Summary
The GroEL-GroES chaperonin system is required for the assisted folding of many essential proteins. The precise nature of this assistance remains unclear, however. Here we show that denatured RuBisCO from Rhodospirillum rubrum populates a stable, nonaggre-gating, and kinetically trapped monomeric state at low temperature. Productive folding of this nonnative intermediate is fully dependent on GroEL, GroES, and ATP. Reactivation of the trapped RuBisCO monomer proceeds through a series of GroEL-induced structural rearrangements, as judged by resonance energy transfer measurements between the amino- and carboxy-terminal domains of RuBisCO. A general mechanism used by GroEL to push large, recalcitrant proteins like RuBisCO toward their native states thus appears to involve two steps: partial unfolding or rearrangement of a nonnative protein upon capture by a GroEL ring, followed by spatial constriction within the GroEL-GroES cavity that favors or enforces compact, folding-competent intermediate states.
Introduction
Many cellular proteins are incapable of folding efficiently on their own (Fayet et al., 1989; Horwich et al., 1993; Houry et al., 1999). The folding landscapes of these proteins seem to be replete with readily populated, non-native states that are highly prone to irreversible aggregation (Dobson, 2003). Aggregation of a critical protein not only deprives a cell of an essential, functioning molecule, but the aggregation process itself, or its tendency to lead to the formation of amyloid fibrils, appears to be the basis of a growing list of human diseases (Dobson, 2001; Horwich, 2002; Koo et al., 1999; Prusiner et al., 1998). One of the essential roles played by molecular chaperones in cellular biology is the prevention or correction of mistakes in folding that lead to the formation of aggregates (Fenton and Horwich, 2003; Hartl and Hayer-Hartl, 2002).
Among the molecular chaperones, the chaperonins (or Hsp60s) deal with error-prone folding in a unique way, capturing and confining nonnative proteins within an enclosed and protected cavity. The GroEL/GroES proteins of Escherichia coli are perhaps the best-studied members of the chaperonin family (Fenton and Horwich, 2003; Hartl and Hayer-Hartl, 2002). GroEL is a large, double-ring oligomer composed of 14 identical 57 kDa subunits arranged in two seven-membered rings, with each ring possessing a large central cavity (Braig et al., 1994). GroES is a smaller ring-shaped oligomer that binds to GroEL, in an ATP-dependent manner, and caps the GroEL cavity (Xu et al., 1997). While years of structural and mechanistic analysis have provided a reasonably detailed picture of the GroEL reaction cycle (Fenton and Horwich, 2003; Sigler et al., 1998), no clear understanding yet exists of how GroEL drives a protein folding reaction that cannot proceed spontaneously.
A more complete picture of GroEL function requires the resolution of the following issue: does GroEL function as a passive aggregation inhibitor or does GroEL actively participate in protein folding, either by unfolding trapped substrate protein or by some other direct action (Wang and Weissman, 1999)? The difficulty in addressing this question stems from the fact that most GroEL-dependent substrates are highly prone to aggregation, and GroEL most certainly functions, at least in part, to prevent irretrievable loss of proteins by aggregation. Whether GroEL additionally acts more directly during a protein folding reaction rests substantially with two questions. First, in the absence of GroEL, how well does a GroEL-dependent substrate fold under conditions where aggregation can be formally excluded? Second, does GroEL modify the conformational states accessible to a folding intermediate at any point along the trajectory of the chaperonin cycle?
Several studies have suggested that GroEL can alter the structure of nonnative proteins (Bhutani and Udgaonkar, 2000; Clark and Frieden, 1999; Gervasoni et al., 1998; Shtilerman et al., 1999; Walter et al., 1996; Zahn et al., 1994, 1996; Zahn and Pluckthun, 1994), though the direct linkage of this action to facilitated folding of strictly dependent GroEL substrate proteins has not been established. However, other studies have shown that some GroEL-dependent substrates can fold in the absence of the chaperonin if aggregation is suppressed through carefully selected solution conditions (Brinker et al., 2001; Schmidt et al., 1994; van der Vies et al., 1992). In most of these cases, though, the presence of inhibitory, low-order aggregates (Ranson et al., 1995; Silow and Oliveberg, 1997) could not be excluded. Significantly, in one case, confinement of a highly GroEL-dependent substrate protein within the GroEL-GroES cavity was found to stimulate folding beyond the observed spontaneous rate (Brinker et al., 2001). These and previous observations (Weissman et al., 1996), as well as several theoretical studies (Baumketner et al., 2003; Takagi et al., 2003), have led to the suggestion that confinement of a protein folding intermediate within the enclosed GroEL-GroES cavity might directly alter the folding landscape of a protein.
To address these questions we have examined the folding of the highly GroEL-dependent protein ribulose-1,5-bisphosphate carboxylase oxygenase (RuBisCO) from Rhodospirillum rubrum. Here we establish solution conditions where RuBisCO is incapable of significant folding and demonstrate that it is stuck in a monomeric, nonnative, nonaggregating, and kinetically trapped state. Reactivation of the trapped RuBisCO intermediate requires GroEL, GroES, and ATP and proceeds through two phases of GroEL-induced structural rearrangement, one upon substrate protein binding to a GroEL ring and the second upon enclosure of the substrate within the GroEL-GroES complex.
Results
A Kinetically Trapped State of RuBisCO Requires GroEL, GroES, and ATP for Productive Folding
The CO2-fixing enzyme RuBisCO from R. rubrum, a model substrate for the GroEL/ES folding reaction, is one of the most strictly GroEL-dependent proteins currently known (Goloubinoff et al., 1989). Native RuBisCO is a dimer of 52 kDa monomers that are highly prone to aggregation upon denaturation (Todd et al., 1994; van der Vies et al., 1992). Even at very low protein concentrations, the extensive and rapid aggregation of RuBisCO can be observed as an explosive increase in light scattering at 25°C when denatured RuBisCO is diluted away from the denaturant. Under these conditions, no reactivation of the RuBisCO enzyme is detectable (Figure 1). However, previous studies have found that certain solution conditions can coax RuBisCO to fold, generally slowly and inefficiently, in the absence of GroEL (e.g., low protein concentration, low temperature, high chloride ion concentrations [Schmidt et al., 1994; van der Vies et al., 1992]). We find that high-order aggregation of 100 nM RuBisCO is completely inhibited at 4°C in low ionic strength buffer (containing no chloride ion), as demonstrated by the elimination of detectable light scattering (Figure 1). However, we detect no refolding of RuBisCO to its enzymatically active native state under these conditions, even after several hours of incubation at 4°C. In the presence of GroEL, GroES, and ATP, however, RuBisCO is rapidly and efficiently refolded, even at 4°C (Figure 1).
Figure 1. Low Temperature Suppresses High-Order Aggregation, but Not GroEL-Mediated Refolding, of Denatured RuBisCO.
(A) Static light scattering of different RuBisCO samples was monitored at 90° and 340 nm. In all cases, the final protein concentration was 100 nM RuBisCO monomer. The arrow indicates the addition of either acid-urea-denatured RuBisCO (“dRub”) or native RuBisCO to standard refolding buffer equilibrated at the indicated temperature.
(B) Spontaneous and GroEL-mediated refolding of RuBisCO at 4°C and 25°C. In all cases, 100 nM acid-urea-denatured RuBisCO was diluted either directly into refolding buffer (“spontaneous”) or into buffer containing 200 nM GroEL (“EL+ES+ATP”). Samples of denatured RuBisCO were also diluted into refolding buffer alone and incubated at the indicated temperature for 10 min, prior to the addition of GroEL (“10 min delay, EL+ES+ATP”). For GroEL-mediated refolding, samples were then supplemented with 400 nM GroES and 1 mM ATP to initiate folding.
