glycolysis during initial activation as well as in effector cells[15*,32]. Furthermore, rapamycin impairs CD8+ effector cell differentiation[33]. Therefore, mTORC1 is critical for both CD4+ and CD8+ effector cell differentiation. As CD8+ T cells transition from effector to memory cells, their metabolism switches from anabolism to catabolism. Several studies have demonstrated a negative role of mTORC1 for CD8+ memory formation. Inhibition of mTORC1 by rapamycin[33,34**,35**] or silencing RAPTOR expression[34**] promotes memory cell generation and function. Importantly, Pearce et al. found that failure to upregulate fatty acid β-oxidation impedes memory T cell formation, which could be ameliorated by mTOR inhibition[35]. These data indicate that mTOR signaling prohibits memory T cell generation, at least partially, by modulating cell metabolism.
Proper immune cell trafficking is critical for a successful immune response and is orchestrated by the expression of chemokine receptors and adhesion molecules[36]. Treatment of CD8+ T cells with rapamycin or deficiency of PI3K or PDK1 prevents TCR or cytokine-induced downregulation of CD62L, CCR7 and S1PR1, molecules important for naïve T cell trafficking[32,37]. Conversely, activation of mTOR through PTEN or TSC1 deficiency is sufficient to downregulate these molecules[23,37]. Mechanistically, mTOR may regulate trafficking molecule expression through KLF2 (Krüppel-like factor 2)[37], FOXO1[38], or HIF1[15*]. Thus, mTOR activation endows T cells with an increased ability to migrate to sites of inflammation instead of retention in secondary lymphoid organs.
mTOR AND REGULATION OF IMMUNE SIGNALS
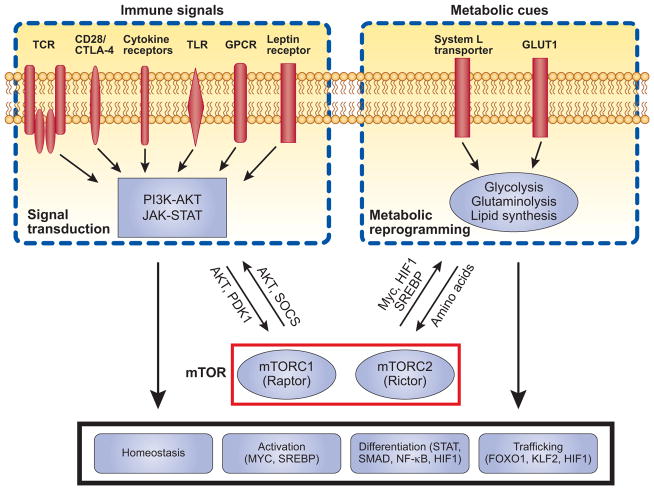
mTOR activity can be positively or negatively regulated by multiple inputs (Figure 2). The classic model posits that three major signals are required for proper T cell activation and differentiation: TCR engagement by antigen-MHC complex, costimulatory signals, and inflammatory cytokines. These signals, together with additional immune-modulatory receptors including Toll-like receptors (TLRs) and G protein-coupled receptors (GPCRs), are sensed and integrated by mTOR; once activated, mTOR can in turn interact with and impinge upon T cell signaling pathways. Thus, the interplay between mTOR and immune signals constitutes an important component to determine the outcome of adaptive immune responses[39].
Figure 2.
mTOR bridges immune signals and metabolic cues to regulate T cell responses. Multiple immune signals regulate mTOR activity, including TCR, costimulatory signals and cytokine receptors, as well as TLR, selective G protein-coupled receptors (GPCRs) and leptin receptors. Nutrients, such as amino acids and glucose, also mediate mTOR activation. These upstream inputs, through PI3K-AKT, JAK-STAT, and additional unidentified pathways, engage mTOR signaling. Once activated, mTOR shapes TCR and cytokine signaling through AKT and SOCS, and probably more importantly, reprograms metabolic activities including glycolysis through transcription factors MYC and HIF1. mTOR-dependent signaling pathways and metabolic programs are important regulators in T cell biology, including homeostasis, activation, lineage differentiation and memory formation. We propose that mTOR serves as a signal integration hub to bridge immune signals and metabolic cues, and the coordinated activation of these two major inputs by mTOR ensures proper execution of adaptive immunity in response to environmental signals
Signaling by TCR and co-stimulation
mTORC1 is activated by TCR engagement and the magnitude of its activation is correlated with antigen dose and the duration of the interaction between T cells and antigen-presenting cells[40,41]. CD28-mediated signal is required for the optimal activation of mTOR[42] and, importantly, for increased glycolytic flux[43]. Importantly, mTORC1 activation by optimal TCR and co-stimulation can occur independently of IL-2[44]. In contrast, ligation of the inhibitory receptors CTLA-4 and PD-1 attenuates mTOR activity and promotes iTreg generation[45,46]. Inhibition of mTOR during T cell activation leads to anergy even in the presence of CD28 signaling and blocking of CTLA-4[47–49].
