Abstract
Human and murine MHC nonclassical class Ib-restricted invariant T (iT) cell subsets, such as invariant natural killer T cells (iNKT) and mucosal-associated invariant T cells, have specialized functions early in immune responses, especially in modulating subsequent adaptive immune responses. Here, we characterize a prominent iT population in the amphibian Xenopus laevis and show the requirement of the class Ib molecule, Xenopus nonclassical gene 10, in its differentiation and function. Using Xenopus nonclassical gene 10 tetramers and RNAi loss of function by transgenesis, we identified a large class Ib-dependent CD8−/CD4− iT subset in unmanipulated frogs and tadpoles. This population is critical for antiviral immunity during early larval stages when classical MHC class Ia function is suboptimal. Furthermore, in young tadpoles with low class Ia expression, deep sequencing revealed additional preponderant invariant T cell receptor (TCR)α rearrangements, implying other iT cell subsets and a predominant selection process mediated by other class Ib molecules. The restriction and requirement of class Ib molecules for development and antiviral immunity of a mammalian iNKT or mucosal-associated invariant T cell counterpart in the amphibian Xenopus show the importance of iT cells in the emergence and evolution of the adaptive immune system.
In jawed vertebrates (Gnathostomes), classical MHC class Ia (class Ia) genes encode highly polymorphic and ubiquitously expressed molecules essential for αβCD8+ T-cell differentiation and function. Gnathostomes also have variable numbers of heterogeneous nonclassical MHC class Ib (class Ib) genes that encode molecules structurally similar to class Ia but usually with more limited tissue distribution and lower polymorphism (1). Some class Ib genes are found within the MHC region, whereas others are located outside the MHC. In mammals, certain class Ib genes, expressed either by thymocytes or other hematopoietic cells, are critically involved in the differentiation and function of distinct subsets of invariant T cells (iT cells) (2–6). These class Ib-restricted T-cell subsets include the CD1d-restricted invariant natural killer T (iNKT) and the MR1-restricted mucosal-associated invariant T (MAIT) cells. Both of these T-cell subsets express a unique semiinvariant T cell receptor (TCR), follow distinct developmental pathways divergent from differentiation of conventional T cells, and have specialized function(s) (4, 6). For example, MAIT cells respond in an MR1-dependent manner to a wide variety of different microbes and have antimicrobial activity, suggesting a role of these cells during microbial infections (7). Similarly, iNKT cells have been implicated in immunity to a variety of bacteria as well as in parasitic infections, viral infections, and fungal disease (8).
Furthermore, certain class Ib genes may play important roles during early development, when class Ia expression is suboptimal (9). However, the full biological significance of iT cells and the importance of nonpolymorphic class Ib molecules for their development are just starting to emerge. Also, nothing is known about the evolutionary conservation of iT cells; their presence in representative species in older classes of vertebrates would identify them, along with conventional T and B cells, as fundamental partners in orchestrating innate and adaptive immunity.
The amphibian Xenopus laevis is an attractive model for developmental immunology owing to the ease of manipulation and visualization of animals at all developmental stages. The immune system is remarkably conserved between mammals and Xenopus, especially intrathymic T-cell development and peripheral T-cell function (10). Unlike mammals, however, the Xenopus immune system and T-cell differentiation in particular are subject to an additional developmental program during metamorphosis (11, 12). Notably, although both larvae and adults are immunocompetent and have CD8+ T cells, the larval thymus lacks significant class Ia protein expression until metamorphosis (13). Conversely, certain class Ib genes are expressed in tadpoles at the onset of thymic organogenesis (14). This expression pattern and the unusually high conservation of certain Xenopus class Ib genes (15) have prompted us to determine whether these class Ib molecules are involved in amphibian iT cell development.
The Xenopus nonclassical gene (XNC) class Ib gene XNC10 is one of X. laevis 30 XNC genes (14–16). XNC10 represents a unique class Ib monogenic lineage phylogenetically distinct from both X. laevis class Ia and other XNC genes but highly conserved among divergent Xenopus species and characterized by interspecies sequence conservation in its putative antigen binding domain (15). Additionally, the XNC10 gene is preferentially expressed by thymocytes as early as 3 d postfertilization (14). We hypothesized that XNC10 is critically involved in Xenopus iT cell biology. Accordingly, we generated an X. laevis class Ib tetramer and developed an effective reverse-genetic loss-of-function approach by combining I-SecI meganuclease-mediated transgenesis with RNAi technology (17).
Results
Specific Binding of XNC10 Tetramer to a CD8−/4− Double Negative Splenic T-Cell Subset in Adult X. laevis.
