Summary
Background
VKORC1 and CYP2C9 are important contributors to warfarin dose variability, but explain less variability for individuals of African descent than for those of European or Asian descent. We aimed to identify additional variants contributing to warfarin dose requirements in African Americans.
Methods
We did a genome-wide association study of discovery and replication cohorts. Samples from African-American adults (aged ≥18 years) who were taking a stable maintenance dose of warfarin were obtained at International Warfarin Pharmacogenetics Consortium (IWPC) sites and the University of Alabama at Birmingham (Birmingham, AL, USA). Patients enrolled at IWPC sites but who were not used for discovery made up the independent replication cohort. All participants were genotyped. We did a stepwise conditional analysis, conditioning first for VKORC1 −1639G→A, followed by the composite genotype of CYP2C9*2 and CYP2C9*3. We prespecified a genome-wide significance threshold of p<5×10−8 in the discovery cohort and p<0·0038 in the replication cohort.
Findings
The discovery cohort contained 533 participants and the replication cohort 432 participants. After the prespecified conditioning in the discovery cohort, we identified an association between a novel single nucleotide polymorphism in the CYP2C cluster on chromosome 10 (rs12777823) and warfarin dose requirement that reached genome-wide significance (p=1·51×10−8). This association was confirmed in the replication cohort (p=5·04×10−5); analysis of the two cohorts together produced a p value of 4·5×10−12. Individuals heterozygous for the rs12777823 A allele need a dose reduction of 6·92 mg/week and those homozygous 9·34 mg/week. Regression analysis showed that the inclusion of rs12777823 significantly improves warfarin dose variability explained by the IWPC dosing algorithm (21% relative improvement).
Interpretation
A novel CYP2C single nucleotide polymorphism exerts a clinically relevant effect on warfarin dose in African Americans, independent of CYP2C9*2 and CYP2C9*3. Incorporation of this variant into pharmacogenetic dosing algorithms could improve warfarin dose prediction in this population.
Funding
National Institutes of Health, American Heart Association, Howard Hughes Medical Institute, Wisconsin Network for Health Research, and the Wellcome Trust.
Introduction
Warfarin is the most widely prescribed oral anticoagulant in the USA, with more than 35 million prescriptions in 2011.1 Complicated by a narrow therapeutic index, warfarin contributes to 33% of hospital admissions related to adverse drug events in individuals aged at least 65 years from the USA.2 Rapid and predictable therapeutic anticoagulation is difficult because dose requirements vary substantially between patients, but is crucial for safe and effective treatment.3,4
Dose variability is affected by single nucleotide polymorphisms (SNPs) in genes encoding cytochrome P450 2C9 (CYP2C9), which metabolises the S-enantiomer of warfarin (more active than the R-enantiomer), and vitamin K epoxide reductase complex 1 (VKORC1), which is the target enzyme for the drug. Both candidate gene studies and genome-wide association studies (GWAS) have consistently shown that VKORC1 and CYP2C9 genotypes explain up to 30% of the total variability in warfarin dose requirements in people of European or Asian origin.5–12 The International Warfarin Pharmacogenetics Consortium (IWPC) has shown that the VKORC1 −1639G→A SNP (rs9923231), CYP2C9*2 (rs1799853), and CYP2C9*3 (rs1057910) can be used with clinical variables to predict warfarin dose.13,14
Unfortunately, these commonly studied genetic variants explain substantially less variability in individuals of African descent than in those of European or Asian origin.5,13,14 African Americans have been absent in previous warfarin GWAS. Importantly, the genomes of people of African ancestry have decreased linkage disequilibrium and increased diversity.15 Therefore, we aimed to identify novel variants contributing to warfarin dose requirements in African Americans.
Methods
Study population
We did a GWAS of two study samples: one from IWPC sites across the USA and one from the University of Alabama at Birmingham (UAB; Birmingham, AL, USA). Together, these samples made up the discovery cohort. Patients enrolled in IWPC sites, but who were not used for discovery (enrolled at local site after samples from that site were sent for genotyping or their local site did not join IWPC until after IWPC samples were sent for genotyping), made up an independent replication cohort. All study participants (discovery and replication cohorts) were adults (aged ≥18 years) of self-reported African ancestry who were taking a stable maintenance dose of warfarin, and for whom clinical and demographic data had been obtained as previously reported.5,13 Definitions of stable dose varied between sites (appendix), but most sites insisted that patients had to have been on a stable dose for at least three consecutive clinic visits, which is indicative of the maintenance phase of warfarin treatment after initial stable dosing has been achieved. Patients provided a DNA sample (whole-blood, saliva, or mouthwash sample). All participants gave written informed consent to participate, and all protocols were approved by local institutional review boards.
Procedures
For the IWPC samples, data curation and quality control procedures were as described previously.13,14 Data for several variables were obtained for participants from IWPC sites in the discovery cohort (appendix). These individuals were genotyped with the Illumina 610 Quad BeadChip (Illumina, San Diego, CA, USA) at the RIKEN Center for Genomic Medicine (Yokohama, Japan).