As for the signaling components involved in the interplay between mTOR and TCR signaling, a notable example is the interaction between PI3K-AKT and mTOR. PI3K-AKT delivers a classical signal for mTORC1 activation, which in turn institutes feedback inhibition of PI3K-AKT[23,24]. In addition, mTORC2 plays a crucial role for the full activation of AKT by phosphorylating the Ser473 residue[13,14]. Thus, mTOR can be both downstream and upstream of PI3K-AKT signaling. Moreover, a PI3K-AKT-independent, but PDK1-dependent pathway has been identified for the activation of mTORC1 in CD8+ T cells[15*,32]. Additional TCR signaling components have also been shown to interact with mTOR. TCR stimulation activates Src family protein kinases LCK and FYN, which phosphorylate and activate ZAP-70. This in turn leads to activation of PLC-γ and ultimately several major signaling and transcriptional pathways: NFAT, NF-κB and MAP kinase (MAPK) cascade[50]. Both LCK and FYN are required for TCR-induced mTOR activation[51], whereas rapamycin impairs LCK and ZAP-70 activation[52], suggesting a crosstalk between TCR proximal signaling and mTOR. TCR-induced mTOR activation also depends on intact MAPK signaling[51,53]. Conversely, inhibition of mTOR can result in MAPK activation in cancer cells[54], although whether this is operative in lymphocytes is unknown. Further, mTOR activation, particularly mTORC2, is required for NF-κB activation through PKC-θ[13,52], but whether NF-κB can directly affect mTOR pathway remains unclear.
Signaling by cytokines and other immunomodulatory factors
Multiple cytokines activate mTOR as an important mechanism to shape T cell homeostasis and activation. For instance, a properly controlled mTOR activity is required for naïve T cell survival and homeostasis in response to IL-7[23,55]. IL-2 activates mTOR to prevent anergy in TCR-stimulated cells[47,56] and to sustain glycolysis, glucose and amino acid uptake in CD8+ effector T cells[15*,57**]. More importantly, the interplay between mTOR and cytokine signaling orchestrates T cell differentiation. IL-12 activates mTOR to facilitate CD8+ effector T cell differentiation[33]. During CD4+ T cell differentiation, mTOR is activated by different polarizing cytokines, such as IL-1 and IL-23 during TH17 differentiation[58,59]. Reciprocally, mTORC1 contributes to STAT4 and STAT3 activation mediated by IL-12 and IL-6, respectively, while IL-4 mediated STAT6 activation requires mTORC2 activation[14**]. This is largely mediated by mTOR-dependent effect on the expression of SOCS family members[14**]. Consistent with a key role for mTOR in cytokine signaling and T cell differentiation, expression of lineage-specific transcription factors T-bet and RORγt is impaired in RHEB-deficient T cells, whereas RICTOR deficiency diminishes GATA3 expression[13,14**]. Moreover, mTOR signaling influences epigenetic modifications that accompany T cell differentiation. Rapamycin treatment during T cell activation leads to increased CpG methylation at promoter regions of Il4 and Ifng[60], but reduces CpG methylation and increases permissive histone methylation at Foxp3 promoter region[30,60]. Interestingly, epigenetic regulation is likely an evolutionarily conserved function of mTOR, as TOR signaling in yeast is linked to histone acetylation[61]. Altogether, various interactions between mTOR and cytokine signaling appear to converge into transcriptional and epigenetic events to dictate T cell fate decisions.
mTOR bridges the interaction between immunomodulatory factors and the pleiotropic cytokine TGF-β, which signals primarily through transcription factors SMAD2 and SMAD3[62]. S1PR1 is a GPCR that mediates lymphocyte trafficking by sensing lipid sphingoshine-1-phosphate (S1P)[63]. S1PR1 activates mTOR signaling to promote TH1 differentiation while suppressing Treg generation through inhibition of SMAD3 activation[31,64]. Conversely, deletion of mTOR or inhibition of mTOR pathway in T cells increases SMAD2/3 phosphorylation and enhances iTreg generation[26**,65]. Neutralization of TGF-β largely blocks the excessive iTreg generation in mTOR-deficient T cells, implicating the obligate role of TGF-β signaling in this process[26]. However, FOXP3 induction by PI3K/mTOR inhibition has also been shown to occur independently of TGF-β[30]. Thus, the extent to which mTOR-mediated regulation of iTreg generation is contingent upon TGF-β signaling remains to be established[66]. A recent study also reveals that the anaphylatoxins C3a and C5a, through complement receptor C3aR and C5aR, activate mTOR and interfere with TGF-β-mediated iTreg generation[67].