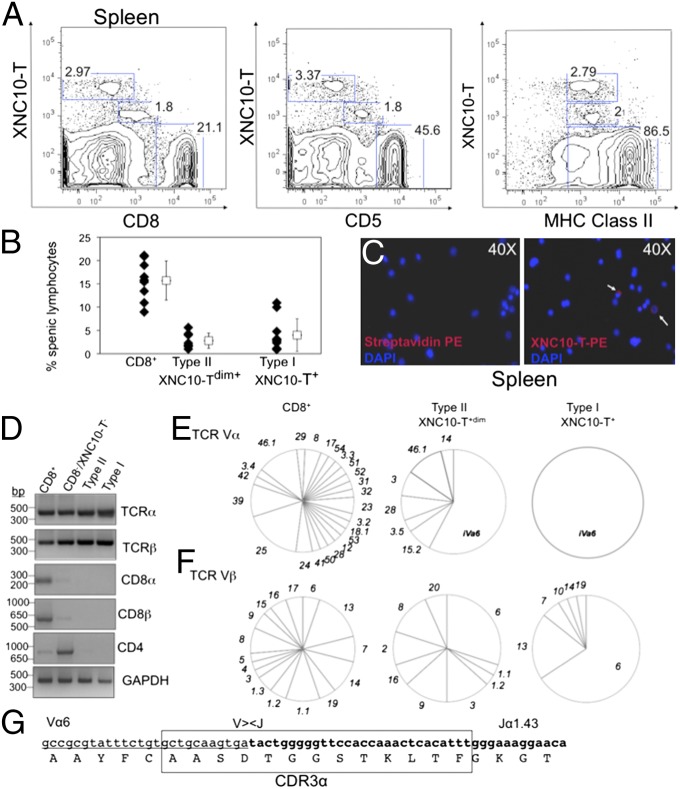
To determine whether class Ib-restricted iT cells are present in nonmammalian vertebrates, we designed a Xenopus class Ib tetramer (XNC10-T) using the methodology used for CD1d tetramers (18) (Fig. S1). We selected XNC10 because of its high degree of evolutionary conservation and its early expression in the developing thymus (14). Flow cytometry analysis of splenic leukocytes from naïve adult frogs revealed two distinct XNC10-T–reactive lymphocyte populations that we classified as types I and II based on differences in phenotype and TCR repertoire (Fig. 1). XNC10-T–reactive cells were also detectable by immunofluorescence staining of total splenocytes (Fig. 1C). Type I bright XNC10-T+ cells stained negative for CD8 and the X. laevis marker CD5 that stains the majority of thymocytes and peripheral T cells (10), whereas type II XNC10-Tdim+ cells displayed an intermediate surface staining for CD8 and CD5 (Fig. 1A). Both types I and II XNC10-T–reactive populations were MHC class IIlow+ (Fig. 1A), which is typical of Xenopus adult T cells (10). XNC10-T–reactive T cells represented a substantial fraction of splenic lymphocytes of healthy frogs (3.5% ± 3.5 for type I XNC10-T+ and 2.5% ± 2.6 for type II XNC10-Tdim+). Higher frequencies of XNC10-T–reactive lymphocytes were observed in some animals (>9%) (Fig. 1B). A substantial fraction of type I XNC10-T+ cells was also detected in peripheral blood leukocytes (1% ± 0.5) (Fig. S2). Because thymocytes express XNC10, we looked for XNC10-T–reactive thymocytes in young adults (>10 mo) when the thymus is well-developed. Although XNC10-T–reactive lymphocytes were consistently present in the spleen of froglets, no significant tetramer staining was detected for thymocytes (Fig. S3).
Fig. 1.
Identification of XNC10-T+ T-cell populations in the spleen of adult Xenopus. (A) Flow cytometry of live spleen leukocytes isolated from adult Xenopus and stained with XNC10-T and CD8, CD5, or MHC class II-specific mAbs. (B) Percent XNC10-Tbright+ (3.5% ± 3.5), XNC10-Tdim+ (2.5% ± 2.6), and CD8+ cells (15.76% ± 3.9) of total live spleen leukocytes (n = 10 adult outbred Xenopus; mean and SD for each population are shown). (C) Immunofluorescence using XNC10-T on adult Xenopus spleen leukocytes is shown in red; nuclei (blue) were stained with DAPI. (Magnification: 40×.) Arrows indicate XNC10-T+ cells. (D) Gene expression profiles of sorted cells from spleen leukocytes. (E and F) TCRα and -β variable segment (V) repertoire (listed by numbers) in sorted populations from inbreed J-strain X. laevis. TCR Vα, n = 35 clones sequences from each population; TCR Vβ, n = 26, 30, and 35 clones, respectively. Data shown are representative of three independent experiments. (G) Nucleotides and amino acids of the invariant CDR3α sequence. Va6 nucleotides are underlined, Jα1.43 nucleotides are bold, and the CDR3α region is boxed.
Furthermore, we sorted splenic XNC10-T+ and XNC10-Tdim+ subsets (>85% purity) and examined the expression of several immune-relevant genes by RT-PCR. In several experiments (Fig. 1 D–G and Fig. S4), XNC10-T+ and XNC10-Tdim+ cells expressed transcripts for functionally rearranged TCR α- and β-chains but were negative for CD4 and CD8β. In addition, type I cells did not express CD8α transcripts, suggesting that these cells are CD8−/CD4− double negative (DN) T cells, whereas CD8α gene expression for XNC10-T+/CD8dim cells varied among individuals (i.e., weak to no expression). Finally, neither type I nor type II cells expressed the promyelocytic leukemia zinc finger, a signature transcription factor of mammalian NKT, iNKT, and MAIT cells (19). Collectively, these results show that splenic XNC10-T+ T cells are mostly DN with some CD8ααlow cells. The fact that XNC10-T specifically identifies a distinct T-cell population suggests that either XNC10 is devoid of ligand in its class I groove or a conserved ligand is available that binds to the XNC10 groove during biosynthesis in the insect cells.
Type I XNC10-T+ DN Cells Express an Invariant TCR α-Chain with a Defined Complementarily Determining Region 3α.
To determine whether XNC10-T–reactive T cells express an invariant TCR, we first analyzed the TCR Vα repertoire of sorted type I XNC10-T+ and type II XNC10-Tdim+ populations by 5′-rapid amplification of cDNA ends (RACE) analysis using Cα reverse primers. Because two different Cα genes (Cαa and Cαb) (20) sharing 76.4% amino acid identity have been identified in X. laevis, consensus Cα reverse primers were designed. Interestingly, no Cαa was found in 61 sequences from types I and II XNC10-T+ cells, whereas it was used at very low frequency (1% of the total transcript analyzed) in XNC10-T−/CD8+ cells. Most notably, type I XNC10-T+ cells displayed a strictly invariant TCRα that consisted of Vα6 rearranged to Jα1.43 (Fig. 1 E and G). In addition, the iVα6-Jα1.43 junction (ASDTG) was conserved in all sequences analyzed from three genetically unrelated individuals, including two outbreds and one MHC homozygous inbred J-strain frog. This feature is reminiscent of the invariant TCR α-chains characteristic but limited to mammalian iNKT and MAIT cells (21, 22). The iVα6-Jα1.43 rearrangement was also detected in the majority (∼58%) of XNC10-Tdim+ transcripts, but there was also a minor representation of six additional Vα and seven additional Jα segments (Fig. 1E and Table S1). In contrast, control XNC10-T−/CD8+ T-cell populations isolated from the same animal expressed a broad TCRα repertoire using many different Vα and Jα segments with no significant expansion for any particular rearrangement (Fig. 1E and Table S2). Finding a diversified TCRα repertoire from RNA of the same animal and experiment rules out potential PCR artifacts.