Information about demographic and clinical variables was obtained for the UAB study sample as previously described.5 Participants from UAB were genotyped at the University of Washington's Department of Genome Sciences (Seattle, WA, USA) with the Illumina Human1M-Duo 3.0 (Illumina, San Diego, CA, USA). SNPs previously associated with warfarin dose were genotyped independently (ie, via single SNP genotyping and at local institutions) in both datasets.
In the replication cohort, samples were tested for a genome-wide significant signal (p<5×10−8). To ensure important signals approaching genome-wide significance were not missed, we identified an additional 12 SNPs with 10−5>p>5×10−8 as being of interest that were also tested in the replication cohort. IWPC replication samples were genotyped at Vanderbilt University's DNA Resources Core (Nashville, TN, USA) with the Sequenom (San Diego, CA, USA) platform, and UAB replication samples at the University of Washington (Seattle, WA, USA) by Sanger sequencing.
To provide insight into potential functional mechanisms of an identified SNP, we assessed the pharmacokinetics of warfarin in an independent cohort of 60 African Americans on stable warfarin doses, as previously described.16 Briefly, venous blood was obtained 12–16 h after a warfarin dose. Warfarin enantiomer concentrations were established by a chiral high-performance liquid chromatography-based method, and the oral clearance of both enantiomers, corrected for body surface area, was calculated.17 Warfarin is almost exclusively cleared by liver metabolism, thus oral clearance approximates the hepatic metabolism of warfarin.
The phenotypic data and a restricted genotypic dataset (eg, VKORC1, CYP2C9, CYP4F2, and rs12777823) are accessible for download at PharmGKB. The IWPC GWAS data are made available under the IWPC Data Usage Agreement and can be accessed at PharmGKB.
Statistical analysis
Conventional GWAS quality control procedures were undertaken, such as checks for sex mismatches, identity by descent testing, deviation from Hardy–Weinberg Equilibrium, and SNP and sample exclusions on the basis of call rates (appendix).
Imputation of additional SNPs was done with hidden Markov models via MACH (version 1.0),18 on the basis of HapMap Phase 2 Release 22 data (appendix). The quality metric (r2) was calculated for all imputed SNPs; no SNPs reported here had a value of less than 0·8.
Global estimates of ancestry (appendix) were established by the first and second principal components, with the first principal component used as a global estimate of African ancestry. All covariates obtained in each dataset (including drugs; appendix) and the first ten principal components obtained through EIGENSTRAT (version 4.2)19 were tested as single covariates in a PLINK (version 1.07) additive model analysis for association with stable warfarin dose. Mean height and weight by sex were used for participants missing these variables in the IWPC cohort.
For all analyses, the primary phenotype (weekly stable warfarin dose) was log transformed to give a normal distribution. We excluded individuals missing data for non-genetic covariates associated with warfarin dose. For the discovery cohort, GWAS analysis was done in PLINK with a linear regression additive genetic model, by use of covariates significantly associated with stable warfarin dose. We combined GWAS results for the IWPC and UAB study samples with fixed effects meta-analysis. We used the same quality-control procedures and analysis approaches in the replication cohort.
Previous warfarin GWAS in individuals of European and Asian ancestry emphasised the importance of conditioning analyses for the well recognised VKORC1 and CYP2C9 SNPs.10,12,20 Therefore, a-priori analysis plans also included a stepwise conditional analysis, conditioning first for VKORC1 −1639G→A, followed by the composite genotype (presence or absence of a variant allele) of CYP2C9*2 and CYP2C9*3 (appendix).
We prespecified a genome-wide significance threshold of p<5×10−8 for the discovery cohort. For the replication cohort, we defined the significance level on the basis of the number of samples tested (13 SNPs). Therefore, significant replication was defined as a Bonferroni-corrected p value of less than 0·05/13 or 0·0038.
Additionally, we looked at a previous GWAS of individuals of European and Asian ancestry9,10,12 to identify any association between the identified SNP and warfarin dose. We tested the SNP in a previously described Egyptian cohort.21
We modelled the residual dose (actual dose minus predicted IWPC dose) with linear regression, with genotypes of a replicated SNP as covariates in participants in the discovery cohort for whom complete data were available for calculation of warfarin dose with the IWPC algorithm. We tested the difference between the models (with and without rs12777823 as a covariate) by ANOVA. All statistical analyses were done with R (version 2.10.1).
Pharmacokinetic variables were compared between genotype groups by ANOVA, and between the GG genotype and both the AA and AG genotypes by the one-sided Student's unpaired t test, with the expected direction defined by the data from the GWAS.
Role of the funding source
The sponsors of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. MAP, LHC, NAL, NJC, RBA, TEK, and JAJ had complete access to the data, and JAJ had final responsibility for the decision to submit for publication.