TLRs recognize pathogen associated molecular patterns and can be expressed on T cells. TLR-2 expressed by CD8+ T cells transduces a costimulatory signal to activate mTOR, increase T-bet expression and enhance CD8+ T cell effector function[68]. MYD88, the essential adaptor molecule for many TLR signaling, is required for IL-1β and IL-23-mediated mTOR activation and TH17 differentiation[59].
mTOR AND REGULATION OF METABOLIC PROGRAMS
mTOR is activated by hormones and nutrients
mTOR signaling is intimately linked with cellular metabolism. Hormones and nutrients are important factors that modulate systemic and cellular metabolism and they feed into the mTOR pathway. Leptin is an adipocyte-derived hormone that controls food intake and metabolism. Recent studies have demonstrated that leptin-induced mTOR activation is critical for effector T cell proliferation[52], whereas it maintains the in vitro anergic status of Treg and negatively controls Treg proliferation[69,70]. Leptin promotes TH1 differentiation and inflammatory cytokine production, but suppresses TH2 differentiation[71]. Moreover, leptin-induced mTOR activation also mediates survival and activation of autoreactive CD4+ T cells[72]. As leptin is a well-characterized “fat-sensor”, these studies establish leptin-mTOR signaling as a direct link between nutritional status and lymphocyte function.
While it is well documented that mTOR senses amino acids, energy level and reactive oxygen species in other cell types[12,73], we are just beginning to appreciate this regulation in T cells. Deprivation of amino acids, glucose or energy through limiting essential amino acids or pharmacological inhibition leads to mTOR inactivation, accompanied by T cell anergy[42] and FOXP3 induction[74]. A recent study demonstrates that amino acid uptake by T cells activates mTORC1. Specifically, TCR engagement induces expression of System L transporter, SLC7A5, which mediates uptake of large neutral amino acids, such as leucine. Furthermore, expression of SLC7A5 is required for MYC expression and MYC-mediated metabolic reprograming. Consequently, SLC7A5-deficient T cells cannot be activated by immunization[57**]. How mTOR senses energy level and other nutrients in T cells awaits further investigation.
mTOR coordinates metabolic programs through key transcription factors
Preceding the metabolic reprogramming of T cell activation is the rapid upregulation of selective transcription factors responsible for the induction of metabolic genes. Among them, the oncogene MYC is critical for T cell activation-induced glycolysis and glutaminolysis[4**]. Acute deletion of MYC impairs upregulation of multiple glycolytic and glutaminolytic enzymes and induction of glutamine antiporter CD98, which modulates amino acid availability and mTOR activation[75]. Consequently, MYC-deficient T cells have severe defects in growth and proliferation. Rapamycin treatment diminishes MYC expression, whereas MYC-deficient T cells display reduced mTOR activation[4**], indicative of an intimate link between MYC and mTOR signaling. Interestingly, the TSC1/2-mTOR and MYC pathways can promote each other’s activity in a feed-forward loop to amplify their oncogenic effects in cancer cells[76]. Hence, just as T cell activation shares many metabolic features with cancer cells, they may adopt similar signaling connections as well.
HIF1α is another transcription factor associated with T cell metabolic reprogramming[4**,77]. Although its deficiency does not impair initial T cell activation or metabolism[4**], it is specifically required for the glycolytic program in TH17 cells[9] and the balance between TH17 and iTreg differentiation[9,78]. HIF1α is preferentially expressed in TH17 cells and its induction is dependent upon mTOR[9,77]. Under TH17-polarizing conditions, HIF1α-deficient T cells have diminished TH17 but increased iTreg differentiation, accompanied by reduced glycolytic activity. This phenotype is recapitulated by pharmacological inhibition of glycolysis, highlighting the importance of metabolic regulation of T cell fates[9]. Furthermore, HIF1α controls these two lineages by directly interacting with lineage-specific transcription factors RORγt and FOXP3 to modulate their function and stability[78]. Therefore, mTOR directs CD4+ T cell metabolic, transcriptional and posttranslational changes partly through HIF1α induction. Recently, Finlay et al. reported an important role of mTOR-induced HIF1 expression in effector CD8+ T cell metabolism, function and trafficking[15*]. HIF1 does not initiate glycolysis in naïve CD8+ T cells, but is crucial for glucose uptake and glycolytic metabolism in effector CD8+ T cells. HIF1β-deficient effector CD8+ T cells have diminished induction of perforin and granzymes, but increased expression of trafficking molecules. Thus, mTOR-induced HIF1 expression controls effector CD8+ T cell differentiation[15*].
