Abstract
The congenital dyserythropoietic anemias are a heterogeneous group of rare disorders primarily affecting erythropoiesis with characteristic morphological abnormalities and a block in erythroid maturation. Mutations in the CDAN1 gene, which encodes Codanin-1, underlie the majority of congenital dyserythropoietic anemia type I cases. However, no likely pathogenic CDAN1 mutation has been detected in approximately 20% of cases, suggesting the presence of at least one other locus. We used whole genome sequencing and segregation analysis to identify a homozygous T to A transversion (c.533T>A), predicted to lead to a p.L178Q missense substitution in C15ORF41, a gene of unknown function, in a consanguineous pedigree of Middle-Eastern origin. Sequencing C15ORF41 in other CDAN1 mutation-negative congenital dyserythropoietic anemia type I pedigrees identified a homozygous transition (c.281A>G), predicted to lead to a p.Y94C substitution, in two further pedigrees of SouthEast Asian origin. The haplotype surrounding the c.281A>G change suggests a founder effect for this mutation in Pakistan. Detailed sequence similarity searches indicate that C15ORF41 encodes a novel restriction endonuclease that is a member of the Holliday junction resolvase family of proteins.
Introduction
The congenital dyserythropoietic anemias (CDAs) are characterized by moderate to severe macrocytic anemia and reticulocytopenia, arising from ineffective intramedullary erythropoiesis which is accompanied in some subtypes by an extravascular hemolytic component.1 Characteristic findings in congenital dyserythropoietic anemia type I (CDA-I, [MIM 224120]), which is inherited recessively, are the presence of macrocytosis, Pappenheimer inclusions and gross aniso-poikilocytosis on peripheral blood smears. Light microscopy of aspirated bone marrow smears shows megaloblastic erythropoiesis, binuclear erythroblasts (<10%) and occasional tri-and tetra-nucleate erythroid cells, basophilic stippling and the relatively specific feature of inter-nuclear bridges between intermediate erythroblasts (1.4–7.9% of total erythroblasts).2 Ultrastructural detail of bone marrow, observed by electron microscopy (EM) shows a pathognomonic pattern of spongy heterochromatin, present in a high proportion (up to 60%) of the intermediate erythroblasts.
In 2002, homozygous mutations in CDAN1, a highly conserved 28-exon gene, were shown to underlie CDA-I in a large Israeli-Arab pedigree.3 Subsequently the majority of individuals with CDA-I have been shown to harbor bi-allelic mutations in CDAN1. To date, approximately 30 CDAN1 mutations have been reported in CDA-I patients. Although the majority of these are missense changes, there are also nonsense, splicing and frameshift mutations.4
A recent epidemiological study reports an incidence of 169 cases of CDA-I from 143 families in Europe5 and in addition there are in excess of 70 cases in a large Bedouin tribe.6 Pooled data on CDAN1 mutations show that approximately 50% of affected patients/families have homozygous or compound heterozygous CDAN1 mutations. However, in 30% only a single CDAN1 mutant allele can be identified7 and it is not known whether such patients harbor cryptic mutations on the second CDAN1 allele or if there may be digenic inheritance. The remaining approximately 20% of families in whom no CDAN1 mutation can be identified may harbor uncharacterized CDAN1 mutations or a second locus may be involved. The latter is supported by analysis of two pedigrees which manifest clinical abnormalities consistent with CDA-I and in which the disease has been shown to segregate independently of the CDAN1 locus on chromosome 15q15.2.7,8
We undertook whole-genome sequencing of an individual from one of these pedigrees, a previously reported consanguineous Middle-Eastern family, with CDA-I disease9,10 and identified a homozygous missense mutation in a gene, C15ORF41, predicted to encode a novel endonuclease. We identified a second missense change in C15ORF41 in two further pedigrees of South-East Asian origin on the same haplotype background, strongly suggesting a founder effect.
Methods
Further information regarding study Design and Methods is available in the Online Supplementary Appendix.
Patients and DNA sequencing
This study involved whole-genome sequencing of an individual affected with CDA-I from a large consanguineous Kuwaiti pedigree that has been reported previously.7,9,10 Ethical approval for the studies presented here was provided by the Oxfordshire Research Ethics Committee (reference: 06/Q1605/3) and informed consent was obtained. The coding region of C15ORF41 was DNA sequenced in 9 further CDA-I families harboring either no CDAN1 missense changes (6 families) or a single CDAN1 change (3 families). Fragments were sequenced on the ABI PRISM 3730 DNA sequencer, employing Big Dye Terminator mix version 3.1 (Applied Biosystems). The human genome hg19 sequence release (February 2009) was used for all analyses and all primer sequences used in this study are shown in Online Supplementary Table S1. For the C15ORF41 transcript, NCBI Reference Sequence: NM_001130010.1 is used for all analyses presented here.
