Abstract
A capability for scanning electron microscopy of wet biological specimens is presented. A membrane that is transparent to electrons protects the fully hydrated sample from the vacuum. The result is a hybrid technique combining the ease of use and ability to see into cells of optical microscopy with the higher resolution of electron microscopy. The resolution of low-contrast materials is ≈100 nm, whereas in high-contrast materials the resolution can reach 10 nm. Standard immunogold techniques and heavy-metal stains can be applied and viewed in the fluid to improve the contrast. Images present a striking combination of whole-cell morphology with a wealth of internal details. A possibility for direct inspection of tissue slices transpires, imaging only the external layer of cells. Simultaneous imaging with photons excited by the electrons incorporates data on material distribution, indicating a potential for multilabeling and specific scintillating markers.
Electron microscopy (EM) has been an indispensable tool for the life and medical sciences since its inception more than half a century ago. Much of the substantial advances in the field were propelled by the need to find methods to best preserve and analyze structures at a state most closely approximating the native state. Little if any attention has been given to wet samples, under the assumption that it was practically impossible. However, an ability to observe fully hydrated samples at room or body temperatures could help eliminate many artifacts of sample preparation and allow routine and reproducible imaging.
Recent progress in adaptation of scanning EM (SEM) for observation of partially hydrated samples relies on technological improvements in differential pumping capabilities and of detectors, which together allow conditions that sustain the sample in a vapor environment [e.g., environmental SEM (1–3)]. However, the goal of imaging wet, fully fluid samples has not been met by these advances until now. The question of whether imaging at acceptable resolution and contrast is at all possible and what can be seen once cells are imaged remained open.
We present here a significant step in this direction, in which wet samples can be maintained in fully physiological conditions and imaged with little loss of resolution compared to standard SEM. Wet SEM relies on a thin, membranous partition that protects the sample from the vacuum while being transparent to the beam electrons. This approach was proposed at the advent of the scanning electron microscope (early attempts are best seen in the work shown in ref. 4) but yielded an unacceptable resolution due to the unavailability of adequate materials at that time. Developments in polymer technology have yielded thin membranes that are practically transparent to energetic electrons yet are tough enough to withstand atmospheric pressure differences. The volume imaged is in close proximity to the membrane, typically probing a few micrometers into the sample. This is ideal for the inspection of fluids or objects that are in close contact with the surface. The presence of fluid helps in preventing charging effects and eliminates the need to coat the sample.
This imaging system enables a number of observations that previously were inaccessible to SEM. First, SEM can now be used to probe the inside of whole cells, giving information on organelles and internal structure. Second, staining and gold immunolabeling can be imaged with no subsequent critical-point drying and coating (5). Third, we show that tissue sections can be viewed, giving structural information on the connectivity and organization of cells and extracellular structures in situ.
Another important advantage is the development of concurrent monitoring of light emitted from molecules inside the sample that are excited by the electron beam (6) [cathodoluminescence (CL)], which gives a complementary view of material distribution, including the ability to distinguish multiple labels.
Ultimately, the resolution available for untreated biological samples is limited by the physical constraints on EM. Because we are imaging the differences in scattering between low atomic number (Z) components such as carbon (cells) and oxygen (water), the contrast is often low and may depend strongly on the local concentration of heavier ions. Although this lowers the resolution, images are still much better than feasible by using optical methods and (as discussed below) are better than what a naive theoretical estimate based on electron scattering in materials gives.
Methods
Chambers. The sample holder centers on a rigid enclosure with a window that consists of a thin, electron-transparent partition membrane. Beam electrons go through the partition membrane, probe the sample, and scatter back to the backscattered electron (BSE) detector placed above the sample. Several membranes have been tested, of which polyimide membranes of 145 nm in thickness were found to be the most appropriate. Variations on the basic design have been developed that allow different modes of insertion of the specimen to close contact with the membrane. For example, adherent cultured cells can be grown directly on the partition medium; bacteria, mammalian, or other cells grown in suspension can be adsorbed or centrifuged onto the membrane before imaging; and tissues may be brought to direct contact with the membrane manually or mechanically. Another design incorporates a small leakage hole to the microscope chamber that is calculated to maintain the sample at low pressures while keeping it fully hydrated. This reduces the strain on the membrane, improves the endurance of the membrane, and enables ultrathin membranes (down to 50 nm) to be used for increased resolution. The polyimide membrane was treated by fibronectin to enhance affinity of cells, whereas bacterial adhesion was achieved with poly-(l-lysine).