The Kinetically Trapped RuBisCO State Is Monomeric
The measurements shown in Figure 1 eliminate high-order, irreversible aggregation as the cause of the RuBisCO monomer’s inability to fold efficiently at 4°C in low ionic strength buffer. However, these measurements do not directly establish the extent to which low-order aggregates, groupings of only a few monomers in a nonnative agglomeration, are populated under these conditions. Because low-order, reversible aggregates are known to slow or block productive protein folding (Silow and Oliveberg, 1997), we sought to determine whether RuBisCO forms low-order, reversible aggregates under solution conditions where neither high-order aggregation nor refolding can be detected.
Previous studies with high-resolution HPLC suggested that, under certain conditions, RuBisCO can populate a stable, nonnative monomeric state (Luo et al., 2001). However, since chromatographic separation might induce nonequilibrium perturbations in a population of unstable, oligomeric states, we turned to fluorescence resonance energy transfer (FRET [Lakowicz, 1999; Van Der Meer et al., 1994]) to probe the extent of RuBisCO aggregation in a homogeneous system. By fluorescently labeling two different populations of RuBisCO, denaturing them together, and then diluting away the denaturant under different conditions, the extent to which the two monomer populations interact with one another could be monitored by FRET. To accomplish this goal, we exploited our previous observations that RuBisCO from R. rubrum can be derivatized with fluorescent dyes without perturbing the enzyme’s stability, activity, or GroEL-dependent refolding (Rye et al., 1997, 1999).
The structure of the RuBisCO dimer (Schneider et al., 1990) is shown in Figure 2, along with the positions that were utilized to attach fluorescent probes through Cys side chain alkylation. Position 58 is a naturally occurring Cys residue and is the only intrinsic RuBisCO Cys residue that is surface exposed and reactive in the native enzyme. An Ala at position 454 was chosen at the opposite end of the monomer as an additional site of modification. Two different RuBisCO samples were expressed and purified: Rub58C, wild-type RuBisCO with a Cys at position 58, and Rub454C, carrying both A454C and C58A mutations. These two RuBisCO variants, each with a single surface-exposed Cys residue, were labeled with different fluorescent dyes (AEDANS as a FRET donor and fluorescein as an acceptor). Dye conjugation was efficient and specific to the Cys sites targeted (see Experimental Procedures). Neither mutation nor fluorescent probe attachment had any significant effect on RuBisCO activity or GroEL-dependent refolding (Figure 2).
Figure 2. GroEL Efficiently Refolds Fluorescent Derivatives of RuBisCO.
(A) Ribbon diagram of the R. rubrum RuBisCO structure (9RUB [Schneider et al., 1990]) derivatized with AEDANS (a FRET donor) and fluorescein (“F”; a FRET acceptor). The fluorescent dyes were modeled into the RuBisCO structure using PyMOL (DeLano Scientific), ChemDraw, and Chem3D (CambridgeSoft). Dye orientations are for reference only and have not been energy minimized.
(B) Refolding of fluorescent RuBisCO variants requires GroEL and GroES and is comparable to the refolding of wild-type, unmodified RuBisCO. Acid-urea-denatured samples of each protein (100 nM final) were mixed with 150 nM GroEL, and folding was then initiated by adding 300 nM GroES and 2 mM ATP.
When two different samples of RuBisCO, one carrying a AEDANS donor and the other carrying a fluorescein acceptor (R0 = 44–48 Å; see Table 1), are mixed together, denatured, and then diluted into buffer (without denaturant) at 25°C, nearly complete quenching of the AEDANS donor fluorescence is observed (Figure 3A), showing extensive aggregate formation under these conditions and consistent with the light scattering data in Figure 1. At 4°C, however, FRET between the two RuBisCO samples is highly dependent on the total monomer concentration. At very low protein concentration (≤100 nM), no significant FRET is detectable when the two denatured RuBisCO monomers are mixed at 4°C (Figure 3A). This steady-state measurement was confirmed by time-domain measurements of the AEDANS intensity decay (see Experimental Procedures). At higher protein concentrations (>150 nM), however, significant FRET is detectable (Figure 3B). We further carried out measurements on several combinations of labeled sites over a range of protein concentrations, using both single acceptor-labeled RuBisCO (e.g., 58-Donor + 58-Acceptor, 58-Donor + 454-Acceptor, 454-Donor + 454-Acceptor; Figure 3B) and double acceptor-labeled RuBisCO (454-Donor + 58-Acceptor/356-Acceptor; see Supplemental Figure S1 at http://www.molecule.org/cgi/content/full/16/1/23/DC1/). In all cases, no significant FRET was detectable when the total RuBisCO concentration was ≤100 nM.
Table 1.
Steady-State and Time-Resolved FRET Distances
| Sample | R0 (Å ) | Steady-State FRET Distance (Å) | Time-Resolved FRET Distance (Å) | Crystal Structure Distance;a Cα → Cα (Å ) |
|---|---|---|---|---|
| Rub58A/454D (native) | 43.8 | 39.8 | 39.0 | 35 |
| dRub58A/454D (4° int) | 43.7 | 39.0 | 37.0 | – |
| EL-Rub58A/454D | 45.2 | 68.3 | ≥70 | – |
| SR1-Rub58A/454D | 47.8 | 46.7 | 46.1 | – |
This distance is measured across the dimer interface, between positions 58 and 454 on opposite subunits (see Figure 2). The intramonomer distance is outside the FRET distance for these positions and this pair of dyes.
Figure 3. The Kinetically Trapped State Populated by RuBisCO at Low Protein Concentration and Low Temperature Is Not a Low-Order Aggregate.
(A) The extent of monomer-monomer contact upon formation of low-order RuBisCO aggregates was monitored by donor-side, steady-state FRET. Samples of unlabeled, donor-only (AEDANS; “58-D”)- and acceptor-only (fluorescein; “58-A”)-labeled RuBisCO were denatured in acid-urea and then mixed together in 1:1 ratios, as indicated. These concentrated, denatured samples were then diluted to 100 nM (final protein concentration) in standard refolding buffer at the indicated temperature.
(B) The dependence of low-order aggregate formation on the total RuBisCO monomer concentration at 4°C is shown. The extent of FRET for each pair of donor- and acceptor-labeled monomers at each final protein concentration was determined by both steady-state (closed symbols) and time-resolved fluorescence measurements (open symbols). Three different pairs of labeled proteins were examined: (triangles) RubC58A/A454C-AEDANS (“454-D”) mixed with RubC58A/A454C-F (“454-A”); (circles) RubC58-AEDANS (“58-D”) mixed with RubC58-F (“58-A”); (squares) RubC58-AEDANS (“58-D”) RubC58A/A454C-F (“454-A”).
We conclude that under these solution conditions, the RuBisCO monomer populates a nonnative conformation (or ensemble of conformations) that is kinetically trapped. This is consistent with earlier observations that RuBisCO folding involves an unstable, partially folded, and aggregation-prone intermediate state (van der Vies et al., 1992). More significantly, however, our analysis demonstrates that this kinetically trapped state is not due to aggregation and that its reversal must involve a direct action of GroEL on the individual RuBisCO monomer structure. This observation raises at least two important questions. First, is the trapped RuBisCO monomer misfolded? Second, how does GroEL reverse this conformational trap and place the protein back on a productive folding manifold?
The Trapped RuBisCO Monomer Is Expanded and Misfolded
In order to characterize the conformational properties of the trapped RuBisCO monomer, we first examined the apparent hydrodynamic volume of this intermediate state by determining its average rotational correlation time. This was accomplished by measuring the time-resolved fluorescence anisotropy decay of a pyrene-labeled RuBisCO sample. The exceptionally long fluorescence lifetime of pyrene (∼100 ns) provides an ideal probe of the rotational correlation times expected for a protein the size of R. rubrum RuBisCO. The measured rotation correlation time for the pyrene-labeled, native RuBisCO dimer at 4°C was found to be 123 ± 6 ns (data not shown), which is consistent with a native, hydrated protein the size (104 kDa) and shape (asymmetric prolate ellipsoid) of a RuBisCO dimer.
