Summary
Chromatin regulators have become attractive targets for cancer therapy, but it is unclear why inhibition of these ubiquitous regulators should have gene-specific effects in tumor cells. Here, we investigate how inhibition of the widely expressed transcriptional coactivator BRD4 leads to selective inhibition of the MYC oncogene in multiple myeloma (MM). BRD4 and Mediator were found to co-occupy thousands of enhancers associated with active genes. They also co-occupied a small set of exceptionally large super-enhancers associated with genes that feature prominently in MM biology, including the MYC oncogene. Treatment of MM tumor cells with the BET-bromodomain inhibitor JQ1 led to preferential loss of BRD4 at super-enhancers and consequent transcription elongation defects that preferentially impacted genes with super-enhancers, including MYC. Super-enhancers were found at key oncogenic drivers in many other tumor cells. These observations have implications for the discovery of cancer therapeutics directed at components of super-enhancers in diverse tumor types.
Introduction
Chromatin regulators are attractive as therapeutic targets for cancer because they are deregulated in numerous cancers (Baylin and Jones, 2011; Elsässer et al., 2011; Esteller, 2008; Feinberg and Tycko, 2004; You and Jones, 2012) and are amenable to small-molecule inhibition (Cole, 2008; Dawson and Kouzarides, 2012; Geutjes et al., 2012). Inhibition of some chromatin regulators has already proven to be efficacious for treatment of certain cancers (Issa and Kantarjian, 2009; Marks and Xu, 2009). Most chromatin regulators, however, are expressed in a broad range of healthy cells and contribute generally to gene expression, so inhibition of these important genome-associated proteins might be expected to adversely affect global gene expression in healthy cells and thus produce highly toxic effects. Nonetheless, inhibitors of some chromatin regulators, such as BRD4, have been shown to selectively inhibit transcription of key oncogenic drivers such as c-MYC (hereafter referred to as MYC) in multiple tumor types (Dawson et al., 2011; Delmore et al., 2011; Mertz et al., 2011; Zuber et al., 2011). It is important to understand how inhibition of a widely expressed, general regulator such as BRD4 can exert a selective effect on the expression of a small number of genes in specific cells.
BRD4 is a member of the bromodomain and extraterminal (BET) subfamily of human bromodomain proteins, which includes BRDT, BRD2, BRD3, and BRD4. These proteins associate with acetylated chromatin and facilitate transcriptional activation (LeRoy et al., 2008; Rahman et al., 2011). BRD4 was first identified as an interaction partner of the murine Mediator coactivator complex (Jiang et al., 1998) and was subsequently shown to associate with Mediator in a variety of human cells (Dawson et al., 2011; Wu and Chiang, 2007). BRD4 is involved in the control of transcriptional elongation by RNA polymerase II (RNA Pol II) through its recruitment of the positive transcription elongation factor P-TEFb (Jang et al., 2005; Yang et al., 2005). Almost all human cells express the BRD4 gene, based on analysis of human tissue expression data across 90 distinct tissue types (human body index - transcriptional profiling, see Extended Experimental Procedures), and BRD4 is found to be associated with a large population of active genes in CD4+ T cells (Zhang et al., 2012). It is not yet clear whether the BRD4 protein is generally involved in the transcription of active genes in tumor cells or if it is selectively associated with a subset of these genes.
Two recently developed bromodomain inhibitors, JQ1 and iBET, selectively bind to the amino-terminal twin bromodomains of BRD4 (Filippakopoulos et al., 2010; Nicodeme et al., 2010). These BET inhibitors cause selective repression of the potent MYC oncogene in a range of tumors, including multiple myeloma (MM), Burkitt's lymphoma (BL), acute myeloid leukemia (AML), and acute lymphoblastic leukemia (ALL) (Dawson et al., 2011; Delmore et al., 2011; Mertz et al., 2011; Ott et al., 2012; Zuber et al., 2011). The inhibition of MYC apparently occurs as a consequence of BRD4 depletion at the enhancers that drive MYC expression (Delmore et al., 2011). Although BRD4 is widely expressed in mouse tissues, mice are reasonably tolerant of the levels of BET bromodomain inhibition that inhibit certain tumors in mouse models (Dawson et al., 2011; Delmore et al., 2011; Filippakopoulos et al., 2010; Mertz et al., 2011; Zuber et al., 2011).
The MM cell line (MM1.S) used to study the effects of JQ1 has an IgH-MYC rearrangement, and MYC gene expression is driven by factors associated with the IgH enhancer (Dib et al., 2008; Shou et al., 2000). Enhancers function through cooperative and synergistic interactions between multiple transcription factors and coactivators (Carey et al., 1990; Giese et al., 1995; Kim and Maniatis, 1997; Thanos and Maniatis, 1995). Cooperative binding and synergistic activation confer increased sensitivity so that small changes in activator concentration can lead to dramatic changes in activator binding and transcription of associated genes (Carey, 1998). Furthermore, enhancers with large numbers of transcription factor binding sites can be more sensitive to small changes in factor concentration than those with smaller numbers of binding sites (Giniger and Ptashne, 1988; Griggs and Johnston, 1991). This concept led us to postulate that some features of the IgH enhancer might account for the selective effect of BRD4 inhibition.
We show here that BRD4 and Mediator are associated with most active enhancers and promoters in MM1.S tumor cells, but exceptionally high levels of these cofactors occur at a small set of large enhancer regions, which we call super-enhancers. Super-enhancers are associated with MYC and other key genes that feature prominently in the biology of MM, including many lineage-specific survival genes. Treatment of MM tumor cells with the BRD4 inhibitor JQ1 caused a preferential loss of BRD4, Mediator, and P-TEFb at super-enhancers and caused preferential loss of transcription at super-enhancer-associated genes, including the MYC oncogene. Tumor cell addiction to high-level expression of these oncogenes may then contribute to their vulnerability to super-enhancer disruption (Chin et al., 1999; Felsher and Bishop, 1999; Jain et al., 2002; Weinstein, 2002). We find super-enhancers in additional tumor types, where they are similarly associated with key oncogenes. Thus, key oncogene drivers of tumor cells are regulated by super-enhancers, which can confer disproportionate sensitivity to loss of the BRD4 coactivator and thus cause selective inhibition of transcription.
Results
BRD4 and Mediator Co-occupy Promoters of Active Genes in Multiple Myeloma
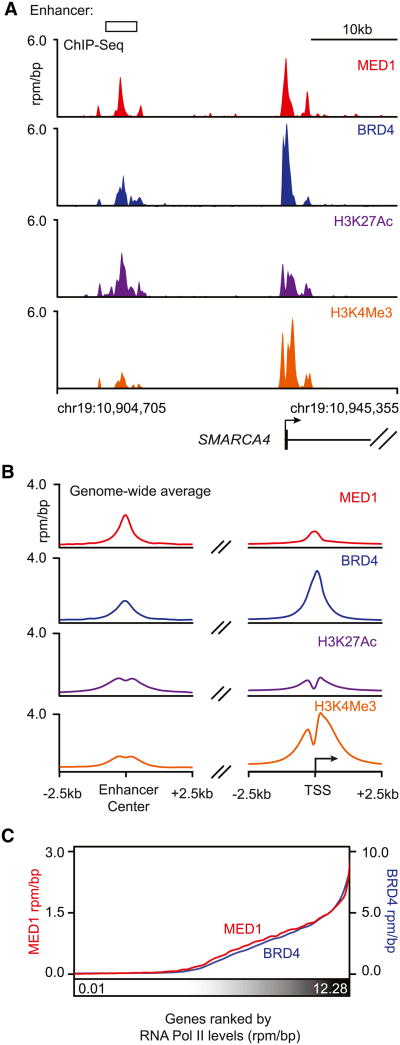
Transcription factors bind to enhancers and recruit the Mediator coactivator, which in turn becomes associated with RNA Pol II at the transcription start site (TSS), thus forming DNA loops between enhancers and core promoters (Kagey et al., 2010). BRD4 is known to associate with Mediator in some mammalian cells (Dawson et al., 2011; Jiang et al., 1998; Wu et al., 2003). To identify active promoter and enhancer elements and to determine how BRD4 and Mediator occupy the genome in MM1.S MM cells, we used chromatin immunoprecipitation coupled to high-throughput sequencing (chromatin immunoprecipitation [ChIP]-seq) with antibodies against the Mediator subunit MED1, BRD4, the enhancer-associated histone modification H3K27Ac, and the TSS-associated histone modification H3K4Me3 (Figure 1). ChIP-seq signals for both Mediator and the histone modification H3K27Ac have previously been shown to occur at both enhancers and TSSs (Creyghton et al., 2010; Heintzman et al., 2009; Rada-Iglesias et al., 2011), and enhancers can be distinguished from TSSs by the absence of TSS annotation and relatively low levels of H3K4Me3. We found that BRD4 co-occupied enhancers and TSSs with MED1 throughout the genome (Figures 1A and 1B) and that the levels of BRD4 and MED1 were strongly correlated (Figure S1 available online).