Staining Methods. Staining was done following standard methods. Unless specified otherwise, cells and tissues were fixed with 3% paraformaldehyde and 2% glutaraldehyde in 0.1 M cacodylate buffer containing 1% sucrose and then rinsed in the same buffer. For uranyl acetate staining, the fixed samples were rinsed extensively in water, treated with 1% tannic acid for 5 min, rinsed in water, and stained with uranyl acetate (0.1–2% solution, pH 3.5) for 15–30 min. Osmium tetroxide staining was performed by washing the fixed samples in water and then incubating for 10–30 min in 1% osmium tetroxide in water. In all cases, the samples were rinsed in water and viewed in the wet state.
Resolution. Theoretically, the resolution of wet SEM depends on the volume sampled by Rutherford scattering of electrons (BSE). This volume depends mainly on the Z of the sample and the acceleration voltage, or energy, of the beam electrons and is approximated by the Kanaya–Okayama radius (5). For biological samples, the Z is low (e.g., carbon Z = 6 and oxygen Z = 8), and the radius of interaction is typically a few micrometers for acceleration voltages of 15–30 kV. Surprisingly, as Fig. 1B shows, the actual resolution is an order of magnitude better, because fat droplets in milk <100 nm can be resolved. This is because the multiply scattered BSEs probe such a large region (on the scale of a few micrometers) that their signal varies only slowly from point to point. The contrast then is obtained from electrons that scatter back after only a few interactions. These probe a much smaller region, on the scale of the width of the beam in the sample, and define a high resolution (similar to that discussed in chapter 4.6.2 of ref. 5). The viscosity inside a cell limits thermal motion such that, theoretically, resolution of unfixed objects above ≈20 nm is possible. Images similar to that shown in Fig. 1C were used to evaluate the resolution numerically (7), giving values in the range of 100–120 nm. We found that emulsions yield a strong signal (and are easier to resolve than polymeric beads), presumably because they deform to wet the membrane and have more material close to the surface.
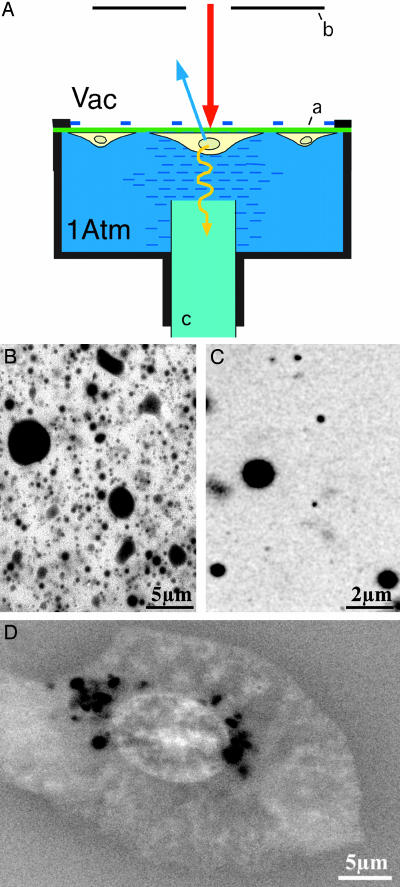
Fig. 1.
Imaging of unstained specimens. (A) Wet-SEM sample holder (schematic). The sample is depicted here as cultured cells grown on the thin partition membrane (a) that separates the vacuum (Vac) from the fluid in the holder. Imaging is done in scanning electron microscope: the electron beam (thick arrow) penetrates into the sample, and BSEs (thin blue arrow) are detected by a BSE detector (b). A grid (dashed line) is used to support and reinforce the membrane. A light guide (c) is introduced at the bottom for CL applications (see text and Fig. 4). The membranes are mounted on a plastic ring and treated with extracellular matrix proteins. Cells in their appropriate growth medium are deposited onto the membrane and incubated. Before observation, the membrane is turned upside down and assembled onto the sample holder. Different microscopes were used: JEOL 6400 SEM, FEI (Hillsboro, OR) XL30 ESEM, and FEI XL30 ESEM-FEG in both low [0.1 torr (1 torr = 133 Pa)] and high (≈10-5 torr) vacuum modes. (B) Low-fat (1.5%) milk (voltage, 12 kV). (C) As described for B. (D) Untreated CHO cell grown on a fibronectin-coated membrane in normal growth medium.