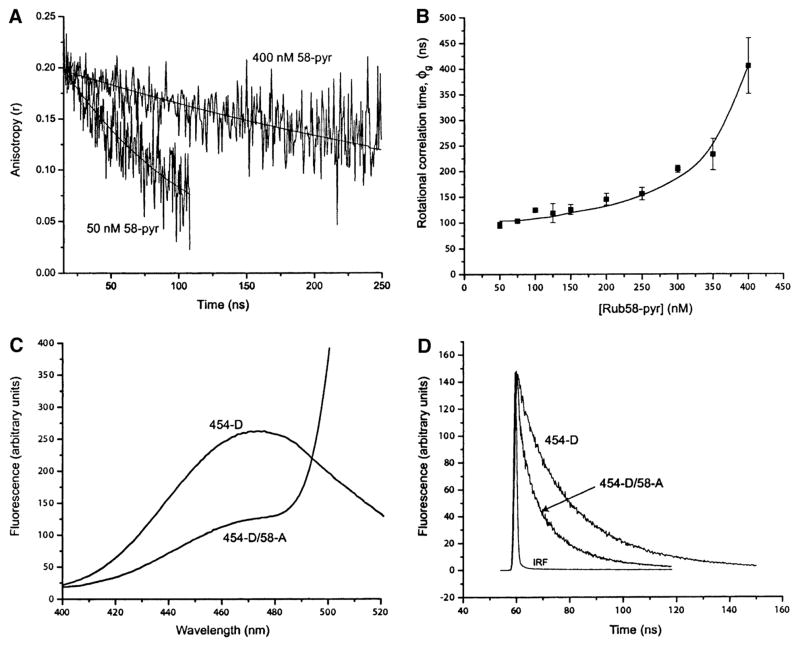
Figure 4A shows a plot of the anisotropy decay of nonnative RuBisCO-pyrene at monomer concentrations of 50 and 400 nM. The apparent rotational correlation times extracted from a series of similar experiments at different protein concentrations are shown in Figure 4B. In all cases, the observed anisotropy decays were well described by a rotational diffusion model with a single correlation time, which represents an average value for the protein states populated under each condition. The measured rotational correlation time increases dramatically as the RuBisCO concentration is raised above 150 nM, suggesting that the measured apparent rotational correlation time becomes increasingly dominated by the formation of low-order aggregates. However, at concentrations below 150 nM, the rotational correlation time remains nearly constant, confirming our observations from the FRET assay that the critical concentration for the onset of low-order aggregate formation is between 150 and 250 nM under these conditions. While only an average picture of the kinetically trapped state, the limiting value of the rotational correlation time from these measurements (∼100 ns) is consistent with an asymmetric, nonnative, and significantly expanded RuBisCO monomer.
Figure 4. The Low-Temperature Kinetically Trapped RuBisCO Monomer Is Expanded and Displays a Shortened, Nonnative Distance between the N- and C-Terminal Domains.
Samples of pyrene-labeled RuBisCO were denatured in acid-urea and then diluted into refolding buffer at 4°C. The anisotropy decays for 50 and 400 nM pyrene-labeled RuBisCO (“58-pyr”) are shown in (A). The global fit of this data to a rotational diffusion model containing a single rotational decay term and two lifetime components is shown. The rotational correlation times associated with overall rotational diffusion (ϕg) were 92 ± 14 ns at 50 nM and 438 ± 52 ns at 400 nM. (B) Measurement of the global rotational correlation time of the denatured, pyrene-labeled RuBisCO monomer at 4°C as a function of total protein concentration. The curve shown is present for illustration purposes only. (C and D) FRET measurements of the kinetically trapped RuBisCO monomer at 4°C. A sample of the donor-only (RubC58A/A454C-AEDANS; “454-D”)- and donor-acceptor (RubC58-F/A454C-AEDANS; “454-D/58-A”)-labeled RuBisCO were denatured in acid-urea and diluted into refolding buffer at 4°C to a final protein concentration of 100 nM. Both steady-state (C) and time-resolved (D) measurements were taken on each sample. The instrument response function (“IRF”) for the lifetime measurement is shown in (D).
We next extended our FRET strategy to directly examine the conformation of the trapped RuBisCO monomer. This approach involved the creation of RuBisCO monomers carrying two fluorescent probes. We first introduced the A454C mutation into the wild-type RuBisCO monomer to generate a variant of the protein with two surface-exposed Cys residues (58C and A454C). We then developed a sequential labeling strategy in which a specific fluorescent probe was attached to either the N- or C-terminal region of the protein (see Experimental Procedures). As with the single labeled RuBisCO variants described above, double labeling had no apparent effect on enzymatic activity, stability, or GroEL-dependent refolding of the protein (Figure 2B).
When the doubly labeled RuBisCO was denatured and diluted into buffer at 4°C, we observed a substantial decrease in the steady-state fluorescence intensity of the donor, a decrease in the donor fluorescence lifetime (Figures 4C and 4D), and a concomitant increase in the steady-state fluorescence intensity of the acceptor (data not shown). These measurements demonstrate that substantial energy transfer takes place between the labeled domains of RuBisCO and give an energy transfer efficiency (E) of 0.66–0.73. This yields an average distance of 37–39 Å between positions 58 and 454 in the kinetically trapped RuBisCO monomer (Table 1). The distance between these positions is approximately 85 Å (Cα to Cα) in the native monomer (Figure 2A). Thus, in the kinetically trapped state, the ends of the RuBisCO monomer appear to be collapsed, with the N- and C-terminal domains much closer together on average than in the native state. The nonnative distance between the N-terminal and C-terminal domains of the RuBisCO monomer strongly suggests that the low-temperature, kinetically trapped RuBisCO monomer is misfolded. Further, how GroEL avoids this conformational trap could now be directly examined with a stringent substrate by following changes in the RuBisCO N- to C-terminal distance during a GroEL reaction cycle.
Binding to Both GroEL and SR1 Causes Large-Scale Stretching of the Misfolded RuBisCO Monomer
We first determined whether simple binding of RuBisCO to a GroEL ring could induce large-scale changes in the conformation of the RuBisCO monomer. This was accomplished by monitoring the energy transfer efficiency of the doubly labeled RuBisCO monomer upon binding to both wild-type, double-ring GroEL and a single ring variant of GroEL (SR1 [Weissman et al., 1995]). A significant decrease in the FRET efficiency is reported upon mixing the kinetically trapped RuBisCO monomer with either GroEL or SR1 (Figure 5). While the overall change in the FRET efficiencies is biphasic, the rate of the fast component was found to be directly proportional to the concentration of GroEL rings present (Figure 5B), yielding an apparent bimolecular rate constant (kbind) of 2–4 × 105 M−1 s−1. This binding rate is, however, considerably slower than previous measurements of acid-denatured RuBisCO binding to GroEL at 25°C (kbind ∼2–5 × 107 M−1 s−1 [Rye et al., 1999]). Because both the temperature and the initial RuBisCO conformational state are significantly different in the experiments described here, we conducted a series of control experiments using donor-labeled RuBisCO and acceptor-labeled GroEL to directly measure the rate of RuBisCO binding to GroEL by FRET (Rye et al., 1999). Under identical, low-temperature conditions, direct FRET measurement of the kinetically trapped RuBisCO monomer binding to GroEL yielded a kbind of 2 × 105 M−1 s−1 (Supplemental Figure S2). This value is in excellent agreement with the indirect measurement of the binding rate from changes in the FRET efficiency of the doubly labeled RuBisCO. These observations strongly suggest that the trapped RuBisCO monomer is subjected to a conformational expansion upon binding to a GroEL ring.