Figure 1. Mediator and BRD4 Co-occupy Promoters of Active Genes in Multiple Myeloma.
(A) Gene tracks of MED1, BRD4, H3K27Ac, and H3K4Me3 ChIP-seq occupancy at the enhancer (left) and promoter (right) of SMARCA4 in MM1.S MM cells. The x axis shows genomic position, and enhancer-containing regions are depicted with a white box. The y axis shows signal of ChIP-seq occupancy in units of reads per million mapped reads per base pair (rpm/bp).
(B) Metagene representation of global MED1, BRD4, H3K27Ac, and H3K4Me3 occupancy at enhancers and promoters. The x axis shows the ±2.5 kb region flanking either the center of enhancer regions (left) or the TSS of active genes (right). The y axis shows the average background subtracted ChIP-seq signal in units of rpm/bp.
(C) Median MED1 and BRD4 levels in the ±1 kb region around the TSSs of actively transcribed genes ranked by increasing RNA Pol II occupancy in MM1.S cells. Levels are in units of rpm/bp, with the left y axis showing levels of MED1 and the right y axis showing levels of BRD4. Promoters were binned (50/bin), and a smoothing function was applied to median levels.
See also Figure S1.
To confirm that BRD4 and Mediator are generally associated with active genes in MM1.S cells, we compared the ChIP-seq data for these regulators with that for RNA Pol II and the histone modification H3K4Me3. The levels of BRD4 and Mediator correlated with the levels of RNA Pol II genome wide (Figure 1C). Signals for BRD4 and Mediator were found together with those for the histone modification H3K4Me3 and RNA Pol II at ∼10,000 annotated TSSs, and these were considered active TSSs (Table S1). Signals for BRD4 and the enhancer-associated histone modification H3K27Ac were found in ∼8,000 Mediator-occupied regions either lacking TSSs or extending beyond the immediate vicinity of the TSS, and these were considered enhancer regions (Table S2, Data S1, and Extended Experimental Procedures).
Super-Enhancers Are Associated with Key Multiple Myeloma Genes
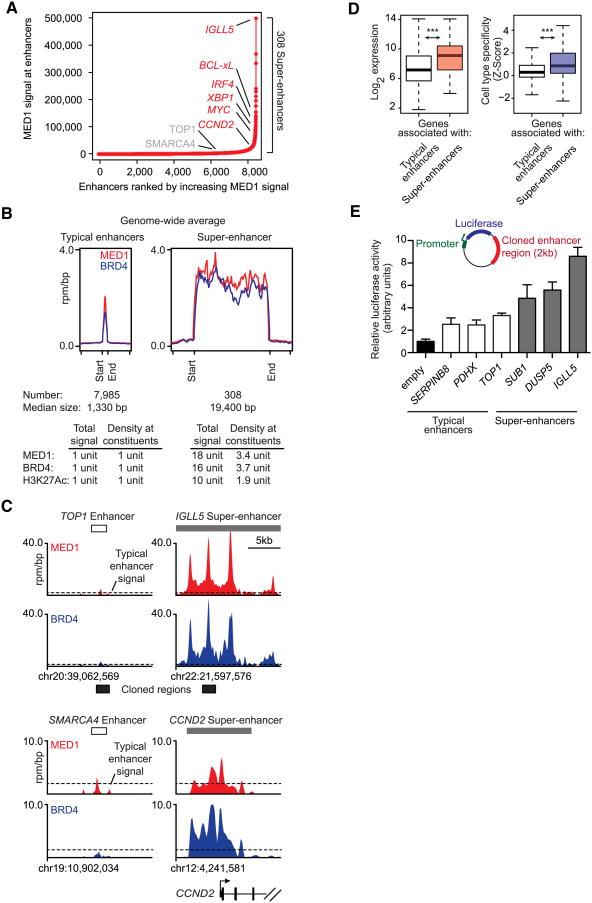
Further analysis of the ∼8,000 enhancer regions revealed that the MED1 signal at 308 enhancers was significantly greater than at all other enhancers and promoters (Figures 2A and S2A and Table S2). These 308 super-enhancers differed from typical enhancers in both size and Mediator levels (Figure 2B). Remarkably, ∼40% of all enhancer-bound Mediator and BRD4 occupied these 308 super-enhancers. Whereas the typical enhancer had a median size of 1.3 kb, the super-enhancers had a median size of 19.4 kb. These super-enhancers were thus 15-fold larger than typical enhancers and were occupied, based on ChIP-seq signal, by 18-fold more Mediator and 16-fold more BRD4. Similarly high levels of H3K27Ac were observed in these large regions (Figure 2B). Examples of gene tracks showing super-enhancers at either end of the spectrum of Mediator occupancy (Figure 2A) are shown in Figure 2C. The largest super-enhancer was found associated with the IGLL5 gene, which encodes an immunoglobulin lambda peptide expressed at high levels in these cells.
Figure 2. Super-Enhancers Identified in Multiple Myeloma.
(A) Total MED1 ChIP-seq signal in units of reads per million in enhancer regions for all enhancers in MM1.S. Enhancers are ranked by increasing MED1 ChIP-seq signal.
(B) Metagene representation of global MED1 (red line) and BRD4 (blue line) occupancy at typical enhancers and super-enhancers. The x axis shows the start and end of the enhancer (left) or super-enhancer (right) regions flanked by ±5 kb of adjacent sequence. Enhancer and super-enhancer regions on the x axis are relatively scaled. The y axis shows the average signal in units of rpm/bp.
(C) Gene tracks of MED1 (top) and BRD4 (bottom) ChIP-seq occupancy at the typical enhancer upstream of TOP1, the super-enhancer downstream of IGLL5, the typical enhancer upstream of SMARCA4, and the super-enhancer overlapping the CCND2 gene TSS. The x axis shows genomic position, and super-enhancer-containing regions are depicted with a gray box. The y axis shows signal of ChIP-seq occupancy in units of rpm/bp.
(D) Left: box plots of expression values for genes with proximal typical enhancers (white) or with proximal super-enhancers (pink). The y axis shows expression value in Log2 arbitrary units. Right: box plots of cell-type specificity values for genes with proximal typical enhancers (white) or with proximal super-enhancers (purple). The y axis shows the Z score of the Jensen-Shannon (JS) divergence statistic for genes, with higher values corresponding to a more cell-type-specific pattern of expression. Changes between expression levels are significant (two-tailed Welch's t test, p < 2 × 10−16), as are changes between cell-type-specificity levels (two-tailed Welch's t test, p = 1 × 10−14).
(E) Bar graph depicting luciferase activity of reporter constructs containing cloned fragments of typical enhancers and super-enhancers in MM1.S cells. 2 kb fragments of three super-enhancers, IGLL5, DUSP5, and SUB1, and three typical enhancers, PDHX, SERPINB8, and TOP1, ranked 1, 129, 227, 2352, 4203, and 4794, respectively, in terms of MED1 occupancy, were cloned into reporter plasmids downstream of the luciferase gene, driven by a minimal MYC promoter. Luciferase activity is represented as fold over empty vector. Error bars represent SD of triplicate experiments.
See also Figure S2 and Data S1.