CL Images. CL images are obtained simultaneously with BSE images. A light guide at the bottom of our sample efficiently collects the light and brings it to the entrance of the photomultiplier of a single photon-counting module. The data are collected into a timer–counter computer card, which also receives digital signals from the microscope when a frame and a new scan line are initiated. Using these three signals, a computer program reconstitutes the CL image in parallel to the image collected in the standard electron channel.
Results
Untreated Samples. Notably, we observe that, even in unstained samples, the differences between water and oil droplets (Fig. 1 B and C) or between different constituents of the cell (Fig. 1D) generate sufficient contrast to distinguish them. In cells, higher-Z materials such as salts, phosphor, and iron, which may be concentrated in different regions, tend to improve the contrast. Fig. 1D shows an untreated Chinese hamster ovary (CHO) cell in normal culture medium at room temperature. The cell outline and the nucleus with its internal structure are clearly visible, as are a series of dark, spherical particles in the cytoplasm. The identification of these particles as lipid droplets is discussed below.
Heavy-Metal Staining. The limits on resolution and contrast when observing low-Z materials suggest that substantial advantages can be achieved by staining the cells with high-Z markers such as electron-dense stains or nanoparticle gold labels. Indeed, we are able to detect gold beads in water down to a diameter of 10 nm by using ultrathin membranes (data not shown).
Differential staining of organelles inside cells using electrondense materials is a standard in EM technique, although not usually applied for SEM. Fig. 2 shows a rich variety of structures inside of cultured cells that were grown directly on the partition membrane and then fixed and stained with uranyl acetate (8). Intracellular organelles are clearly visible, including internal details of mitochondria (Fig. 2C), actin stress fibers (Fig. 2D), and complex tubular and cytoskeletal structures (Fig. 2 A and B). The higher beam energy in Fig. 2C probes internal structure, whereas at lower energy (Fig. 2D), the surface close to the membrane is visualized. Thus, different energies can be used to obtain three-dimensional information.
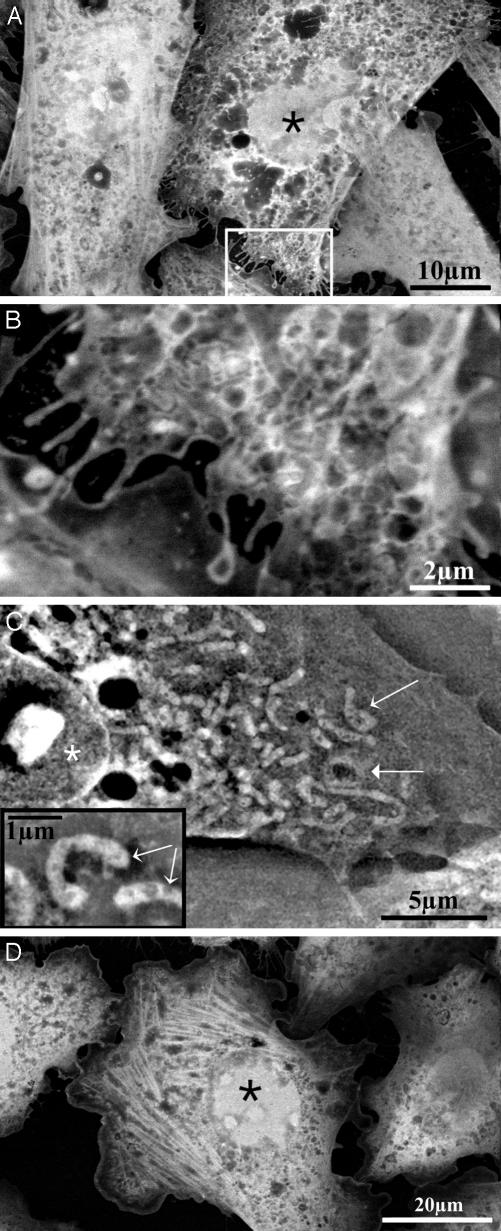
Fig. 2.
