Abstract
Parasite infection impacts population dynamics through effects on fitness and fecundity of the individual host. In addition to the known roles of environmental factors, host susceptibility to parasites has a genetic basis that has not been well characterized. We previously mapped quantitative trait loci (QTL) for susceptibility to rat tapeworm (Hymenolepis diminuta) infection in Tribolium castaneum using dominant AFLP markers; however, the resistance genes were not identified. Here, we refined the QTL locations and increased the marker density in the QTL regions using new microsatellite markers, sequence-tagged site markers, and single-strand conformational polymorphism markers. Resistance QTL in three linkage groups (LG3, LG6, and LG8) were each mapped to intervals <1.0 cM between two codominant markers. The effects of 21 genes in the three QTL regions were investigated by using quantitative RT-PCR analysis, and transcription profiles were obtained from the resistant TIW1 and the susceptible cSM strains. Based on transcription data, eight genes were selected for RNA interference analysis to investigate their possible roles in H. diminuta resistance, including cytochrome P450 (LOC657454) and Toll-like receptor 13 (TLR13, LOC662131). The transcription of P450 and TLR13 genes in the resistant TIW1 strains was reduced more than ninefold relative to the control. Moreover, the effects of gene knockdown of P450 and TLR13 caused resistant beetles to become susceptible to tapeworm infection, which strongly suggests an important role for each in T. castaneum resistance to H. diminuta infection.
Keywords: parasite resistance gene, high-resolution mapping, quantitative trait loci, Hymenolepis diminuta, Tribolium castaneum
PARASITES exert negative effects on host survivorship and reproductive success, and in turn hosts develop resistance to parasites by reducing susceptibility to infection. Despite the importance to medicine and agriculture of host resistance to parasitism, and the fact that host resistance is genetically determined, the genetics of parasite resistance in insect hosts is not well known. Infection of the red flour beetle, Tribolium castaneum, by the rat tapeworm, Hymenolepis diminuta, has been well characterized with regard to host–parasite interactions (Keymer and Anderson 1979; Zhong et al. 2003, 2005). The system also represents an excellent model to study the evolutionary genetics of resistance to parasite infection, as infection can be easily controlled and monitored in the laboratory. Serving as an intermediate host, infection of Tribolium by H. diminuta occurs upon ingestion of the parasite eggs in rat feces. The eggs then hatch and develop into cysticercoids that are capable of infecting the mammalian host and amplifying when ingested, but cannot be horizontally or vertically transmitted.
Previously, we used AFLP markers to identify three major quantitative trait loci (QTL) that affected T. castaneum beetle susceptibility to H. diminuta on linkage groups LG3 [hds 4(3, L1B1.69)], LG6 [hds(6, L1A16.141)], and LG8 [hds(8, L6B2.100)]. The gene action at QTL hds(3, L1B1.69) and hds(8, L6B2.100) was overdominance in the resistant TIW1 strain. In contrast, the gene action at QTL hds(6, L1A16.141) was overdominance in the resistant TIW1 strain. In contrast, the gene action at QTL hds[6, L1A16.141] was underdominance or recessive in resistant TIW1 strain (Zhong et al. 2003, 2005). Fine-mapping of QTL can be achieved by increasing marker density within the chromosomal region of interest, increasing the number of individuals for which phenotypic information can be obtained, or by increasing the accuracy of assigning QTL genotypes (Nezer et al. 2003). QTL should be defined by informative, codominant markers within a 1- to 2-cM range or less to facilitate candidate gene identification (Yu et al. 2006). However, the QTL that we identified were positioned within large marker intervals due to the dominant AFLP markers used in the linkage analysis (Adarichev et al. 2003). In addition, the genomic regions containing the QTL were too large for positional cloning. Thus, the resistance genes could not be mapped with a reasonable degree of certainty, nor could sufficient sequence information be obtained.
To decrease the size of the marker intervals obtained in our previous study and facilitate the identification of the T. castaneum resistance genes, we applied the advanced intercross lines (AIL) method for high-resolution fine-mapping of QTL (Darvasi and Soller 1995). An AIL is generated by random intercross breeding of two inbred strains for several generations. As such, AIL can accumulate many more recombination events than the conventional methods. AIL has been used to refine multiple proximally located QTL (Iraqi et al. 2000; Wang et al. 2003; Jagodic et al. 2004). Recent advances in Tribolium genome resources, including the development of a high-resolution bacterial artificial chromosome (BAC) fingerprint map that integrates genetic, physical, and comparative mapping information and genome sequence data (Tribolium Genome Sequencing Consortium 2008), provide a powerful means of identifying new high-density markers for QTL analysis. Utilizing this technology, we increased the codominant marker density in the QTL regions associated with resistance to H. diminuta and refined the map position of the parasite resistance QTL. We conducted high-resolution QTL analysis in the 15th generation (G15) of an advanced intercross line, focusing specifically on the three previously identified QTL (Zhong et al. 2003). Based on the results of the QTL analysis, transcription profiles were obtained for selected candidate genes in resistant and susceptible populations, and RNA interference (RNAi) was used to investigate the function of putative resistance genes with regard to the resistant and susceptible phenotypes.
Materials and Methods
Mapping populations
Two T. castaneum strains, cSM and TIW1, were used to set up segregating populations for QTL high-resolution mapping. Both were standard laboratory strains that have been used in genetic linkage mapping and other ecological and evolutionary genetics studies. TIW1 is less susceptible to tapeworm infection than is cSM, but there is considerable intrastrain variability in beetle susceptibility to parasites. The two strains have been reared in the laboratory for >15 years. The origin of the two strains and their F1 and F2 segregation populations have been published (Zhong et al. 2003, 2005). The first population was generated from pairwise mating between a TIW1 male and a cSM female and an F1 intercross (cross 1). The second was from pairwise mating between a cSM male and a TIW1 female and an F1 intercross (cross 2). Two AIL G15 (after 15 generations of inbreeding in cross 1 or cross 2) segregation populations were generated by random intercross breeding in this study. Beetles were raised in 8-dram shell vials (25 mm × 95 mm) containing ∼5 g standard medium (95% by weight fine-sifted whole-wheat flour and 5% dried powdered brewer’s yeast). Experimental vials were maintained in a dark incubator regulated at 29° and 70% relative humidity. All beetles used for QTL fine-mapping studies were raised under the same conditions.