Amplification of C15ORF41 from cDNA
Total RNA was isolated from transformed B lymphocytes and in vitro cultured erythroblasts11 from healthy individuals and cDNA generated using oligo (dT) primers (RevertAid, Fermentas). cDNA was amplified with primers homologous to the 3′- and 5′-untranslated regions of C15ORF41 (see Online Supplementary Table S1 for primer sequences and Figure 2 for locations).12
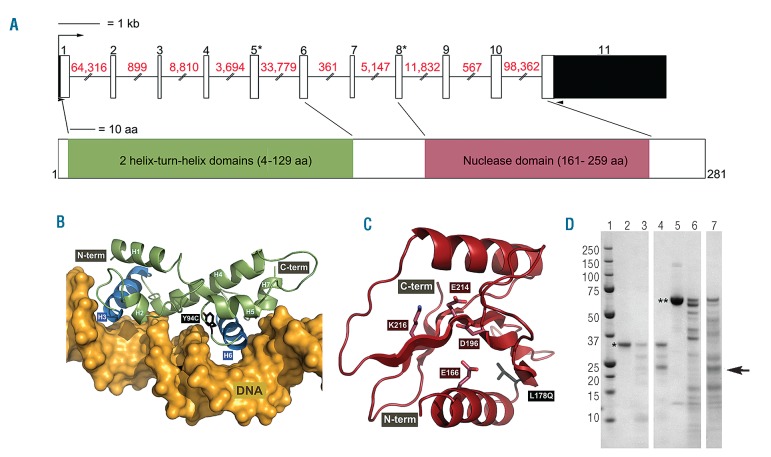
Figure 2.
C15ORF41 gene and protein structure. (A) Schematic representation of the C15ORF41 gene, exons are shown to scale with coding sequence shown in white and UTRs in black. Red numbers above lines indicate intron sizes (not to scale); numbers above exons indicate exon number; asterisks indicate the two exons in which mutations have been identified. Black arrow heads indicate locations of primers used to amplify the C15ORF41 transcript. The lower section shows the C15ORF41 protein with annotated domains shown to scale. (B,C) Predicted tertiary structure of the conserved domains identified in C15ORF41. Missense changes found in CDA-I patients are shown using sticks (Y94C and L178Q) and labeled in black. (B) Two helix-turn-helix domains predicted for the N-terminal of C15ORF41 (amino acids 4–129). Helices are numbered and putative DNA interaction helices are shown in blue (H3 and H6). The displayed DNA molecule was extracted from Rhee et al.12 (C) The PD-(D/E)XK nuclease domain predicted for the C-terminal region of C15ORF41 (amino acids 161–259). Highly conserved residues in the PD-(D/E)XK nuclease superfamily that form part of its active centre are labeled in red and side chains are shown using sticks. (D) Purified recombinant C15ORF41 protein was treated with varying concentrations of trypsin (lanes 2–4) for 1 hour at 37°C and digests were analyzed by SDS-PAGE. As a control, another recombinant protein (human RECQ1) was similarly digested (lanes 5–7). Lane 1: size markers. Lanes 2, 5: no trypsin. Lanes 3, 6: 4 μg/mL trypsin. Lanes 4, 7: 100 μg/mL trypsin. The arrow indicates the location of trypsin, * is C15ORF41, ** is RECQ1.
Sequence and structural analyses
C15ORF41 structural models were created using Modeller.13,14 Models are presented using Pymol (http://www.pymol.org). Secondary-structure predictions were performed using PsiPred.14
Expression and analysis of C15ORF41 protein
The open reading frame of C15ORF41 was cloned into a baculovirus transfer vector and converted to recombinant baculoviruses as previously described.15,16 Protein purification is detailed in the Online Supplementary Appendix. Partial trypsin and chymotrypsin digestion was performed over concentrations from 0.8–100 μg/mL from 10 min to 3 h. Reaction mixes were analyzed by mass spectrometry and SDS-PAGE.