T cell activation is accompanied by rapid upregulation of lipid biosynthesis and its gene expression program[79]. The sterol regulatory element-binding proteins SREBP1 and SREBP2 are transcription factors that activate cholesterol and fatty acid biosynthesis. Inhibition of mTOR by either rapamycin or PI3K inhibitor treatment prevents the processing of full-length SREBP and subsequent accumulation of mature SREBPs in the nucleus[80*]. Thus, mTOR promotes T cell lipogenesis through activation of SREBPs.
The estrogen-related receptor-alpha (ERRα) is another T cell activation-induced transcription factor that mediates effector T cell metabolism and differentiation. ERRα-deficient T cells have impaired metabolism and proliferation, associated with altered mTOR activity. Further, increasing mTOR activity through TSC2 deficiency renders T cell independent of ERRα, suggesting a potential interaction between ERRα and mTOR signaling[81].
Crosstalk between mTOR-controlled metabolism and immune signaling
Many posttranslational modifications, such as methylation, acetylation, glycosylation and prenylation, are mediated by metabolites. Emerging evidence suggests that metabolite-sensitive protein modifications can modulate signal transduction[82,83], with several examples identified in lymphocytes. For instance, UDP-N-acetylglucosamine (UDP-GlcNAc), a metabolite in the hexosamine biosynthetic branch of glucose metabolism, is required for IL-3 receptor (IL-3R) glycosylation and its membrane presentation. Absence of glucose downregulates membrane IL-3R expression and cell growth but these defects are reversible by supplement of GlcNAc, suggesting that glucose metabolism through the hexosamine biosynthetic pathway directly influences lymphocyte signaling[84]. Protein prenylation and palmitoylation were also found to influence TCR signaling and T cell differentiation[85–87]. Currently, no evidence is available that directly links mTOR-mediated metabolic programs to lymphocyte signaling. However, given the central role of mTOR in cell metabolism, it is conceivable that mTOR could influence immune signaling through metabolite production or availability. We propose that mTOR is uniquely situated in T cells to bridge immune signals and metabolic cues, and this could be partly mediated by mTOR-associated nutrients and metabolites (Figure 2).
CONCLUSION
T cells are at the center of adaptive immunity that protects the body from pathogen infections, or mediates self-destructive autoimmune diseases. mTOR integrates immune signals and metabolic cues to direct T cell homeostatic and functional fates, and this is shaped by the extensive interplay between mTOR signaling and cell metabolism. Despite the recent remarkable advances, a number of questions remain to be answered. We have yet to fully understand mTOR-associated upstream signal inputs and downstream effector pathways in T cells. Also, how mTOR-mediated metabolism interacts with immune signaling is another fascinating question. Further, modulation of mTOR activity with newly developed inhibitors, or direct targeting of specific metabolic pathways, holds promises as novel strategies for therapeutic intervention of immune-mediated diseases.
Highlights.
Different T cell activation states have distinct metabolic profiles and mTOR activity
mTOR orchestrates T cell quiescence, functional activation, and fate decisions
mTOR is activated by and impinge upon antigen receptor and other immune signaling
mTOR senses metabolic cues and coordinates T cell metabolic reprograming
Acknowledgments
We acknowledge the large number of researchers who have contributed to this field whose work was not cited owing to space limitations. The authors’ research is supported by US National Institutes of Health (R01 NS064599 and R21 AI094089), National Multiple Sclerosis Society, Lupus Research Institute, and the American Lebanese Syrian Associated Charities (H.C.).