Imaging of stained cells. Asterisks denote nuclei, and thin arrows denote mitochondria. (A) HeLa cells grown on the membrane in normal growth medium, then fixed with paraformaldehyde, and stained with uranyl acetate (imaged at 12 kV). (B) Magnification of the marked rectangle shown in A. (C) CHO cell fixed with glutaraldehyde and paraformaldehyde, stained with uranyl acetate, and maintained in water (imaged at 30 kV). The thick arrow denotes a mitochondrion that surrounds a lipid droplet. (Inset) Higher magnification showing mitochondria (thin arrows). (D) Actin fibers in stained CHO cells. Treatment and imaging are as described for A.
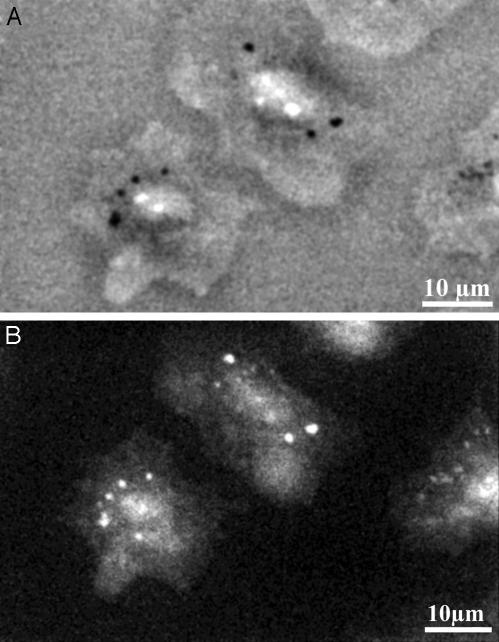
Nanoparticle Gold Markers. High resolution and specificity of detection can be achieved by binding of antibodies or other ligands. As with other EM methods, labeling is done most conveniently by using gold nanoparticles. In contrast with transmission EM (TEM), which samples only very thin and often arbitrary sections, wet SEM allows a quick switching between the local and global view of labeling all over the cell. Fig. 3 A and B show how 40-nm nanoparticle gold-labeled monoclonal antiepidermal growth factor receptor antibodies were used for labeling the epidermal growth factor receptors on intact A431 cancer cells. Fixation is often useful and convenient even in wet SEM and may facilitate the labeling as well as the transfer to the microscope. The cells presented here are also stained with uranyl acetate, which allows alignment of the labeled receptors with the general structure of the cells. The high contrast and uniform size of the nanoparticles enable unambiguous identification and localization. Knowing the precise localization of single receptor molecules opens the possibility of measuring events such as epidermal growth factor-induced receptor dimerization. Staining of the cells is not needed for the labeling, and we have included an image of unstained, labeled cells (Fig. 7, which is published as supporting information on the PNAS web site). Also shown is an image demonstrating that marking of internal parts of the cell is possible (Fig. 8, which is published as supporting information on the PNAS web site), in this case, the marking of mitochondria with gold beads. Labeling of actin filaments inside of cells has also been achieved by using subnanometer gold particles, followed by silver enhancement (data not shown).
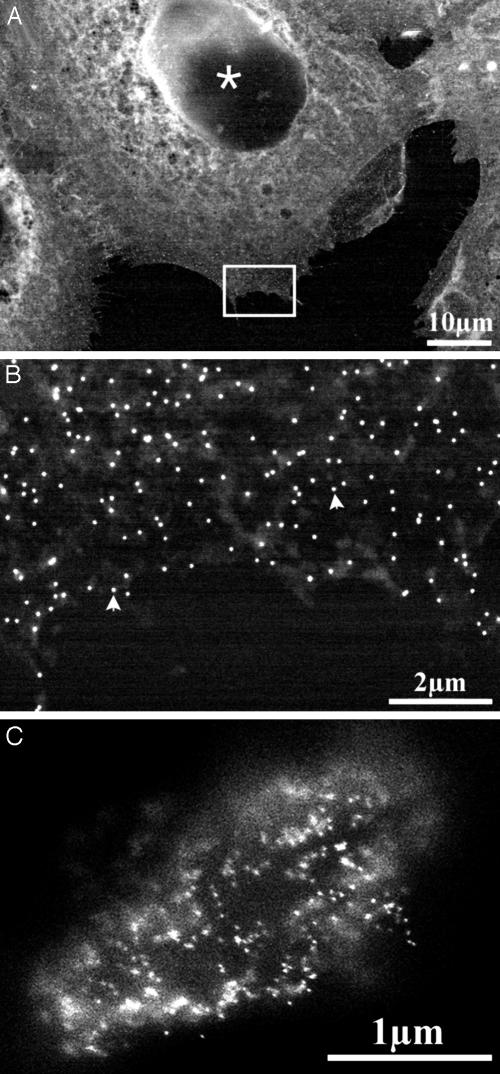
Fig. 3.