Results and Discussion
This study was prompted by a report of a family with CDA-I in whom the disease was not linked to the CDAN1 locus on chromosome 15q15.2.7 The pedigree (Figure 1, Family 1) includes 3 surviving affected siblings born to first-cousin-once-removed parents who also have 10 normal children of both sexes. Affected siblings manifested typical hematologic features of CDA-I: megaloblastic erythropoiesis with severe dyserythropoietic changes, bi- and multi-nuclear erythroblasts and inter-nuclear chromatin bridges. All 3 patients were severely affected requiring transfusion support during childhood9,10 (Online Supplementary Table S1).
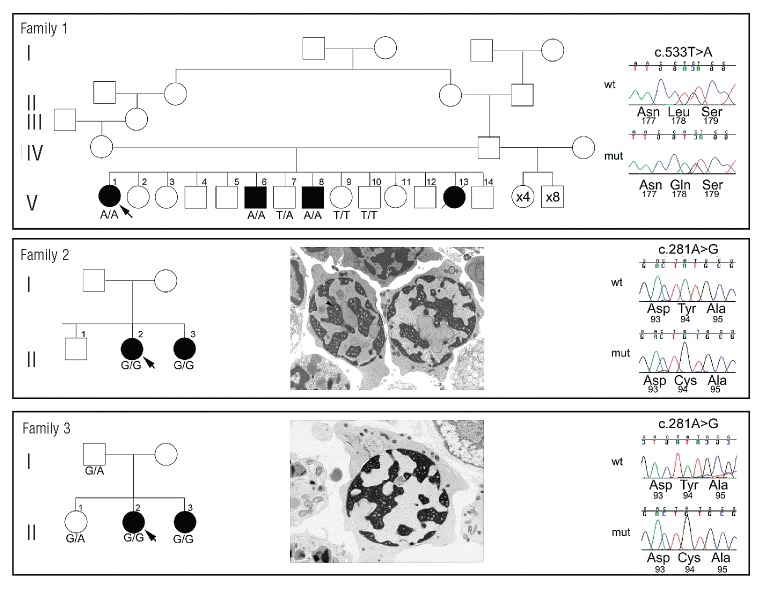
Figure 1.
Pedigrees of three families with novel non-synonymous variants of C15ORF41. Black filled symbols indicate individuals with CDA-I. Chromatograms from a control individual (upper) and the proband (lower) show sequence changes identified, together with the amino acids encoded by the change and by adjacent codons. Positions of nucleotide changes are shown above the chromatogram and are given according to numbering of the base within the open reading frame in the C15ORF41 cDNA (ENST00000566621). The lower two panels also show electron micrographs of intermediate erythroblasts from each proband clearly showing the spongy heterochromatin (arrowed) indicative of CDA-I. Probands are indicated by arrows. Below each symbol the status of that individual for the change identified in the proband is shown.
On the basis of the consanguinity in this family, coupled with the exclusion of CDAN1, and because none of the 12 half-siblings of the affected individuals manifested CDA-I, we hypothesized that the proband had a different, autosomal-recessive basis for her condition. DNA isolated from a lymphoblastoid cell line derived from the proband was sequenced (see Methods). We initially identified 4,274,834 predicted variants that we prioritized by removing intergenic variants and those present in dbSNP (build 136) (http://www.ncbi.nlm.nih.gov/projects/SNP/); this left 172,445 predicted variants. We next selected only variants in regions shown to be homozygous in the proband by analysis of the array data (Online Supplementary Methods); after this filter, 3,255 predicted variants remained. To further narrow our search, we selected only variants predicted by ANNOVAR17 to alter protein coding sequences; specifically, insertions or deletions predicted to alter the reading frame, non-synonymous amino acid changes or loss or gain of a stop codon. This filter left 59 predicted homozygous coding changes. Visual inspection of these sequence calls, using a customized GBrowse database,18 revealed 19 to be invalid, their presence most likely owing to low coverage or to result from misalignment of a highly similar sequence. To allow further filtering of novel homozygous coding changes, we conducted segregation analysis using variation data (see Methods) from the 2 affected siblings of the proband (Figure 1, Family 1 V-6 and V-8) and identified only 9 homozygous coding variants predicted to be shared between all 3 affected individuals. We verified these by DNA sequencing and checked segregation in the 3 affected and 3 unaffected individuals for whom samples were available. Of the 9 variant sequence calls, 6 were reproducible by Sanger sequencing and only one segregated with the CDA-I disease in this pedigree (Figure 1). This was a T to A transversion in exon 8 (c.533T>A) of C15ORF41, an uncharacterized gene, leading to an L178Q substitution altering a highly conserved hydrophobic leucine to a polar glutamine.
