Abstract
Hepatitis C virus (HCV) replication is limited by cyclophilin inhibitors but it remains unclear how viral genetic variations influence susceptibility to cyclosporine (cyclosporine A, CsA), a cyclophilin inhibitor. In this study HCV from liver transplant patients was sequenced before and after CsA exposure. Phenotypic analysis of NS5A sequence was performed by using HCV sub genomic replicon to determine CsA susceptibility. The data indicates an atypical proline at position 328 in NS5A causes increases CsA sensitivity both in the context of genotype 1a and 1b residues. Point mutants mimicking other naturally occurring residues at this position also increased (Ala) or decreased (Arg) replicon sensitivity to CsA relative to the typical threonine (genotype 1a) or serine (genotype 1b) at this position. This work has implications for treatment of HCV by cyclophilin inhibitors.
Keywords: Hepatitis C virus, Transplantation, Cyclophilin inhibitor, Cyclosporine, Antiviral
Introduction
Hepatitis C virus (HCV) is a genetically diverse virus that exists as a quasispecies in a single host, and has multiple variants between different infected individuals at least in part because of adaptation to the host's specific adaptive immune system. HCV infects over 200 million people, and current strategies to eradicate the virus are compromised by toxicity, cost, and efficacy. Newer multidrug approaches are currently in development, including the recently approved protease inhibitors polymerase inhibitors, as well as cyclophilin inhibitors among others. HCV has 6 recognized genotypes which express at least 10 different proteins, some of which interact with cyclophilin A and modulate/inhibit HCV replication (Ciesek et al., 2009; Fernandes et al., 2007; Foster et al., 2011; Goto et al., 2009; Tang, 2010; Verdegem et al., 2011; Watashi et al., 2005).
HCV is the most common indication currently for liver transplant in the US. After a liver transplant almost all liver recipients receive a calcineurin inhibitor immunosuppressant, either CsA or tacrolimus and all HCV infected patient's new liver gets reinfected by residual viremia in the patient. CsA and its nonimmunosuppressive analogs, DEBIO-25 (Alisporivir), SCY-635, and NIM811 are inhibitors of cellular prolyl-peptidyl isomerases called cyclophilins (Cyps), which are necessary for HCV replication (Fischer et al., 2010; Flisiak et al., 2008; Hanoulle et al., 2009; Kaul et al., 2009; Ma et al., 2006; Vermehren and Sarrazin, 2011). The anti-HCV impact of CsA in patients with HCV remains controversial (Flisiak et al., 2008). Since cyclophilin inhibitors are also being developed as a component of multidrug therapy directed against HCV, the effect of CsA in HCV infected liver transplant recipients represents an ideal cohort to study the impact of cyclophilin inhibitor “mono” therapy on HCV. However, at the moment tacrolimus is more commonly used than CsA in liver transplantation. In order to study whether or not CsA had any effect on the evolution of HCV post-transplant, we took advantage of a unique cohort of HIV/HCV infected patients [HIV and transplant study, (Terrault et al., 2012)]. These samples were banked during the study and later used to address if CsA selected viral variants.
The multi-domain (domains I, II and III) nonstructural protein 5A (NS5A) (Tellinghuisen et al., 2008) is particularly genetically diverse. It is unclear if the naturally occurring variation in NS5A alters cyclophilin dependance, and arguments against NS5A altering cyclophilin dependance have been made (Chatterji et al., 2010), but amino acid variation could explain heterogeneity in a putative antiviral effect of CsA post-transplant. Earlier we reported laboratory acquired mutations within domains II and III region of genotype 1b NS5A that conferred resistance to CsA (Fernandes et al., 2007, 2010). For that reason, we sought to determine if CsA exposure selected for NS5A mutations in patients who received CsA but not other antivirals. We performed consensus sequencing of NS5A and NS5B of patients before and during CsA exposure. In most of the serial viral sequences we did not find compelling evidence for selection by CsA, but in one patient we were able to correlate selection at amino acid 328 in NS5A with altered CsA susceptibility in cell culture.