Tapeworm infection and DNA extraction
Fresh rat feces mixed with H. diminuta eggs were obtained from Carolina Biological Supply Company (Burlington, NC), which has maintained rats infected with H. diminuta for the past 40 years. To identify the tapeworm parasite genotypes, the DNA of H. diminuta eggs was extracted, and the PCR fragments of the mitochondrial cytochrome c oxidase subunit I (COI) gene and ribosomal internal transcribed spacer 2 (ITS2) gene were sequenced using the PCR primers (COI gene: 5′-CGGGTATTGGCTGAACATTT-3′ and 5′-ACACTCGACGAGGTAAACCA-3′; ITS2 gene: 5′-GAACTGTATGCGGTGGATCA-3′ and 5′-AAGTTCAGCGGGTAATCACG-3′). Three haplotypes of the COI gene sequence were identified with one to two mutations for the 957-bp PCR fragment (GenBank accession nos. KC990401–KC990403), and seven unique ITS2 sequences were detected with three microsatellite polymorphic regions for the PCR fragment 756–769 bp (GenBank accession nos. KC990404–KC990410). Phylogenetic analysis of three COI gene sequences indicated that the tapeworm parasites are most closely related to the USA strain (GenBank accession no. AF314223). Therefore, the parasites used in the present study are mixed genotypes.
The parental TIW1 and cSM populations, F1 and G15 individuals from reciprocal crosses of TIW1 and cSM strains, were evaluated for tapeworm susceptibility using the infection and dissection methods described previously (Pai and Yan 2003). A total of 300 female and 300 male beetles of each cross from the G15 segregating population were exposed to tapeworm eggs following the previously established infection protocol (Pai and Yan 2003). Large sample size is critical for placing the resistance genes in relation to closely linked molecular markers with high confidence. Two weeks post infection, 200 live beetles of each sex were selected and dissected in a cold DNA extraction buffer to determine infection intensity. The beetle carcasses were collected for DNA extraction and genotyping, and the parasite tissues were discarded (Zhong et al. 2003).
Screening and development of molecular markers
Three major QTL on LG3 [hds(3, L1B1.69)], LG6 [hds(6, L1A16.141)], and LG8 [hds(8, L6B2.100)] were selected for QTL high-resolution mapping. The primer sequences for microsatellite and sequence-tagged site (STS) markers previously developed in T. castaneum (Demuth et al. 2007; Lorenzen et al. 2005) were used to screen for polymorphism between TIW1 and cSM parent strains surrounding the three QTL regions of ∼10 megabases. To increase the marker density, we developed new microsatellite and single-strand conformational polymorphism (SSCP) markers in the QTL region. The microsatellite markers were developed based on the Tribolium genome sequences using the online software WebSat (http://wsmartins.net/websat/). The SSCP markers were designed based on the sequences of the intron region of genes surrounding the QTL regions using the Primer3 software (http://frodo.wi.mit.edu/primer3/). The sequences of forward and reverse primers are listed in Supporting Information, Table S1. Marker polymorphism was screened using the method described previously (Zhong et al. 2004, 2006) and the Li-Cor model 4300 automated DNA analyzer (Li-Cor, Lincoln, NE). The Gene ImagIR 4.33 software (Li-Cor) program was used to quantify allele size based on the pattern of the height of signal peaks.
Genotyping of AIL mapping population
The polymorphic markers between parents were selected to genotype two G15 segregation populations. The detection of PCR products and PCR conditions were described previously (Zhong et al. 2004, 2006). A total of 800 individuals were genotyped in the two G15 populations using 54 polymorphic markers (Table S1). PCR products were resolved on denaturizing polyacrylamide gels and were detected by using the Li-Cor Model 4300 automated DNA analyzer (Li-Cor). Genotypes were evaluated manually and double-checked by the same individual..
Double-stranded RNA synthesis and RNAi analysis
Double-stranded RNA (dsRNA) was synthesized using the Ambion MEGAscript high-yield transcription kit as described previously (Tomoyasu et al. 2008). The sequences of gene-specific primers (marked with footnote a in Table 1) for dsRNA synthesizing and the size of the dsRNA are listed in Table 1. Total RNA was extracted from cSM and TIW1 adult beetles. The dsRNA of genes on LG3 and LG8 were synthesized from the total RNA extract from the TIW1 strain (dsRNA-T), while the dsRNA of genes on LG6 were synthesized from the total RNA extract from the cSM strain (dsRNA-C). Approximately 300–500 ng of dsRNA was injected into each beetle pupa (dsRNA-T for the TIW1 strain, dsRNA-C for cSM strain). For each RNAi experiment, 100 female pupae were injected with dsRNA and another 100 female pupae were injected with injection buffer as control. Injected pupae were then kept in a dark incubator at 29° and 70% relative humidity. Two weeks later, half the adult beetles were evaluated for tapeworm susceptibility using the infection and dissection methods previously described (Pai and Yan 2003). The other half of the beetles was used for gene expression analysis.