Imaging of nanoparticle gold markers on cells. The arrowheads denote gold nanoparticles. (A) Epidermal growth factor receptors immunolabeled with 40-nm gold nanoparticles on A431 cells and counterstained with uranyl acetate (imaged at 30 kV). (B) Magnification of the marked rectangle shown in E. (C) Putative gastrin receptors on H. pylori bacterium, after incubation with complexed biotinylated gastrin on streptavidin-coated 20-nm gold particles, followed by glutaraldehyde and sedimentation onto the poly-(l-lysine)-coated membrane (imaged at 20 kV).
An example of gold labeling on bacteria is given in Fig. 3C. Here, the putative gastrin receptor of Helicobacter pylori (9) is detected by incubation with complexes of biotinylated gastrin and streptavidin-coated 20-nm gold nanoparticles. Administering the biotinylated gastrin before that of the streptavidin-coated gold yielded almost no attachment of nanoparticles to the bacterium. This result was obtained independently of the time between the two steps, suggesting that the uptake of gastrin into H. pylori is almost immediate. The fact that complexed gastrin did not enter the cells points to the possibility of defining the limits of the size of particles that can be internalized by the bacteria.
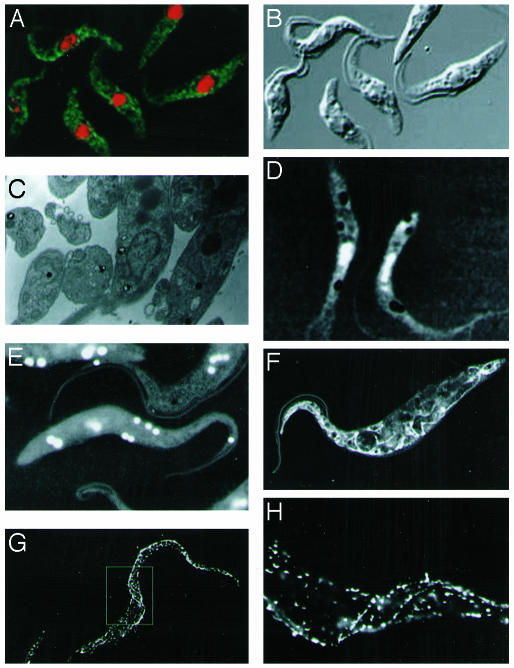
Comparison of Imaging Techniques (Trypanosoma brucei). It is useful to compare this technology with existing imaging techniques using a single-model system. Such a study is detailed in Fig. 4, using the parasite T. brucei procyclic form that propagates in the midgut of the tsetse fly. The first three images display existing modes of imaging: optical fluorescence microscopy (Fig. 4A), differential interference contrast microscopy (Fig. 4B), and TEM (Fig. 4C). The strength of existing techniques is apparent: localized marking in fluorescence and detailed sections in TEM. The untreated optical (Fig. 4B) and wet-SEM (Fig. 4D) images delineate the borders of the cell but are low in contrast and lack detail. Both emphasize the nucleus, but other organelles that appear in the differential interference contrast image are not clearly identified, whereas the wet-SEM image brings out the lipid droplets to an advantage. Osmium tetroxide staining (Fig. 4E) is not as useful in this system, emphasizing mostly the lipid droplets and some unidentified inner organelles. The strongest imaging is seen in Fig. 4F, for which uranyl acetate staining was used. The advantage of intact imaging with which the full cell can be seen (along with its fine internal structures that can be observed in detail) is apparent, especially in comparison to TEM imaging (Fig. 4C). Finally, wet-SEM imaging of whole-cell-specific marking with antibodies against the major glycosylphosphatidylinositol-anchored surface coat protein EP procyclin (10) is shown in Fig. 4 G and H. Interestingly, EP procyclin is known to cover the whole cell uniformly, but the observed signal is clustered. We attribute this to the tendency of the underlying lipid rafts to cluster under the influence of antibodies, a process that fixation in formaldehyde is unable to prevent. The gold markers also elucidate the three-dimensional conformation of the cell in a manner that is reminiscent of what standard SEM can image (an excellent, representative SEM image of T. brucei can be found at http://tryps.rockefeller.edu/crosslab_intro.html).