Results
Development of cyclosporine resistance in NS5A in a patient exposed to cyclosporine
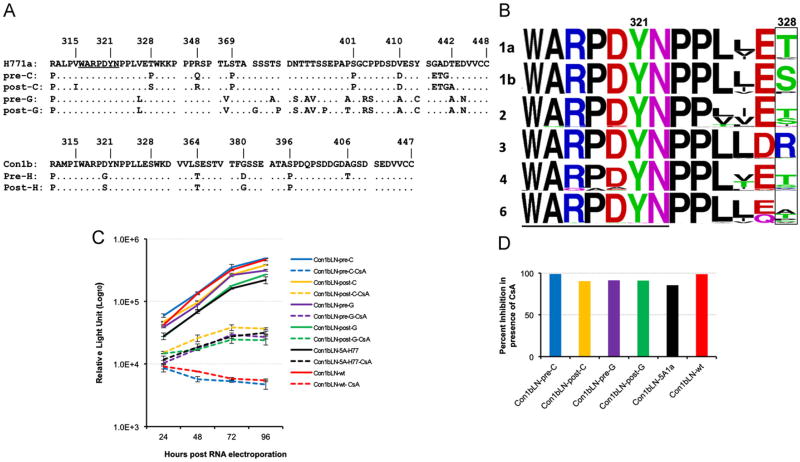
The HIV and transplant study (HIV--TR, www.HIVtransplant.com) evaluated transplantation of 89 HCV/HIV infected patients and compared CsA and tacrolimus as initial immunosuppression and found no clear benefit for either inhibitors (Terrault et al., 2012) despite cell culture data from multiple groups demonstrating CsA inhibition of HCV (Fernandes et al., 2010; Fischer et al., 2010; Tang, 2010). As genotype 1 was both the most prevalent and the most interferon resistant, we limited ourselves to the 61 genotype 1 infected patients. Only 15 of these were initially treated with CsA and did not receive anti-HCV interferon based therapy. In order to specifically examine if CsA selected resistant HCV variants we limited ourselves to this smaller subset of patients and attempted to amplify both NS5A and NS5B regions spanning from PQLPG of NS5A (residue 29) to GGDIYHS of NS5B (residue 563). From nine of these patients either a post-transplant or a pre-transplant sample or both were missing or full amplification of HCV was unsuccessful. Three of these nine patients were considered spontaneous cures (Haque et al., 2010; Terrault et al., 2012). We were able to amplify consensus sequencing of above mentioned NS5A carboxy terminal region from six genotype 1 infected patients (A–F) pre- and post-transplant. All six patients were continuously CsA exposed during the interval between pre-transplant sequencing and 6 months to 2 years post-transplant and received no anti-HCV therapy during this time. One of these six was a genotype 1b (patient H), and the rest were genotype 1a. One of these patients (patient C) though was noted to have an atypical pre-transplant proline at amino acid 328 of NS5A (Fig. 1A), which mutated to a consensus serine post-transplant rather than the more typical genotype 1a threonine (Fig. 1A and B).While a serine is the consensus amino acid for ∼91% of genotype 1b strains, it represents only 2% of genotype 1a strains a difference that is statistically significant (p value (p<0.0001) based on 358 genotype 1b and 224 genotype 1a sequences retrieved from European database. The 328 Pro is near lab selected resistance mutations that we and others have mapped (Fernandes et al., 2010; Grise et al., 2012; Tang, 2010). When the carboxy terminus of NS5A (from amino acids FARALPV to MSYTWT) was amplified from the pre-C cDNA and cloned into a genotype 1b replicon, the CsA sensitivity of the resultant replicon was similar to the parental genotype 1b replicon (Fig. 1C, compare solid and dashed blue lines of patient C with solid and dashed red lines of parental 1b replicon). The replicon with genotype 1a laboratory strain H77 (Con1bLN-5AH77) though was relatively more cell culture resistant to CsA than the pre-transplant patient C replicon (Fig. 1C, black solid and dotted lines). Furthermore we cloned the corresponding fragment of the carboxy terminus of NS5A from the same patient (patient C) post-transplant (6 months later, post-C) and found the replicon less sensitive to CsA (Fig. 1C) (dashed blue suppressed ∼100 fold while dashed yellow suppressed only ∼10 fold (Fig. 1D)). No naturally occurring NS5A fragment (genotype 1b or 1a, pre or post-transplant) was found to be as sensitive to CsA as the pre-transplant patient C sequence (data not shown). We could detect no obvious clinical benefit accrued to patient C. The earliest post-transplant HCV viremia performed was at one year and was over 8 million IU/ml, which is typical of HCV infected post-transplant patients.
Fig. 1.