Figure 5. Direct Binding of the Trapped RuBisCO Intermediate to a GroEL Ring Results in a Stretching of the N- to C-Terminal Domain Distance.
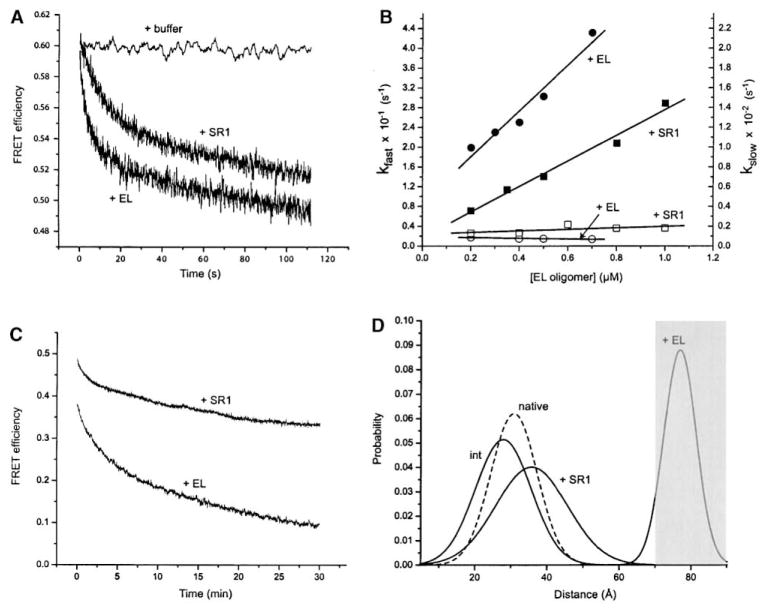
(A) Binding of the kinetically trapped RuBisCO monomer to both wild-type GroEL and SR1 induces a significant decrease in the FRET efficiency of the labeled monomer. Samples of labeled RuBisCO were denatured in acid-urea, diluted into refolding buffer at 4°C (100 nM), and then loaded into a stopped-flow device thermally jacketed at 4°C. The kinetically trapped monomer samples were rapidly mixed 1:1 with either buffer only (“+ buffer”) or with wild-type GroEL (“+ EL”; 400 nM final concentration) or SR1 (“+ SR1”; 400 nM). The observed change in the FRET efficiency upon binding to both wild-type GroEL and SR1 required two exponential fitting components (kfast and kslow).
(B) The rate of the fast change in the FRET signal (kfast) is directly proportional to the concentration of GroEL rings present for both wild-type GroEL (closed circles) and SR1 (closed squares).
(C) Example of a manual mixing experiment, with GroEL and SR1 present at molar ratios of 4:1 relative to RuBisCO. The rate of the slower FRET change for both SR1 and GroEL requires two exponential fitting components (kslow,1 = 0.0094 ± 0.0005 s−1 and kslow,2 = 0.0008 ± 0.0001 s−1 for GroEL; kslow,1 = 0.015 ± 0.0035 s−1 and kslow,2 = 0.0016 ± 0.0004 s−1 for SR1), neither of which was dependent on the chaperonin concentration. The dominant component of the slow phase (∼95%) is shown at different GroEL ([B]; open circles) or SR1 ([B]; open squares) concentrations.
(D) The intraprobe distance distribution between the N- and C-terminal domains of RuBisCO expands upon binding to SR1. The apparent distance distributions consistent with time-resolved FRET measurements of the native RuBisCO dimer (“native”), the kinetically trapped intermediate (“int”), and the SR1 (“+ SR1”) and wild-type GroEL (“+ EL”) binary complexes are shown. The apparent distributions were obtained by global fitting of the time-resolved FRET data to a distributed distance model (see Experimental Procedures). Note that the distance distribution measured for the native RuBisCO dimer is measured across the dimer interface between the sites on different monomers, while the other distributions are measured for intrasite distances within individual monomers. The apparent distribution recovered for the GroEL binary complex is outside the reliable distance range measurable with the AEDANS-fluorescein energy transfer pair, as indicated by the shaded area.
The rate of the slower FRET change for both SR1 and wild-type GroEL is much slower than the rate of RuBisCO binding (Figures 5B and 5C). This change is not dependent on GroEL concentration (Figure 5B) and most likely represents additional expansion of the Ru-BisCO intermediate following the initial binding event. Note that the end-point of the structural change for SR1 and wild-type GroEL are not the same, with GroEL capable of inducing a very large distance change in the bound intermediate over 30 min. To more carefully examine the extent of structural rearrangement, a set of steady-state and time-resolved FRET measurements were made on each binary complex (see Table 1 and Supplemental Figure S3). Upon binding to both wild-type GroEL and SR1, the FRET efficiency dropped dramatically, suggesting a significant expansion of the distance between the ends of the nonnative protein. The final apparent distance between the N- and C-terminal domains of the bound RuBisCO monomer was 46–47 Å for SR1 and ≥70 Å for wild-type GroEL at both 4°C and 25°C.
While the distance measurements outlined above provide a clear picture of the average conformational behavior of the RuBisCO intermediate along one principal axis, the nonnative RuBisCO monomer is likely to populate an ensemble of conformational states around that average. Time-resolved FRET measurements can, in principal, provide an estimate of this conformational heterogeneity (Haas et al., 1978, 1988; Navon et al., 2001). We therefore globally fit time-resolved FRET data from each RuBisCO sample to a distributed distance model (Figure 5D). The apparent distribution for the native RuBisCO dimer demonstrated a half-width of ∼7 Å. This value is consistent with both the length of the fluorescent probe linkers and with previous measurements of intra-site distance distributions in another native protein made with this same FRET pair (Lillo et al., 1997). Remarkably, the apparent distribution of the kinetically trapped state, while showing a decrease in the mean distance as expected, was only slightly wider than that observed for the native protein. Upon binding to SR1, the distance distribution was found to expand in width and shift to a larger mean value. The distance distribution observed with wild-type GroEL, however, displayed an even greater mean value, consistent with the average distance measurements noted above. The apparent distribution width observed with wild-type GroEL, while appearing narrower, is substantially outside of the reliable distance range accessible with the AEDANS-fluorescein FRET pair (the maximum measurable distance is given by R0 + 1/2 R0 ≈ 70 Å) and is probably not a reliable measure of the conformational heterogeneity of this state.
Binding of ATP and GroES to an SR1-RuBisCO Binary Complex Induces a Compaction of the Encapsulated Monomer
In addition to structural rearrangements induced by direct binding, further modification of the intermediate structure might be imposed by the ATP-dependent binding of GroES to form the enclosed GroEL-ATP-GroES complex (the so-called cis ternary complex [Shtilerman et al., 1999]). In order to examine this possibility, we monitored the energy transfer efficiency of the doubly labeled RuBisCO monomer upon the rapid addition of GroES and ATP to an SR1-RuBisCO binary complex (Figure 6A). Strikingly, we observed a significant increase in the transfer efficiency. This change in the transfer efficiency strongly suggests that binding of GroES to the RuBisCO-SR1 binary complex induces a significant and transient compaction of the bound folding intermediate. Following this collapse, the distance between the N-and C-terminal domains slowly increases on the inside of the GroEL-GroES cavity with a time constant (t1/2 = 3 min) that matches the rate-limiting, GroEL-dependent folding rate of the RuBisCO monomer inside the SR1-GroES complex (Rye et al., 1997). As a control, complementary experiments were conducted with wild-type GroEL (data not shown). While the formation of both cis and trans substrate complexes make quantitative interpretations of experiments with double-ring GroEL difficult, the qualitative changes in FRET efficiency were very similar to those observed in Figure 6A.