Results
For the IWPC study sample, eight institutions contributed genetic and clinical data for 345 African-American individuals, although 18 were subsequently excluded because of missing data or quality-control issues. The UAB study sample consisted of 206 African Americans; no participant was excluded from analysis because of quality control issues. Therefore, the discovery cohort contained 533 participants after quality control (table 1). The replication cohort consisted of 432 African Americans enrolled from six IWPC sites, including UAB (table 1; appendix).
Table 1.
Baseline characteristics
|
Discovery cohort |
Replication cohort (n=432) | |||
|---|---|---|---|---|
| International Warfarin Pharmacogenetics Consortium (n=327) | University of Alabama at Birmingham (n=206) | |||
| Stable warfarin dose (mg/wk) | 45·6 (18·6) | 42·2 (18·4) | 45·7 (23·8) | |
| International normalised ratio | 2·46 (0·44) | 2·41 (0·40) | 2·48 (0·31) | |
| Age (years) | 57·1 (14·8) | 59·1 (15·3) | 58·6 (16·0) | |
| Height (cm) | 169·4 (10·1) | 170·9 (10·5) | 170·4 (27·2) | |
| Weight (kg) | 94·4 (10·1) | 89·6 (22·9) | 93·5 (27·8) | |
| Sex | ||||
| Male | 121 (37%) | 92 (45%) | 168 (39%) | |
| Female | 206 (63%) | 114 (55%) | 264 (61%) | |
| Primary warfarin indication* | ||||
| Pulmonary embolism or deep vein thrombosis | 153 (47%) | 97 (47%) | 186 (59%) | |
| Atrial fibrillation or flutter | 81 (25%) | 68 (33%) | 67 (21%) | |
| Cardiomyopathy or left ventricle dilation | 5 (2%) | 0 | 0 | |
| Heart valve replacement | 26 (8%) | 0 | 7 (2%) | |
| Stroke | 28 (9%) | 21 (10%) | 29 (9%) | |
| Other | 34 (10%) | 20 (10%) | 27 (9%) | |
| Current smoker | 53 (16%) | 34 (17%) | NA | |
| Amiodarone use | 16 (5%) | 7 (3%) | 21 (5%) | |
| Aspirin use | 92 (28%) | 93 (45%) | 149 (34%) | |
| Genotype† | ||||
| CYP2C9*2 | 12 (2%) | 6 (1%) | 19 (2%) | |
| CYP2C9*3 | 5 (1%) | 6 (1%) | 13 (2%) | |
| VKORC1 −1639G→A | 59 (10%) | 38 (10%) | 77 (11%) | |
Data are mean (SD) or n (%). NA=not available.
Primary indication was not available for 116 individuals in the replication cohort.
Reported as number of people carrying one or two variant alleles; percentage is minor allele frequency.
Participants in both the IWPC and UAB cohorts clustered between the HapMap CEU (northern and western European ancestry) and YRI (African ancestry) samples, as was expected (appendix). Mean proportion of African ancestry in the discovery cohort was 85% (SD 9%) in participants from IWPC sites and 86% (8%) from UAB. The proportion of African ancestry was not associated with warfarin dose in participants from IWPC sites (p=0·08) or from UAB (p=0·944). 557 286 SNPs passed quality-control checks in the IWPC samples (mean call rate 99·96%) and 950 007 in the UAB samples (99·95%).
Age, weight, height, aspirin use, and amiodarone use were associated with warfarin dose in the discovery cohort (p<0·05 for all in both IWPC and UAB samples; appendix). A glomerular filtration rate of less than 30 mL/min in UAB samples was also associated with warfarin dose (p<0·05).
None of the SNPs reported here that were associated with stable warfarin dose significantly deviated from Hardy–Weinberg equilibrium. SNPs on chromosome 16 reached genome-wide significance, including VKORC1 –1639G→A (rs9923231; p=2·08×10−9; figure 1, appendix). Associations with SNPs in linkage disequilibrium with rs9923231 on chromosome 16 were also significant (appendix).
Figure 1.