To determine whether the XNC10-T+ invariant complementarily determining region 3α (CDR3α) region underwent n-nucleotide additions, we performed BLAST analysis on iVα6-Jα1.43 cDNA sequences of an inbred J-strain adult against the partially annotated X. laevis J-strain genome (http://www.xenbase.org/genomes/static/laevis.jsp). The rearranged iVα6-Jα1.43 CDR3α region was exclusively encoded by germ-line sequences with no evidence of n-nucleotide additions. The invariant junctional aspartic acid (D) was always encoded by 2 nt derived from the Vα6 segment and 1 nt from the Jα1.43 gene segment. Comparatively, CDR3α regions of type II XCN10-Tdim+ cells (excluding the iVα6-Jα1.43) were more diverse, ranging from 10 to 14 residues in length, with n-nucleotides identified in 63.6% of nonredundant sequences (Table S1). The CDR3α of CD8+/XNC10-T− cells was highly diverse; 62% of junctional regions contained 1–4 nongerm line-encoded nt additions (Table S2). We next examined types I and II CDR3α regions for amino acid conservation. However, with the exception of a glycine at position 110 (23) in the majority of types I and II CDR3α regions and the positively charged amino acid lysine at position 115 followed by a leucine at position 116, no additional conserved residues were identified. All of these conserved CDR3α residues exclusively originated from germ-line sequences.
Additional analysis of the X. laevis genomic scaffolds revealed that Vα6 is located ∼81.7 kbp 5′ of the Cαb region, with the interspersing region containing 5 additional functional Vα gene segments and at least 55 unique Jα segments (Table S3). In addition, both Vα6 and Jα1.43 genes were flanked by canonical recombination signal sequence motifs highly conserved among different Vα and Jα segments. RT-PCR of XNC10-T−/CD8+ populations and total splenocytes using Vα6-specific forward primers with Cαb reverse primers showed that Vα6 can rearrange to different Jα segments, suggesting that the predominance of Vα6-Jα1.43 in XNC10-T–reactive cells results, at least partially, from a selection process rather than genetic programming.
Type I XNC10-T+ DN Cells Predominantly Use TCR Vβ6/Vβ13, Dβ1.2, and Jβ13/Jβ3 Gene Segments.
To examine the degree of TCR invariance of XNC10-T–reactive T cells, we characterized their TCRβ repertoire by 5′-RACE-PCR (Fig. 1F). Type I XNC10-T+ cells showed a restricted TCRβ repertoire with predominant use of Vβ6-Jβ13 (∼66%) and to a lesser extent, Vβ13/Vβ7 gene segments (20% and 5%, respectively) (Table S4). Notably, Vβ6 and Vβ7 shared identical CDR2β regions but divergent CDR1β regions (Fig. S5). Likewise, type II XNC10-Tdim+ cells predominantly used Vβ6-Jβ13 segments (30%), with a minor representation of eight additional Vβ segments. (Table S1). The limited TCRβ use displayed by these XNC10-T–reactive T cells is again a typical feature found in mammalian MAIT and iNKT cells.
We next addressed D segment use. To date, two putative Dβ segments have been identified in X. laevis (24). To determine the precise VD and DJ junctions and degree of n-nucleotide contribution in the CDR3β regions, we searched the X. laevis genome and identified two Dβ (Dβ1.1 and Dβ1.2) genes. Both Dβ1.1 and Dβ1.2 are flanked by canonical recombination signal sequences. The predominant Vβ6-Jβ13 rearrangement of type I XNC10-T+ cells exclusively used Dβ1.2, and the CDR3β region was only encoded by germ line-derived sequences. Interestingly, one of the Vβ7 containing transcripts consisted of a CDR3 region identical to CDR3 of the predominant Vβ6-Jβ13. Moreover, CDR3β regions of Vβ10, Vβ13, Vβ14, and Vβ19 all contained a Dβ1.2 core segment. However, in contrast to the predominant Vβ6-Jβ13 junctional region, the CDR3β regions consisted of germ line- and nongerm line-encoded nucleotides at both the Vβ-Dβ and Dβ-Jβ junctions. Moreover, with the exception of a serine residue at position 107 (25) and a glutamine located three to four residues upstream of the conserved GXG motif in the majority (66.6%) of sequences, no apparent conserved motifs were detected in the type I XNC10-T+ cell CDR3β regions. Thus, the type I XNC10-T+ cells display a limited TCRβ repertoire diversity, albeit more variable than TCRα.
Concerning type II XNC10-Tdim+ CDR3β regions, all transcripts analyzed except the Vβ6-Jβ13 consisted of germ-line and nongerm-line sequences displaying both Dβ1.1 (3.3%) and Dβ1.2 (96.7%) segments (Table S1). Control XNC10-T−/CD8+ populations isolated from the same animal showed widely diversified Vβ/Jβ use and CDR3β regions (Fig. 1F and Table S5).
XNC10-T+ Cells Are Present in Premetamorphic Tadpoles.
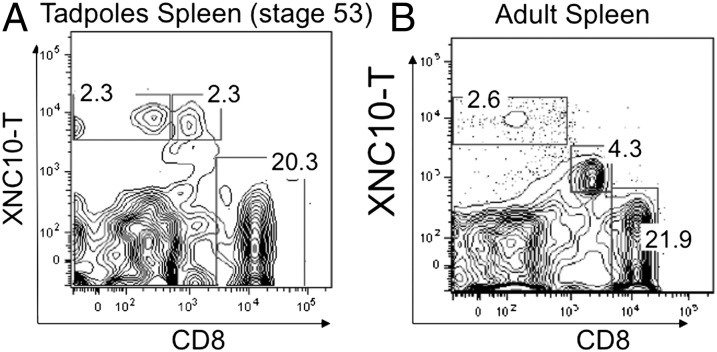
Unlike adult frogs, where cell surface class Ia molecules are ubiquitously expressed, larvae have weak and limited class I surface protein expression until the onset of metamorphosis (10–13). Because class Ib gene expression, specifically XNC10, is detected from early thymus organogenesis, class Ib expression has been postulated to compensate for the low class Ia expression during early ontogeny (14). To investigate this possibility, we first determined whether XNC10-T+–reactive cells were present in the spleens of premetamorphic tadpoles (stage 53, 25–26 d postfertilization) by flow cytometry. Because of low cell numbers, splenocytes from four tadpoles were pooled before staining. As in adults, two distinct populations of XNC10-T+–reactive cells were found in the tadpole: a type I XNC10-T+/CD8− and a type II XNC10-T+/CD8dim+ cell subset, representing 2% ± 0.8 and 2.3% ± 0.5 of total splenic lymphocytes, respectively (Fig. 2). In contrast to adults, the XNC10-T fluorescent signal intensity of the type II XNC10-T+/CD8dim+ larval subset was higher than adult, suggesting an increase in surface expression of the TCR bound by XNC10-T. As in young adults, no consistent XNC10-T staining was detected on larval thymocytes.