Table 1. Genes selected for RNAi assay.
| No. | Locusa | LG | Location (cM) | Gene name | Forward primer | Reverse primer | Size (bp) |
|---|---|---|---|---|---|---|---|
| 1 | LOC100142401 | 3 | 13.7 | Similar to discoidin domain receptor CG33531-PA | ATCTATGGGTGTCGCTGGAC | TGTTCTTACGCGTGTCGTTC | 244 |
| 2 | LOC657454a | 3 | 14.1 | Similar to probable cytochrome P450 | ACTCGCTGAAAGACCGAAAA | AAGTGTTCTTCCGGGTGTTG | 277 |
| 3 | LOC100141631a | 3 | 14.2 | Similar to AGAP006113-PA | CCTGGAGGACACCTCGAATA | CGTCACCATATTGACGCAAC | 232 |
| 4 | LOC657788 | 3 | 15.2 | Hypothetical protein LOC657788 | TCTCCACTGCAGACACCAAG | AACCCAATTTTTCCGATTCC | 387 |
| 5 | LOC659770 | 3 | 15.4 | Similar to CG5642-PA | TGGATTCCATCCCTGATGAT | AGCTCTGAGCCACTCTCCAG | 310 |
| 6 | LOC660033 | 3 | 15.5 | Similar to CG1221-PA, isoform A | AGTGAACGAGGAGCCAAGAA | GTTGGGCTTCAGTTTGTCGT | 243 |
| 7 | LOC654972 | 3 | 16.7 | Similar to CG5547-PD, isoform D | GCGCTTTTGATCTCTTCCAC | TCAAACTCCAGCCTGTTCCT | 406 |
| 8 | LOC663292 | 6 | 7.8 | Similar to CG4225-PA | CTTGGTTCAGTCCCGTTTGT | CAGAAGGGAAAAGGGGTAGG | 172 |
| 9 | LOC663441a | 6 | 7.9 | Similar to CG1512-PB, isoform B | CAGAAACTGTCCGATGCTGA | AAGAATTGTGCCCCTGACTG | 216 |
| 10 | LOC663587a | 6 | 8.1 | Similar to CG2052-PB, isoform B | CAGCATCTCAGAAACCACGA | ATTGGGGTCCTTGTGTGTGT | 234 |
| 11 | LOC663602a | 6 | 8.7 | Similar to CG7332-PA | CCGAAACTAAAACCGACCAA | GCACCAATGAAAACACATCG | 249 |
| 12 | LOC655450 | 6 | 9.6 | Similar to leucine-rich repeat containing 47 | CCCTCCGGAAATTAACCAAT | CCTTTTGTTCGCTCTTTTGC | 239 |
| 13 | LOC656106 | 6 | 9.7 | Similar to ubiquitin-specific protease 2 | TAACCGAAGACGGCATAACC | AGTTGGCCGGTGAGTATTTG | 228 |
| 14 | LOC661773 | 8 | 10.5 | Similar to potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | TGGTGGCAATGAGCAAAATA | CATAGCGGAACACCCTGTTT | 231 |
| 15 | LOC661984 | 8 | 10.9 | Similar to CG10889-PA | AGTGCACATACGGCAACAAA | ACACCACGACATGTTGCACT | 195 |
| 16 | LOC662131a | 8 | 11.0 | Similar to Toll-like receptor 13 | ACGGTTTGGACAACTTGGAG | CCCTCAGGTGCGTATTTGTT | 228 |
| 17 | LOC662235a | 8 | 11.2 | Similar to CG17839-PB, isoform B | CTGTTGTTTTGGCCCTTGTT | GTTCTTCCAGCCTTGCACTC | 245 |
| 18 | LOC662348a | 8 | 11.4 | Similar to CG8503-PA | AGAAGCAGTTCTTGCCCAAA | TCGTGTCCATTGACCTGAAA | 208 |
| 19 | LOC662521 | 8 | 11.8 | Similar to myeloid leukemia factor (myelodysplasia-myeloid leukemia factor) | ACCGCATGAGCAACTCTCTT | TAATACCCCCAGGAGCTGTG | 198 |
| 20 | LOC663023 | 8 | 12.0 | Similar to glyceraldehyde-3-phosphate dehydrogenase II | CAAAGTTATCCCGGCTTTGA | AAATCGACGAGTGCGTATCC | 230 |
| 21 | LOC663134 | 8 | 12.1 | Similar to endothelin-converting enzyme 2 | TTTTTACCGCTCCTGCCTTA | AAAATCAAATTCCCCGGTTC | 237 |
Locus name, location, product size, and primer sequence of qRT-PCR of the candidate genes in the QTL regions in T. castaneum.
The dsRNA of genes on LG3 and LG8 were synthesized from the total RNA extract from the TIW1 strain (dsRNA-T), while the dsRNA of genes on LG6 were synthesized from the total RNA extract from the cSM strain (dsRNA-C).
Quantitative RT-PCR analysis
To test the expression difference between the two parental strains (resistant TIW1 beetles and susceptible cSM strains), we used quantitative real-time PCR (qRT-PCR) analysis and selected the genes surrounding the QTL regions. A total of 21 genes in the three QTL regions were selected (Table 1). Total RNA was extracted from a pool of 20 resistant TIW1 beetles and 20 susceptible cSM beetles (2-week-old female adults) in the absence of parasite infection, using the RNeasy Mini Kit (Qiagen Inc., Valencia, CA). The RNA was then treated with DNase I. Similarly, total RNA was extracted from 20 dsRNA-injected beetles and 20 control beetles at 14 days post infection. The poly(A+) messenger RNA (mRNA) was isolated from the total RNA by the Oligotex mRNA kit (Qiagen) according to the Qiagen protocol. The purified mRNA samples were used for reverse transcription, using the iScriptTM cDNA synthesis kit (Bio-Rad, Hercules, CA). The qRT-PCR primers for each candidate gene were designed using Primer 3.0, based on the corresponding complementary DNA sequences obtained from GenBank. The qRT-PCR analysis was performed using the method described previously (Wang et al. 2010). The ribosomal protein RPS3 and RPS18 genes were used as the standard for expression normalization for each gene because of their stable expression in T. castaneum after exposure to bacteria (Lord et al. 2010). The qRT-PCR assays were conducted in triplicate, and the average value of the triplicate was used for analysis of gene expression differences.