Figure 6. Binding of GroES over the Top of a GroEL Bound RuBisCO Monomer Induces a Significant and Transient Compaction of the Distance between the N- and C-Terminal Domains.
A binary complex between SR1 (400 nM) and either donor-only- or donor-acceptor-labeled RuBisCO (100 nM) was prepared and loaded into a stopped-flow device thermally jacketed at 25°C. The binary complex was then rapidly mixed (1:1) with either buffer only, ATP only (2 mM; “ATP”), or ATP and GroES (2 mM ATP and 800 nM GroES; “ATP + GroES”). Using both EDANS-fluorescein (A) and fluorescein-rhodamine (B) as the FRET pair, a significant (∼4–5 Å) and rapid (t1/2 = 0.5 s) increase in FRET is observed as ATP and GroES bind and encapsulate the SR1-captured RuBisCO. Subsequently, the FRET signal shows a slow decrease (t1/2 = 175 s). The inset is an expansion of the first 20 s of each data set. The preshot before the arrow shows the FRET efficiency of the SR1-RuBisCO binary complex mixed only with buffer. The data after the arrow are the result of mixing either ATP only or ATP and GroES with the SR1 binary complex.
To confirm these observations and eliminate other trivial photophysical explanations, we conducted a series of additional control experiments. To eliminate any effect of temperature on the measured RuBisCO compression, experiments were conducted with SR1-RuBisCO complexes at 4°C. These experiments showed that assembly of the SR1-ATP-ES complex at 4°C resulted in the same extent of RuBisCO compaction as at 25°C (data not shown). Second, we repeated the SR1 rapid mixing experiment with a different pair of energy transfer dyes (Figure 6B). In this case, we utilized fluorescein as the donor and rhodamine as the acceptor. As with the AEDANS-fluorescein pair, we see a dramatic increase in the FRET efficiency upon GroES binding to the SR1-RuBisCO binary complex. Because a dramatic change in fluorescent probe mobility upon GroES binding could contribute to the apparent FRET signal, we further monitored the steady-state anisotropy of each dye in control experiments (Supplemental Figure S4). In all cases, the probe anisotropies did not change significantly upon GroES binding. Together, these data demonstrate that N- and C-terminal domains of the nonnative RuBisCO monomer collapse toward one another by 4–5 Å, on average, upon cis complex formation.
Discussion
GroEL and GroES Do Not Function as a Passive Folding Box
A variety of mechanisms have been proposed to explain how GroEL facilitates protein folding. We have shown that GroEL functions as a molecular chaperone, at least in part, through the active rearrangement of protein conformational states. In contrast, purely passive models of GroEL action, for example the Anfinsen cage model (Ellis, 1994), envision GroEL’s essential role as simply suppressing nonproductive protein aggregation. Here, however, we have shown that a kinetic trap that blocks folding of RuBisCO results from stable, intrachain mis-folding and not from aggregation. Significantly, this mis-folded but monomeric RuBisCO cannot fold spontaneously over the course of several hours, though GroEL and GroES can drive refolding with a half time of approximately 20 min under the same conditions. These observations demonstrate that GroEL-mediated refolding of the kinetically trapped RuBisCO cannot be due to simple, passive inhibition of aggregation.
The corrective action taken by GroEL occurs at two levels. First, the gross structure of the nonnative RuBisCO intermediate is rearranged upon binding to a GroEL ring. Subsequently, binding of ATP and GroES to a RuBisCO-occupied GroEL ring induces a dramatic compaction of the folding intermediate as it is sealed into the enclosed GroEL-GroES-ATP cis complex. These observations support and unify two active models of GroEL-mediated folding: (1) substrate unfolding and (2) conformational restriction.
The Unfolding of Substrate Proteins by GroEL
Though mechanistically controversial, unfolding of substrate proteins by GroEL has been previously observed. Unfolding of a target protein by GroEL has been suggested to occur by two mechanisms: (1) binding-induced unfolding and (2) forced unfolding. Binding-induced unfolding has been observed with several nonstringent GroEL substrate proteins (Bhutani and Udgaonkar, 2000; Clark and Frieden, 1999; Gervasoni et al., 1998; Walter et al., 1996; Zahn et al., 1994, 1996; Zahn and Pluckthun, 1994). In general, GroEL seems capable of driving extensive unfolding of many proteins through the selection and stable binding of less folded conformational states. Forced unfolding, by contrast, has been suggested to occur upon formation of the GroEL-ES cis cavity. In essence, a mechanical unfolding force is imagined to disrupt the structure of a bound substrate protein as the GroEL apical domains rearrange to accommodate the binding of GroES (Shtilerman et al., 1999). While tritium exchange studies with RuBisCO were consistent with this idea (Shtilerman et al., 1999), similar experiments with MDH found no evidence for forced unfolding upon cis complex formation (Chen et al., 2001). More importantly, extensive unfolding of strictly dependent GroEL substrates has not been demonstrated. Indeed, several studies have found little evidence of global structural destabilization of several proteins upon binding to GroEL (Chen et al., 2001; Goldberg et al., 1997; Gross et al., 1996; Robinson et al., 1994). These divergent observations have left open the question of whether substrate unfolding plays any significant, mechanistic role in GroEL-mediated folding.
Our demonstration that RuBisCO folding involves rearrangement of a kinetically trapped folding intermediate strongly suggests that substrate unfolding by GroEL can be of central importance in facilitated protein folding. However, the data presented in the current study provide no evidence for forced unfolding. We detect no dramatic stretching of the RuBisCO monomer upon cis complex formation. Indeed, we observe quite the opposite. Rather than a forced expansion of RuBisCO upon GroES binding, we observe a dramatic compression of the RuBisCO structure. While these observations are not consistent with forced unfolding, the single, average distance measurements made in the current study cannot exclude the possibility that some unfolding accompanies the compaction of the RuBisCO structure upon GroES binding. Indeed, previous experiments monitoring the fluorescence anisotropy of the RuBisCO tryptophan residues showed that cis complex formation induces significant internal changes in the bound RuBisCO structure (Rye et al., 1997).
The expansion and unfolding we observed appears to occur during and following binding of the RuBisCO monomer to an open GroEL ring. However, the maximum unfolding GroEL is capable of delivering to RuBisCO does not appear to be required for fully productive folding. While extensive modification of the RuBisCO intermediate can occur on an open GroEL ring during a typical reaction cycle (10–20 s), the maximum rearrangement we observe is too slow (∼30 min) to be obligatory (Figure 5). In addition, despite the fact that SR1 does not cause as large a structural disruption as wild-type GroEL (Figure 5), both GroEL and SR1 are equally efficient at folding RuBisCO (Rye et al., 1997). It therefore remains unclear how much unfolding is necessary to trigger productive folding. Remarkably, binding-induced unfolding may provide enough conformational modification for some productive folding of RuBisCO to occur following release from the trans ring (Farr et al., 2003). This trans-ring folding mode is significantly less efficient at folding RuBisCO than is GroEL-assisted folding that involves GroES encapsulation, however. In addition, RuBisCO folding is maximally productive with SR1 after only a single round of binding-induced unfolding, provided the intermediate is maintained within the enclosed SR1-GroES complex (Rye et al., 1997). These observations suggest that, in addition to unfolding, the encapsulation of a folding intermediate within the cis complex is an essential additional level of assistance provided by GroEL and GroES.