Fig. 2.
Identification of XNC10-T+ cells in the spleens of early developmental stage 53 tadpoles (3 wk). Flow cytometry of spleen leukocytes pooled from either (A) four individuals or (B) one adult and double stained with XNC10-T and CD8 mAb. Data shown are representative of three different experiments.
XNC10 Loss of Function by RNAi in Vivo Results in Impaired iVα6-Jα1.43 Expression, XNC10-T+ Cell Deficiency, and Weakened Larval Resistance to Viral Infection.
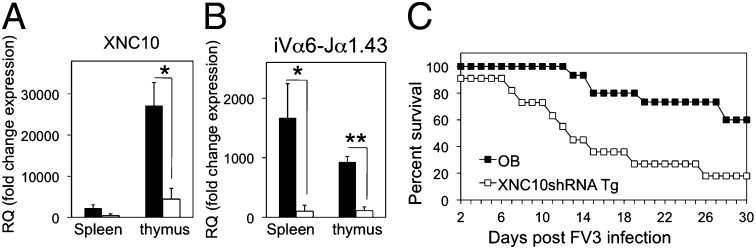
Given the XNC10 gene expression by larval thymocytes at the onset of thymic organogenesis and the presence of splenic XNC10-T+–reactive cells during early development, we hypothesized that XNC10 is required for iVα6T cell differentiation. To test this hypothesis, we used our recently developed reverse-genetic loss-of-function approach, which combines RNAi and I-SceI meganuclease-mediated transgenesis (17). A key advantage of this technique is that the high efficiency and nonmosaic transgene expression permit the direct use of F0 animals (17). Outbred F0 transgenic tadpoles (stage 53, 25 d postfertilization) were screened for uniform nonmosaic GFP expression, and XNC10 knockdown was assessed by quantitative PCR (qPCR) compared with age-matched controls (Fig. 3A). Dejellying was performed before microinjection on experimental and control eggs to remove the protective membranes that surrounds the embryo. Notably, XNC10 silencing in transgenic animals resulted in a drastic decreased expression of the invariant iVα6 rearrangement in both spleens (P = 0.0396) and thymuses (P = 0.00712) compared with age-matched controls (Fig. 3B), suggesting that successful differentiation of the iVα6 T-cell lineage is XNC10-dependent. Furthermore, nested RT-PCR revealed a high frequency of nonproductive Vα6 rearrangements in the spleens of transgenic animals (37.1% ± 15, n = 3) compared with controls (8.7% ± 14.4, n = 3) (Fig. S6). Finally, flow cytometric analysis of an F0 transgenic adult showed a marked reduction of both splenic types I and II XNC10-T+ cells compared with age-matched dejellied controls (Fig. 4). No obvious differences were observed in the total numbers of CD8+ cells, indicating that XNC10 silencing specifically effected the iVα6 T-cell populations but not other T-cell pools. Collectively, these results provide convincing evidence of the restriction and requirement of XNC10 for the iVα6 T-cell lineage development, which further argues for functional similarity to mammalian MAIT and iNKT cells.
Fig. 3.
XNC10shRNA Tg Xenopus tadpoles have reduced expression of iVα6. Expression of (A) XNC10 or (B) iVα6 in Tg developmental stage 53/54 tadpoles (white box) or age-matched dejellied controls (black box). Results are normalized to an endodgenous control and presented as fold change in expression compared with expression of XNC10 or iVα6 in the skin of Tg tadpoles. All results are presented as mean ± SE (n = 4). *P < 0.05 and **P < 0.005 denote significant differences (Student t test). (C) F0 Tg tadpoles (stage 53, n = 12) or control larvae (stage 53, n = 15) were i.p. infected with 1 × 104 pfu FV3, and survival was monitored daily. Active FV3 infection was verified postmortem by RT-PCR using viral polymerase II on kidney-derived genomic DNA. OB, outbred animals; RQ, relative quantification.
Fig. 4.
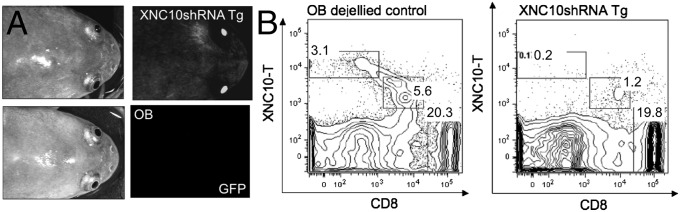
XNC10shRNA Tg animals have fewer XNC10-T+ cells. (A) Representative image of GFP expression in F0 Tg adult frogs (<10 mo). (B) Flow cytometry analysis of spleen from Tg animal and age-matched dejellied control using XNC10-T and CD8 mAb. Percent XNC10-T+, XNC10-Tdim+, and CD8+ cells are shown in the plot.
To determine whether XNC10-dependent iVα6 T-cell deficiency has immunological consequences, we infected F0 transgenic tadpoles (stage 53, 25 d postfertilization) with the ranavirus frog virus 3 (FV3), a natural amphibian pathogen (26). XNC10 deficiency resulted in a marked increase in susceptibility to FV3 infection, especially at early stages of infection, compared with nontransgenic controls of the same parent (Fig. 3C). The median survival time on infection of transgenic tadpoles was significantly shorter than controls (12.5 versus 36.5 d, P > 0.001).