Linkage analysis and statistics
Linkage analysis between molecular markers and resistance QTL was performed using GNU R 2.6.0 with the QTL package version 1.07–12 (R/qtl) (Broman et al. 2003). The physical map is derived from the National Center for Biotechnology Information (NCBI) Tribolium genome sequence at http://www.ncbi.nlm.nih.gov. Marker positions were obtained from the NCBI Tribolium genome Build 2.1 statistics. Data were analyzed by implementing a nonparametric model for quantitative traits. Confidence intervals (C.I.’s) for QTL were defined as the region within the maximum logarithm of odds (LOD). To further evaluate identified QTL, a multiple QTL model test was performed using R/qtl software. Analysis was performed for the two reciprocal crosses of G15 populations separately and also in a combined analysis to increase statistical power. Separate analyses were performed with the multiple imputation method (normal model) with 64 simulations (step = 2, ndraws = 64) in R/qtl. Because different populations were used in the combined cross analysis, marker regression with 64 simulations was used. Only physical positions for the markers were used in the combined analysis since they are constant between populations in contrast to genetic positions. The experiment-wise significance threshold levels were determined by the permutation method in R/qtl using 10,000 permutations (Broman et al. 2003).
Gene expression data obtained from qRT-PCRs for 21 loci in both TIW1 and cSM strains were analyzed as a one-way ANOVA. To assess differences in tapeworm infection intensity with or without dsRNA injection (RNAi), the gene expression ratio data obtained from qRT-PCRs in eight selected loci in the TIW1 or cSM strain were also explored using one-way ANOVA. We used pairwise Student’s t-tests to detect statistically significant differences in the absolute average infection intensity of tapeworm between dsRNA and buffer injections. JMP software package (SAS Institute Inc., Cary, NC) was used to perform the statistical analysis.
Results
Phenotypic variability in susceptibility to tapeworm parasites in AIL
Infection intensity of the cSM and TIW1 parental and the resultant F1 progeny was reported previously, with the cSM and F1 populations being significantly more susceptible to tapeworm parasite infection than the TIW1 (Zhong et al. 2003, 2005). In the present study, the individuals of two G15 AIL populations were exposed to feces obtained from H. diminuta-infected rats. A total of 400 G15 AIL beetles in each cross were infected with tapeworm parasites. The average infection intensity of cross 1 AIL populations was 5.3 ± 0.7 (range 0–28; n = 400) and 3.85 ± 0.5 (range 0–22; n = 400) for cross 2. There was no difference in the parasite infection intensity between male and female beetles for cross 1 and cross 2, which suggests that sex is not a factor for susceptibility to tapeworm infection.
High-resolution mapping of the QTL [hds(3, L1B1.69)] region on linkage group LG3
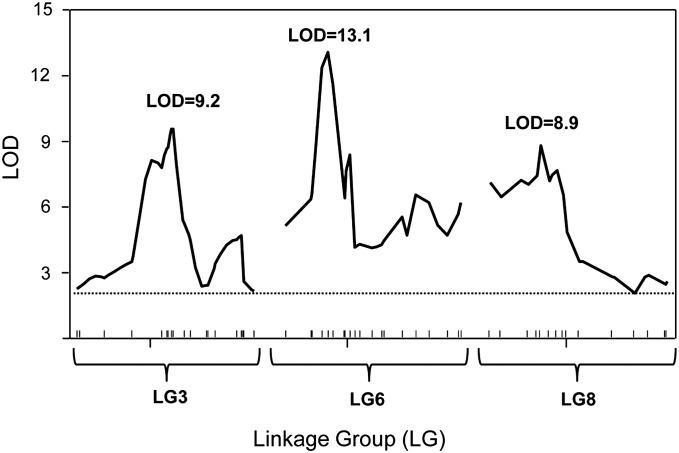
A 14.17-cM (∼5.0-Mb) region of the QTL LG3 [hds(3, L1B1.69)] was selected for high-resolution mapping (map position: 6.48–20.65 cM). Previously described markers (Zhong et al. 2004; Demuth et al. 2007) were used to screen for polymorphism between cSM and TIW1 parent strains. Of the 42 markers previously reported on LG3, 9 were polymorphic between the two parental strains. Ninety-two new primers were subsequently designed based on Tribolium genome sequences in the QTL region on LG3; 10 of these were polymorphic between the two parental strains. Thus, a total of 19 markers were used to genotype the two AIL populations (Table S1). Linkage analysis was performed in two AIL populations separately and confirmed the locus with a high-LOD score (>8) in the two AILs. The combined population linkage analysis indicated an LOD score of 9.2 at an interval of the two markers Tca3.4924 and Tca3.4970 for the QTL (Figure 1, Figure S1A, and Table S1). The QTL region was narrowed to a genomic region of ∼0.13 cM (46 kb) within a 95% C.I. Based on the Tribolium genomic database, this region contains four predicted genes, including a candidate gene, cytochrome P450 (GenBank accession no. XM_963914) (Figure S1A), an important enzyme system involved in insecticide metabolism.
Figure 1.
Fine-mapping QTL-associated beetle susceptibility to tapeworm parasite in AIL (G15). LOD scores are presented on the y-axis and physical positions (in centimorgans) of the linkage group are given on the x-axis. LOD score values were determined using R/qtl software (Broman et al. 2003). The physical position of the markers was retrieved from the T. castaneum genome database at http://www.ncbi.nlm.nih.gov/genome/guide/beetle/index.html.
High-resolution mapping of the QTL [hds(6, L1A16.141)] region on linkage group LG6
A 14.31-cM (∼5.0-Mb) region of the QTL LG6 [hds(6, L1A16.141)] was selected for high-resolution mapping by the same method used for the analysis on LG3. Of 17 markers previously reported in the region (Demuth et al. 2007), 9 markers identified polymorphisms in the parental strains and 11 new markers were then developed, which identified additional polymorphisms in this region. A total of 20 markers were used to genotype the two AIL populations (Table S1). Linkage analysis was performed in two AIL populations separately, and the locus was confirmed with a high-LOD score of >10 in the two AILs. The combined population linkage analysis indicated an LOD score of 13.1 at an interval of the two markers Tca6.2726 and Tca6.2926 for the QTL (Figure 1, Figure S1B, and Table S1). The QTL region was narrowed to a genomic region of ∼0.55 cM (192 kb) within a 95% C.I. Based on the Tribolium genomic database, this region contains 11 predicted genes of unknown function (Figure S1B).