Substrate Protein Confinement and Compaction
Two simple mechanisms can explain the rapid compression of the GroEL bound RuBisCO intermediate upon cis complex formation. Compaction could result from either (1) internal collapse of the expanded folding intermediate upon cis complex formation and release into the cavity, or (2) a directed, compressive action delivered to the expanded folding intermediate as it is stuffed into the cis cavity upon GroES binding. At the level of our current analysis, we cannot distinguish between these mechanisms. The compaction we observe might be a transient, compressive impulse delivered directly to the bound protein that is necessary to induce additional structural alteration in the folding intermediate and thus trigger folding. Alternately, if the compaction we observe is due to relaxation of the stretched folding intermediate upon release into the cis cavity, it is striking that the average distance between N- and C-terminal domains in the first few seconds following release (∼42 Å) is measurably different from the average distance between these domains in the kinetically trapped state (∼37 Å). This observation strongly suggests that confinement within the cis complex explicitly blocks population of intermediate states of the RuBisCO monomer that are kinetically nonproductive in free solution.
Our observations thus support an active role for GroEL in protein folding beyond simple unfolding of misfolded intermediate states. Essentially, GroEL appears to edit the conformational space available to a folding protein by compacting and confining it within the spatially constricted GroEL-GroES cavity. Several additional observations also support such a model of GroEL action. One recent study found that encapsulation of the RuBisCO monomer within the GroEL-GroES cavity accelerates folding beyond the rate achievable in free solution under permissive conditions (Brinker et al., 2001). It was suggested that confinement of the RuBisCO folding intermediate within the cis complex might therefore modify the folding landscape of a protein and stimulate productive folding. Several theoretical treatments have lent support to this idea (Baumketner et al., 2003; Takagi et al., 2003). The stimulatory effect of confinement would, however, be predicted to only remain significant while a folding intermediate was maintained inside the GroEL-GroES cavity. Upon release, that subset of molecules that have reached a committed state would go on to complete folding, while those that relapse into kinetically trapped states would require recapture, unfolding, and compaction.
In summary, the folding mechanism utilized by GroEL to stimulate productive folding seems to operate on three levels (Figure 7). First, binding of GroEL to aggregation-prone folding intermediates serves to directly halt the progression of irreversible aggregation, particularly at the concentrations of protein present within a living cell. Second, the hydrophobic, multivalent GroEL ring is directly capable of significant structural rearrangement of the folding intermediates that it captures. However, efficient folding of proteins like RuBisCO further requires the binding of GroES over the top of a captured folding intermediate, resulting in the compaction of the folding intermediate upon formation of the cis cavity. While the extent to which compression and confinement operate on various substrate proteins is not fully clear, it does seem certain that confinement is important for some stringent GroEL substrates. Graded levels of assistance in GroEL-mediated folding could permit this chaperone machine to work effectively on different proteins with a variety of requirements.
Figure 7. Three-Stage Model for GroEL-Dependent RuBisCO Refolding.

Denaturation of a RuBisCO dimer results in the formation of a misfolded, kinetically trapped monomer. The misfolded monomer is highly prone to aggregation, which in its early stages is reversible. Binding of GroEL to the aggregation-prone monomer can block the progression of aggregation. Fully productive refolding of RuBisCO requires two additional stages of directed structural modification of the misfolded monomer. First, binding of the RuBisCO monomer to a GroEL ring results in expansion or stretching of the distance between the N and C-terminal domains. Additional conformational alteration of the RuBisCO intermediate, attendant with the observed N-to-C terminal expansion, is probable. Subsequent encapsulation of the RuBisCO monomer upon binding of ATP and GroES results in a compaction of the folding intermediate inside the GroEL-GroES cavity.
Experimental Procedures
Proteins
GroEL, SR1, and GroES were expressed in Escherichia coli and purified as previously described (Rye et al., 1997; Weissman et al., 1995). The A454C and C58A/A454C variants of Rhodospirillum rubrum RuBisCO were created using standard site-directed muta-genesis methods. All RuBisCO molecules were expressed in Escherichia coli and purified as previously described (Rye et al., 1997, 1999).
RuBisCO Refolding
RuBisCO denaturation and refolding were carried out essentially as previously described (Rye et al., 1997). In brief, samples of RuBisCO were first denatured in acid-urea and then diluted into either buffer alone or into buffer containing GroEL. For GroEL-containing samples, folding was initiated by the addition of GroES and ATP. See Supplemental Experimental Procedures for a more a detailed description.
Light Scattering
The extent of light scattering from various RuBisCO samples was examined at a single angle (90°) at 340 nm (4 nm excitation slits; 4 nm detection slits) using a PTI photon counting spectrofluorometer (Photon Technology International). See Supplemental Experimental Procedures for a more a detailed description.
Labeling of RuBisCO with Fluorescent Dyes
The thiol-reactive dyes used in this study were 5-iodoacetamidofluorescein (5-IAF), 5-(2-acetamidoethyl) aminonaphthalene-1-sulfonate (IAEDANS), tetramethylrhodamine-5-iodoacetamide (TMRIA), and N-1-pyrene-maleimide (pyr). All dyes were obtained from Molecular Probes Inc. and were prepared fresh from dry powder in anhydrous DMF immediately prior to use.
Single-Site Labeling of RuBisCO Variants
RuBisCO variants containing single surface Cys residues were derivatized with reactive dyes essentially as described (Rye, 2001; Rye et al., 1999). Specific labeling of the targeted site was confirmed by both tryptic digestion and reverse phase chromatography (C18, Vydac) and mass spectrometry. In some cases, additional purification of the labeled proteins by ion exchange was conducted.
Double Labeling of the 58C/A454C RuBisCO Variant
Taking advantage of a significant difference in reaction rates between the 58C and 454C sites, a sample of the 58C/A454C RuBisCO variant was prepared and treated as above to completely label the 58C position. Under typical conditions, this resulted in simultaneous modification of roughly 50% of the 454C sites. The fraction of the protein labeled only at position 58 was separated from unmodified and 454-reacted molecules using high-resolution ion exchange chromatography (Mono-Q, Pharmacia). Following purification, the singly labeled 58C-dye/A454C protein was then carried through the basic labeling protocol to derivatize the 454C position with a fluorescent dye. Sample purity and extent and specificity of labeling were assessed as described above.
Steady-State Resonance Energy Transfer
Steady-state fluorescence emission spectra were acquired with a PTI photon-counting spectrofluorometer equipped with a temperature-jacketed cuvette holder. See Supplemental Experimental Procedures for a more detailed description of instrument settings and energy transfer calculations.
Time-Resolved Donor Lifetime Measurement and Resonance Energy Transfer
Donor-excited state lifetimes were measured in the time domain using a PTI TimeMaster Fluorescence Lifetime Spectrometer. The lifetimes of donor-only and donor-plus-acceptor samples were measured using a pulsed, N2 laser-pumped dye laser coupled to an optical boxcar detector (James et al., 1992). See Supplemental Experimental Procedures for a more a detailed description of instrument settings and energy transfer calculations.
Time-Resolved Fluorescence Anisotropy Decay and Rotational Correlation Times
Fluorescence anisotropy decay measurements were performed with the PTI TimeMaster Lifetime instrument using film polarizers for selection of vertically (V) and horizontally (H) polarized excitation and emission components. A set of time-resolved fluorescence polarization decays (VV and VH) and their associated background curves were collected for each RuBisCO sample. Simultaneous analysis of vertical and horizontal intensity decays and extraction of experimental anisotropy decays and rotational correlation times was performed using Globals WE (Laboratory of Fluorescence Dynamics, University of Illinois). See Supplemental Experimental Procedures for a more a detailed description of instrument settings and data analysis.