We next sought to identify the complete set of MM1.S genes that are most likely associated with super-enhancers. Enhancers tend to loop to and associate with adjacent genes in order to activate their transcription (Göndör and Ohlsson, 2009; Lelli et al., 2012; Ong and Corces, 2011; Spitz and Furlong, 2012). Most of these interactions occur within a distance of ∼50 kb of the enhancer (Chepelev et al., 2012). Using a simple proximity rule, we assigned all transcriptionally active genes (TSSs) to super-enhancers within a 50 kb window, a method shown to identify a large proportion of true enhancer/promoter interactions in embryonic stem cells (Dixon et al., 2012). This identified 681 genes associated with super-enhancers (Table S3), and 307 of these had a super-enhancer overlapping a portion of the gene, as shown for CCND2 in Figure 2C.
Super-enhancer-associated genes were generally expressed at higher levels than genes with typical enhancers and tended to be specifically expressed in MM1.S cells (Figure 2D). To test whether components of super-enhancers confer stronger activity compared to typical enhancers, we cloned representative super-enhancer or typical enhancer fragments of similar size into luciferase reporter constructs and transfected these into MM1.S cells. Cloned sequence fragments from super-enhancers generated 2- to 3-fold higher luciferase activity compared to typical enhancers of similar size (Figure 2E and Extended Experimental Procedures). These results are consistent with the notion that super-enhancers help to activate high levels of transcription of key genes that regulate and enforce the MM1.S cancer cell state.
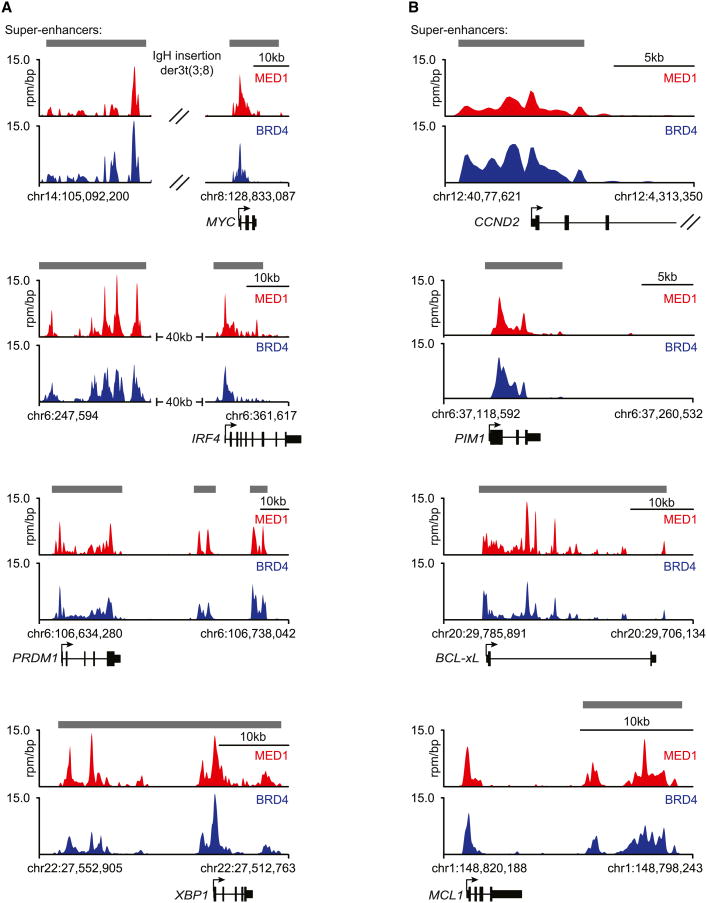
The super-enhancer-associated genes included most genes that have previously been shown to have important roles in MM biology, including MYC, IRF4, PRDM1/BLIMP-1, and XBP1 (Figure 3A). MYC is a key oncogenic driver in MM (Chng et al., 2011; Dib et al., 2008; Holien et al., 2012; Shou et al., 2000), and the MM1.S MYC locus contains a chromosomal rearrangement that places MYC under the control of the IgH enhancer, which qualifies as a super-enhancer in MM1.S cells. The IRF4 gene encodes a key plasma cell transcription factor that is frequently deregulated in MM (Shaffer et al., 2008). PRDM1/BLIMP-1 encodes a transcription factor that is considered a master regulator of plasma cell development and is required for the formation of plasma cell tumors in a mouse model (Shapiro-Shelef et al., 2003; Turner et al., 1994). XBP1 encodes a basic-region leucine zipper (bZIP) transcription factor of the CREB-ATF family that governs plasma cell differentiation (Reimold et al., 2001). XBP1 is frequently overexpressed in human MM and can drive the development of MM in a mouse model (Carrasco et al., 2007; Claudio et al., 2002).
Figure 3. Super-Enhancers Are Associated with Key Multiple Myeloma Genes.
(A and B) Gene tracks of MED1 and BRD4 ChIP-seq occupancy at super-enhancers near genes with important roles in MM biology (A) or genes with important roles in cancer (B). Super-enhancers are depicted in gray boxes over the gene tracks. The x axis shows genomic position, and super-enhancer-containing regions are depicted with a gray box. The y axis shows signal of ChIP-seq occupancy in units of rpm/bp.
Super-enhancers were associated with many additional genes that have important roles in cancer pathogenesis more generally (Figure 3B). Cyclin D2 (CCND2) is deregulated in many human cancers, including MM (Bergsagel et al., 2005; Musgrove et al., 2011). The PIM1 kinase has been implicated in the biology of many different cancers (Shah et al., 2008). MCL1 and BCL-xL, members of the BCL-2 family of apoptosis regulators, are frequently deregulated in cancer, promoting cell survival and chemoresistance (Beroukhim et al., 2010). We conclude that super-enhancers are frequently associated with genes that feature prominently in the biology of MM and other human cancers.
Inhibition of BRD4 Leads to Displacement of BRD4 Genome Wide
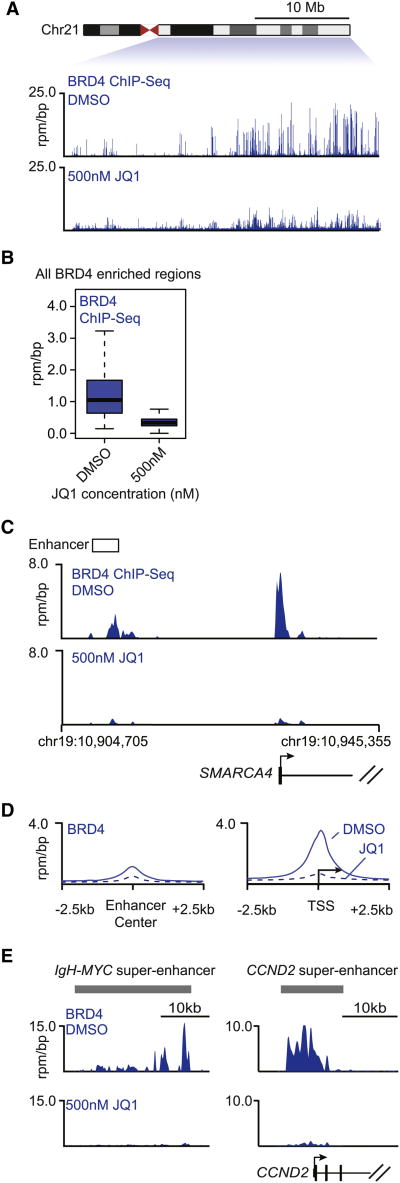
BRD4 interacts with chromatin-associated proteins such as transcription factors, the Mediator complex, and acetylated histones (Dawson et al., 2011; Dey et al., 2003; Jang et al., 2005; Jiang et al., 1998; Wu and Chiang, 2007; Wu et al., 2013). Previous studies have shown that treatment of MM1.S cells with JQ1 leads to reduced levels of BRD4 at the IgH enhancer that drives MYC expression (Delmore et al., 2011), but it is not clear whether such treatment causes a general reduction in the levels of BRD4 associated with the genome. We found that treatment of MM1.S cells with 500 nM JQ1 for 6 hr reduced the levels of BRD4 genome wide by ∼70% (Figures 4A and 4B). This reduction in BRD4 occupancy was evident both by inspection of individual gene tracks (Figure 4C) and through global analysis of the average effects at enhancers and TSSs (Figure 4D). JQ1 treatment led to ∼60% reduction in BRD4 signal at enhancers and ∼90% reduction at promoters (Figure 4D). The reduction in BRD4 was more profound at super-enhancers such as those associated with IgH-MYC and CCND2 (Figure 4E), where the loss of BRD4 was nearly complete. We conclude that BET bromodomain inhibition of BRD4 leads to reduced levels of BRD4 at enhancers and promoters throughout the genome in MM1.S cells.