High-resolution mapping of the QTL [hds(8, L6B2.100)] region on linkage group LG8
A 9.93-cM (∼3.48-Mb) region of the QTL LG8 [hds(8, L6B2.100)] was selected for high-resolution mapping. Of 20 markers previously reported in the region (Demuth et al. 2007), seven markers identified polymorphisms in the parental strains, and eight newly developed markers identified additional polymorphisms (Table S1), resulting in a total of 15 markers that were used to genotype the two AIL populations. The linkage analysis was performed separately, and the locus was confirmed with a high-LOD score of >8 in the two AILs. The combined population linkage analysis indicated an LOD score of 8.9 at an interval of the two markers Tca8.3781 and Tca8.4007 for the QTL (Figure 1, Figure S1C, and Table S1). The QTL region was narrowed to a genomic region of ∼0.70 cM (245 kb) with a 95% C.I. This region contains 12 predicted genes, including a candidate gene, Toll-like receptor 13 (TLR13; GenBank accession no. XM_968248) (Figure S1C), which has been implicated in innate immunity and endotoxin susceptibility in mice (Roach et al. 2005; Mishra et al. 2008; Shi et al. 2009).
Gene expression differences between resistant and susceptible strains
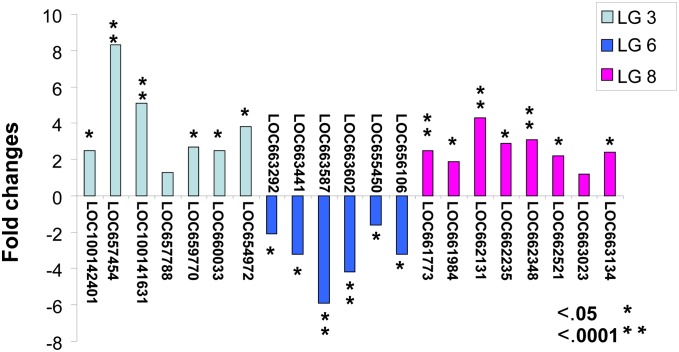
The transcription profiles of genes associated with the loci identified in the resistant TIW1 and susceptible cSM strains (2-week-old female adults) were examined using qRT-PCR. A total of 21 genes surrounding the three QTL regions were selected for qRT-PCR analysis: seven genes on LG3, six genes on LG6, and 8 genes on LG8 (Table 1, Figure 2). The range of expression level between the resistant and susceptible strain was a 1.3- to 8.3-fold difference for LG3, 1.6- to 5.9-fold for LG6, and 1.2- to 4.3-fold for LG8 (Figure 2). Nineteen genes of the 21 selected genes showed significant differences in gene expression (P < 0.05) between resistant and susceptible strains. The resistant TIW1 strain had a higher expression level for the genes on LG3 and LG8 whereas a lower expression level was found for the genes on LG6 (Figure 2). These results are consistent with the overdominance of QTL hds[3, L1B1.69] on LG3 and hds[8, L6B2.100] on LG8 and underdominance or recessive of QTL hds[6, L1A16.141] on LG6 in the resistant TIW1 strain (Zhong et al. 2003, 2005).
Figure 2.
Gene expression difference (in fold) between the resistant TIW1 strain and the susceptible cSM strain in the absence of parasite infection. A positive value indicates that the resistant strain had a higher mRNA level, while a negative value indicates that the susceptible strain had higher mRNA level.
RNAi effect on parasite infection intensity and gene expression level changes
A total of eight genes were selected for RNAi analysis (Table 2). RNAi was performed by injecting the beetle pupae and by subsequently examining the parasite infection intensity and comparing the mRNA levels with those of buffer-injected controls. Significant differences in parasite infection densities were observed for genes on LG3 and LG8, but not on LG6 (Table 2). The dsRNA injected beetles had significantly more parasites compared to control beetles (buffer-injected) in the resistant TIW1 strain, but not in the susceptible cSM strain. Transcription levels for the selected genes were significantly altered after RNAi treatment for both the resistant and the susceptible strains (Table 3). Both genes (LOC657454 and LOC100141631) on LG3 had significantly reduced transcription (10- to 13-fold changes, P < 0.001). Similar to the two genes on LG3, the three genes used for RNAi analysis on LG8 had a significant gene knockdown effect (3- to 9-fold changes, P < 0.01). The TLR13 gene (LOC662131) had the highest gene knockdown effect (9.49-fold change, P < 0.001) in the resistant TIW1 strain (Table 3). While no significant difference in parasite infection intensity was observed for genes on LG6 as a result of RNAi treatment, the transcription levels of the three selected LG6 genes were significantly altered.
Table 2. RNAi effect on parasite infection intensity.
| Strain/treatment (injection) | LG | n | Infection intensity (mean ± SE) | Significancea |
|---|---|---|---|---|
| TIW1 (dsRNA-T) | ||||
| LOC657454 (dsRNA) | 3 | 44 | 5.30 ± 0.93 | A |
| LOC662131 (dsRNA) | 8 | 44 | 4.48 ± 0.60 | A |
| LOC100141631 (dsRNA) | 3 | 38 | 2.42 ± 0.40 | B |
| LOC662348 (dsRNA) | 8 | 40 | 1.85 ± 0.41 | BC |
| LOC662235 (dsRNA) | 8 | 45 | 1.58 ± 0.32 | BC |
| Control (injection buffer) | 48 | 0.60 ± 0.15 | C | |
| cSM (dsRNA-C) | ||||
| LOC663441 (dsRNA) | 6 | 43 | 7.74 ± 1.03 | A |
| LOC663587 (dsRNA) | 6 | 42 | 8.81 ± 1.08 | A |
| LOC663602 (dsRNA) | 6 | 44 | 7.91 ± 1.10 | A |
| Control (injection buffer) | 47 | 8.60 ± 1.07 | A |
Student’s t-test was performed to compare the difference. n, number of beetles.
RNAi effect on parasite infection. Means with the same letter are not significantly different.