Footnotes
Competing interest statement
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krauss S, Brand MD, Buttgereit F. Signaling takes a breath--new quantitative perspectives on bioenergetics and signal transduction. Immunity. 2001;15:497–502. doi: 10.1016/s1074-7613(01)00205-9. [DOI] [PubMed] [Google Scholar]
- 3.Jones RG, Thompson CB. Revving the engine: signal transduction fuels T cell activation. Immunity. 2007;27:173–178. doi: 10.1016/j.immuni.2007.07.008. [DOI] [PubMed] [Google Scholar]
- **4.Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. This paper described MYC-mediated metabolic reprograming during T cell activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 6.Doughty CA, Bleiman BF, Wagner DJ, Dufort FJ, Mataraza JM, Roberts MF, Chiles TC. Antigen receptor-mediated changes in glucose metabolism in B lymphocytes: role of phosphatidylinositol 3-kinase signaling in the glycolytic control of growth. Blood. 2006;107:4458–4465. doi: 10.1182/blood-2005-12-4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: energy metabolism and the T-cell response. Nature reviews Immunology. 2005;5:844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- 8.Pearce EL. Metabolism in T cell activation and differentiation. Curr Opin Immunol. 2010;22:314–320. doi: 10.1016/j.coi.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **9.Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. The Journal of experimental medicine. 2011;208:1367–1376. doi: 10.1084/jem.20110278. The authors demonstrated that different helper T cell lineages have distinct glycolytic activities and HIF1 reciprocally mediates TH17 vs iTreg differentiation through metabolic reprograming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. Journal of immunology. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang R, Green DR. Metabolic checkpoints in activated T cells. Nature immunology. 2012;13:907–915. doi: 10.1038/ni.2386. [DOI] [PubMed] [Google Scholar]
- 12.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **13.Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, Boothby M. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **14.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nature immunology. 2011;12:295–303. doi: 10.1038/ni.2005. These studies showed that mTORC1 and mTORC2 are both important for helper T cell lineage differentiation, but with distinct contributions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *15.Finlay DK, Rosenzweig E, Sinclair LV, Feijoo-Carnero C, Hukelmann JL, Rolf J, Panteleyev AA, Okkenhaug K, Cantrell DA. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. The Journal of experimental medicine. 2012;209:2441–2453. doi: 10.1084/jem.20112607. This study showed that mTOR, through the transcription factor HIF1, mediates metabolism, effector function and migration in CD8+ T cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouyang W, Li MO. Foxo. in command of T lymphocyte homeostasis and tolerance. Trends in immunology. 2011;32:26–33. doi: 10.1016/j.it.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hedrick SM, Hess Michelini R, Doedens AL, Goldrath AW, Stone EL. FOXO transcription factors throughout T cell biology. Nature reviews Immunology. 2012;12:649–661. doi: 10.1038/nri3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nature reviews Immunology. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annual review of immunology. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Limon JJ, Fruman DA. Akt and mTOR in B Cell Activation and Differentiation. Frontiers in immunology. 2012;3:228. doi: 10.3389/fimmu.2012.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Brien TF, Gorentla BK, Xie D, Srivatsan S, McLeod IX, He YW, Zhong XP. Regulation of T-cell survival and mitochondrial homeostasis by TSC1. European journal of immunology. 2011;41:3361–3370. doi: 10.1002/eji.201141411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Q, Liu Y, Chen C, Ikenoue T, Qiao Y, Li CS, Li W, Guan KL, Zheng P. The tuberous sclerosis complex-mammalian target of rapamycin pathway maintains the quiescence and survival of naive T cells. Journal of immunology. 2011;187:1106–1112. doi: 10.4049/jimmunol.1003968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nature immunology. 2011;12:888–897. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **26.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. This study provided the first genetic evidence that mTOR signaling promotes effector T cell differentiation, but negatively impacts iTreg generation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurebayashi Y, Nagai S, Ikejiri A, Ohtani M, Ichiyama K, Baba Y, Yamada T, Egami S, Hoshii T, Hirao A, et al. PI3K-Akt-mTORC1-S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORgamma. Cell reports. 2012;1:360–373. doi: 10.1016/j.celrep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *29.