Stopped-Flow Fluorescence and Fluorescence Anisotropy
Stopped-flow measurements were conducted with an SFM-400 rapid mixing unit (BioLogic) equipped with a custom designed, two-channel, photon-counting detection system similar to a previously reported design (Rye et al., 1997). Stopped flow anisotropy measurements were taken by adding film polarizers (Meadowlark Optics) to the excitation and emission optical paths. See Supplemental Experimental Procedures for a more detailed description of instrument settings and data analysis.
For the binding experiments described in Figure 5, the stopped-flow apparatus was first precooled to 4°C, and the cuvette exterior and detection optics were purged with dry nitrogen. The low-temperature RuBisCO folding intermediate was prepared by diluting denatured samples of RuBisCO to 100 nM in degassed buffer at 4°C. These samples were loaded into one prechilled stopped-flow syringe, followed by loading of wild-type GroEL or SR1 into a separate syringe. The denatured RuBisCO was rapidly mixed (1:1) with either buffer alone or chaperonin for a final RuBisCO concentration of 50 nM.
For the refolding experiments described in Figure 6, the stopped-flow apparatus was maintained at 25°C. Binary complexes of RuBisCO and SR1 were prepared by mixing 100 nM RuBisCO with a 4-fold excess of SR1. The binary complex was loaded into one syringe of the stopped-flow and buffer only, ATP, or ATP and GroES were loaded in another syringes. The binary complex was then mixed 1:1 with buffer, ATP, or ATP and GroES.
Distance Calculation by FRET
The average distance between donor and acceptor probes (<r >) was calculated from the FRET efficiency as per (Lakowicz, 1999):
| (1) |
A value of R0 was calculated for each donor-acceptor pair in each state examined (Table 1). The value of κ2 was assumed to be 2/3 for all distance measurements. See Supplemental Experimental Procedures for a more detailed description of the values and procedures used to determine R0.
Modeling of energy transfer distance distributions was accomplished through global analysis (Globals WE) of the donor-only and donor-acceptor data using a probability-weighted (Gaussian) energy transfer model (Haas et al., 1978, 1988; Lillo et al., 1997). We did not attempt to model intrasite diffusion (Beechem and Haas, 1989), and the very short, low amplitude lifetime component of the AEDANS decay was assumed to not participate in the energy transfer process (Lillo et al., 1997).
Supplementary Material
Acknowledgments
We would like to thank Chavela Carr, Frederick Hughson, and members of the Rye lab for helpful discussions and critical comments on the manuscript. We would also like to thank Krystyna Furtak for help in constructing the A454C and C58A/A454C RuBisCO clones, Saw Kyin for mass spectrometry analysis of labeled protein samples, and Enrico Gratton for assistance with data analysis using Globals. This work was supported by grants from the Beckman Foundation through a Beckman Young Investigator Award and the National Institutes of Health.
References
- Baumketner A, Jewett A, Shea JE. Effects of confinement in chaperonin assisted protein folding: rate enhancement by decreasing the roughness of the folding energy landscape. J Mol Biol. 2003;332:701–713. doi: 10.1016/s0022-2836(03)00929-x. [DOI] [PubMed] [Google Scholar]
- Beechem JM, Haas E. Simultaneous determination of intramolecular distance distributions and conformational dynamics by global analysis of energy transfer measurements. Biophys J. 1989;55:1225–1236. doi: 10.1016/S0006-3495(89)82918-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Udgaonkar JB. A thermodynamic coupling mechanism can explain the GroEL-mediated acceleration of the folding of barstar. J Mol Biol. 2000;297:1037–1044. doi: 10.1006/jmbi.2000.3648. [DOI] [PubMed] [Google Scholar]
- Braig K, Otwinowski Z, Hegde R, Boisvert DC, Joachimiak A, Horwich AL, Sigler PB. The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature. 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- Brinker A, Pfeifer G, Kerner MJ, Naylor DJ, Hartl FU, Hayer-Hartl M. Dual function of protein confinement in chaperonin-assisted protein folding. Cell. 2001;107:223–233. doi: 10.1016/s0092-8674(01)00517-7. [DOI] [PubMed] [Google Scholar]
- Chen J, Walter S, Horwich AL, Smith DL. Folding of malate dehydrogenase inside the GroEL-GroES cavity. Nat Struct Biol. 2001;8:721–728. doi: 10.1038/90443. [DOI] [PubMed] [Google Scholar]
- Clark AC, Frieden C. The chaperonin GroEL binds to late-folding non-native conformations present in native Escherichia coli and murine dihydrofolate reductases. J Mol Biol. 1999;285:1777–1788. doi: 10.1006/jmbi.1998.2403. [DOI] [PubMed] [Google Scholar]
- Dobson CM. The structural basis of protein folding and its links with human disease. Philos Trans R Soc Lond B Biol Sci. 2001;356:133–145. doi: 10.1098/rstb.2000.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- Ellis RJ. Molecular chaperones. Opening and closing the Anfinsen cage. Curr Biol. 1994;4:633–635. doi: 10.1016/s0960-9822(00)00140-8. [DOI] [PubMed] [Google Scholar]
- Farr GW, Fenton WA, Chaudhuri TK, Clare DK, Saibil HR, Horwich AL. Folding with and without encapsulation by cis- and trans-only GroEL-GroES complexes. EMBO J. 2003;22:3220–3230. doi: 10.1093/emboj/cdg313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayet O, Ziegelhoffer T, Georgopoulos C. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J Bacteriol. 1989;171:1379–1385. doi: 10.1128/jb.171.3.1379-1385.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton WA, Horwich AL. Chaperonin-mediated protein folding: fate of substrate polypeptide. Q Rev Biophys. 2003;36:229–256. doi: 10.1017/s0033583503003883. [DOI] [PubMed] [Google Scholar]
- Gervasoni P, Gehrig P, Pluckthun A. Two conformational states of beta-lactamase bound to GroEL: a biophysical characterization. J Mol Biol. 1998;275:663–675. doi: 10.1006/jmbi.1997.1481. [DOI] [PubMed] [Google Scholar]
- Goldberg MS, Zhang J, Sondek S, Matthews CR, Fox RO, Horwich AL. Native-like structure of a protein-folding intermediate bound to the chaperonin GroEL. Proc Natl Acad Sci USA. 1997;94:1080–1085. doi: 10.1073/pnas.94.4.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloubinoff P, Gatenby AA, Lorimer GH. GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature. 1989;337:44–47. doi: 10.1038/337044a0. [DOI] [PubMed] [Google Scholar]
- Gross M, Robinson CV, Mayhew M, Hartl FU, Radford SE. Significant hydrogen exchange protection in GroEL-bound DHFR is maintained during iterative rounds of substrate cycling. Protein Sci. 1996;5:2506–2513. doi: 10.1002/pro.5560051213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas E, Katchalski-Katzir E, Steinberg IZ. Brownian motion of the ends of oligopeptide chains in solution as estimated by energy transfer between chain ends. Biopolymers. 1978;17:11–31. [Google Scholar]