Table 3. RNAi effect on gene expression.
| Strain/locus name | LG | Expression changes (RNAi to control) | ANOVA (P-value) |
|---|---|---|---|
| TIW1 | |||
| LOC657454 | 3 | −13.21 | 0.0002 |
| LOC662131 | 8 | −9.49 | 0.0008 |
| LOC100141631 | 3 | −10.31 | <0.0001 |
| LOC662348 | 8 | −3.63 | 0.0015 |
| LOC662235 | 8 | −5.00 | 0.0074 |
| cSM | |||
| LOC663441 | 6 | −6.56 | 0.0006 |
| LOC663587 | 6 | −15.00 | <0.0001 |
| LOC663602 | 6 | −5.43 | 0.0046 |
The gene expression between RNAi and control groups is compared. ANOVA analysis for significance is listed.
Discussion
To refine the QTL associated with T. castaneum susceptibility to H. diminuta infection, we generated two AIL populations from resistant TIW1 and susceptible cSM strains. Using 29 newly developed microsatellite and STS markers, together with 25 previously published microsatellite markers, we were able to identify three tapeworm-resistant QTL in genome regions of ∼0.13 cM on LG3, 0.55 cM on LG6, and 0.70 cM on LG8. The QTL on LG3 contain sequences homologous to cytochrome P450 (CPYIXF2), a family of enzymes involved in insecticide activation and detoxification. The QTL on LG8 contain gene TLR13, a novel member of the Toll-like receptor family (Shi et al. 2009), which plays a key role in the innate immune system. Because these QTL regions may include genes that play a role in inhibiting parasite development in the beetle host, we designated these genes as candidate genes for resistance to H. diminuta infection.
A biological basis for the involvement of the candidate genes in resistance to tapeworm infection may exist. Cytochrome P450s constitute the largest gene superfamily found in nature, displaying a wide variety of functions. However, only a small subset of these cytochrome P450 genes is involved in insecticide metabolism and the innate immune response. Insect cytochrome P450s are known to play an important role in detoxifying insecticides, such as pyrethroids (Feyereisen 2005; Karunker et al. 2009). The most important character of insect cytochrome P450s is that they are constitutively overexpressed in the insecticide-resistant phenotype, causing enhanced metabolic detoxification of insecticides (Feyereisen 2005; Zhu et al. 2008). In Tribolium, >200 sequences of P450 genes have been registered in the GenBank database, but the physiological functions of P450s remain largely unknown. Zhu et al. (2010) identified a P450 gene, CYP6BQ9 responsible for the majority of deltamethrin resistance that showed a >200-fold increase in expression in the deltamethrin-resistant QTC279 strain when compared with a deltamethrin-susceptible Lab-S strain.
In our present study, we found that the resistant TIW1 strain had significantly higher transcription of CPYIXF2 (LOC657454, probable cytochrome P450) compared to the susceptible cSM strain in the absence of parasite infection. Our analysis showed that injection of dsRNA corresponding to CYPIXF2 sequences in the resistant TIW1 strain (LOC657454) resulted in a significant increase in infection intensity that was accompanied by a significant decrease in the transcription of CYPIXF2. These data strongly suggest that this P450 gene is involved in resistance to tapeworm infection.
Toll-like receptors (TLRs) are an important family of pattern-recognition receptors that play a key role in the innate immune system (Roach et al. 2005). The Toll receptor was initially identified in Drosophila melanogaster for its role in embryonic development and was recognized as a key regulator of immune response (Leulier and Lemaitre 2008). TLRs have been identified in many animal species, including pigs (Shinkai et al. 2006), chickens (Fukui et al. 2001), fish (Tsujita et al. 2004), and insects (Tauszig et al. 2000; Imamura and Yamakawa 2002; Aronstein and Saldivar 2005). It has been estimated that most mammalian species have between 10 and 15 types of Toll-like receptors (Roach et al. 2005). Insect TLRs and mammalian TLRs are evolutionarily conserved and share characteristic domain organization (Gangloff et al. 2003; Roach et al. 2005). It has been demonstrated that experimental infection of humans with the malaria parasite Plasmodium falciparum can enhance TLR-mediated responses (Franklin et al. 2009). In T. castaneum, Zou et al. (2007) reported nine genes (TLRs 1–4, TLRs 6–10) that encode Toll and Toll homologs. Zhang et al. (2004) demonstrated that TLR11 recognizes urinary pathogenic Escherichia coli. TLR13 expression was evident in brain cells of mice, both infected and uninfected with the Mesocestoides corti parasite, and parasite infection caused a several-fold increase in mRNA and protein levels of TLR13 (Mishra et al. 2008). In our study, the TLR13 gene of the resistant TIW1 strain exhibited a significantly higher TLR13 mRNA level than the susceptible cSM strain in the absence of parasite infection. RNAi analysis showed that injection of dsRNA corresponding to the TLR13 gene sequences (LOC662131) resulted in a significant gene knockdown effect in the resistant TIW1 strain, as evidenced by the presence of a significantly higher number of parasites than in the buffer-injected controls. Therefore, these data strongly suggest the role of TLR13 in T. castaneum resistance to H. diminuta infection.
Only a few invertebrate parasite-susceptibility QTL-mapping studies have been completed to date, and all have reported multiple regions involving parasite susceptibility, such as QTL affecting malaria-parasite and filarial-parasite infection in Aedes aegypti (Beerntsen et al. 1995; Severson et al. 1995, 1999; Morlais et al. 2003; Aronstein and Saldivar 2005) and in Anopheles gambiae (Menge et al. 2006). The polygenic, quantitative genetic patterns that we have observed in these experiments are supported by earlier studies that approached parasite resistance evolution as a quantitative character rather than a single genetic factor (or gene). Characterization of the Tribolium beetle in response to tapeworm parasite infection will provide valuable insights into the molecular basis of host resistance to parasites and coevolution between beetle host and tapeworm parasites. This will have important implications for the development of novel strategies for Tribolium pest control. The knowledge and tools developed in such studies are useful in analyzing natural host–parasite systems.