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. The Journal of experimental medicine. 2008;205:565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *30.Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, Spivakov M, Knight ZA, Cobb BS, Cantrell D, O’Connor E, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7797–7802. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *31.Liu G, Burns S, Huang G, Boyd K, Proia RL, Flavell RA, Chi H. The receptor S1P1 overrides regulatory T cell-mediated immune suppression through Akt-mTOR. Nature immunology. 2009;10:769–777. doi: 10.1038/ni.1743. Reference 29–31 demonstrated that AKT-mTOR pathway negatively regulates Treg differentiation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macintyre AN, Finlay D, Preston G, Sinclair LV, Waugh CM, Tamas P, Feijoo C, Okkenhaug K, Cantrell DA. Protein kinase B controls transcriptional programs that direct cytotoxic T cell fate but is dispensable for T cell metabolism. Immunity. 2011;34:224–236. doi: 10.1016/j.immuni.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **34.Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **35.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. References 34 and 35 were the first to show that memory CD8 T cell differentiation is negatively regulated by mTOR signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finlay D, Cantrell D. Phosphoinositide 3-kinase and the mammalian target of rapamycin pathways control T cell migration. Annals of the New York Academy of Sciences. 2010;1183:149–157. doi: 10.1111/j.1749-6632.2009.05134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinclair LV, Finlay D, Feijoo C, Cornish GH, Gray A, Ager A, Okkenhaug K, Hagenbeek TJ, Spits H, Cantrell DA. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nature immunology. 2008;9:513–521. doi: 10.1038/ni.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, DePinho RA, Hedrick SM. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nature immunology. 2009;10:176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powell JD, Delgoffe GM. The mammalian target of rapamycin: linking T cell differentiation, function, and metabolism. Immunity. 2010;33:301–311. doi: 10.1016/j.immuni.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner MS, Kane LP, Morel PA. Dominant role of antigen dose in CD4+Foxp3+ regulatory T cell induction and expansion. Journal of immunology. 2009;183:4895–4903. doi: 10.4049/jimmunol.0901459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katzman SD, O’Gorman WE, Villarino AV, Gallo E, Friedman RS, Krummel MF, Nolan GP, Abbas AK. Duration of antigen receptor signaling determines T-cell tolerance or activation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18085–18090. doi: 10.1073/pnas.1010560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng Y, Delgoffe GM, Meyer CF, Chan W, Powell JD. Anergic T cells are metabolically anergic. Journal of immunology. 2009;183:6095–6101. doi: 10.4049/jimmunol.0803510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, Elstrom RL, June CH, Thompson CB. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–777. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 44.Colombetti S, Basso V, Mueller DL, Mondino A. Prolonged TCR/CD28 engagement drives IL-2-independent T cell clonal expansion through signaling mediated by the mammalian target of rapamycin. Journal of immunology. 2006;176:2730–2738. doi: 10.4049/jimmunol.176.5.2730. [DOI] [PubMed] [Google Scholar]
- 45.Karman J, Jiang JL, Gumlaw N, Zhao H, Campos-Rivera J, Sancho J, Zhang J, Jiang C, Cheng SH, Zhu Y. Ligation of cytotoxic T lymphocyte antigen-4 to T cell receptor inhibits T cell activation and directs differentiation into Foxp3+ regulatory T cells. J Biol Chem. 2012;287:11098–11107. doi: 10.1074/jbc.M111.283705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, Sharpe AH. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. The Journal of experimental medicine. 2009;206:3015–3029. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng Y, Collins SL, Lutz MA, Allen AN, Kole TP, Zarek PE, Powell JD. A role for mammalian target of rapamycin in regulating T cell activation versus anergy. Journal of immunology. 2007;178:2163–2170. doi: 10.4049/jimmunol.178.4.2163. [DOI] [PubMed] [Google Scholar]
- 48.Vanasek TL, Khoruts A, Zell T, Mueller DL. Antagonistic roles for CTLA-4 and the mammalian target of rapamycin in the regulation of clonal anergy: enhanced cell cycle progression promotes recall antigen responsiveness. Journal of immunology. 2001;167:5636–5644. doi: 10.4049/jimmunol.167.10.5636. [DOI] [PubMed] [Google Scholar]
- 49.Powell JD, Lerner CG, Schwartz RH. Inhibition of cell cycle progression by rapamycin induces T cell clonal anergy even in the presence of costimulation. Journal of immunology. 1999;162:2775–2784. [PubMed] [Google Scholar]