Figure 4. Inhibition of BRD4 Leads to Loss of BRD4 Genome Wide.
(A) Tracks showing BRD4 ChIP-seq occupancy on the 35 Mb right arm of chromosome 21 after DMSO (top) or 500 nM JQ1 (bottom) treatment. The chromosome 21 ideogram is displayed above the gene tracks with the relevant region highlighted in blue. The x axis of the gene tracks shows genomic position, and the y axis shows BRD4 ChIP-seq signal in units of rpm/bp.
(B) Box plot showing the distributions of BRD4 ChIP-seq signal at BRD4-enriched regions after DMSO (left) or 500 nM JQ1 (right) treatment. BRD4-enriched regions were defined in MM1.S cells treated with DMSO. The y axis shows BRD4 ChIP-seq signal in units of rpm/bp. The loss of BRD4 occupancy at BRD4-enriched regions after JQ1 is highly significant (p value < 1 × 10−16, Welch's t test).
(C) Gene tracks of BRD4 ChIP-seq occupancy at the enhancer (left) and promoter (right) of SMARCA4 in MM1.S cells after DMSO (top) or 500 nM JQ1 (bottom) treatment for 6 hr. The x axis shows genomic position, and enhancer-containing regions are depicted with a white box. The y axis shows signal of ChIP-seq occupancy in units of rpm/bp.
(D) Metagene representation of global BRD4 occupancy at enhancers and promoters after DMSO (solid line) or 500 nM JQ1 (dotted line) treatment. The x axis shows the ±2.5 kb region flanking either the center of enhancer regions (left) or the TSS of active genes. The y axis shows the average background subtracted ChIP-seq signal in units of rpm/bp.
(E) Gene tracks of BRD4 binding at super-enhancers after DMSO (top) or 500 nM JQ1 (bottom) treatment. The x axis shows genomic position, and super-enhancer-containing regions are depicted with a gray box. The y axis shows signal of ChIP-seq occupancy in units of rpm/bp.
Transcription of Super-Enhancer-Associated Genes Is Highly Sensitive to BRD4 Inhibition
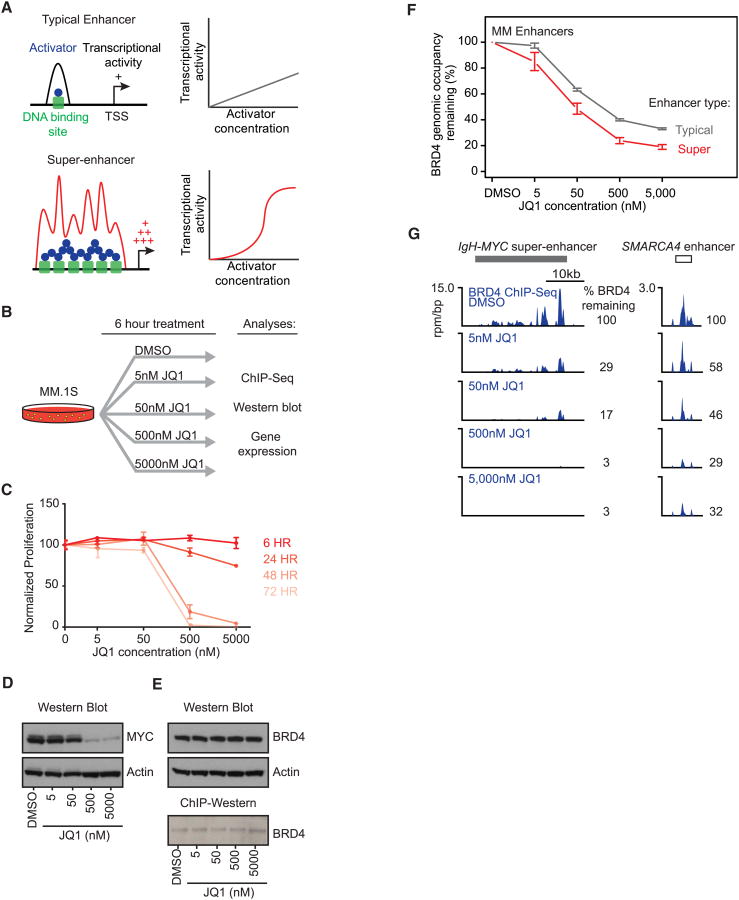
Enhancers are formed through cooperative and synergistic binding of multiple transcription factors and coactivators (Carey, 1998; Carey et al., 1990; Giese et al., 1995; Kim and Maniatis, 1997; Thanos and Maniatis, 1995). As a consequence of this binding behavior, enhancers bound by many cooperatively interacting factors lose activity more rapidly than enhancers bound by fewer factors when the levels of enhancer-bound factors are reduced (Giniger and Ptashne, 1988; Griggs and Johnston, 1991). The presence of super-enhancers at MYC and other key genes associated with MM led us to consider the hypothesis that super-enhancers are more sensitive to reduced levels of BRD4 than typical enhancers and that genes associated with super-enhancers might then experience a greater reduction of transcription than genes with average enhancers when BRD4 is inhibited (Figure 5A).
Figure 5. BRD4 Occupancy at Super-Enhancers Is Highly Sensitive to Bromodomain Inhibition.
(A) Schematic example of how cooperative interactions of enhancer-associated factors at super-enhancers lead to both higher transcriptional output and increased sensitivity to factor concentration.
(B) Measuring the effects of various concentrations of JQ1 genome wide on BRD4 occupancy. Schematic depicting the experimental procedure.
(C) Short-term JQ1 treatment (6 hr) has little effect on MM1.S cell viability. JQ1 sensitivity of MM1.S cells by measurement of ATP levels (CellTiterGlo) after 6, 24, 48, and 72 hr of treatment with JQ1 (5, 50, 500, or 5,000 nM) or vehicle (DMSO, 0.05%). Error bars represent the SD of triplicate experiments.
(D) Western blot of relative MYC levels after 6 hr of JQ1 or DMSO treatment.
(E) Western blot of relative BRD4 levels after 6 hr of JQ1 or DMSO treatment. ChIP-western blot of the relative levels of immunoprecipitated BRD4 after 6 hr of JQ1 or DMSO treatment.
(F) Line graph showing the percentage of BRD4 occupancy remaining after 6 hr treatment at various JQ1 concentrations for typical enhancers (gray line) or super-enhancers (red line). The y axis shows the fraction of BRD4 occupancy remaining versus DMSO. The x axis shows different JQ1 concentrations (DMSO [none], 5 nM, 50 nM, and 500 nM). Error bars represent 95% confidence intervals of the mean (95% CI).
(G) Gene tracks of BRD4 ChIP-seq occupancy after various concentrations of JQ1 treatment at the IgH-MYC-associated super-enhancer (left) and the SMARCA4-associated typical enhancer (right). The x axis shows genomic position, and gray boxes depict super-enhancer regions. The y axis shows signal of ChIP-seq occupancy in units of rpm/bp. The percent of BRD4 remaining after each concentration of JQ1 treatment is annotated to the right of the gene tracks.