Gene expression analysis was performed on female beetles only; the effects of host sex and age on their immune response and resistance to parasite infection were not examined. Freitak et al. (2012) found that 54% of microRNAs exhibited gender-specific expression patterns upon exposure to environmental stress in T. castaneum. When we examined the between-sex variation in the expression of 29 immune-related genes in T. castaneum, we found significant among-strain variations in the response of the immune-related genes (Zhong et al. 2013). Furthermore, despite the fact that the tapeworm parasites used in the study have been maintained in the Carolina Supply Company for 40 years, we identified three genotypes by COI gene, suggesting that tapeworm parasites were not genetically homogenous. This is consistent with natural situations in which parasite populations are genetically heterogeneous. More studies are needed to determine the effects of interactions between parasite and host genotypes on host infectivity.
In summary, the tapeworm parasite resistance QTL of T. castaneum beetles were finely mapped to genomic regions in T. castaneum using AILs and high-density molecular markers. Two genes were identified as candidates for resistance genes to H. diminuta infection (CPYIXF2 and TLR13) using transcription analysis and RNAi. This work not only lays a foundation for identification and cloning of tapeworm parasite resistance genes in the Tribolium host, but also improves the recognition of function in resistance genes to the indirectly transmitted macroparasites.
Supplementary Material
Acknowledgments
We thank Sarah May K. Daguplo, Amy Gillen, Christina Costa, Varma Penumetsa, and Monica Karsay-Klein for technical assistance. Mariangela Bonizzoni provided helpful advice. Two anonymous reviewers provided constructive criticisms and helpful suggestions. This research was supported by a grant from the National Science Foundation (DEB 0716275).
Footnotes
Communicating editor: C. D. Jones
Literature Cited
- Adarichev V. A., Valdez J. C., Bardos T., Finnegan A., Mikecz K., et al. , 2003. Combined autoimmune models of arthritis reveal shared and independent qualitative (binary) and quantitative trait loci. J. Immunol. 170: 2283–2292. [DOI] [PubMed] [Google Scholar]
- Aronstein K., Saldivar E., 2005. Characterization of a honey bee Toll related receptor gene Am18w and its potential involvement in antimicrobial immune defense. Apidologie (Celle) 36: 3–14. [Google Scholar]
- Beerntsen B. T., Severson D. W., Klinkhammer J. A., Kassner V. A., Christensen B. M., 1995. Aedes aegypti: a quantitative trait locus (QTL) influencing filarial worm intensity is linked to QTL for susceptibility to other mosquito-borne pathogens. Exp. Parasitol. 81: 355–362. [DOI] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Darvasi A., Soller M., 1995. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 141: 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth J. P., Drury D. W., Peters M. L., Van Dyken J. D., Priest N. K., et al. , 2007. Genome-wide survey of Tribolium castaneum microsatellites and description of 509 polymorphic markers. Mol. Ecol. Notes 7: 1189–1195. [Google Scholar]
- Feyereisen, R., 2005 Insect cytochrome P450, pp. 1–77 in Comprehensive Molecular Insect Science, edited by L. I. Gilbert, K. Iatrou, and S. S. Gill. Elsevier, Amsterdam; New York. [Google Scholar]
- Franklin B. S., Parroche P., Ataide M. A., Lauw F., Ropert C., et al. , 2009. Malaria primes the innate immune response due to interferon-γ induced enhancement of toll-like receptor expression and function. Proc. Natl. Acad. Sci. USA 106: 5789–5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitak D., Knorr E., Vogel H., Vilcinskas A., 2012. Gender- and stressor-specific microRNA expression in Tribolium castaneum. Biol. Lett. 8: 860–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui A., Inoue N., Matsumoto M., Nomura M., Yamada K., et al. , 2001. Molecular cloning and functional characterization of chicken Toll-like receptors. J. Biol. Chem. 276: 47143–47149. [DOI] [PubMed] [Google Scholar]
- Gangloff M., Weber A. N., Gibbard R. J., Gay N. J., 2003. Evolutionary relationships, but functional differences, between the Drosophila and human Toll-like receptor families. Biochem. Soc. Trans. 31: 659–663. [DOI] [PubMed] [Google Scholar]
- Imamura M., Yamakawa M., 2002. Molecular cloning and expression of a Toll receptor gene homologue from the silkworm, Bombyx mori. Biochim. Biophys. Acta 1576: 246–254. [DOI] [PubMed] [Google Scholar]
- Iraqi F., Clapcott S. J., Kumari P., Haley C. S., Kemp S. J., et al. , 2000. Fine mapping of trypanosomiasis resistance loci in murine advanced intercross lines. Mamm. Genome 11: 645–648. [DOI] [PubMed] [Google Scholar]
- Jagodic M., Becanovic K., Sheng J. R., Wu X., Backdahl L., et al. , 2004. An advanced intercross line resolves Eae18 into two narrow quantitative trait loci syntenic to multiple sclerosis candidate loci. J. Immunol. 173: 1366–1373. [DOI] [PubMed] [Google Scholar]
- Karunker I., Morou E., Nikou D., Nauen R., Sertchook R., et al. , 2009. Structural model and functional characterization of the Bemisia tabaci CYP6CM1vQ, a cytochrome P450 associated with high levels of imidacloprid resistance. Insect Biochem. Mol. Biol. 39: 697–706. [DOI] [PubMed] [Google Scholar]
- Keymer A. E., Anderson R. M., 1979. The dynamics of infection of Tribolium confusum by Hymenolepis diminuta: the influence of infective-stage density and spatial distribution. Parasitology 79: 195–207. [DOI] [PubMed] [Google Scholar]
- Leulier F., Lemaitre B., 2008. Toll-like receptors: taking an evolutionary approach. Nat. Rev. Genet. 9: 165–178. [DOI] [PubMed] [Google Scholar]
- Lord J. C., Hartzer K., Toutges M., Oppert B., 2010. Evaluation of quantitative PCR reference genes for gene expression studies in Tribolium castaneum after fungal challenge. J. Microbiol. Methods 80: 219–221. [DOI] [PubMed] [Google Scholar]