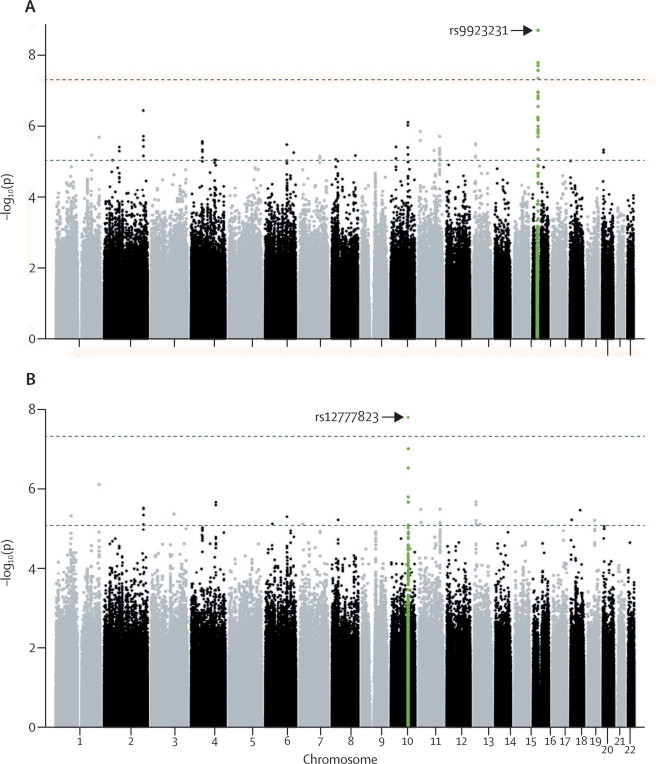
Manhattan plots showing associations between single nucleotide polymorphisms and stable warfarin dose in the discovery cohort
(A) Without and (B) with prespecified conditioning. The top line shows p=5×10−8. Green points indicate genome-wide significant regions.
After the prespecified conditioning on the VKORC1 locus, CYP2C9*2, and CYP2C9*3, the most significant SNP association with warfarin dose was rs12777823 on chromosome 10 (p=1·51×10−8; figure 1, appendix). The minor allele frequency was about 25% in the discovery cohort (appendix). Other top signals after conditioning are shown in the appendix.
The SNP that achieved genome-wide significance in the discovery cohort after conditioning (rs12777823) was also significantly associated with stable warfarin dose in the replication cohort after conditioning (p=5·04×10−5; figure 2, appendix). This p value is well below the threshold of 0·0038 that had been defined for significant replication. A combined analysis of the discovery and replication cohorts for rs12777823 leads to a combined p value of 4·5×10−12. None of the 12 SNPs of interest were associated with stable warfarin dose (appendix). rs12777823 was also not associated with warfarin dose in individuals of European and Japanese ancestry, or Egyptian individuals (data not shown).
Figure 2.
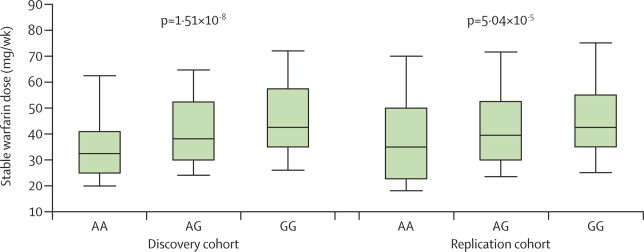
Stable warfarin dose by rs12777823 genotype
Lines within boxes represent medians; lower and upper boundaries represent IQRs; and whiskers represent tenth and 90th percentiles. p values calculated after conditioning on the VKORC1 locus, CYP2C9*2, CYP2C9*3, and non-genetic covariates associated with warfarin dose.
The IWPC warfarin dosing equation is less predictive for African Americans than for other populations.13 We modelled the residual dose with linear regression in 504 participants from the discovery cohort. Addition of rs12777823 genotype to the IWPC algorithm improved the ability to predict actual therapeutic warfarin dose with the IWPC algorithm by an absolute 5% (relative 21%; table 2). Individuals heterozygous or homozygous for the rs12777823 A allele have reduced stable warfarin doses (table 2).
Table 2.
Effects of novel CYP2C variation on warfarin dose prediction with the IWPC dosing algorithm
| Coefficient* | p value | Adjusted model R2† | |
|---|---|---|---|
| IWPC dosing equation Predicted Dose | .. | <2×10−16 | 0·2193 |
| rs12777823 AG | −6·92 | 6·76×10−6 | .. |
| rs12777823 AA | −9·34 | 0·00050 | .. |
| IWPC dosing equation and rs1277823 | .. | 1·7×106 | 0·2666 |
IWPC=International Warfarin Pharmacogenetics Consortium.
Represents change in warfarin dose (mg/week).
Increase indicates increase in amount of variability explained.
In the independent cohort of 60 African Americans, the hepatic metabolism of the S-enantiomer of warfarin (as estimated by oral clearance) differed significantly with rs12777823 genotype (table 3). Additionally, the ratio of the R-enantiomer and S-enantiomer differed significantly by rs12777823 genotype, consistent with genotypic differences in metabolism of the S-enantiomer, but not of the R-enantiomer (table 3).
Table 3.