Fig. 4.
Multimodal imaging of T. brucei. The typical length of a cell is 20 μm in length and 3 μm in width. (A) Confocal fluorescence microscopy. Anti-EP procyclin antibodies (Cedarline) were used, and the binding was detected with anti-mouse linked to fluorescein isothiocyanate (green). Staining of the nucleus and kinetoplast was with propidium iodide (red). (B) Optical (differential interference contrast) imaging. (C) TEM sections of a cell, followed by uranyl acetate staining, revealing internal organelles. (D) Wet-SEM imaging of whole, hydrated, unstained cells. (E) As described for D using osmium tetroxide staining. (F) As described for D using uranyl acetate staining. (G) Nanoparticle gold markers linked to anti-mouse antibodies, showing the location of the surface EP procyclin protein and enabling at the same time a three-dimensional view. (H) Higher magnification of the midsection of the cell shown in G, elucidating an area of three-dimensional helicity in the cell.
Obviously, TEM will have better resolution, in general, than wet SEM, but the images presented in the example of T. brucei for comparison are “typical” images obtained in the same laboratory. Because wet SEM is different in its utility than TEM and optical microscopy, in T. brucei it can be seen as an important complementary capability, and one can expect that the full picture would be given by a combination of optical, TEM, and wet-SEM technique.
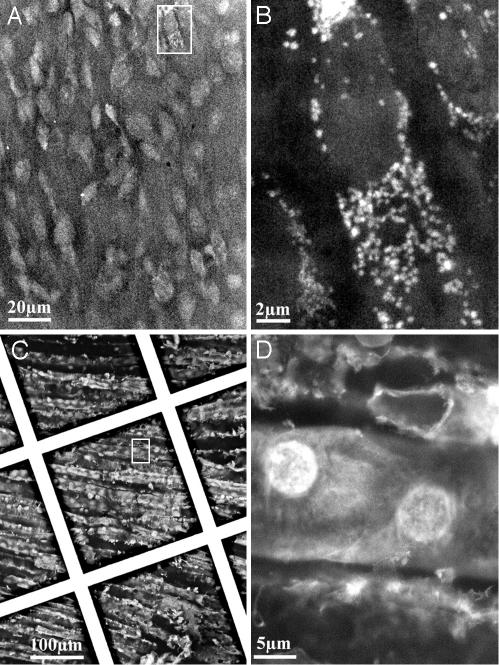
Tissue Sections. Wet SEM can be used for thick specimens such as tissue fragments by simply mounting the specimen against the membrane. The limited sampling depth of BSEs allows imaging of a “virtual section,” extending from the surface into a defined depth of up to a few micrometers without the need for thin sectioning.
Fig. 5 A and B shows the direct visualization of untreated tissues from mouse. Fig. 5A is a small-magnification image of cardiac tissue. The organization of the muscle cells is apparent. Zooming onto one cell (Fig. 5B) gives a vivid view of the nucleus and intracellular organelles, possibly mitochondria, and their dispersion in the cell. As shown above for cultured cells, staining of tissues is advantageous but not imperative and can enhance the visualization of many features. Fig. 5 C and D shows a region of the renal cortex of a rat. The staining (potassium ferricyanide) accentuates the overall organization and cell contacts within the epithelia of the renal tubules. Careful adjustment of staining and imaging conditions can reveal different and complementary information such as tissue architecture; e.g., tissues stained with uranyl acetate yield a clear image of the extracellular matrix (data not shown).
Fig. 5.
Imaging of tissues. (A) Mouse heart, untreated, imaged directly in the sample holder at 30 kV. (B) Higher magnification of the rectangle shown in A. (C) Rat kidney was dissected, cut, and fixed in formalin for 24 h. The tissue then was stained with 0.1% uranyl acetate for 10 min. C and D show epithelial cells within the inner surface of the tubules of the medullary rays. The grid supporting the membrane is visible in this picture. (D) Magnification of the box shown in C.