To test this hypothesis, we first examined the effects of various concentrations of JQ1 on BRD4 occupancy genome wide (Figure 5B). JQ1 had little effect on MM1.S cell viability when treated for 6 hr at these various concentrations, whereas at later time points, JQ1 had a significant antiproliferative effect (Figure 5C). As expected, MYC protein levels were significantly depleted by exposure of MM1.S cells to 50 nM or greater doses of JQ1 for 6 hr (Figure 6D) (Delmore et al., 2011). In contrast, JQ1 did not affect total BRD4 protein levels within the cells and did not significantly reduce ChIP efficiency (Figure 5E). When BRD4 occupancy was examined genome wide in cells exposed to increasing concentrations of JQ1, it was evident that super-enhancers showed a greater loss of BRD4 occupancy than typical enhancer regions (Figure 5F). For example, the IgH super-enhancer showed significantly greater reduction in BRD4 occupancy in cells treated with 5 nM or 50 nM JQ1 than typical enhancer regions such as that upstream of SMARCA4 (Figure 5G). Ultimately, virtually all BRD4 occupancy was lost at the IgH super-enhancer (97% reduction versus DMSO control) after treatment with 500 nM JQ1, whereas loss of BRD4 occupancy at the typical enhancer for SMARCA4 was less pronounced (71% reduction versus DMSO control) (Figure 5G).
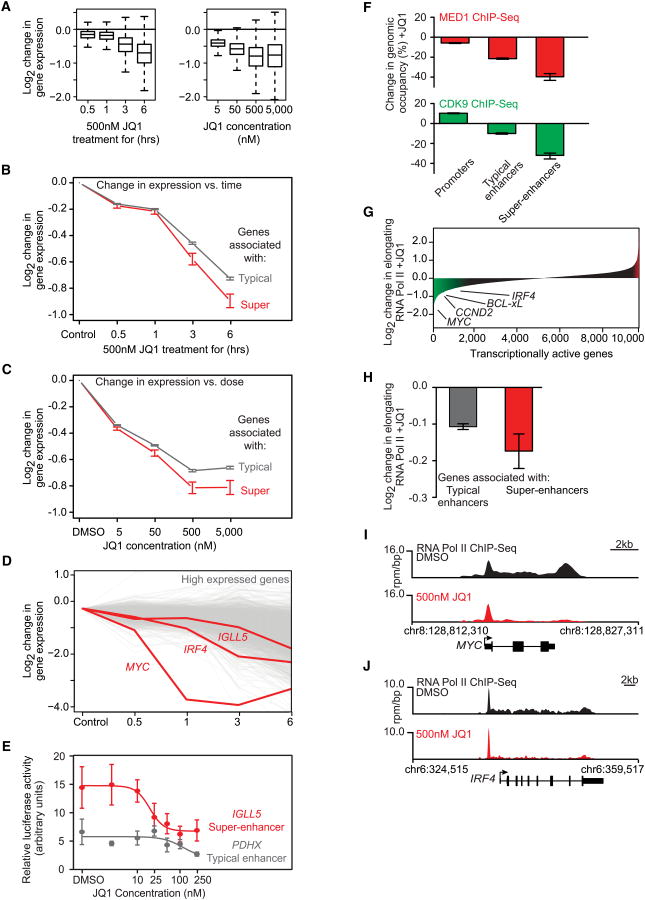
Figure 6. JQ1 Causes Disproportionate Loss of Transcription at Super-Enhancer Genes.
(A) Box plots showing the Log2 change in gene expression for all actively transcribed genes in JQ1-treated versus control cells for a time course of cells treated with 500 nM JQ1 (left) or for a concentration course of cells treated for 6 hr with varying amounts of JQ1 (right). The y axis shows the Log2 change in gene expression versus untreated control cells (left graph) or control cells treated with DMSO for 6 hr (right graph).
(B and C) Line graph showing the Log2 change in gene expression versus control cells after JQ1 treatment in a time- (B) or dose (C)-dependent manner for genes associated with typical enhancers (gray line) or genes associated with super-enhancers (red line). The y axis shows the Log2 change in gene expression of JQ1 treated versus untreated control cells. The x axis shows time of 500 nM JQ1 treatment (B) or JQ1 treatment concentration at 6 hr (C). Error bars represent 95% confidence intervals of the mean (95% CI).
(D) Graph showing the Log2 change in gene expression after JQ1 treatment over time for genes ranked in the top 10% of expression in MM1.S cells. Each line represents a single gene, with the MYC and IRF4 genes drawn in red. The y axis shows the Log2 change in gene expression of JQ1-treated versus untreated control cells. The x axis shows time of 500 nM JQ1 treatment.
(E) Line graph showing luciferase activity after JQ1 treatment at various concentrations for luciferase reporter constructs containing either a fragment from the IGLL5 super-enhancer (red line) or the PDHX typical enhancer (gray line). The y axis represents relative luciferase activity in arbitrary units. The x axis shows JQ1 concentrations. Error bars are SEM.
(F) Bar graphs showing the percentage loss of either MED1 (top, red) or CDK9 (bottom, green) at promoters, typical enhancers, and super-enhancers. Error bars represent 95% CI.
(G) Graph of loss of RNA Pol II density in the elongating gene body region for all transcriptionally active genes in MM1.S cells after 6 hr of 500 nM JQ1 treatment. Genes are ordered by decrease in elongating RNA Pol II in units of Log2 fold loss. Genes with a greater than 0.5 Log2 fold change in elongating RNA Pol II are shaded in green (loss) or red (gain). The amount of RNA Pol II loss is indicated for select genes.
(H) Bar graph showing the Log2 fold change in RNA Pol II density in elongating gene body regions after 6 hr of 500 nM JQ1 treatment for genes with typical enhancers (left, gray) or genes with super-enhancers (red, right). Error bars represent 95% confidence intervals of the mean (95% CI).
(I and J) Gene tracks of RNA Pol II ChIP-seq occupancy after DMSO (black) or 500 nM JQ1 treatment (red) at the super-enhancer proximal MYC gene (I) and IRF4 gene (J). The y axis shows signal of ChIP-seq occupancy in units of rpm/bp.
See also Figure S3.
We next investigated whether genes associated with super-enhancers might experience a greater reduction of transcription than genes with average enhancers when BRD4 is inhibited. As expected, treatment of MM1.S cells with 500 nM JQ1 led to progressive reduction in global messenger RNA (mRNA) levels over time (Figures 6A and S3A). Similarly, treatment with increasing concentrations of JQ1 caused progressive reductions in global mRNA levels (Figures 6A and S3B). There was a selective depletion of mRNAs from super-enhancer-associated genes that occurred in both temporal (Figure 6B) and concentration-dependent manners (Figure 6C). Notably, MYC and IRF4 mRNA levels were more rapidly depleted than other mRNAs that are expressed at similar levels (Figure 6D). The levels of transcripts from super-enhancer-associated genes were somewhat more affected than those from genes that have multiple typical enhancers bound by BRD4 (Figures S3C and S3D). Thus, BET bromodomain inhibition preferentially impacts transcription of super-enhancer-driven genes.
To further test the model that super-enhancers are responsible for the special sensitivity to BRD4 inhibition, we transfected MM1.S cells with luciferase reporter constructs containing super-enhancer and typical enhancer fragments and examined the effects of various JQ1 concentrations on luciferase activity. Upon treatment with JQ1, MM1.S cells transfected with a super-enhancer reporter experienced a greater reduction in luciferase activity than those transfected with a typical enhancer reporter (Figure 6E). Interestingly, the dose-response curve observed for luciferase activity of the super-enhancer construct is consistent with that expected for enhancers that are bound cooperatively by multiple factors (Figure 5A) (Giniger and Ptashne, 1988; Griggs and Johnston, 1991). These results are also consistent with the model that super-enhancers are responsible for the special sensitivity of gene transcription to BRD4 inhibition.
BRD4 Inhibition and Transcription Elongation
At active genes, enhancers and core promoters are brought into close proximity, so factors associated with enhancers can act on the transcription apparatus in the vicinity of TSSs and thereby influence initiation or elongation. BRD4 is known to interact with Mediator and P-TEFb and to be involved in the control of transcriptional elongation by RNA Pol II (Conaway and Conaway, 2011; Dawson et al., 2011; Jang et al., 2005; Krueger et al., 2010; Rahman et al., 2011; Yang et al., 2005). This suggests that the preferential loss of BRD4 from super-enhancers might affect the levels of Mediator and P-TEFb at these sites and, furthermore, that the reduced levels of mRNAs from super-enhancer-associated genes might be due to an effect on transcription elongation.