Pharmacokinetic parameters of warfarin R-enantiomer and S-enantiomer by rs12777823 genotype
| AA (n=6) | AG (n=24) | GG (n=30) | |
|---|---|---|---|
| Oral clearance of S-enantiomer (mL/min/m2)* | 1·20 (0·69) | 1·38 (0·48) | 1·80 (0·83) |
| Plasma concentration ratio of R-enantiomer to S-enantiomer† | 1·29 (0·50) | 1·42 (0·43) | 1·76 (0·68) |
Data are mean (SD). Clearance values are normalised for body surface area.
p=0·042 for association between the three genotype groups (one-way ANOVA); p=0·049 for comparison between AA and GG genotypes, and p=0·016 for comparison between AG and GG genotypes (one-sided Student's unpaired t test).
p=0·049 for association between the three genotype groups (one-way ANOVA); p=0·039 for comparison between AA and GG genotypes, and p=0·018 for comparison between AG and GG genotypes (one-sided Student's unpaired t test).
CYP2C9*2 and CYP2C9*3 occurred at low frequency (table 1) and were not significantly associated with warfarin dose variability (CYP2C9*2 nominal p=0·834; CYP2C9*3 nominal p=0·023). Of the CYP2C9 non-synonymous polymorphisms specific to individuals of African descent, only CYP2C9*11 was imputed in the meta-analysis and had a significant association with warfarin dose (p=0·0001). The CYP2C9*5, CYP2C9*6, and CYP2C9*8 alleles were not included on the GWAS arrays and could not be imputed. Analyses with additional CYP2C9 alleles (eg, CYP2C9*5, CYP2C9*6, and CYP2C9*8) had low power, because few participants were genotyped for these alleles independent of the GWAS. Notably, rs12777823 is in low linkage disequilibrium with CYP2C9*2, CYP2C9*3, and CYP2C9*11 (all r2<0·05), on the basis of the imputed genotypes for these alleles in the GWAS cohorts. Additionally, on the basis of participants independently genotyped for CYP2C9*5, CYP2C9*6, and CYP2C9*8, rs12777823 is also in low linkage disequilibrium with these SNPs (all r2<0·10), suggesting that the rs12777823 signal is independent of previously studied CYP2C9 polymorphisms. The CYP4F2 Val433Met variant (rs2108622) that has been previously associated with warfarin dose in those of European and Asian descent10,12 was not associated with warfarin dose in this study (p=0·07).
Discussion
Our results show that VKORC1 genotype is the primary genetic determinant of variability in warfarin dose requirements in African Americans, which is consistent with previous GWAS data from European and Japanese populations.9,10,12 After adjustment for VKORC1 −1639G→A, CYP2C9*2, and CYP2C9*3, we identified a novel association between rs12777823 and warfarin dose variability; we replicated this finding in an independent African American cohort.
In the context of other genotypes and clinical factors in the IWPC model, the rs12777823 genotype was associated with a reduction in dose of almost 7 mg/week in heterozygotes and of 9 mg/week in homozygotes—an effect similar to that of the CYP2C9*2 allele in the IWPC dosing algorithm.13 Thus, the effect is consistent with a polymorphism that is well accepted to be clinically relevant for individuals. The effect is also important at a population level, because of both the effect size and the frequency of the polymorphism. Specifically, rs12777823 explains 5% of the variability in warfarin dose in African Americans, which is consistent with the variability explained by CYP2C9*2 and CYP2C9*3 in individuals of European ancestry,8 and greatly exceeds the variability explained by CYP2C9*2 and CYP2C9*3 in African Americans (1–2%).5,22
Our findings have important implications for prediction of warfarin maintenance dose in individuals of African ancestry, for whom current warfarin pharmacogenetic algorithms are less predictive.13,14 On the basis of our data, the IWPC algorithm could be used to estimate warfarin dose, and then the rs12777823 variant in African Americans could be accounted for with dose reductions (panel). Further improvement in dose estimation might eventually be possible for African Americans when additional SNPs specific to individuals with African ancestry are identified.
Panel. Research in context.
Systematic review
We searched PubMed for reports of studies and meta-analyses published in any language before Dec 1, 2012. We used the search terms “warfarin” and “genome wide association study”. We identified three genome-wide association studies of warfarin dose requirement9,10,12 and one of acenocoumarol.20 All previous studies were done in either European or Asian populations. They all showed that that VKORC1 –1639G→A (rs9923231), CYP2C9*2 (rs1799853), CYP2C9*3 (rs1057910), and CYP4F2 (rs2108622) are the main genetic determinants of drug dose. The study of acenocoumarol20 identified a single nucleotide polymorphism (SNP) upstream of CYP2C18, although it did not reach genome-wide significance after conditioning for CYP2C9*2 and CYP2C9*3. To identify previous studies testing genetic variation associated with warfarin dose requirements in African Americans, we did another search of PubMed with the same date criteria and no language restrictions, but used the search terms “warfarin” and “African American”. We identified five studies,22–26 all of which used a candidate-gene approach.