Simultaneous Photon Collection (CL). BSE detection in wet SEM can be complemented by the simultaneous collection of photons. The scanning electron beam excites molecules in the sample, which then may emit light at characteristic wavelengths (CL). The light intensity then is used to derive an image of the distribution of scintillating molecules, either endogenous to the cell or labels that can be introduced extraneously. This image is obtained simultaneously with the imaging by BSE at a resolution limited by electron–matter interactions and not by light diffraction (6). We inserted a light guide into the fluid below the sample, which guides the emitted light to a photomultiplier. This setup achieves good coupling and efficient collection of the photons excited by the electron beam without compromising the efficiency of concurrent BSE detection.
Fig. 6 shows untreated CHO cells, visualized simultaneously by BSE and photons. In Fig. 6A, the cell borders are clearly outlined in the electron-imaging mode, and the nucleus (along with its nucleoli) are prominent, as are the dark ≈1-μm spots around the nucleus. These appear in the cytoplasm of all eukaryotic cells we investigated, and although their number varies, they usually are dispersed around the nucleus. In the CL image shown in Fig. 6B, the cell outline and nucleus are clearly defined, indicative of a distribution of scintillating molecules (similar to autofluorescence) in the cell. In this mode, the same spots are characterized by a high emission of photons. The combination of a high cathodoluminescent signal and high carbon content is typical of lipid droplets (11), which participate in the energy storage of the cell (12). An independent test of adding 200 μM oleic acid caused a strong proliferation of these droplets in the cell (data not shown), consistent with the notion that the observed spots are lipid droplets.
Fig. 6.
Simultaneous imaging with backscattered and electron beam-excited photons (CL). (A) Unstained CHO cell, imaged by using BSEs as described for Fig. 1D. (B) Emitted light image taken simultaneously with that shown in A.
Wet SEM has an advantage over standard SEM techniques in the preservation of lipids, especially in large aggregates such as lipid droplets. Eliminating the need for drying saves the lipids from organic solvents that may dissolve them. Although osmium treatment will preserve some lipid bilayers, in larger aggregates the osmium quickly reacts to create an external crust that does not let osmium in, leaving the aggregates vulnerable to subsequent organic-solvent treatment. In cryogenic preparations, the tendency of the cleavage process to crack along lipid–water boundaries is also a hazard to large lipid structures.
High-resolution imaging of markers is a highly desirable capability of the CL detection mode, requiring the development of labels for specific molecules in cells. In Fig. 9, which is published as supporting information on the PNAS web site, we demonstrate that standard fluorescent beads 0.2 μm in diameter emit photons intensely under the electron beam and can be imaged with a resolution of ≈100 nm (7). Imaging of smaller beads is limited by low brightness, entailing the development of specialized scintillation beads at small diameter [similar to the larger particles used in scintillation proximity assays (13)]. Because the mechanism for excitation of scintillation is strongest when the beam is directly impinging on the bead, we expect that this can extend the resolution of fluorescent imaging an order of magnitude beyond that available with optical microscopy.
Discussion
The wet-SEM technique presents, in many respects, a unique imaging modality. First, the complete separation of the sample from the vacuum allows direct imaging of fully hydrated, whole-mount samples in an electron microscope acting at moderate beam energies. Second, the use of BSE detection and the elimination of charging allow the visualization of the internal structure of cells in a scanning electron microscope. Third, the geometry of the sample holder allows high-efficiency, simultaneous detection of CL, which opens the possibility for analysis of light emission at a resolution exceeding the limits of light microscopy.
The method presents some additional beneficial features, some of which were not wholly anticipated. We obtain resolutions between 10 and 100 nm, an order of magnitude smaller than could be predicted from the volume of interaction of the electron beam with an aqueous sample. Contrast between low-Z materials such as carbon and oxygen can be readily detected. Thick samples such as tissue biopsies can be seen without thin sectioning. Both the global and high-resolution distribution of colloidal gold labels on cells can be readily determined.
Under the intense radiation of the electron microscope (0.1–1 electron per Å2), the question of viability of cells naturally arises, because the amount of radiation absorbed during highmagnification imaging is sufficient to cause cell death (14). However, no obvious structural damage was apparent, and several repeated scans gave the same images. At longer times, on the scale of an hour after the initial imaging, some signs of deterioration such as cell shrinkage were visible.