Proline is an atypical residue for genotype 1a or 1b at amino acid 328 NS5A, but was present pre-transplant in patient C and was associated with more CsA susceptibility than H771a. (A) Alignment of H771a, patient C and patient G pre and post-transplant amino acids sequences along with alignment of Con1b, patient H pre and post-transplant amino acids sequences are presented. (B) The Logo analysis of genotypes 1a, 1b, 2, 3, 4 and 6 sequences near the highly conserved WARPDYN motif (underlined) associated with laboratory aquired CsA resistance. The amino acid at position 328 is boxed and sequences derived from different genotypes are labeled. (C) Transient replication of Con1bLN-pre-C, Con1bLN-post-C, Con1bLN-Pre-G, Con1bLN-post-G, Con1bLN-5AH77 and Con1bLN-wt replicons and their sensitivity in presence (dashed lines) andin absence (solid lines) of CsA at different time points. (D) Percent inhibition of HCV replicons are presented at 96 h post RNA electroporation by comparing CsA treated vs. untreated luciferase data as in 1C.
The post-transplant sequences from a seventh patient (patient G), with a divergent genotype 1a sequence from H77, who also developed multiple mutations in this region were compared to the pre-transplant sequence (Fig. 1A, pre-G and post-G) and its sensitivity towards CsA was tested after cloning into the 1b replicon. Unlike patient C, we did not observe a more CsA resistant phenotype despite having acquired two additional prolines and few other changes post-transplant (Fig. 1C, pre and post G are purple and green). Similar to the Con1bLN-5AH77 chimeric replicon, the patient G replicon was relatively resistant both pre and post-transplant compared to the pre-transplant patient C chimeric replicon (Fig. 1D). All the chimeric replicons had similar growth kinetics in absence of CsA (Fig. 1C, solid lines).
Mutational analysis demonstrates proline at 328 confers CsA relative susceptibility
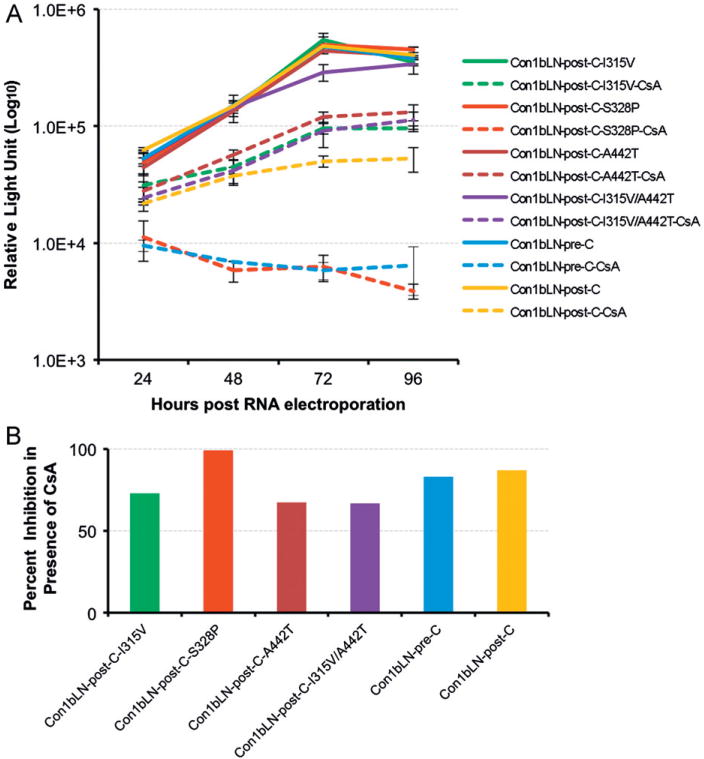
The replicon derived from patient C pre-transplant and post-transplant differed by 4 amino acids (Fig. 1A). The Con1bLN-post-C replicon was mutated to contain single amino acid substitutions at Val315, Pro328 and Thr442 and a double amino acids substitution at Val315 and Thr442. Replicons containing Val or Thr mutations either in single or in combination replicated similar to Con1bLN-post-C replicon containing all three mutations (Fig. 2A and B). However, mutating only Ser to Pro at position 328 rendered Con1bLN-post-C-S328P replicon CsA sensitive to the level of Con1bLN-pre-C replicon (compare dashed red with dashed blue line). In a converse experiment we mutated Con1bLN-pre-C replicon to contain Ser at position 328 and this change was sufficient enough to shift the CsA susceptibility similar to post-replicon confirming the involvement of amino acid 328 in CsA susceptibility (Fig. 3A and B, dashed purple line).