To test these predictions, we carried out ChIP-seq for the Mediator component MED1 and the catalytic subunit of the P-TEFb complex CDK9 in MM1.S cells treated with DMSO or 500 nM JQ1 for 6 hr. In control cells, MED1 and CDK9 were found at enhancers and promoters of active genes throughout the MM genome, as expected (Figures 1A, 1B, and S3E). In cells treated with JQ1, reduced levels of MED1 and CDK9 were observed primarily at enhancers, with the greatest loss at super-enhancers (Figure 6F). As many super-enhancers span contiguous regions that encompass or overlap the TSS, we analyzed MED1 and CDK9 loss in either TSS proximal or TSS distal regions of super-enhancers and again observed loss of MED1 and CDK9 predominantly at TSS distal regions (Figure S3F). We conclude that inhibition of BRD4 genomic binding leads to a marked reduction in the levels of Mediator and P-TEFb at genomic regions distal to TSSs, with the greatest reduction occurring at super-enhancers.
To determine whether reduced levels of BRD4 lead to changes in transcription elongation, we quantified changes in transcription elongation by performing ChIP-seq of RNA Pol II before and after treatment of MM1.S cells with 500 nM JQ1. We then calculated the fold loss of RNA Pol II occupancy in the gene body regions for all transcriptionally active genes and found that more than half of these genes show a decrease in elongating RNA Pol II density after JQ1 treatment (Figure 6G). Importantly, genes associated with super-enhancers showed a greater decrease of RNA Pol II in their elongating gene body regions compared to genes associated with typical enhancers (Figures 6H and S3G). Inspection of individual gene tracks revealed pronounced elongation defects at super-enhancer-associated genes such as MYC and IRF4, with the greatest effects observed with MYC (Figures 6I and 6J). Thus, the selective effects of JQ1 on the transcription of MYC and other super-enhancer-associated genes can be explained, at least in part, by the sensitivity of super-enhancers to reduced levels of BRD4, which leads to a pronounced effect on pause release and transcription elongation.
Super-Enhancers Are Associated with Disease-Critical Genes in Other Cancers
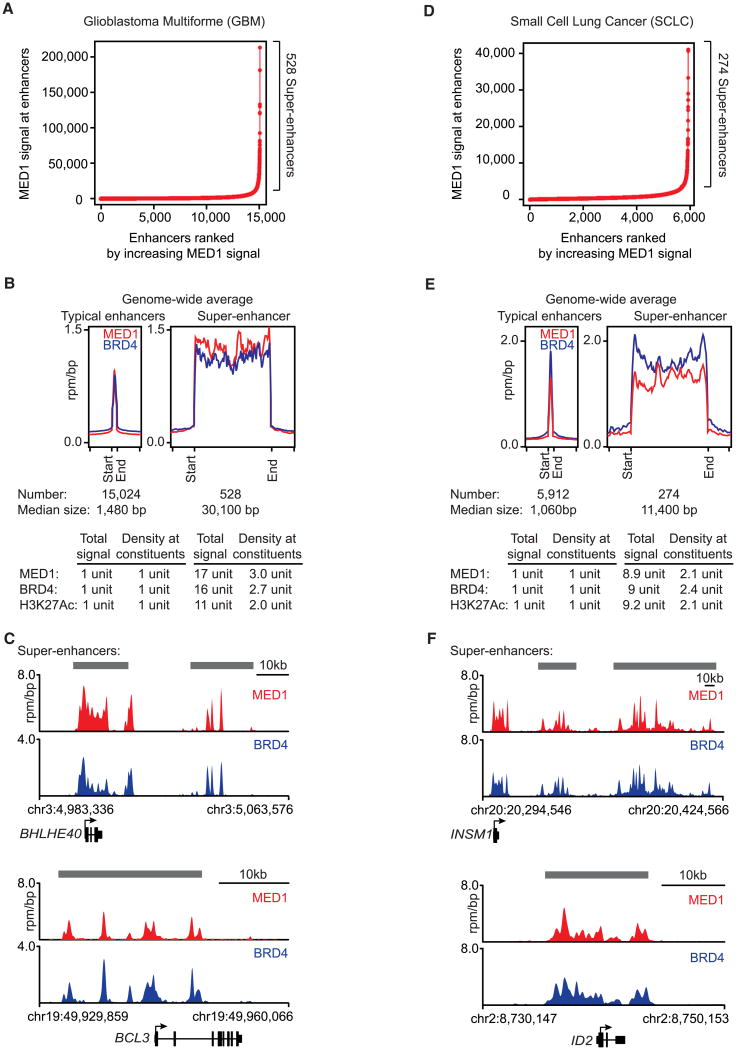
To map enhancers and to determine whether super-enhancers occur in additional tumor types, we investigated the genome-wide occupancy of Mediator (MED1), BRD4, and the enhancer-associated histone modification H3K27Ac using ChIP-seq in glioblastoma multiforme (GBM) and small-cell lung cancer (SCLC) (Figure 7). Mediator (MED1) occupancy was used to identify enhancer elements because enhancer-bound transcription factors bind directly to Mediator (Borggrefe and Yue, 2011; Conaway and Conaway, 2011; Kornberg, 2005; Malik and Roeder, 2010; Taatjes, 2010) and because it has proven to produce high-quality evidence for enhancers in mammalian cells (Kagey et al., 2010). Global occupancy of BRD4 and H3K27Ac was used as corroborative evidence to identify enhancer elements (Figure S4 and Table S4). Analysis of the regions occupied by Mediator revealed that, as in MM1.S cells, large genomic domains were occupied by this coactivator in both GBM and SCLC (Figures 7A, 7B, 7D, and 7E). The median super-enhancer was 30 kb in GBM cells and 11 kb in SCLC cells (Figures 7B and 7E). As in MM1.S cells, these GBM and SCLC super-enhancers were an order or magnitude larger and showed a commensurate increase in MED1, BRD4, and H3K27Ac levels when compared to normal enhancers (Figures 7B and 7E).
Figure 7. Super-Enhancers Are Associated with Key Genes in Other Cancers.
(A and D) Total MED1 ChIP-seq signal in units of reads per million in enhancer regions for all enhancers in (A) the GBM cell line U-87 MG or (D) the SCLC cell line H2171. Enhancers are ranked by increasing MED1 ChIP-seq signal.
(B and E) Metagene representation of global MED1 and BRD4 occupancy at (B) typical GMB enhancers and super-enhancers or (E) typical SCLC enhancers and super-enhancers. The x axis shows the start and end of the enhancer (left) or super-enhancer (right) regions flanked by ±5 kb of adjacent sequence. Enhancer and super-enhancer regions on the x axis are relatively scaled. The y axis shows the average signal in units of rpm/bp.
(C and F) Gene tracks of MED1 and BRD4 ChIP-seq occupancy at (C) super-enhancers near BHLHE40 and BCL3, genes with important roles in GBM, or at (F) super-enhancers near INSM1 and ID2, genes with important roles in SCLC. Super-enhancers are depicted in gray boxes over the gene tracks.
See also Figure S4.
The super-enhancers in GBM and SCLC were found to be associated with many well-known tumor-associated genes (Figures 7C and 7F and Table S5). In GBM, super-enhancers were associated with genes encoding three transcription factors (RUNX1, FOSL2, and BHLHE40) critical for mesenchymal transformation of brain tumors (Carro et al., 2010); the super-enhancers associated with BHLHE40 are shown in Figure 7C. BCL3, which associates with NF-κB and is deregulated in many blood and solid tumor types, is associated with a super-enhancer in GBM (Figure 7C) (Maldonado and Melendez-Zajgla, 2011). In SCLC, a super-enhancer is associated with the INSM1 gene, which encodes a transcription factor involved in neuronal development that is highly expressed in neuroendocrine tissue and tumors such as SCLC (Figure 7F) (Pedersen et al., 2003). A super-enhancer is also associated with the ID2 gene, which is highly expressed in SCLCs and encodes a protein that interacts with the well-known retinoblastoma tumor suppressor (Figure 7F) (Pedersen et al., 2003; Perk et al., 2005). These results indicate that super-enhancers are likely to associate with critical tumor oncogenes in diverse tumor types.