Interpretation
As far as we are aware, ours is the first genome-wide association study to investigate warfarin dose requirements in African Americans. Our results identified a SNP upstream of CYP2C18, rs12777823, that is significantly associated with warfarin dose requirement in African Americans and is independent of previous associations with VKORC1 and CYP2C9. Patients carrying this SNP have a significantly lower stable dose of warfarin than do those without this variant, and the addition of this SNP improved the International Warfarin Pharmacogenetics Consortium algorithm by 21%. We have also shown that this SNP significantly alters warfarin clearance. To our knowledge, no other studies of warfarin dose requirement have identified this SNP.
rs12777823 is located within the CYP2C gene cluster, upstream from CYP2C18 on chromosome 10q23, and includes the CYP2C9, CYP2C8, CYP2C18, and CYP2C19 genes. In a GWAS of clopidogrel antiplatelet effect in Amish individuals,27 rs12777823 was the strongest signal, although it was in strong linkage disequilibrium (r2=0·87) with CYP2C19*2 (rs4244285), which produces a non-functional truncated CYP2C19 protein and is believed to be the causative variant. We reported low linkage disequilibrium (r2=0·5) between rs12777823 and CYP2C19*2, and the association between warfarin dose requirements and CYP2C19*2 was not significant at the genome-wide level and was absent when conditioned on rs12777823. Therefore, the effect of rs12777823 in African Americans is probably independent of CYP2C19*2.
The functional basis for the reported association between warfarin dose and rs12777823 is unknown, but the pharmacokinetic data indicate an effect on metabolism of the S-enantiomer. The S-enantiomer is metabolised primarily by CYP2C9, and the stereoselective nature of the genetic association suggests rs12777823 affects CYP2C9 activity. Importantly, the low linkage disequilibrium between this SNP and other known functional SNPs in CYP2C9 suggest that it has an independent and novel genetic effect on CYP2C9 metabolism, which could have clinically important implications for many other drugs that are CYP2C9 substrates.
rs12777823 is common in African Americans: in our study, the minor allele frequency was 25% in both the discovery and replication cohorts. Although common in European (minor allele frequency 14%) and Japanese (32%) populations, rs12777823 did not have a significant association with warfarin dose in previous GWAS,9,10,12 or when we interrogated the European and Japanese GWAS datasets for this SNP. Similarly, this SNP did not have an association with warfarin dose in about 200 individuals from Egypt. Collectively, these data suggest that the association in African Americans might not be due to rs12777823 itself, but a causal SNP in linkage disequilibrium with rs12777823 in African Americans but not in other populations.
Although the CYP2C9*2 and CYP2C9*3 alleles contribute to warfarin dose variability in populations of European descent,9,10 we recorded no genome-wide significant association in the GWAS of African Americans. The low frequency of these alleles in this population might have contributed to this finding.
GWAS has limitations because not all variants are interrogated—eg, a novel VKORC1 variant (rs61162043) identified through resequencing and associated with increased warfarin dose requirements in African Americans22 is not captured on the GWAS genotyping platforms, nor is it in HapMap.28 CYP2C9*5, CYP2C9*6, CYP2C9*8, and CYP2C9*11 have been previously associated with warfarin dose variability,5,22,29 but were not included on the GWAS genotyping platforms; only CYP2C9*11 was available through imputation. Because few sites independently genotyped for these variants, we could not reliably estimate their contribution to warfarin dose requirements in our study. However, we could rule out the CYP2C9*2, CYP2C9*3, and CYP2C9*11 alleles as causes of the recorded association with the rs12777823 variant on the basis of conditional analyses and the computed pairwise linkage disequilibrium between these SNPs. Additionally, linkage disequilibrium between rs12777823 and CYP2C9*5, CYP2C9*6, and CYP2C9*8 was low in participants for whom these data were available.
Although use of new oral anticoagulants is expected to increase with time, warfarin will probably be widely used for the foreseeable future. A study reported in 20121 suggested that dabigatran use has risen consistently since its approval; 8·1% of oral anticoagulant prescriptions in the USA in late 2011 were for this drug. In Sweden, the proportion of prescriptions for oral anticoagulants that were for new drugs rose from 3·0% in 2011, to 4·7% in 2012.30 Thus, although new oral anticoagulant use is increasing, warfarin remains the predominant oral anticoagulant. Additionally, in December, 2012, the US Food and Drugs Administration issued a safety notice that outlined that dabigatran is contraindicated in patients with mechanical heart valves, meaning new oral anticoagulants will not be used for all the indications for which warfarin is presently used.
Importantly, because of cost issues, warfarin will probably remain the mainstay oral anticoagulant for the uninsured or underinsured in the USA and individuals in developing countries—particularly in Africa—for many years to come. Therefore, identification of a way to achieve therapeutic anticoagulation efficiently with warfarin is a priority. Pharmacogenetic warfarin dosing algorithms, such as the IWPC algorithm, could reduce the time needed to reach stable therapeutic dosing;8,13,31 genotype-guided warfarin dosing better predicts warfarin maintenance dose than does standard dosing, and reduces out-of-range international normalised ratio values.31 Secondary analyses in a previous study31 also suggested the potential for improved outcomes via a pharmacogenetic approach. Randomised controlled trials are assessing the clinical utility of genotype-guided warfarin dosing.32,33 If the trials show benefits for pharmacogenetic approaches to warfarin, then future consensus guidelines will probably support warfarin pharmacogenetics in the clinical setting.