Fig. 2.
Replicons with proline at amino acid 328 have increased CsA susceptibility than serine in both genotype 1a and 1b lineages. (A) Transient replication of Con1bLN-post-C replicon in presence (dashed line) and in absence (solid line) of CsA after mutating amino acids at position 315, 328, and 442 similar to patient C pre-transplant sequences. (B) Percent inhibition of HCV replicons are presented at 96 h post RNA electroporation by comparing CsA treated vs untreated luciferase data as in 2A.
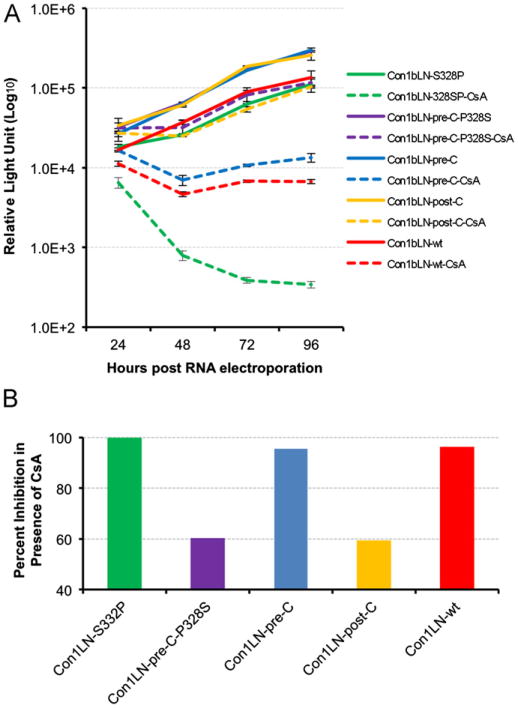
Fig. 3.
The Con1bLN-wt replicon with 328P is more sensitive to CsA that Con1bLN-wt. (A) Transient replication of Con1bLN-S328P and Con1bLN-pre-C-P328S replicons were analyzed along with Con1bLN-pre-C, Con1bLN-post-C and Con1bLN-wt replicon in presence (dashed line) and in absence (solid line) of CsA at different time points. (B) Percent inhibition of HCV replicons are presented at 96 h post RNA electroporation by comparing CsA treated vs. untreated luciferase data as in 3A.
Interestingly the consensus amino acid at position 328 is quite distinct between the genotype 1a lineage (90% threonine), and the genotype 1b (∼90% serine), but a proline can occur in either lineage and is approximately twice as frequent for genotype 1b as for genotype 1a (Fig. 1B). Therefore a proline was engineered into the Con1bLN-wt (1b nonchimeric) replicon. Substitution of Ser to Pro increased the susceptibility of the Con1bLN-S328P replicon to CsA just as it did for the Con1bLN-pre-C replicon (Fig. 3A and B, dotted green line) suggesting the effect is not an artifact of chimeric replicons and residue 328 is relevant to the CsA susceptibilities of both genotypes 1b and 1a.
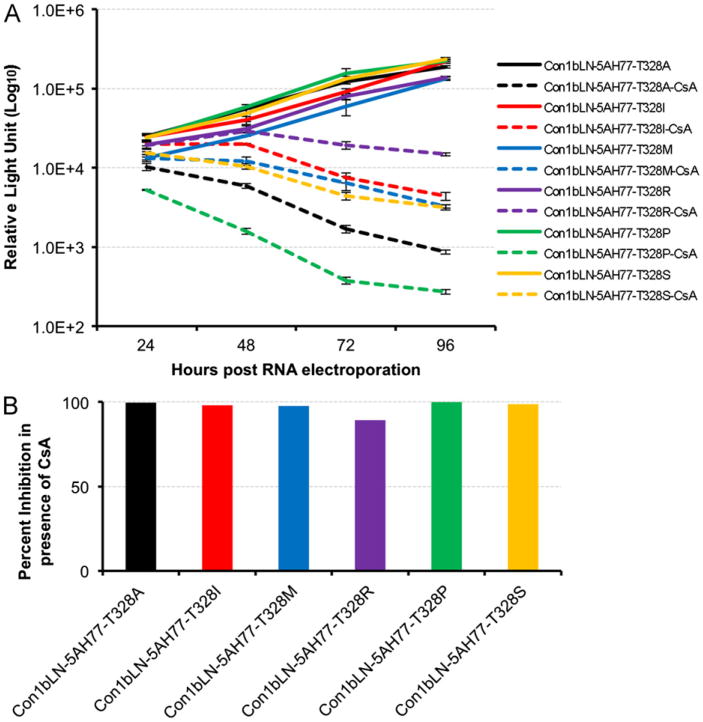
While proline is the most common variant other than Ser (genotype 1b) or Thr (genotype 1a) there are other more rare variants reported in other HCV genotypes (Fig. 1B). To investigate the effect of these variants on CsA susceptibility we mutated Thr 328 in the context of Con1bLN-5AH77 chimeric replicon to Ala, Ser, Met, Ile and Arg residues which are present in the different HCV genotypes (Fig. 1B). All mutated replicons displayed at least some sensitivity to CsA, but proline conferred the most susceptibility. Ser, Met, Ala and Ile displayed intermediate susceptible phenotypes, while Arg was the least susceptible to CsA treatment in our transient replication model (Fig. 4A and B).