Future directions of developing and applying wet SEM are manifold. The easy processing of multiple samples suggests assays involving multiple time points or treatments. The system is easy to automate, as was previously done for the inspection of semiconductors (15), and this opens the possibility for drug screening based on high-resolution imaging of the treated cells. The development of specific and localized labeling in the cell, such as scintillating markers and recombinant metal-binding proteins (e.g., ferritin), is promising. Finally, the direct imaging of tissues opens opportunities for investigative and medical histopathology.
The results presented here show the broad potential and large scope for applications of our technique. We believe that wet SEM will eventually find a place alongside that of optical and standard EM imaging. We further expect that systems will be found where wet SEM will reveal totally new information.
In summary, wet SEM combines several advantages: the ability to probe into cells similar to optical microscopy or TEM, a simplicity of sample preparation comparable with optical microscopy, and the resolution of standard SEM along with its ease of use and ability of zooming in.
Supplementary Material
Acknowledgments
We thank Nathan Ezov for help with tissue samples; Sini Ezer, Miri Horowitz, Yiftach Karni, Eugenia Klein, Trisha Rice, Alon Sabban, Anya Vainshtein, and Orna Yeger for support and assistance; and Iris Barshack, Deborah Brown, Benny Geiger, David Joy, Abraham Nyska, and Daniel Zajfman for helpful advice and discussions. We thank Vered Ozeri and Li Liu for the help in the trypanosome project. This work was supported partially by the Segre Research Grant and the European Commission under project HPRN-CT-2002-00312.
Abbreviations: EM, electron microscopy; SEM, scanning EM; CL, cathodoluminescence; Z, atomic number; BSE, backscattered electron; CHO, Chinese hamster ovary; TEM, transmission EM.
References
- 1.Robinson, V. N. (1975) J. Microsc. 103, 71-77. [DOI] [PubMed] [Google Scholar]
- 2.Danilatos, G. D. (1991) J. Microsc. 162, 391-402. [Google Scholar]
- 3.Philips Corporation (1996) Environmental Scanning Electron Microscope (Robert Johnson Assoc., El Dorado Hills, CA).
- 4.Thornley, R. F. M. (1960) Ph.D. thesis (Univ. of Cambridge, Cambridge, U.K.).
- 5.Goldstein, J. I., Newbury, D. E., Echlin, P., Joy, D. C., Romig, A. D., Lyman, C. E., Fiori, C. & Lifshin, E. (1992) Scanning Electron Microscopy and X-Ray Microanalysis (Plenum, New York), 2nd Ed.
- 6.Boyde, A. & Reid, S. A. (1983) Nature 302, 522-523. [DOI] [PubMed] [Google Scholar]
- 7.Joy, D., Ko, Y. U. & Hwu, J. (2000) Proc. SPIE 3998, 108-115; also available at http://web.utk.edu/~srcutk. [Google Scholar]
- 8.Levenberg, S., Katz, B.-Z., Yamada, K. M. & Geiger, B. (1998) J. Cell Sci. 111, 347-357. [DOI] [PubMed] [Google Scholar]
- 9.Chowers, M. Y., Keller, N., Tal, R., Barshack, I., Lang, R., Bar-Meir, S. & Chowers, Y. (1999) Gastroenterology 117, 1113-1118. [DOI] [PubMed] [Google Scholar]
- 10.Liu, L, Liang, X. H., Uliel, S., Unger, R., Ullu, E. & Michaeli, S. (2002) J. Biol. Chem. 277, 47348-47367. [DOI] [PubMed] [Google Scholar]
- 11.Ning, G., Fugimoto, T., Koike, H. & Ogawa, K. (1993) Cell Tissue Res. 271, 217-225. [DOI] [PubMed] [Google Scholar]
- 12.Brown, D. A. (2001) Curr. Biol. 11, R446-R449. [DOI] [PubMed] [Google Scholar]
- 13.Bosworth, N. & Towers, P. (1989) Nature 341, 167-168. [DOI] [PubMed] [Google Scholar]
- 14.National Research Council (1990) Health Effects of Exposure to Low Levels of Ionizing Radiation, Biological Effects of Ionizing Radiation Report V (Natl. Acad. Press, Washington, DC).
- 15.Somekh, S. (1999) Semiconductor Fabtech, 10, 39-43. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.