In conclusion, we have shown that VKORC1 is the major gene affecting warfarin dose variability in African Americans, with the CYP2C locus exerting influence independent of well known CYP2C9*2, CYP2C9*3, and CYP2C19*2 polymorphisms. Our findings suggest that warfarin dose variability is affected by variants other than the well established VKORC1 and CYP2C9 ones in African Americans; these new variants could improve dose prediction in these individuals.
Acknowledgments
Acknowledgments
MAP (K23 HL089808), NAL (K23 NS45598; RO1 HL092173; UL1 TR000165), NL (R01GM081488), MT and JKB (UL1 TR000427), SAS (KL2 TR000069), DMR (U19 HL 065962; RC2 GM092618), NJC (U19 GM61393; U19 HL065962), RBA and TEK (R24 GM061374), and JAJ (U01 GM074492) were supported by National Institutes of Health grants. LHC was supported by an American Heart Association Grant-In-Aid (10GRNT3750024). RD was supported by the Howard Hughes Medical Institute. MT and JKB were also supported by the Wisconsin Network for Health Research. PD was supported by the Wellcome Trust (grant 098051).
Contributors
MAP, LHC, NAL, PD, MJW, DMR, RBA, TEK, and JAJ did the literature search. MAP, LHC, NAL, DCC, RJD, SIK, SAL, MJR, SAS, AHBW, JKB, PD, MJW, TM, DMR, NJC, RBA, TEK, YN, and JAJ designed the study. MAP, LHC, NAL, DCC, NT, HT, YB, BMB, JLH, TYL, EAN, MO, MHS, SRP, HS, MT, KEW, MJR, SAS, MJW, TM, and MK gathered data and generated laboratory data. MAP, LHC, NAL, ERG, AK, and AP prepared figures. MAP, LHC, NAL, ERG, AK, RD, AP, DCC, JW, NL, NT, SB, HT, MJR, MK, and NJC analysed data. MAP, LHC, NAL, ERG, AK, RD, AP, DCC, JL, NL, NT, SB, HT, SAS, AHBW, JKB, PD, MJW, TM, DMR, NJC, RBA, TEK, YN, and JAJ interpreted data. MAP, LHC, NAL, RBA, TEK, and JAJ drafted the report. All authors contributed to revision of the report.
Conflicts of interest
NAL, RJD, JLH, JAJ are principal investigators; TYL, SAS, and JKB are members of the genotyping committee; and JAJ is a member of the executive committee for the National Institutes of Health Clarification of Optimal Anticoagulation through Genetics trial. KEW is an unpaid consultant for Roche Diagnostics and a paid consultant for the Gentris Corporation. JKB is a site principal investigator for the Iverson Warfarin Trial and holds a patent pending for CYP4F2 use for warfarin dosing. RBA is founder, equity holder, and consultant for Personalis. The other authors declare that they have no conflicts of interest.
Supplementary Material
References
- 1.Kirley K, Qato DM, Kornfield R, Stafford RS, Alexander GC. National trends in oral anticoagulant use in the United States, 2007 to 2011. Circ Cardiovasc Qual Outcomes. 2012;5:615–621. doi: 10.1161/CIRCOUTCOMES.112.967299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Budnitz DS, Lovegrove MC, Shehab N, Richards CL. Emergency hospitalizations for adverse drug events in older Americans. N Engl J Med. 2011;365:2002–2012. doi: 10.1056/NEJMsa1103053. [DOI] [PubMed] [Google Scholar]
- 3.Hylek EM, Go AS, Chang Y. Effect of intensity of oral anticoagulation on stroke severity and mortality in atrial fibrillation. N Engl J Med. 2003;349:1019–1026. doi: 10.1056/NEJMoa022913. [DOI] [PubMed] [Google Scholar]
- 4.Hylek EM, Evans-Molina C, Shea C, Henault LE, Regan S. Major hemorrhage and tolerability of warfarin in the first year of therapy among elderly patients with atrial fibrillation. Circulation. 2007;115:2689–2696. doi: 10.1161/CIRCULATIONAHA.106.653048. [DOI] [PubMed] [Google Scholar]
- 5.Limdi NA, Arnett DK, Goldstein JA. Influence of CYP2C9 and VKORC1 on warfarin dose, anticoagulation attainment and maintenance among European-Americans and African-Americans. Pharmacogenomics. 2008;9:511–526. doi: 10.2217/14622416.9.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferder N, Eby CS, Deych E. Ability of VKORC1 and CYP2C9 to predict therapeutic warfarin dose during the initial weeks of therapy. J Thromb Haemost. 2010;8:95–100. doi: 10.1111/j.1538-7836.2009.03677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rieder MJ, Reiner AP, Gage BF. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 8.Gage B, Eby C, Johnson J. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84:326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper GM, Johnson JA, Langaee TY. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeuchi F, McGinnis R, Bourgeois S. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Limdi NA, Beasley TM, Crowley MR. VKORC1 polymorphisms, haplotypes and haplotype groups on warfarin dose among African-Americans and European-Americans. Pharmacogenomics. 2008;9:1445–1458. doi: 10.2217/14622416.9.10.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cha PC, Mushiroda T, Takahashi A. Genome-wide association study identifies genetic determinants of warfarin responsiveness for Japanese. Hum Mol Genet. 2011;19:4735–4744. doi: 10.1093/hmg/ddq389. [DOI] [PubMed] [Google Scholar]