- 50.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annual review of immunology. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salmond RJ, Emery J, Okkenhaug K, Zamoyska R. MAPK, phosphatidylinositol 3-kinase, and mammalian target of rapamycin pathways converge at the level of ribosomal protein S6 phosphorylation to control metabolic signaling in CD8 T cells. Journal of immunology. 2009;183:7388–7397. doi: 10.4049/jimmunol.0902294. [DOI] [PubMed] [Google Scholar]
- 52.Procaccini C, De Rosa V, Galgani M, Carbone F, Cassano S, Greco D, Qian K, Auvinen P, Cali G, Stallone G, et al. Leptin-induced mTOR activation defines a specific molecular and transcriptional signature controlling CD4+ effector T cell responses. Journal of immunology. 2012;189:2941–2953. doi: 10.4049/jimmunol.1200935. [DOI] [PubMed] [Google Scholar]
- 53.Gorentla BK, Wan CK, Zhong XP. Negative regulation of mTOR activation by diacylglycerol kinases. Blood. 2011;117:4022–4031. doi: 10.1182/blood-2010-08-300731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. The Journal of clinical investigation. 2008;118:3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rathmell JC, Farkash EA, Gao W, Thompson CB. IL-7 enhances the survival and maintains the size of naive T cells. Journal of immunology. 2001;167:6869–6876. doi: 10.4049/jimmunol.167.12.6869. [DOI] [PubMed] [Google Scholar]
- 56.Dure M, Macian F. IL-2 signaling prevents T cell anergy by inhibiting the expression of anergy-inducing genes. Mol Immunol. 2009;46:999–1006. doi: 10.1016/j.molimm.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **57.Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, Cantrell DA. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nature immunology. 2013;14:500–508. doi: 10.1038/ni.2556. The authors identified System L transporter, SLC7A5, as the crucial mediator of amino acid uptake and mTOR activation during T cell activation, thereby promoting metabolic reprograming and adaptive immune responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gulen MF, Kang Z, Bulek K, Youzhong W, Kim TW, Chen Y, Altuntas CZ, Sass Bak-Jensen K, McGeachy MJ, Do JS, et al. The receptor SIGIRR suppresses Th17 cell proliferation via inhibition of the interleukin-1 receptor pathway and mTOR kinase activation. Immunity. 2010;32:54–66. doi: 10.1016/j.immuni.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang J, Burkett PR, Borges CM, Kuchroo VK, Turka LA, Chang CH. MyD88 is essential to sustain mTOR activation necessary to promote T helper 17 cell proliferation by linking IL-1 and IL-23 signaling. Proceedings of the National Academy of Sciences of the United States of America. 2013 doi: 10.1073/pnas.1206048110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomasoni R, Basso V, Pilipow K, Sitia G, Saccani S, Agresti A, Mietton F, Natoli G, Colombetti S, Mondino A. Rapamycin-sensitive signals control TCR/CD28-driven Ifng, Il4 and Foxp3 transcription and promoter region methylation. European journal of immunology. 2011;41:2086–2096. doi: 10.1002/eji.201041130. [DOI] [PubMed] [Google Scholar]
- 61.Rohde JR, Cardenas ME. The tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol Cell Biol. 2003;23:629–635. doi: 10.1128/MCB.23.2.629-635.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell. 2008;134:392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spiegel S, Milstien S. The outs and the ins of sphingosine-1-phosphate in immunity. Nature reviews Immunology. 2011;11:403–415. doi: 10.1038/nri2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu G, Yang K, Burns S, Shrestha S, Chi H. The S1P(1)-mTOR axis directs the reciprocal differentiation of T(H)1 and T(reg) cells. Nature immunology. 2010;11:1047–1056. doi: 10.1038/ni.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao YG, Wang Y, Guo Z, Gu AD, Dan HC, Baldwin AS, Hao W, Wan YY. Dihydroartemisinin Ameliorates Inflammatory Disease by Its Reciprocal Effects on Th and Regulatory T Cell Function via Modulating the Mammalian Target of Rapamycin Pathway. Journal of immunology. 2012;189:4417–4425. doi: 10.4049/jimmunol.1200919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thomson AW, Turnquist HR, Raimondi G. Immunoregulatory functions of mTOR inhibition. Nature reviews Immunology. 2009;9:324–337. doi: 10.1038/nri2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strainic MG, Shevach EM, An F, Lin F, Medof ME. Absence of signaling into CD4(+) cells via C3aR and C5aR enables autoinductive TGF-beta1 signaling and induction of Foxp3(+) regulatory T cells. Nature immunology. 2012;14:162–171. doi: 10.1038/ni.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Geng D, Zheng L, Srivastava R, Asprodites N, Velasco-Gonzalez C, Davila E. When Toll-like receptor and T-cell receptor signals collide: a mechanism for enhanced CD8 T-cell effector function. Blood. 