TCRα Repertoire in CD8dim+ and CD8− T-Cell Populations in Tadpole Is Limited with Several Dominant Distinct Invariant Rearrangements.
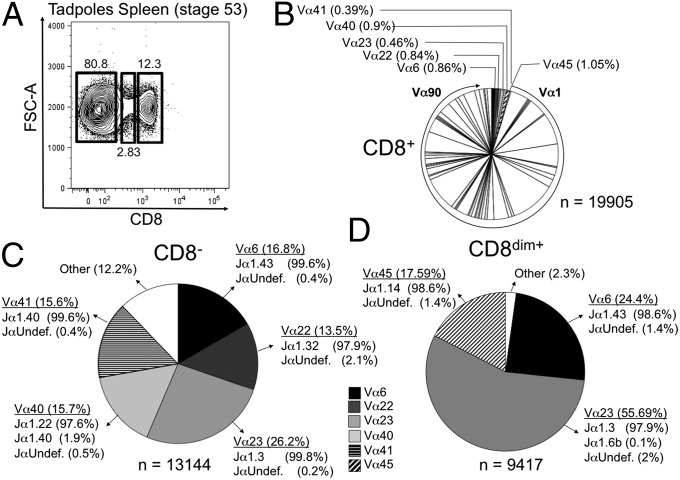
Given the suboptimal class Ia gene expression during early ontogeny, especially in the thymus (10–13), we have proposed that certain class Ib genes, such as XNC10, may play a more preponderant role in T-cell differentiation during early development. Accordingly, one would predict a general bias in the TCR repertoire at an early larval stage. To examine the nontetramer-biased global TCRα repertoire, we adapted our 5′-RACE analysis to 454-pyrosequencing technology. Using the inbred J strain to avoid allelic variation, we sorted CD8+, CD8dim+, and CD8− populations from splenocytes pooled of 167 stage 50/51 tadpoles (15–16 d postfertilization), when the first mature T cells begin to populate the spleen. After selection for a minimum size of 250 bp, a total of 42,466 high-quality functional rearranged TCRα sequences was analyzed, including 19,905 CD8+, 9,417 CD8dim+, and 13,144 CD8− sequences. Based on a >85% nucleotide identity cutoff, the different sequences were grouped into clusters of distinctive Vα families irrespective of Jα segments (Fig. 5). The Jα segment used within each cluster was then determined by using all 60 known genomic Jα segments.
Fig. 5.
TCRα repertoire in stage 50 tadpoles is limited with several dominant invariant TCR α-chains. (A) CD8+, CD8dim+, and CD8− populations were sorted from spleens of stage 50 inbred J-strain tadpoles (n = 167); potential TCRα repertoire in each population was determined using 5′RACE-PCR and next generation 454 pyrosequencing. The frequency of different Vα families based on >85% nucleotide identity for (B) CD8+, (C) CD8dim, and (D) CD8− is show with n = total reads. The frequency and number of Vα6, Vα22, Vα23, Vα40, Vα41, and Vα45 are indicated in each population. For CD8+, each slice represents a unique Vα family starting with Vα1 and moving clockwise to Vα90. The total CD8+ Vα family repertoire is summarized in Table S6. (C and D) Each of the dominant Vα family clusters was reanalyzed with a >95% nucleotide identity, and Jα gene segment use within each cluster is indicated. Undef indicates no match with >95% sequence identity to any previously identified Jα. FSC-A, forward scatter.
Larval CD8+ T cells exhibited a wide variety of distinct Vα and Jα segments indicative of a diversified TCRα repertoire already present at this early developmental stage at the population level (if not the individual level) (Fig. 5A and Table S6). Additional analysis using a higher stringency (>95%) and basic local alignment tool searches against the X. laevis genome confirmed the diversification by a diverse Jα use in each cluster. In contrast, when the same criteria were applied to the CD8dim+- and CD8−-derived sequences, we found a marked overrepresentation of five distinct Vα families. The dominant clusters in each of these populations were reclustered using a >95% sequence identity cutoff (Fig. 5 C and D), and each cluster was analyzed for Jα use and n-nucleotide additions. We identified three highly overrepresented invariant Vα rearrangements with no n-nucleotide additions in the CD8dim+ population (Vα6-Jα1.43, Vα45-Jα1.14, and Vα23-Jα1.3). Two of these rearrangements, Vα6-Jα1.43 and Vα23-Jα1.3, were also overrepresented in the CD8− population together with three additional invariant Vα rearrangements (Vα22-Jα1.32, Vα40-Jα1.22, and Vα41-Jα1.40), comprising from 13.5% to 55.6% of the total Vα repertoire in these populations (Fig. S7). Collectively, our findings indicate that, during early development, the TCRα repertoire is highly biased by at least six distinct dominant invariant TCR Vα rearrangements, one of which (Vα6-Jα1.43) is strictly class Ib-dependent (XNC10).
Discussion
Our study provides compelling evolutionary evidence not limited to mammals of the biological importance of iT cells and the requirement of class Ib in their differentiation and function. The identification of a distinct class Ib-restricted iT cell subset in a cold-blooded vertebrate suggests the conservation throughout jawed vertebrates of T-cell subsets with characteristics similar to mammalian iNKT and MAIT cells. We have extensively characterized (in the amphibian X. laevis,) a distinct and sizable class Ib (XNC10) -dependent type I DN iT cell subset that expresses the invariant TCR Vα6/Jα1.43 rearrangement without terminal deoxynucleotidyl transferase-mediated n diversification and a limited TCRβ repertoire. These iT cells comprise a significant fraction of splenocytes and circulating blood cells of healthy Xenopus adults and larvae. We also identified an additional type II CD8dim+ (presumably CD8α/α) XNC10-reactive iT subset with a predominant iVα6 bias, albeit with a more diverse TCRα and -β repertoire than the type I subset. It is unclear whether this partial TCR diversity results from contamination because of the slight overlap between the XNC10-Tdim+/CD8dim+ and XNC10−/CD8+ populations or whether type II XNC10-T+ cells represent a distinct XNC10-T+–reactive subset reminiscent of mammalian type II NKT cells that are (like the type I iNKT) CD1d-restricted but do not express the canonical TCR α-chain (27). In either case, the identification of iT cells controlled by class Ib molecules in a species as evolutionarily distant from mouse and human as Xenopus constitutes a strong argument for the functional relevance of these cells. This observation has strong significance, because the biological role of this system in immunity is neither fully understood nor fully appreciated, despite marked progress in characterization of mammalian iT cells.