- 13.Klein TE, Altman RB, Eriksson N. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Limdi NA, Wadelius M, Cavallari L. Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood. 2010;115:3827–3834. doi: 10.1182/blood-2009-12-255992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tishkoff SA, Dietzsch E, Speed W. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 1996;271:1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Jeong H, Takahashi H. Decreased warfarin clearance associated with the CYP2C9 R150H (*8) polymorphism. Clin Pharmacol Ther. 2012;91:660–665. doi: 10.1038/clpt.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi H, Kashima T, Nomizo Y. Metabolism of warfarin enantiomers in Japanese patients with heart disease having different CYP2C9 and CYP2C19 genotypes. Clin Pharmacol Ther. 1998;63:519–528. doi: 10.1016/S0009-9236(98)90103-5. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 20.Teichert M, Eijgelsheim M, Rivadeneira F. A genome-wide association study of acenocoumarol maintenance dosage. Hum Mol Genet. 2009;18:3758–3768. doi: 10.1093/hmg/ddp309. [DOI] [PubMed] [Google Scholar]
- 21.Shahin MH, Khalifa SI, Gong Y. Genetic and nongenetic factors associated with warfarin dose requirements in Egyptian patients. Pharmacogenet Genomics. 2011;21:130–135. doi: 10.1097/FPC.0b013e3283436b86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perera MA, Gamazon E, Cavallari LH. The missing association: sequencing-based discovery of novel SNPs in VKORC1 and CYP2C9 that affect warfarin dose in African Americans. Clin Pharmacol Ther. 2011;89:408–415. doi: 10.1038/clpt.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez AH, Shi Y, Schildcrout JS. Predicting warfarin dosage in European-Americans and African-Americans using DNA samples linked to an electronic health record. Pharmacogenomics. 2012;13:407–418. doi: 10.2217/pgs.11.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cavallari LH, Perera M, Wadelius M. Association of the GGCX (CAA)16/17 repeat polymorphism with higher warfarin dose requirements in African Americans. Pharmacogenet Genomics. 2012;22:152–158. doi: 10.1097/FPC.0b013e32834f288f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schelleman H, Brensinger CM, Chen J, Finkelman BS, Rieder MJ, Kimmel SE. New genetic variant that might improve warfarin dose prediction in African Americans. Br J Clin Pharmacol. 2010;70:393–399. doi: 10.1111/j.1365-2125.2010.03709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voora D, Koboldt DC, King CR. A polymorphism in the VKORC1 regulator calumenin predicts higher warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87:445–451. doi: 10.1038/clpt.2009.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shuldiner AR, O'Connell JR, Bliden KP. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gamazon ER, Ziliak D, Im HK. Genetic architecture of microRNA expression: implications for the transcriptome and complex traits. Am J Hum Genet. 2012;90:1046–1063. doi: 10.1016/j.ajhg.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavallari LH, Langaee TY, Momary KM. Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87:459–464. doi: 10.1038/clpt.2009.223. [DOI] [PubMed] [Google Scholar]
- 30.The National Board of Health and Welfare Läkemedelsstatistik. http://192.137.163.49/sdb/lak/val.aspx (accessed May 10, 2013).
- 31.Anderson JL, Horne BD, Stevens SM. A randomized and clinical effectiveness trial comparing two pharmacogenetic algorithms and standard care for individualizing warfarin dosing (CoumaGen-II) Circulation. 2012;125:1997–2005. doi: 10.1161/CIRCULATIONAHA.111.070920. [DOI] [PubMed] [Google Scholar]
- 32.French B, Joo J, Geller NL. Statistical design of personalized medicine interventions: the Clarification of Optimal Anticoagulation through Genetics (COAG) trial. Trials. 2010;11:108. doi: 10.1186/1745-6215-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Schie RM, Wadelius MI, Kamali F. Genotype-guided dosing of coumarin derivatives: the European pharmacogenetics of anticoagulant therapy (EU-PACT) trial design. Pharmacogenomics. 2009;10:1687–1695. doi: 10.2217/pgs.09.125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