Fig. 4.
CsA sensitivity of natural variants at position 328 present in different HCV genotypes. (A) The Con1bLN-5AH77 chimeric replicon was mutated to contain Ala, Ser, Met, Ile and Arg at position 328 and their replication efficiency in presence (dotted line) and in absence (solid line) of CsA at different time points. (B) Percent inhibition of HCV replicons are presented at 96 h post RNA electroporation by comparing CsA treated vs. untreated luciferase data as in (A).
Discussion
Antiviral development for HCV is improving rapidly, but many patients can still not be cured. Furthermore, many viral sequences are not replication competent in cell culture, and therefore it is difficult to know if a specific viral sequence is susceptible to a specific antiviral agent. Through the use of NS5A chimeric replicons generated from patients exposed to CsA, we directly compared the CsA susceptibility of specific NS5A consensus sequences without the confounding effects of differences in other parts of the genome. Prior work has shown that NS5A derived from genotypes 1a, 1b, 2a, and 2b binds cyclophilin. This cyclophilin binding and the strong conservation of the WARPDYN binding site for CypA identified by NMR (Hanoulle et al., 2009) has been used to argue that cyclophilin inhibitors are “pangenotypic” and the heterogeneity of NS5A does not correlate with cyclophilin inhibition (Chatterji et al., 2010). Cyclophilin inhibitors do block the NS5A:CypA interaction in vitro (Fernandes et al., 2010; Hopkins et al., 2012). Our data is consistent with most if not all HCV being susceptible to cyclophilin inhibitors, but suggests that NS5A polymorphisms outside the conserved DYN sequence also influence the degree of CsA susceptibility. While HCV exists as a swarm of closely related quasispecies, we did not detect a 328 threonine containing sequence in patient C pre-transplant via clonal analysis (data not shown). Clearly isolates from different patients vary at this and many positions in NS5A Deep sequencing may have shown cyclosporine selection at other positions besides 328, or in other patients but we only have consensus sequencing data for these patients. We did not find any variant that has been described from cell culture selection with cyclophilin inhibitors including D320E or Y321N (Puyang et al., 2010; Yang et al., 2010).
The DYN amino acids 320-322 is embedded among multiple prolines that biochemical data suggest are targets of cyclophilin (Fernandes et al., 2010; Hanoulle et al., 2009; Tang, 2010). Genetic differences between genotypes in NS5A could alter how susceptible HCV is in patients and do alter it in cell culture (Ansari and Striker, 2012) but it is unclear that these changes in cell culture susceptibility are clinically significant. This case series shows no antiviral effect of CsA in most patients but suggests in patient C selection of resistant HCV can occur. Moreover, this data lends confirmatory in vivo evidence that NS5A is one of the critical targets of cyclophilin inhibitors. Both NS2 and NS5B have also been hypothesized as targets of cyclophilin, but we sequenced most of NS5B as well as NS5A and could not confirm any selective pressure on NS5B. There were other consensus mutations that varied between pre- and post-sequences in NS5B and NS5A, but there was no obvious pattern. In cell culture, multiple mutations have been found to be required for significant decreases in CsA cell culture susceptibility (Garcia-Rivera et al., 2012), but our mutational analysis within the naturally occurring NS5A sequence suggests the bulk of the decrease CsA susceptibility for our patient samples came from the proline to serine mutation. Strikingly, the only genotype 1b patient had pre-transplant GYN rather than the typical DYN consensus sequence that mutated to SYN post-transplant (patient H), but replicon data refuted that this variation altered cell culture sensitivity (data not shown). Perhaps other sequence variation outside of the cloned region obscured our ability to associate this change with relative CsA resistance. Since neither patient B nor C received interferon or ribavirin, these drugs cannot explain the selection of consensus mutants near known positions of laboratory required CsA resistance. While proline 328 is the consensus residue in only 5–10% of HCV infected patients it is possible that patients with a proline present explains some of the benefit anecdotally seen with switching to CsA (Lorho et al., 2005). In patient C, the only viral load data available to us was 6 months after we detected the threonine mutant and one year after transplantation so it is difficult to known whether CsA delayed or partially suppressed the patient's viremia or not.