- Haas E, McWherter CA, Scheraga HA. Conformational unfolding in the N-terminal region of ribonuclease A detected by nonradiative energy transfer: distribution of interresidue distances in the native, denatured, and reduced-denatured states. Biopolymers. 1988;27:1–21. doi: 10.1002/bip.360270102. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Horwich A. Protein aggregation in disease: a role for folding intermediates forming specific multimeric interactions. J Clin Invest. 2002;110:1221–1232. doi: 10.1172/JCI16781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich AL, Low KB, Fenton WA, Hirshfield IN, Furtak K. Folding in vivo of bacterial cytoplasmic proteins: role of GroEL. Cell. 1993;74:909–917. doi: 10.1016/0092-8674(93)90470-b. [DOI] [PubMed] [Google Scholar]
- Houry WA, Frishman D, Eckerskorn C, Lottspeich F, Hartl FU. Identification of in vivo substrates of the chaperonin GroEL. Nature. 1999;402:147–154. doi: 10.1038/45977. [DOI] [PubMed] [Google Scholar]
- James DR, Siemiarczuk A, Ware WR. Stroboscopic optical boxcar technique for the determination of fluorescence lifetimes. Rev Sci Instrum. 1992;63:1710–1716. [Google Scholar]
- Koo EH, Lansbury PT, Jr, Kelly JW. Amyloid diseases: abnormal protein aggregation in neurodegeneration. Proc Natl Acad Sci USA. 1999;96:9989–9990. doi: 10.1073/pnas.96.18.9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz . Fluorescence anisotropy. In: Lakowicz JR, editor. Principles of Fluorescence Spectroscopy. New York: Kluwer Academic/Plenum Publishers; 1999. pp. 368–391. [Google Scholar]
- Lillo MP, Beechem JM, Szpikowska BK, Sherman MA, Mas MT. Design and characterization of a multisite fluorescence energy-transfer system for protein folding studies: a steady-state and time-resolved study of yeast phosphoglycerate kinase. Biochemistry. 1997;36:11261–11272. doi: 10.1021/bi9707887. [DOI] [PubMed] [Google Scholar]
- Luo S, Wang ZY, Kobayashi M, Nozawa T. The dimerization of folded monomers of ribulose 1,5-bisphosphate carboxylase/oxygenase. J Biol Chem. 2001;276:7023–7026. doi: 10.1074/jbc.M006838200. [DOI] [PubMed] [Google Scholar]
- Navon A, Ittah V, Landsman P, Scheraga HA, Haas E. Distributions of intramolecular distances in the reduced and denatured states of bovine pancreatic ribonuclease A. Folding initiation structures in the C-terminal portions of the reduced protein. Biochemistry. 2001;40:105–118. doi: 10.1021/bi001946o. [DOI] [PubMed] [Google Scholar]
- Prusiner SB, Scott MR, DeArmond SJ, Cohen FE. Prion protein biology. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- Ranson NA, Dunster NJ, Burston SG, Clarke AR. Chaperonins can catalyse the reversal of early aggregation steps when a protein misfolds. J Mol Biol. 1995;250:581–586. doi: 10.1006/jmbi.1995.0399. [DOI] [PubMed] [Google Scholar]
- Robinson CV, Gross M, Eyles SJ, Ewbank JJ, Mayhew M, Hartl FU, Dobson CM, Radford SE. Conformation of GroEL-bound alpha-lactalbumin probed by mass spectrometry. Nature. 1994;372:646–651. doi: 10.1038/372646a0. [DOI] [PubMed] [Google Scholar]
- Rye HS. Application of fluorescence resonance energy transfer to the GroEL-GroES chaperonin reaction. Methods. 2001;24:278–288. doi: 10.1006/meth.2001.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rye HS, Burston SG, Fenton WA, Beechem JM, Xu Z, Sigler PB, Horwich AL. Distinct actions of cis and trans ATP within the double ring of the chaperonin GroEL. Nature. 1997;388:792–798. doi: 10.1038/42047. [DOI] [PubMed] [Google Scholar]
- Rye HS, Roseman AM, Chen S, Furtak K, Fenton WA, Saibil HR, Horwich AL. GroEL-GroES cycling: ATP and non-native polypeptide direct alternation of folding-active rings. Cell. 1999;97:325–338. doi: 10.1016/s0092-8674(00)80742-4. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Buchner J, Todd MJ, Lorimer GH, Viitanen PV. On the role of groES in the chaperonin-assisted folding reaction. Three case studies. J Biol Chem. 1994;269:10304–10311. [PubMed] [Google Scholar]
- Schneider G, Lindqvist Y, Lundqvist T. Crystallographic refinement and structure of ribulose-1,5-bisphosphate carboxylase from Rhodospirillum rubrum at 1.7 A resolution. J Mol Biol. 1990;211:989–1008. doi: 10.1016/0022-2836(90)90088-4. [DOI] [PubMed] [Google Scholar]
- Shtilerman M, Lorimer GH, Englander SW. Chaperonin function: folding by forced unfolding. Science. 1999;284:822–825. doi: 10.1126/science.284.5415.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigler PB, Xu Z, Rye HS, Burston SG, Fenton WA, Horwich AL. Structure and function in GroEL-mediated protein folding. Annu Rev Biochem. 1998;67:581–608. doi: 10.1146/annurev.biochem.67.1.581. [DOI] [PubMed] [Google Scholar]
- Silow M, Oliveberg M. Transient aggregates in protein folding are easily mistaken for folding intermediates. Proc Natl Acad Sci USA. 1997;94:6084–6086. doi: 10.1073/pnas.94.12.6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi F, Koga N, Takada S. How protein thermodynamics and folding mechanisms are altered by the chaperonin cage: molecular simulations. Proc Natl Acad Sci USA. 2003;100:11367–11372. doi: 10.1073/pnas.1831920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd MJ, Viitanen PV, Lorimer GH. Dynamics of the chaperonin ATPase cycle: implications for facilitated protein folding. Science. 1994;265:659–666. doi: 10.1126/science.7913555. [DOI] [PubMed] [Google Scholar]
- Van Der Meer BW, Coker GI, Simon Chen SY. Resonance Energy Transfer: Theory and Data. New York: VCH Publishers, Inc.; 1994. [Google Scholar]
- van der Vies SM, Viitanen PV, Gatenby AA, Lorimer GH, Jaenicke R. Conformational states of ribulosebisphosphate carboxylase and their interaction with chaperonin 60. Biochemistry. 1992;31:3635–3644. doi: 10.1021/bi00129a012. [DOI] [PubMed] [Google Scholar]
- Walter S, Lorimer GH, Schmid FX. A thermodynamic coupling mechanism for GroEL-mediated unfolding. Proc Natl Acad Sci USA. 1996;93:9425–9430. doi: 10.1073/pnas.93.18.9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Weissman JS. Thinking outside the box: new insights into the mechanism of GroEL-mediated protein folding. Nat Struct Biol. 1999;6:597–600. doi: 10.1038/10636. [DOI] [PubMed] [Google Scholar]
- Weissman JS, Hohl CM, Kovalenko O, Kashi Y, Chen S, Braig K, Saibil HR, Fenton WA, Horwich AL. Mechanism of GroEL action: productive release of polypeptide from a sequestered position under GroES. Cell. 1995;83:577–587. doi: 10.1016/0092-8674(95)90098-5. [DOI] [PubMed] [Google Scholar]
- Weissman JS, Rye HS, Fenton WA, Beechem JM, Horwich AL. Characterization of the active intermediate of a GroEL-GroES-mediated protein folding reaction. Cell. 1996;84:481–490. doi: 10.1016/s0092-8674(00)81293-3. [DOI] [PubMed] [Google Scholar]
- Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- Zahn R, Pluckthun A. Thermodynamic partitioning model for hydrophobic binding of polypeptides by GroEL. II GroEL recognizes thermally unfolded mature beta-lactamase. J Mol Biol. 1994;242:165–174. doi: 10.1006/jmbi.1994.1567. [DOI] [PubMed] [Google Scholar]
- Zahn R, Spitzfaden C, Ottiger M, Wuthrich K, Pluckthun A. Destabilization of the complete protein secondary structure on binding to the chaperone GroEL. Nature. 1994;368:261–265. doi: 10.1038/368261a0. [DOI] [PubMed] [Google Scholar]
- Zahn R, Perrett S, Stenberg G, Fersht AR. Catalysis of amide proton exchange by the molecular chaperones GroEL and SecB. Science. 1996;271:642–645. doi: 10.1126/science.271.5249.642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.