Discussion
Chromatin regulators have become attractive targets for cancer therapy, but many of these regulators are expressed in a broad range of healthy cells and contribute generally to gene expression. Thus, it is unclear how inhibition of a global chromatin regulator such as BRD4 might produce selective effects, such as at the MYC oncogene (Delmore et al., 2011). We have found that key regulators of tumor cell state in MM1.S cells are associated with large enhancer domains, characterized by disproportionately high levels of BRD4 and Mediator. These super-enhancers are more sensitive to perturbation than typical enhancers, and the expression of the genes associated with super-enhancers is preferentially affected. Thus, the preferential loss of BRD4 at super-enhancers associated with the MYC oncogene and other key tumor-associated genes can explain the gene-selective effects of JQ1 treatment in these cells.
BRD4 is an excellent example of a chromatin regulator that is expressed in a broad range of healthy cells and contributes generally to gene expression. Most cell types for which RNA-seq data are available express the BRD4 gene. ChIP-seq data revealed that BRD4 generally occupies the enhancer and promoter elements of active genes with the Mediator coactivator in MM1.S cells (Figure 1). These results eliminate the model that BRD4 is exclusively associated with a small set of genes that are thereby rendered inactive by the BRD4 inhibitor JQ1 and instead suggest that the gene-specific effects of the small molecule have other causes.
We have found that ∼3% of the enhancers in MM1.S cells are exceptionally large and are occupied by remarkably high amounts of BRD4 and Mediator. These super-enhancers are generally an order of magnitude larger and contain an order of magnitude more BRD4, Mediator, and histone marks associated with enhancers (H3K27Ac) than typical enhancers. Our results suggest that super-enhancers are collections of closely spaced enhancers that can collectively facilitate high levels of transcription from adjacent genes. Importantly, the super-enhancers are associated with the MYC oncogene and additional genes such as IGLL5, IRF4, PRDM1/BLIMP-1, and XBP1 that feature prominently in MM biology.
Cooperative and synergistic binding of multiple transcription factors and coactivators occurs at enhancers. Enhancers bound by many cooperatively interacting factors can lose activity more rapidly than enhancers bound by fewer factors when the levels of enhancer-bound factors are reduced (Giniger and Ptashne, 1988; Griggs and Johnston, 1991). The presence of super-enhancers at MYC and other key genes associated with MM led us to test the hypothesis that super-enhancers are more sensitive to reduced levels of BRD4 than average enhancers. We found that treatment of these tumor cells with the BET-bromodomain inhibitor JQ1 leads to preferential loss of BRD4 at super-enhancers. In addition, this decrease in BRD4 occupancy is accompanied by a corresponding loss of MED1 and CDK9 at super-enhancers. Consequent transcription elongation defects and mRNA decreases preferentially impact super-enhancer-associated genes, with an especially profound effect at the MYC oncogene.
Super-enhancers are not restricted to MM cells. We have identified super-enhancers in two additional tumor types, small-cell lung cancer and glioblastoma multiforme. Super-enhancers identified in these cell types have characteristics similar to those found in MM1.S; they span large genomic regions and contain exceptional amounts of Mediator and BRD4. These super-enhancers are also associated with important tumor genes in both cell types. In GBM cells, BHLHE40 and BCL3 are known to be important in tumor biology and are each associated with super-enhancers in this cell type. In H2171 SCLC cells, super-enhancers are associated with INSM1 and ID2, which are frequently overexpressed in SCLC. In fact, super-enhancers are not restricted to tumor cells and have been identified in several additional cell types in which they similarly associate with key cell identity genes (Whyte et al., 2013 [this issue of Cell]).
Our results demonstrate that super-enhancers occupied by BRD4 regulate critical oncogenic drivers in MM and show that BRD4 inhibition leads to preferential disruption of these super-enhancers. This insight into the mechanism by which BRD4 inhibition causes selective loss of oncogene expression in this highly malignant blood cancer may have implications for future drug development in oncology. Tumor cells frequently become addicted to oncogenes, thus becoming unusually reliant on high-level expression of these genes (Cheung et al., 2011; Chin et al., 1999; Felsher and Bishop, 1999; Garraway and Sellers, 2006; Garraway et al., 2005; Jain et al., 2002; Weinstein, 2002). Thus, preferential disruption of super-enhancer function may be a general approach to selectively inhibiting the oncogenic drivers of many tumor cells.
Experimental Procedures
Cell Culture
MM1.S MM cells (CRL-2974 ATCC) and U-87 MG glioblastoma cells (HTB-14 ATCC) were purchased from ATCC. H2171 small-cell lung carcinoma cells (CRL-5929 ATCC) were kindly provided by John Minna, UT Southwestern. MM1.S and H2171 cells were propagated in RPMI-1640 supplemented with 10% fetal bovine serum and 1% GlutaMAX (Invitrogen, 35050-061). U-87 MG cells were cultured in Eagle's minimum essential medium (EMEM) modified to contain Earle's Balanced Salt Solution, nonessential amino acids, 2 mM L-glutamine, 1 mM sodium pyruvate, and 1,500 mg/l sodium bicarbonate. Cells were grown at 37°C and 5% CO2.
For JQ1 treatment experiments, cells were resuspended in fresh media containing JQ1 (5 nM, 50 nM, 500 nM, and 5,000 nM) or vehicle (DMSO, 0.05%) and treated for a duration of 6 hr, unless otherwise indicated.
ChIP-Seq
ChIP was carried out as described in Lin et al. (2012). Additional details are provided in Extended Experimental Procedures. Antibodies used are as follows: total RNA Pol II (Rpb1 N terminus), Santa Cruz sc-899 lot K0111; MED1, Bethyl Labs A300-793A lot A300-793A-2; BRD4, Bethyl Labs A301-985A lot A301-985A-1; CDK9, Santa Cruz Biotechnology sc-484, lot D1612. ChIP-seq data sets of H3K4Me3 and H3K27Ac in MM1.S and MED1 and H3K27Ac in U-87 MG and H2171 were previously published (Lin et al., 2012).
Luciferase Reporter Assays
A minimal Myc promoter was amplified from human genomic DNA and cloned into the SacI and HindIII sites of the pGL3 basic vector (Promega). Enhancer fragments were likewise amplified from human genomic DNA and cloned into the BamHI and SalI sites of the pGL3-pMyc vector. All cloning primers are listed in Table S6. Constructs were transfected into MM1.S cells using Lipofectamine 2000 (Invitrogen). The pRL-SV40 plasmid (Promega) was cotransfected as a normalization control. Cells were incubated for 24 hr, and luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega). For the JQ1 concentration course, cells were resuspended in fresh media containing various concentrations of JQ1 24 hr after transfection and were incubated for an additional 6 hr before harvesting. Luminescence measurements were made using the Dual-Luciferase Reporter Assay System (Promega) on a Wallac EnVision (Perkin Elmer) plate reader.
Cell Viability Assays
Cell viability was measured using the CellTiterGlo assay kit (Promega, G7571). MM1.S cells were resuspended in fresh media containing JQ1 (5 nM, 50 nM, 500 nM, and 1,000 nM) or vehicle (DMSO, 0.05%) and then plated in 96-well plates at 10,000 cells/well in a volume of 100 μl. Viability was measured after 6, 24, 48, and 72 hr incubations by addition of CellTiter Glo reagent and luminescence measurement on a Tecan Safire2 plate reader.
Western Blotting
Western blots were carried out using standard protocols. Antibodies used are as follows: c-Myc (Epitomics, category: 1472-1), BRD4 (Epitomics, category: 5716-1) or β-actin (Sigma, clone AC-15, A5441).
Data Analysis
All ChIP-seq data sets were aligned using Bowtie (version 0.12.9) (Langmead et al., 2009) to build version NCBI36/HG18 of the human genome. Individual data set GEO accession IDs and background data sets used can be found in Table S7.
ChIP-seq read densities in genomic regions were calculated as in Lin et al. (2012). We used the MACS version 1.4.2 (model-based analysis of ChIP-seq) (Zhang et al., 2008) peak finding algorithm to identify regions of ChIP-seq enrichment over background. A p value threshold of enrichment of 1 × 10−9 was used for all data sets.