To gather further genetic evidence that mutations in C15ORF41 underlie CDA-I we undertook DNA sequencing of the coding region of this gene including the intron/exon boundaries in 9 additional CDA-I patients, both familial and sporadic in origin. In 6 of these patients, no CDAN1 mutations had been previously identified despite sequencing of the coding region, while 3 patients had been found to harbor only a single deleterious CDAN1 allele. We identified a homozygous A>G transition in C15ORF41 in exon 5 at position 281 (c.281A>G), leading to a p.Y94C missense change, in 2 CDAN1 mutation-negative patients from unrelated consanguineous South-East Asian pedigrees (Figure 1, Families 2 and 3). We found no likely pathogenic changes in the remaining 7 pedigrees suggesting the presence of at least a further causative locus. Clinical findings in Family 2 have been previously reported8 as showing hematologic results typical of CDA-I (Online Supplementary Table S2) and a substantial proportion of the erythroblasts showing spongy heterochromatin upon EM (Figure 1). Blood indices of affected members of Family 3 indicate anemia (Online Supplementary Table S2) and EM of erythroblasts from individual II-2 shows the characteristic pattern of spongy heterochromatin (Figure 1). We were able to demonstrate segregation of the c.281A>G homozygous change in all available samples with CDA-I in both pedigrees (Figure 1). Although residue Y94 is not as well conserved as L178, this p.Y94C missense change alters a hydrophobic tyrosine to cysteine which could form covalent cross-links via a disulphide bond, thereby disrupting tertiary structure.
Both changes are extremely rare as neither is listed in dbSNP136 (http://www.ncbi.nlm.nih.gov/projects/SNP/) nor in more than 11,800 alleles from African and European Americans listed in the Exome Variant Server (EVS) (http://evs.gs.washington.edu/EVS/) (Online Supplementary Appendix), and may be specific to Middle-Eastern and South-East Asian populations. In addition, we excluded the c.533T>A change from 41 unrelated ethnically matched Saudi Arabian and Jordanian control individuals by DNA sequencing, further suggesting this variant to be disease associated. Taken together these data signal C15ORF41 as a second disease gene for CDA-I.
To investigate the origin of these mutations we assayed 2 informative microsatellites and 8 single nucleotide polymorphisms in probands within a ~335 kb region around the missense changes in C15ORF41 that contains no recombination hotspots (defined as ≥10 cM/Mb) according to the International HapMap Consortium (http://hapmap.ncbi.nlm.nih.gov/). The unrelated South-East Asian patients (both families are of Pakistani descent) shared the same haplotype over C15ORF41 (Online Supplementary Table S3) suggesting a founder effect of this missense mutation in Pakistan. It is, therefore, possible that this mutation causes other cases of CDA-I in this population. However, further screening will be required to confirm this. Establishing the prevalence of this haplotype in the normal Pakistani population may also shed light on the age of any founder effect.
C15ORF41 is an uncharacterized gene located in chromosomal region 15q14 and comprises 11 exons. Data from gene expression arrays show that C15ORF41 is widely transcribed although expression appears to be elevated in B lymphoblasts, CD34+ cells, cardiomyocytes and fetal liver suggesting a specific requirement in hematopoiesis.19 To verify that C15ORF41 generates a spliced transcript we designed oligonucleotides complementary to the 5- and 3-untranslated regions (UTRs) (see Figure 2 for primer locations). Using these we amplified an 1013 bp product spanning 11 exons, corresponding to RefSeq transcript NM_001130010.1 (Ensembl transcript ENST00000566621) which encodes a 281 aa protein, from cDNA generated from a lymphoblastoid cell line and from intermediate stage in vitro cultured erythroblasts, both derived from healthy individuals. There are a number of predicted isoforms of C15ORF41 that we attempted to amplify from cDNA using specific primers. However, we could only detect the single isoform described above in both cell types tested (Online Supplementary Figure S1A). Global gene expression analysis throughout erythropoiesis reveals that C15ORF41 is uniformly expressed during erythroid differentiation,20 suggesting a constant requirement for this protein.