Importantly, XNC10 silencing in vivo resulted in a marked decrease in the expression of iVα6 TCR and a loss of XNC10-T–reactive cells as well as a more than twofold increase in susceptibility to FV3 infection in young tadpoles. Although Xenopus tadpoles are not as resistant to FV3 infection as adults, they do mount antiviral immune responses (26). We previously reported that beta 2 microglobulin (b2m) silencing, which impairs both class Ia and XNCs surface expression, increased susceptibility to FV3 infection in young tadpoles (17). We interpreted these data as evidence for a critical role of class Ib in antiviral immunity during early ontogeny, when class Ia expression is suboptimal (17). The marked increase in susceptibility to FV3 infection resulting in a rapid increase in mortality within a few days postinfection obtained by specifically silencing one single class Ib gene, XNC10, shows that class Ib molecules play a role in antiviral immunity in larvae. Furthermore, this increase in viral susceptibility indicates that XNC10-dependent iVa6 T cells are essential during early viral immunity, a finding that has fundamental relevance for the biological role of iT cells in all vertebrates. Although effector function of iT cells in Xenopus remains to be characterized, their importance in viral immunity has parallels in humans and mice, where some viruses have been shown to target CD1d expression (28). Similar to our study in Xenopus, CD1−/− and Jα18−/− mice have impaired viral clearance of herpes simplex viruses 1 and 2. Likewise, CD1d−/− but not Ja1.18−/− mice show increased mortality to another herpes virus, cytomegalovirus (29). Effector functions of mammalian iNKT cells are still not fully understood and likely to vary with the type of viral infection. For example, cytomegalovirus induces murine iNKT cells to produce IFN-γ, which in turn, amplifies the natural killer cell response (30, 31).
The genes encoding class Ib molecules have been subjected to rapid evolution; thus, their phylogeny remains unclear, and species-specific adaptations are common. In this context, XNC10 is remarkable, because it is a monogenic family conserved across the entire Xenopodinae subfamily corresponding to an evolutionary history of at least 65 million y, implying an important and nonredundant function for this gene. It should be emphasized that this degree of evolutionary conservation of a class Ib pattern of evolution is in stark contrast to mammals, where although few class Ib genes (e.g., CD1 and MR1) have defined evolutionary relationships, most class Ib genes show no orthologous relationships among closely related species (25). Similarly, conserved functional equivalents among class Ib molecules are limited in mammals. Importantly, our reverse-genetic loss-of-function approach reveals that XNC10 is essential for the development of iVα6T cells. The fact that XNC10 silencing had no noticeable effect on the total CD8+ T-cell compartment or the occurrence of other TCR Vα supports the idea that XNC10 is specialized to the differentiation and function of iVα6T cells. This characteristic is reminiscent of CD1d requirement for the development and function of iNKT cells and MR1 for MAIT cells (3, 6). Our data strongly suggest that, despite this evolutionary incertitude, important specialized class Ib functions and presumably underlying mechanisms have been conserved. As such, Xenopus provides a powerful comparative model system to gather additional insight into class Ib-mediated iT cell biology, including their tissue localization and potential role in mucosal immunity.
Although to date, the nature of the XNC10 ligand is unknown, the fact that our XNC10 tetramer, lacking experimentally loaded antigens, can nonetheless specifically bind a defined population of iVα6T cells suggests that either the XNC10 interaction with the iVαT is antigen-independent or purified XNC10 monomers are preloaded with insect-derived antigens conserved enough to the true ligand to facilitate iVαT-cell interactions. There are several observations that support the idea of a conserved antigen. First, XNC10 has a unique putative peptide binding domain distinct from all other X. laevis class Ib as well as class Ia genes (15). The putative peptide-docking residues of XNC10 are highly conserved with those identified from the X. tropicalis homolog SNC10, especially in the C terminus, suggesting that XNC10 presents common or conserved (either endogenous or exogenous) antigenic motif. In this context, it is interesting to note that, based on Southern blot cross-hybridization, the XNC10 gene seems to be conserved and diploidized in all species of the Xenopodinae subfamily, regardless of their ploidy (diploid to dodecaploid) (15). Second, although the direct interaction between XNC10 and the invariant TCR expressed on iVα6T cells has not yet been shown, it represents a likely target. Because the CDR3α region of the invariant Vα chain is maintained among animals of different genetic backgrounds and exclusively encoded by germ-line sequences with no n-nucleotide additions, it is likely that the resulting TCR binds a conserved motif. In humans and mice, predominant use of germ-line sequences in the CDR3α region has been shown in both the canonical type I iNKT and MAIT TCR α-chain, with the MAIT TCR α-chain exhibiting a conserved CDR3α length but displaying some junctional variability in two codons (4, 6, 21).