2010;116:3494–3504. doi: 10.1182/blood-2010-02-268169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Rosa V, Procaccini C, Cali G, Pirozzi G, Fontana S, Zappacosta S, La Cava A, Matarese G. A key role of leptin in the control of regulatory T cell proliferation. Immunity. 2007;26:241–255. doi: 10.1016/j.immuni.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 70.Procaccini C, De Rosa V, Galgani M, Abanni L, Cali G, Porcellini A, Carbone F, Fontana S, Horvath TL, La Cava A, et al. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity. 2010;33:929–941. doi: 10.1016/j.immuni.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lord GM, Matarese G, Howard JK, Baker RJ, Bloom SR, Lechler RI. Leptin modulates the T-cell immune response and reverses starvation-induced immunosuppression. Nature. 1998;394:897–901. doi: 10.1038/29795. [DOI] [PubMed] [Google Scholar]
- 72.Galgani M, Procaccini C, De Rosa V, Carbone F, Chieffi P, La Cava A, Matarese G. Leptin modulates the survival of autoreactive CD4+ T cells through the nutrient/energy-sensing mammalian target of rapamycin signaling pathway. Journal of immunology. 2010;185:7474–7479. doi: 10.4049/jimmunol.1001674. [DOI] [PubMed] [Google Scholar]
- 73.Yalcin S, Marinkovic D, Mungamuri SK, Zhang X, Tong W, Sellers R, Ghaffari S. ROS-mediated amplification of AKT/mTOR signalling pathway leads to myeloproliferative syndrome in Foxo3(−/−) mice. EMBO J. 2010;29:4118–4131. doi: 10.1038/emboj.2010.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cobbold SP, Adams E, Farquhar CA, Nolan KF, Howie D, Lui KO, Fairchild PJ, Mellor AL, Ron D, Waldmann H. Infectious tolerance via the consumption of essential amino acids and mTOR signaling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12055–12060. doi: 10.1073/pnas.0903919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmidt EV, Ravitz MJ, Chen L, Lynch M. Growth controls connect: interactions between c-myc and the tuberous sclerosis complex-mTOR pathway. Cell Cycle. 2009;8:1344–1351. doi: 10.4161/cc.8.9.8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakamura H, Makino Y, Okamoto K, Poellinger L, Ohnuma K, Morimoto C, Tanaka H. TCR engagement increases hypoxia-inducible factor-1 alpha protein synthesis via rapamycin-sensitive pathway under hypoxic conditions in human peripheral T cells. Journal of immunology. 2005;174:7592–7599. doi: 10.4049/jimmunol.174.12.7592. [DOI] [PubMed] [Google Scholar]
- 78.Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen HR, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bensinger SJ, Bradley MN, Joseph SB, Zelcer N, Janssen EM, Hausner MA, Shih R, Parks JS, Edwards PA, Jamieson BD, et al. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 2008;134:97–111. doi: 10.1016/j.cell.2008.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *80.Kidani Y, Elsaesser H, Hock MB, Vergnes L, Williams KJ, Argus JP, Marbois BN, Komisopoulou E, Wilson EB, Osborne TF, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nature immunology. 2013;14:489–499. doi: 10.1038/ni.2570. This study showed that PI3K-mTOR pathway is required for T cell activation-induced SREBP processing and lipogenic gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Michalek RD, Gerriets VA, Nichols AG, Inoue M, Kazmin D, Chang CY, Dwyer MA, Nelson ER, Pollizzi KN, Ilkayeva O, et al. Estrogen-related receptor-alpha is a metabolic regulator of effector T-cell activation and differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18348–18353. doi: 10.1073/pnas.1108856108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Metallo CM, Vander Heiden MG. Metabolism strikes back: metabolic flux regulates cell signaling. Genes & development. 2010;24:2717–2722. doi: 10.1101/gad.2010510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13:270–276. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- 84.Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW, Rabinowitz JD, Coller HA, Thompson CB. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes & development. 2010;24:2784–2799. doi: 10.1101/gad.1985910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dunn SE, Youssef S, Goldstein MJ, Prod’homme T, Weber MS, Zamvil SS, Steinman L. Isoprenoids determine Th1/Th2 fate in pathogenic T cells, providing a mechanism of modulation of autoimmunity by atorvastatin. The Journal of experimental medicine. 2006;203:401–412. doi: 10.1084/jem.20051129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kagami S, Owada T, Kanari H, Saito Y, Suto A, Ikeda K, Hirose K, Watanabe N, Iwamoto I, Nakajima H. Protein geranylgeranylation regulates the balance between Th17 cells and Foxp3+ regulatory T cells. International immunology. 2009;21:679–689. doi: 10.1093/intimm/dxp037. [DOI] [PubMed] [Google Scholar]
- 87.Ladygina N, Martin BR, Altman A. Dynamic palmitoylation and the role of DHHC proteins in T cell activation and anergy. Advances in immunology. 2011;109:1–44. doi: 10.1016/B978-0-12-387664-5.00001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]