C15ORF41 is widely conserved with orthologs broadly distributed in eukaryotes; there are also identifiable homologs in members of the archaea and in viruses (see Online Supplementary Appendix for details of alignments). The consistency of the secondary structure predictions and corroboration by profile-to-profile comparison methods, provide strong evidence that the C15ORF41 protein contains 2 N-terminal AraC/XylS-like wHtH domains followed by a PD-(D/E)XK nuclease domain (Figure 2 and Online Supplementary Figure S1B and C) suggesting C15ORF41 encodes a divalent metal-ion dependent restriction endonuclease. Each of the two mutated residues contributes to the hydrophobic cores of their respective domains, and are both predicted to affect protein stability (Figure 2 and Online Supplementary Figure S1B and C), which is supported by the very similar abnormalities present in patients harboring mutations in both functional domains. Biological functions performed by this family include DNA damage repair, Holliday junction resolution and RNA processing.21 In some members of the PD-(D/E)XK nuclease superfamily this combination of domains underlies protein-protein interactions (usually dimerization) and may establish additional DNA interactions, thereby improving DNA specificity. It is unknown if wHTH domains in C15ORF41 are performing one or both such functions. As none of the commercially available antibodies cross-reacted with C15ORF41 in our hands, we are currently raising an antibody to address this question.
To examine the structure and activity of C15ORF41 protein, we expressed the full-length protein fused to a histidine tag. Four chromatographic steps yielded a purified protein and removed all non-specific nuclease activity. To test the structural integrity and identify possible subdomains, we performed partial proteolysis with trypsin and chymotrypsin. Multiple experiments showed that the C15ORF41 is unusually resistant to proteolysis under native conditions (Figure 2D). Mass spectrometry indicated that only the tag sequence was susceptible to proteolysis. This biochemical data support the prediction of well-ordered domains in C15ORF41 and the absence of general nuclease activity suggests it may exhibit sequence- or structure-specific activity. A recent report suggests C15ORF41 interacts with Asf1b.22 This is significant as Codanin-1 has been proposed to play a role in the transport of histones through interaction with Asf1b and supports the hypothesis that the primary defect in CDA-I is in DNA replication and chromatin assembly.23
Lesions of both C15ORF41 and CDAN1 cause similar lineage-specific phenotypic abnormalities that result in the clinical presentation of CDA-I. In cases of CDA-I caused by CDAN1 mutations the severity of the anemia varies within and between families,24,25 and in addition, there is variation in the iron overload arising as a complication.24,26 The severity of CDA-I caused by C15ORF41 lesions also varies and, in the 3 pedigrees reported here, is comparable with that caused by CDAN1 mutations. Patients with CDA-I caused by C15ORF41 mutations show significant hematologic response to interferon-α, with improved Hb levels and decreased dyserythropoiesis. The patients homozygous for C15ORF41 mutations reported in this study are unresponsive to interferon-α suggesting a subtly different pathogenic mechanism, although the numbers involved are too small to determine whether this is a distinguishing feature. The biochemical basis of the response of the anemia to interferon is currently unknown; therefore, it is still not possible to determine the basis of any differential response in patients.
The mutations identified in C15ORF41 may affect the predicted nuclease activity of this protein thereby disrupting the intrinsic connection between cell cycle dynamics and the instigation of terminal erythroid differentiation.27 An endonuclease involved in DNA repair may be critical in this context; whilst slowly dividing stem cells are able to undertake extensive DNA repair, rapidly dividing erythroid progenitor cells may be particularly susceptible to deficiencies in repair pathways.28
In summary, we have identified mutations in a second causative gene underlying CDA-I and demonstrated a founder effect for one of the mutations. Provocatively, we could not identify likely causative mutations of C15ORF41 in 7 of the 9 CDAN1-mutation-negative CDA-I families we screened, strongly suggesting the existence of at least a further causative locus. We show C15ORF41, previously an uncharacterized gene, produces a spliced transcript in cultured erythroblasts encoding a structurally compact protein with homology to the Holliday junction resolvases.
Acknowledgments
The authors would like to thank Helena Ayyub for DNA preparation, Raffaele Renella, Chris Fisher, Noemi Roy, Andrew Wilkie and Stephen Twigg for stimulating discussions and Tim Rostron, John Frankland and Katalin Di Gleria, Jackie Sloane-Stanley and Sue Butler for technical assistance. The authors would also like to thank the NHLBI GO Exome Sequencing Project and its ongoing studies which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010).
Footnotes
The online version of this article has s Supplementary Appendix.
Funding
This work was supported by the Medical Research Council in CPP and VJB laboratories. The WGS500 project is funded by Wellcome Trust, Oxford NIHR Biomedical Research Centre and Illumina.