Active enhancers were defined as regions of ChIP-seq enrichment for the mediator complex component MED1 outside of promoters (e.g., a region not contained within ±2.5 kb region flanking the promoter). In order to accurately capture dense clusters of enhancers, we allowed MED1 regions within 12.5 kb of one another to be stitched together. To identify super-enhancers, we first ranked all enhancers by increasing total background subtracted ChIP-seq-occupancy of MED1 (x axis) and plotted the total background subtracted ChIP-seq occupancy of MED1 in units of total rpm (y axis). This representation revealed a clear inflection point in the distribution of MED1 at enhancers. We geometrically defined the inflection point and used it to establish the cutoff for super-enhancers (see Extended Experimental Procedures).
Supplementary Material
Acknowledgments
We thank Zi Peng Fan for bioinformatics support; Michael R. McKeown for help with cell viability assays; Jun Qi for providing JQ1; Tom Volkert, Jennifer Love, Sumeet Gupta, and Jeong-Ah Kwon at the Whitehead Genome Technologies Core for Solexa sequencing; and members of the Young, Bradner, and Vakoc labs for helpful discussion. This work was supported by a Swedish Research Council Postdoctoral Fellowship VR-B0086301 (J.L.), the Damon-Runyon Cancer Research Foundation (J.E.B.), a Burroughs-Welcome CAMS award and NCI Cancer Center Support Grant Development Fund CA45508 (C.R.V.), and National Institutes of Health grants HG002668 (R.A.Y.) and CA146445 (R.A.Y., T.I.L.). R.A.Y. and J.E.B. are founders, J.L. and D.A.O. have become employees, and C.R.V. is a scientific advisor of Syros Pharmaceuticals.
Footnotes
Accession Numbers: The GEO accession number for the ChIP-seq and gene expression data reported in this paper is GSE44931 (http://www.ncbi.nlm.nih.gov/geo/).
Supplemental Information: Supplemental Information includes Extended Experimental Procedures, four figures, one data file, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.03.036.
References
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsagel PL, Kuehl WM, Zhan F, Sawyer J, Barlogie B, Shaughnessy J., Jr Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood. 2005;106:296–303. doi: 10.1182/blood-2005-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe T, Yue X. Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin Cell Dev Biol. 2011;22:759–768. doi: 10.1016/j.semcdb.2011.07.022. [DOI] [PubMed] [Google Scholar]
- Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- Carey M, Leatherwood J, Ptashne M. A potent GAL4 derivative activates transcription at a distance in vitro. Science. 1990;247:710–712. doi: 10.1126/science.2405489. [DOI] [PubMed] [Google Scholar]
- Carrasco DR, Sukhdeo K, Protopopova M, Sinha R, Enos M, Carrasco DE, Zheng M, Mani M, Henderson J, Pinkus GS, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11:349–360. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 2012;22:490–503. doi: 10.1038/cr.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci USA. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O'Hagan R, Pantginis J, Zhou H, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- Chng WJ, Huang GF, Chung TH, Ng SB, Gonzalez-Paz N, Troska-Price T, Mulligan G, Chesi M, Bergsagel PL, Fonseca R. Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia. 2011;25:1026–1035. doi: 10.1038/leu.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudio JO, Masih-Khan E, Tang H, Gonçalves J, Voralia M, Li ZH, Nadeem V, Cukerman E, Francisco-Pabalan O, Liew CC, et al. A molecular compendium of genes expressed in multiple myeloma. Blood. 2002;100:2175–2186. doi: 10.1182/blood-2002-01-0008. [DOI] [PubMed] [Google Scholar]
- Cole PA. Chemical probes for histone-modifying enzymes. Nat Chem Biol. 2008;4:590–597. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. Function and regulation of the Mediator complex. Curr Opin Genet Dev. 2011;21:225–230. doi: 10.1016/j.gde.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib A, Gabrea A, Glebov OK, Bergsagel PL, Kuehl WM. Characterization of MYC translocations in multiple myeloma cell lines. J Natl Cancer Inst Monogr. 2008;39:25–31. doi: 10.1093/jncimonographs/lgn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsässer SJ, Allis CD, Lewis PW. Cancer. New epigenetic drivers of cancers. Science. 2011;331:1145–1146. doi: 10.1126/science.1203280. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Sellers WR. Lineage dependency and lineagesurvival oncogenes in human cancer. Nat Rev Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Geutjes EJ, Bajpe PK, Bernards R. Targeting the epigenome for treatment of cancer. Oncogene. 2012;31:3827–3844. doi: 10.1038/onc.2011.552. [DOI] [PubMed] [Google Scholar]
- Giese K, Kingsley C, Kirshner JR, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- Giniger E, Ptashne M. Cooperative DNA binding of the yeast transcriptional activator GAL4. Proc Natl Acad Sci USA. 1988;85:382–386. doi: 10.1073/pnas.85.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göndör A, Ohlsson R. Chromosome crosstalk in three dimensions. Nature. 2009;461:212–217. doi: 10.1038/nature08453. [DOI] [PubMed] [Google Scholar]
- Griggs DW, Johnston M. Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc Natl Acad Sci USA. 1991;88:8597–8601. doi: 10.1073/pnas.88.19.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holien T, Våtsveen TK, Hella H, Waage A, Sundan A. Addiction to c-MYC in multiple myeloma. Blood. 2012;120:2450–2453. doi: 10.1182/blood-2011-08-371567. [DOI] [PubMed] [Google Scholar]
- Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009;15:3938–3946. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg CD, Bishop JM, Felsher DW. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Jiang YW, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway JW, Conaway RC, Kornberg RD. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl Acad Sci USA. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Krueger BJ, Varzavand K, Cooper JJ, Price DH. The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK. PLoS ONE. 2010;5:e12335. doi: 10.1371/journal.pone.0012335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelli KM, Slattery M, Mann RS. Disentangling the many layers of eukaryotic transcriptional regulation. Annu Rev Genet. 2012;46:43–68. doi: 10.1146/annurev-genet-110711-155437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado V, Melendez-Zajgla J. Role of Bcl-3 in solid tumors. Mol Cancer. 2011;10:152. doi: 10.1186/1476-4598-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks PA, Xu WS. Histone deacetylase inhibitors: Potential in cancer therapy. J Cell Biochem. 2009;107:600–608. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–572. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, Rodig SJ, Kung AL, Bradner JE, Weinstock DM. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120:2843–2852. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N, Mortensen S, Sørensen SB, Pedersen MW, Rieneck K, Bovin LF, Poulsen HS. Transcriptional gene expression profiling of small cell lung cancer cells. Cancer Res. 2003;63:1943–1953. [PubMed] [Google Scholar]
- Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, Howley PM. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N, Pang B, Yeoh KG, Thorn S, Chen CS, Lilly MB, Salto-Tellez M. Potential roles for the PIM1 kinase in human cancer - a molecular and therapeutic appraisal. Eur J Cancer. 2008;44:2144–2151. doi: 10.1016/j.ejca.2008.06.044. [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- Shou Y, Martelli ML, Gabrea A, Qi Y, Brents LA, Roschke A, Dewald G, Kirsch IR, Bergsagel PL, Kuehl WM. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc Natl Acad Sci USA. 2000;97:228–233. doi: 10.1073/pnas.97.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- Weinstein IB. Cancer. Addiction to oncogenes—the Achilles heal of cancer Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(this issue):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- Wu SY, Zhou T, Chiang CM. Human mediator enhances activator-facilitated recruitment of RNA polymerase II and promoter recognition by TATA-binding protein (TBP) independently of TBP-associated factors. Mol Cell Biol. 2003;23:6229–6242. doi: 10.1128/MCB.23.17.6229-6242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Lee AY, Lai HT, Zhang H, Chiang CM. Phospho switch triggers brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell. 2013;49:843–857. doi: 10.1016/j.molcel.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Prakash C, Sum C, Gong Y, Li Y, Kwok JJ, Thiessen N, Pettersson S, Jones SJ, Knapp S, et al. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J Biol Chem. 2012;287:43137–43155. doi: 10.1074/jbc.M112.413047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.