In summary, for six of our seven patients, even consensus sequencing and phenotypic analysis could not detect an effect of CsA on viral evolution, or antiviral benefit. The only patient in which we could find evidence for selection from the antiviral effect of CsA had an atypical consensus sequence in the region of NS5A that binds cyclophilin. Likely all, if not most HCV is susceptible to nonimmunosuppressive cyclophilin inhibitors including alisporivir and SCY635, but ∼5–10% of genotype 1 strains that have this proline 328 variant may be even more susceptible. At least one genotype 3 infected patient was cured by a short duration alisporivir monotherapy (Patel and Heathcote, 2011). While the approval of protease inhibitors has greatly increased the possibility of curing HCV, small molecule inhibitors quickly select resistance in HCV unless given in combination with other antivirals with different mechanisms of action. Clinical studies pairing of cyclophilin inhibitors with other NS5A and nonNS5A acting antivirals may benefit from looking at viral genetic differences between HCV isolates.
Material and methods
Clinical trial and study design
The HIV–TR study included pre-transplant and post-transplant serum samples and a detailed protocol post-transplant follow-up (Terrault et al., 2012). The decision to treat HCV infected patients with interferon or not was left to the discretion of the patient care team. This resulted in only 15 patients who were exposed to CsA immediately post-transplant for 6 months or more and we attempted to sequence all of those patients pre and post-transplant for which there were samples available.
RT-PCR and nucleotide sequencing
The viral RNA was isolated from serum samples and used for RT-PCR. Consensus sequencing and analysis of amplified HCV was performed from amino acid 29 in NS5A until amino acid 563 of NS5B as described before (Kuntzen et al., 2007). The nucleotide sequences of 6 cloned fragments derived from 6 individual patients are deposited in GenBank (JX457830-35). The Institutional Review Board of the University of Wisconsin approved this study and all study participants provided informed consent in accordance with the Institutional Review Boards of the participating centers.
Genetic manipulation of Con1bLN replicon
The PCR fragment spanning from amino acids 312 of NS5A to amino acids 5 of NS5B (FARALPV to MSYTWT) was amplified from the above described original cDNA fragment using primers containing XhoI and BstZI restriction sites. This fragment was cloned in TOPO vector and sequenced again to confirm the mutations. The desired fragment was subcloned directionally in Con1bLN replicon (Fernandes et al., 2010) using XhoI and BstZ17I restriction enzymes. All the replicons were sequenced to confirm mutations.
RNA transcription and transient replication assay
The Huh7.5 cells were maintained as before (Fernandes et al., 2010). The CsA was purchased from sigma. The RNA transcription and electroporation was performed as described before (Fernandes et al., 2010). In brief, approximately 2 million cells were electroporated with 6 μg of in vitro transcribed RNA and electroporated cells were divided into two sets into twenty–four well plates. After 6 h of incubation the media was replaced but only one set of plates received 0.5 μg/ml of CsA. The cells were further incubated and renilla luciferase activity was monitored every 24 h as described before. In all the luciferase assays an average of three independent assays was presented along with standard deviations.
Logo analysis
The amino acid sequences of different HCV genotypes 1a (224 strains), 1b (358 strains), 2 (49 strains), 3 (30 strains), 4(36 strains) and 6 (48strains) were retrieved from The European HCV Database (http://euhcvdb.ibcp.fr/euHCVdb/). A small stretch of amino acids encompassing the region of interest (aa 316–328) was subjected to logo analysis (Crooks et al., 2004) using web based program (www.biovirus.org).