Using global TCRα repertoire analysis by deep sequencing, we offer compelling evidence that not only XNC10-dependent iVα6 T cells but also, several additional distinct iT cell subsets are preponderant during early development, when class Ia expression is suboptimal. This biased usage suggests an alternative, possibly class Ib-dependent selection process. This observation is of fundamental relevance, because Xenopus tadpoles represent something of an immunological enigma; they have very limited surface expression of class Ia molecules until the onset of metamorphosis (12, 13). Unlike mammalian embryos, which reside in a relatively antigen-free uterus, Xenopus larvae (like other ectothermic vertebrates) hatch in the surrounding antigen-rich water. As such, within 2 wk postfertilization, the Xenopus immune system is under pressure to develop quickly and produce a diversified lymphocyte receptor repertoire with a very small numbers of lymphocytes (15,000–20,000 T cells). Because the potential TCRα/β repertoire in Xenopus far exceeds the number of lymphocytes, additional mechanisms may have evolved to produce a functional but more limited lymphocyte repertoire during early ontogeny. In this context, surface class Ia expression is first detected on erythrocytes and splenocytes at metamorphic stages (reviewed in ref. 10). Similarly, no class Ia protein is detected by immunofluorescence microscopy, and there was no none-to-weak class Ia and large multi functional peptidase 7 mRNA expression as determined by Northern blot analysis (12, 13). However, we have shown that low levels of class Ia expression are detectable by RT-PCR in the thymus of tadpoles as early as stage 39. Because b2m is also expressed at this stage, the possibility remains that a very low level of class Ia (below Ab detection level) is expressed in the larval thymus. Alternatively, class Ib genes expressed at this early developmental stage may counterbalance the class Ia-deficiency. These two possibilities need not be mutually exclusive. Nevertheless, it is noteworthy that, despite a suboptimal class Ia expression, Xenopus larvae have CD8+ T cells and can reject MHC incompatible grafts, albeit with slower rejection kinetics than adults (32). This ability suggests the presence of a T-cell compartment functionally distinct from T cells of postmetamorphic animals.
Our results indicate that 2-wk-old tadpoles (stage 50) with spleens that are just beginning to be colonized by mature T cells already have, on the one hand, a CD8+ T-cell population with a diversified TCRα repertoire and on the other hand, CD8dim+ and CD8− populations with a highly biased TCRα repertoire largely composed of six different dominant invariant clonotypes, including the XNC10-dependent Vα6/Jα1.43 iT cell rearrangement. The relative fraction of each putative iT cell population expressing these different overrepresented rearrangements in the spleen of individual animals is hard to estimate, because we cannot exclude that the predominant expression of the dominant transcripts is because of a high level of expression in a few cells as opposed to an expanded population. However, for the iVα6 rearrangement, we have shown, using XNC10 tetramers, that these iVα6 T cells represent a substantial fraction (∼5% of total lymphocytes), which reinforces the idea of a biased TCRα repertoire during early development with an overrepresentation of dominant TCR Vα rearrangements.
The availability of XNC10-deficient transgenic animals combined with an XNC10 tetramer as well as efficient technology to generate tetramers and transgenic clones with loss of function for other class Ib genes will contribute to furthering our understanding of the ontogeny and functions of iT cells.
Materials and Methods
Experimental Animals.
Outbreed and inbreed J strains of X. laevis were from the Xenopus laevis Research Resource for Immunology at the University of Rochester (http://www.urmc.rochester.edu/smd/mbi/xenopus/index.htm). Transgenic X. laevis was generated as described in SI Materials and Methods. All animals were handled under strict laboratory and University Committee on Animal Resources regulations (100577/2003–151), and discomfort was minimized at all times.
Production of b2m-XNC10 Tetrameric Complexes.
The X. laevis b2m-linker-XNC10 recombinant protein was produced in a eukaryotic expression system using protocols modified from ref. 18 as described in SI Materials and Methods.
Flow Cytometry and Immunofluorescence Staining.
All anti-X. laevis mAbs were from the Xenopus laevis Research Resource (https://www.urmc.rochester.edu/mbi/resources/Xenopus/). For flow cytometry, cells (0.25 × 106) were stained with 5 μg XNC10-T-allophycocyanin (APC) for 30 min at 4 °C followed by incubation with mAbs as described in SI Materials and Methods. Splenocytes were FACS sorted (F-Aria-18; BD Bioscience). For microscopy, cells were fixed and stained with 5 μg XNC10-T-phycoerythrin plus DNA dye Hoechst-33258, mounted in antifade medium, and visualized with a Leica DMIRB inverted fluorescent microscope.
qPCR, RT-PCR, and 5′-RACE.
RNA and cDNA, SMARTer RACE, and qPCR were performed using standard protocols as described in SI Materials and Methods. All primers used are described in Table S7.
FV3 Infection.
FV3 (Iridoviridae) was grown as previously described. Transgenic tadpoles (stage 53) and age-matched dejellied controls from the same parents were i.p. infected with 1 × 104 pfu, and cumulative mortality was monitored daily over a 30-d period.
TCRα Repertoire Sequence Analysis.
RNAs from FACS-sorted splenic CD8+, CD8dim, and CD8− populations of 167 pooled stage 50/51 inbred J tadpoles were used for 5′-RACE analysis using unique bar-coded primers and sequencing using a GS Junior Sequencer (Roche). The resulting 454 GS junior reads were screened for the Cα anchoring region using BLASTn and clustered using uSEarch version 6.0.307 with a >85% cutoff as described in SI Materials and Methods.
Statistical Analysis.
All quantitative data were analyzed using a one-way ANOVA and the Vassar Stat software (www.vassarstats.net).
Supplementary Material
Acknowledgments
We thank David Albright and Tina Martin for the expert animal husbandry and Drs. Erin Adams, Martin Flajnik, and Nicholas Cohen for discussions and critical reading of the manuscript. This research was supported by National Institutes of Health Grant R25-GM064133 (to L.-M.A.S.), R24-AI-059830, National Sciences Foundation IOS-0923772 and Kesel Fund Award 20115123 (to E.-S.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309840110/-/DCSupplemental.