- Lorenzen M. D., Doyungan Z., Savard J., Snow K., Crumly L. R., et al. , 2005. Genetic linkage maps of the red flour beetle, Tribolium castaneum, based on bacterial artificial chromosomes and expressed sequence tags. Genetics 170: 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menge D. M., Zhong D., Guda T., Gouagna L., Githure J., et al. , 2006. Quantitative trait loci controlling refractoriness to Plasmodium falciparum in natural Anopheles gambiae mosquitoes from a malaria-endemic region in western Kenya. Genetics 173: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra B., Gundra U., Teale J., 2008. Expression and distribution of Toll-like receptors 11–13 in the brain during murine neurocysticercosis. J. Neuroinflammation 5: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlais I., Mori A., Schneider J. R., Severson D. W., 2003. A targeted approach to the identification of candidate genes determining susceptibility to Plasmodium gallinaceum in Aedes aegypti. Mol. Genet. Genomics 269: 753–764. [DOI] [PubMed] [Google Scholar]
- Nezer C., Collette C., Moreau L., Brouwers B., Kim J.-J., et al. , 2003. Haplotype sharing refines the location of an imprinted quantitative trait locus with major effect on muscle mass to a 250-kb chromosome segment containing the porcine IGF2 gene. Genetics 165: 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai A., Yan G., 2003. Effects of tapeworm infection on male reproductive success and mating vigor in the red flour beetle, Tribolium castaneum. J. Parasitol. 89: 516–521. [DOI] [PubMed] [Google Scholar]
- Roach J. C., Glusman G., Rowen L., Kaur A., Purcell M. K., et al. , 2005. The evolution of vertebrate Toll-like receptors. Proc. Natl. Acad. Sci. USA 102: 9577–9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson D. W., Thathy V., Mori A., Zhang Y., Christensen B. M., 1995. Restriction fragment length polymorphism mapping of quantitative trait loci for malaria parasite susceptibility in the mosquito Aedes aegypti. Genetics 139: 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson D. W., Zaitlin D., Kassner V. A., 1999. Targeted identification of markers linked to malaria and filarioid nematode parasite resistance genes in the mosquito Aedes aegypti. Genet. Res. 73: 217–224. [DOI] [PubMed] [Google Scholar]
- Shi Z., Cai Z., Wen S., Chen C., Gendron C., et al. , 2009. Transcriptional regulation of the novel Toll-like receptor TLR13. J. Biol. Chem. 284: 20540–20547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai H., Muneta Y., Suzuki K., Eguchi-Ogawa T., Awata T., et al. , 2006. Porcine Toll-like receptor 1, 6, and 10 genes: complete sequencing of genomic region and expression analysis. Mol. Immunol. 43: 1474–1480. [DOI] [PubMed] [Google Scholar]
- Tauszig S., Jouanguy E., Hoffmann J. A., Imler J.-L., 2000. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc. Natl. Acad. Sci. USA 97: 10520–10525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu Y., Miller S., Tomita S., Schoppmeier M., Grossmann D., et al. , 2008. Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol. 9: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing Consortium , 2008. The genome of the model beetle and pest Tribolium castaneum. Nature 452: 949–955. [DOI] [PubMed] [Google Scholar]
- Tsujita T., Tsukada H., Nakao M., Oshiumi H., Matsumoto M., et al. , 2004. Sensing bacterial flagellin by membrane and soluble orthologs of Toll-like receptor 5 in rainbow trout (Onchorhynchus mikiss). J. Biol. Chem. 279: 48588–48597. [DOI] [PubMed] [Google Scholar]
- Wang M.-H., Marinotti O., James A. A., Walker E., Githure J., et al. , 2010. Genome-wide patterns of gene expression during aging in the African malaria vector Anopheles gambiae. PLoS ONE 5: e13359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Le Roy I., Nicodeme E., Li R., Wagner R., et al. , 2003. Using advanced intercross lines for high-resolution mapping of HDL cholesterol quantitative trait loci. Genome Res. 13: 1654–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Bauer K., Wernhoff P., Koczan D., Moller S., et al. , 2006. Fine mapping of collagen-induced arthritis quantitative trait loci in an advanced intercross line. J. Immunol. 177: 7042–7049. [DOI] [PubMed] [Google Scholar]
- Zhang D., Zhang G., Hayden M. S., Greenblatt M. B., Bussey C., et al. , 2004. A Toll-like receptor that prevents infection by uropathogenic bacteria. Science 303: 1522–1526. [DOI] [PubMed] [Google Scholar]
- Zhong D., Pai A., Yan G., 2003. Quantitative trait loci for susceptibility to tapeworm infection in the red flour beetle. Genetics 165: 1307–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong D., Pai A., Yan G., 2004. AFLP-based genetic linkage map for the red flour beetle (Tribolium castaneum). J. Hered. 95: 53–61. [DOI] [PubMed] [Google Scholar]
- Zhong D., Pai A., Yan G., 2005. Costly resistance to parasitism: evidence from simultaneous quantitative trait loci mapping for resistance and fitness in Tribolium castaneum. Genetics 169: 2127–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong D., Temu E. A., Guda T., Gouagna L., Menge D., et al. , 2006. Dynamics of gene introgression in the African malaria vector Anopheles gambiae. Genetics 172: 2359–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong D., Wang M.-H., Pai A., Yan G., 2013. Transcription profiling of immune genes during parasite infection in susceptible and resistant strains of the flour beetles (Tribolium castaneum). Exp. Parasitol. 134: 61–67. [DOI] [PubMed] [Google Scholar]
- Zhu F., Feng J. N., Zhang L., Liu N., 2008. Characterization of two novel cytochrome P450 genes in insecticide-resistant house-flies. Insect Mol. Biol. 17: 27–37. [DOI] [PubMed] [Google Scholar]
- Zhu F., Parthasarathy R., Bai H., Woithe K., Kaussmann M., et al. , 2010. A brain-specific cytochrome P450 responsible for the majority of deltamethrin resistance in the QTC279 strain of Tribolium castaneum. Proc. Natl. Acad. Sci. USA 107: 8557–8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z., Evans J., Lu Z., Zhao P., Williams M., et al. , 2007. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 8: R177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.