Acknowledgments
We gratefully acknowledge the patients and other investigators involved in the solid organ transplantation in HIV (HIV–TR Study AI052748) and the Project Manager, Rodney Rogers. This work was supported by the American Cancer Society Research Scholar Grant (07-077-01) to R.S., by the Office of Research and Development, Biomedical Laboratory R&D Service, Department of Veterans Affairs. Patient samples were obtained via the solid organ transplantation in HIV: Multi-site study (HIV–TR study AI052748) funded by the National Institute of Allergy and Infectious Diseases.
We also thank John Tavis for playing a critically helpful role in the early phases of this project and Lindsey Moser for critical reading of the manuscript.
References
- Ansari IUH, Striker RT. Subtype specific differences in NS5A domain II reveals involvement of proline at position 310 in cyclosporine susceptibility of Hepatitis C virus. Viruses. 2012;4:3303–3315. doi: 10.3390/v4123303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji U, Lim P, Bobardt MD, Wieland S, Cordek DG, Vuagniaux G, Chisari F, Cameron CE, Targett-Adams P, Parkinson T, Gallay PA. HCV resistance to cyclosporin A does not correlate with a resistance of the NS5A-cyclophilin A interaction to cyclophilin inhibitors. J Hepatol. 2010;53:50–56. doi: 10.1016/j.jhep.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesek S, Steinmann E, Wedemeyer H, Manns MP, Neyts J, Tautz N, Madan V, Bartenschlager R, von Hahn T, Pietschmann T. Cyclosporine A inhibits hepatitis C virus nonstructural protein 2 through cyclophilin A. Hepatology. 2009;50:1638–1645. doi: 10.1002/hep.23281. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes F, Ansari IUH, Striker R. Cyclosporine inhibits a direct interaction between cyclophilins and hepatitis C NS5A. PLoS One. 2010;5:e9815. doi: 10.1371/journal.pone.0009815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes F, Poole DS, Hoover S, Middleton R, Andrei AC, Gerstner J, Striker R. Sensitivity of hepatitis C virus to cyclosporine A depends on nonstructural proteins NS5A and NS5B. Hepatology. 2007;46:1026–1033. doi: 10.1002/hep.21809. [DOI] [PubMed] [Google Scholar]
- Fischer G, Gallay P, Hopkins S. Cyclophilin inhibitors for the treatment of HCV infection. Curr Opin Invest Drugs. 2010;11:911–918. [PubMed] [Google Scholar]
- Flisiak R, Horban A, Gallay P, Bobardt M, Selvarajah S, Wiercinska-Drapalo A, Siwak E, Cielniak I, Higersberger J, Kierkus J, Aeschlimann C, Grosgurin P, Nicolas-Metral V, Dumont JM, Porchet H, Crabbe R, Scalfaro P. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology. 2008;47:817–826. doi: 10.1002/hep.22131. [DOI] [PubMed] [Google Scholar]
- Foster TL, Gallay P, Stonehouse NJ, Harris M. Cyclophilin A interacts with domain II of hepatitis C virus NS5A and stimulates RNA binding in an isomerase-dependent manner. J Virol. 2011;85:7460–7464. doi: 10.1128/JVI.00393-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rivera JA, Bobardt M, Chatterji U, Hopkins S, Gregory MA, Wilkinson B, Lin K, Gallay PA. Multiple mutations in hepatitis C virus NS5A domain II are required to confer a significant level of resistance to alisporivir. Antimicrob Agents Chemother. 2012;56:5113–5121. doi: 10.1128/AAC.00919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Watashi K, Inoue D, Hijikata M, Shimotohno K. Identification of cellular and viral factors related to anti-hepatitis C virus activity of cyclophilin inhibitor. Cancer Sci. 2009;100:1943–1950. doi: 10.1111/j.1349-7006.2009.01263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grise H, Frausto S, Logan T, Tang H. A conserved tandem cyclophilinbinding site in hepatitis C virus nonstructural protein 5A regulates alisporivir susceptibility. J Virol. 2012;86:4811–4822. doi: 10.1128/JVI.06641-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanoulle X, Badillo A, Wieruszeski JM, Verdegem D, Landrieu I, Bartenschlager R, Penin F, Lippens G. Hepatitis C virus NS5A protein is a substrate for the peptidyl–prolyl cis/trans isomerase activity of cyclophilins A and B. J Biol Chem. 2009;284:13589–13601. doi: 10.1074/jbc.M809244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque M, Hashim A, Greanya ED, Steinbrecher UP, Erb SR, Yoshida EM. Spontaneous clearance of hepatitis C infection post-liver transplant: a rare but real phenomenon? A case report and review of the literature. Ann Hepatol. 2010;9:202–206. [PubMed] [Google Scholar]