References
- 1.Flajnik MF, Kasahara M. Comparative genomics of the MHC: Glimpses into the evolution of the adaptive immune system. Immunity. 2001;15(3):351–362. doi: 10.1016/s1074-7613(01)00198-4. [DOI] [PubMed] [Google Scholar]
- 2.Bendelac A, et al. CD1 recognition by mouse NK1+ T lymphocytes. Science. 1995;268(5212):863–865. doi: 10.1126/science.7538697. [DOI] [PubMed] [Google Scholar]
- 3.Matsuda JL, Gapin L. Developmental program of mouse Valpha14i NKT cells. Curr Opin Immunol. 2005;17(2):122–130. doi: 10.1016/j.coi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Kronenberg M, Gapin L. The unconventional lifestyle of NKT cells. Nat Rev Immunol. 2002;2(8):557–568. doi: 10.1038/nri854. [DOI] [PubMed] [Google Scholar]
- 5.Treiner E, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422(6928):164–169. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- 6.Le Bourhis L, et al. Mucosal-associated invariant T cells: Unconventional development and function. Trends Immunol. 2011;32(5):212–218. doi: 10.1016/j.it.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Le Bourhis L, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. 2010;11(8):701–708. doi: 10.1038/ni.1890. [DOI] [PubMed] [Google Scholar]
- 8.Sköld M, Behar SM. Role of CD1d-restricted NKT cells in microbial immunity. Infect Immun. 2003;71(10):5447–5455. doi: 10.1128/IAI.71.10.5447-5455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houlihan JM, Biro PA, Fergar-Payne A, Simpson KL, Holmes CH. Evidence for the expression of non-HLA-A,-B,-C class I genes in the human fetal liver. J Immunol. 1992;149(2):668–675. [PubMed] [Google Scholar]
- 10.Robert J, Ohta Y. Comparative and developmental study of the immune system in Xenopus. Dev Dyn. 2009;238(6):1249–1270. doi: 10.1002/dvdy.21891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flajnik MF, Kaufman JF, Du Pasquier L. Changes in the immune system during metamorphosis of Xenopus. Trends Immunol. 1987;8(2):58–64. doi: 10.1016/0167-5699(87)90240-4. [DOI] [PubMed] [Google Scholar]
- 12.Flajnik MF, et al. Major histocompatibility complex-encoded class I molecules are absent in immunologically competent Xenopus before metamorphosis. J Immunol. 1986;137(12):3891–3899. [PubMed] [Google Scholar]
- 13.Salter-Cid L, Nonaka M, Flajnik MF. Expression of MHC class Ia and class Ib during ontogeny: High expression in epithelia and coregulation of class Ia and lmp7 genes. J Immunol. 1998;160(6):2853–2861. [PubMed] [Google Scholar]
- 14.Goyos A, Ohta Y, Guselnikov S, Robert J. Novel nonclassical MHC class Ib genes associated with CD8 T cell development and thymic tumors. Mol Immunol. 2009;46(8-9):1775–1786. doi: 10.1016/j.molimm.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goyos A, Sowa J, Ohta Y, Robert J. Remarkable conservation of distinct nonclassical MHC class I lineages in divergent amphibian species. J Immunol. 2011;186(1):372–381. doi: 10.4049/jimmunol.1001467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flajnik MF, et al. A novel type of class I gene organization in vertebrates: A large family of non-MHC-linked class I genes is expressed at the RNA level in the amphibian Xenopus. EMBO J. 1993;12(11):4385–4396. doi: 10.1002/j.1460-2075.1993.tb06123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nedelkovska H, Edholm ES, Haynes N, Robert J. Effective RNAi-mediated β2-microglobulin loss of function by transgenesis in Xenopus laevis. Biol Open. 2013;2(3):335–342. doi: 10.1242/bio.20133483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidobre S, Kronenberg M. CD1 tetramers: A powerful tool for the analysis of glycolipid-reactive T cells. J Immunol Methods. 2002;268(1):107–121. doi: 10.1016/s0022-1759(02)00204-1. [DOI] [PubMed] [Google Scholar]
- 19.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29(3):391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haire RN, Kitzan Haindfield MK, Turpen JB, Litman GW. Structure and diversity of T-lymphocyte antigen receptors alpha and gamma in Xenopus. Immunogenetics. 2002;54(6):431–438. doi: 10.1007/s00251-002-0474-4. [DOI] [PubMed] [Google Scholar]
- 21.Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med. 1993;178(1):1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4-8- T cells in mice and humans. J Exp Med. 1994;180(3):1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefranc MP, et al. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev Comp Immunol. 2003;27(1):55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- 24.Chretien I, Marcuz A, Fellah J, Charlemagne J, Du Pasquier L. The T cell receptor beta genes of Xenopus. Eur J Immunol. 1997;27(3):763–771. doi: 10.1002/eji.1830270327. [DOI] [PubMed] [Google Scholar]
- 25.Hansen TH, Huang S, Arnold PL, Fremont DH. Patterns of nonclassical MHC antigen presentation. Nat Immunol. 2007;8(6):563–568. doi: 10.1038/ni1475. [DOI] [PubMed] [Google Scholar]
- 26.Chinchar VG, Hyatt A, Miyazaki T, Williams T. Family Iridoviridae: Poor viral relations no longer. Curr Top Microbiol Immunol. 2009;328:123–170. doi: 10.1007/978-3-540-68618-7_4. [DOI] [PubMed] [Google Scholar]
- 27.Behar SM, Podrebarac TA, Roy CJ, Wang CR, Brenner MB. Diverse TCRs recognize murine CD1. J Immunol. 1999;162(1):161–167. [PubMed] [Google Scholar]
- 28.Diana J, Lehuen A. NKT cells: Friend or foe during viral infections? Eur J Immunol. 2009;39(12):3283–3291. doi: 10.1002/eji.200939800. [DOI] [PubMed] [Google Scholar]
- 29.Grubor-Bauk B, Simmons A, Mayrhofer G, Speck PG. Impaired clearance of herpes simplex virus type 1 from mice lacking CD1d or NKT cells expressing the semivariant V alpha 14-J alpha 281 TCR. J Immunol. 2003;170(3):1430–1434. doi: 10.4049/jimmunol.170.3.1430. [DOI] [PubMed] [Google Scholar]
- 30.Wesley JD, Tessmer MS, Chaukos D, Brossay L. NK cell-like behavior of Valpha14i NK T cells during MCMV infection. PLoS Pathog. 2008;4(7):e1000106. doi: 10.1371/journal.ppat.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyznik AJ, et al. Cutting edge: The mechanism of invariant NKT cell responses to viral danger signals. J Immunol. 2008;181(7):4452–4456. doi: 10.4049/jimmunol.181.7.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barlow EH, Cohen N. The thymus dependency of transplantation allotolerance in the metamorphosing frog Xenopus laevis. Transplantation. 1983;35(6):612–619. doi: 10.1097/00007890-198306000-00018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.