- Hopkins S, Bobardt M, Chatterji U, Garcia-Rivera JA, Lim P, Gallay PA. The cyclophilin inhibitor SCY-635 disrupts hepatitis C virus NS5A-cyclophilin A complexes. Antimicrob Agents Chemother. 2012;56:3888–3897. doi: 10.1128/AAC.00693-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul A, Stauffer S, Berger C, Pertel T, Schmitt J, Kallis S, Zayas M, Lohmann V, Luban J, Bartenschlager R. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009;5:e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntzen T, Timm J, Berical A, Lewis-Ximenez LL, Jones A, Nolan B, Schulze zur Wiesch J, Li B, Schneidewind A, Kim AY, Chung RT, Lauer GM, Allen TM. Viral sequence evolution in acute hepatitis C virus infection. J Virol. 2007;81:11658–11668. doi: 10.1128/JVI.00995-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorho R, Turlin B, de Lajarte-Thirouard AS, Camus C, Lakehal M, Compagnon P, Meunier B, Boudjema K, Messner M. Improved liver function and decreased hepatitis C viral load after tacrolimus was replaced by cyclosporine. Transplant Proc. 2005;37:2871–2872. doi: 10.1016/j.transproceed.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Ma S, Boerner JE, TiongYip C, Weidmann B, Ryder NS, Cooreman MP, Lin K. NIM811, a cyclophilin inhibitor, exhibits potent in vitro activity against hepatitis C virus alone or in combination with alpha interferon. Antimicrob Agents Chemother. 2006;50:2976–2982. doi: 10.1128/AAC.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel H, Heathcote EJ. Sustained virological response with 29 days of Debio 025 monotherapy in hepatitis C virus genotype 3. Gut. 2011;60:879. doi: 10.1136/gut.2010.217323. [DOI] [PubMed] [Google Scholar]
- Puyang X, Poulin DL, Mathy JE, Anderson LJ, Ma S, Fang Z, Zhu S, Lin K, Fujimoto R, Compton T, Wiedmann B. Mechanism of resistance of hepatitis C virus replicons to structurally distinct cyclophilin inhibitors. Antimicrob Agents Chemother. 2010;54:1981–1987. doi: 10.1128/AAC.01236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. Cyclophilin inhibitors as a novel HCV therapy. Viruses. 2010;2:1621–1634. doi: 10.3390/v2081621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellinghuisen TL, Foss KL, Treadaway JC, Rice CM. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J Virol. 2008;82:1073–1083. doi: 10.1128/JVI.00328-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrault NA, Roland ME, Schiano T, Dove L, Wong MT, Poordad F, Ragni MV, Barin B, Simon D, Olthoff KM, Johnson L, Stosor V, Jayaweera D, Fung J, Sherman KE, Subramanian A, Michael Millis J, Slakey D, Berg CL, Carlson L, Ferrell L, Stablein DM, Odim J, Fox L, Stock PG. Outcomes of liver transplantation in HCV–HIV coinfected recipients. Liver Transplant 2012 [Google Scholar]
- Verdegem D, Badillo A, Wieruszeski JM, Landrieu I, Leroy A, Bartenschlager R, Penin F, Lippens G, Hanoulle X. Domain 3 of NS5A protein from the hepatitis C virus has intrinsic alpha-helical propensity and is a substrate of cyclophilin A. J Biol Chem. 2011;286:20441–20454. doi: 10.1074/jbc.M110.182436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermehren J, Sarrazin C. New HCV therapies on the horizon. Clin Microbiol Infec. 2011;17:122–134. doi: 10.1111/j.1469-0691.2010.03430.x. [DOI] [PubMed] [Google Scholar]
- Watashi K, Ishii N, Hijikata M, Inoue D, Murata T, Miyanari Y, Shimotohno K. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell. 2005;19:111–122. doi: 10.1016/j.molcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Yang F, Robotham JM, Grise H, Frausto S, Madan V, Zayas M, Bartenschlager R, Robinson M, Greenstein AE, Nag A, Logan TM, Bienkiewicz E, Tang H. A major determinant of cyclophilin dependence and cyclosporine susceptibility of hepatitis C virus identified by a genetic approach. PLoS Pathog. 2010;6:e1001118. doi: 10.1371/journal.ppat.1001118. [DOI] [PMC free article] [PubMed] [Google Scholar]