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Heimpel H, Wendt F. Congenital dyserythropoietic anemia with karyorrhexis and multinuclearity of erythroblasts. Helvetica Medica Acta. 1968;34(2):103–15 [PubMed] [Google Scholar]
- 2.Heimpel H, Kellermann K, Neuschwander N, Hogel J, Schwarz K. The morphological diagnosis of congenital dyserythropoietic anemia: results of a quantitative analysis of peripheral blood and bone marrow cells. Haematologica. 2010;95(6):1034–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dgany O, Avidan N, Delaunay J, Krasnov T, Shalmon L, Shalev H, et al. Congenital dyserythropoietic anemia type I is caused by mutations in codanin-1. Am J Hum Genet. 2002; 71(6):1467–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renella R, Wood WG. The congenital dyserythropoietic anemias. Hematol Oncol Clin North Am. 2009;23(2):283–306 [DOI] [PubMed] [Google Scholar]
- 5.Heimpel H, Matuschek A, Ahmed M, Bader-Meunier B, Colita A, Delaunay J, et al. Frequency of congenital dyserythropoietic anemias in Europe. Eur J Haematol. 2010;85(1):20–5 [PubMed] [Google Scholar]
- 6.Tamary H, Shalev H, Luria D, Shaft D, Zoldan M, Shalmon L, et al. Clinical features and studies of erythropoiesis in Israeli Bedouins with congenital dyserythropoietic anemia type I. Blood. 1996;87(5):1763–70 [PubMed] [Google Scholar]
- 7.Ahmed MR, Zaki M, Sabry MA, Higgs D, Vyas P, Wood W, et al. Evidence of genetic heterogeneity in congenital dyserythropoietic anaemia type I. Br J Haematol. 2006; 133(4):444–5 [DOI] [PubMed] [Google Scholar]
- 8.Ahmed MR, Chehal A, Zahed L, Taher A, Haidar J, Shamseddine A, et al. Linkage and mutational analysis of the CDAN1 gene reveals genetic heterogeneity in congenital dyserythropoietic anemia type I. Blood. 2006;107(12):4968–9 [DOI] [PubMed] [Google Scholar]
- 9.Zaki M, Hassanein A, Daoud A. Congenital Dyserythropoietic Anaemia Type I in a Brother and a Sister. Med Princ Pract. 1992;3:57–9 [Google Scholar]
- 10.Sabry MA, Zaki M, al Awadi SA, al Saleh Q, Mattar MS. Non-haematological traits associated with congenital dyserythropoietic anaemia type 1: a new entity emerging. Clin Dysmorphol. 1997;6(3):205–12 [DOI] [PubMed] [Google Scholar]
- 11.Fibach E, Manor D, Oppenheim A, Rachmilewitz EA. Proliferation and maturation of human erythroid progenitors in liquid culture. Blood. 1989;73(1):100–3 [PubMed] [Google Scholar]
- 12.Rhee S, Martin RG, Rosner JL, Davies DR. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc Natl Acad Sci USA. 1998;95(18):10413–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815 [DOI] [PubMed] [Google Scholar]
- 14.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292(2):195–202 [DOI] [PubMed] [Google Scholar]
- 15.Savitsky P, Bray J, Cooper CD, Marsden BD, Mahajan P, Burgess-Brown NA, et al. High-throughput production of human proteins for crystallization: the SGC experience. J Struct Biol. 2010;172(1):3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shrestha B, Smee C, Gileadi O. Baculovirus expression vector system: an emerging host for high-throughput eukaryotic protein expression. Methods Mol Biol. 2008; 439:269–89 [DOI] [PubMed] [Google Scholar]
- 17.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12(10):1599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004; 101(16):6062–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merryweather-Clarke AT, Atzberger A, Soneji S, Gray N, Clark K, Waugh C, et al. Global gene expression analysis of human erythroid progenitors. Blood. 2011; 117(13):e96–108 [DOI] [PubMed] [Google Scholar]
- 21.Laganeckas M, Margelevicius M, Venclovas C. Identification of new homologs of PD-(D/E)XK nucleases by support vector machines trained on data derived from profile-profile alignments. Nucleic Acids Res. 2011;39(4):1187–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007; 3:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ask K, Jasencakova Z, Menard P, Feng Y, Almouzni G, Groth A. Codanin-1, mutated in the anaemic disease CDAI, regulates Asf1 function in S-phase histone supply. EMBO J. 2012;31(8):2013–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wickramasinghe SN. Congenital dyserythropoietic anaemias: clinical features, haematological morphology and new biochemical data. Blood Rev. 1998;12(3):178–200 [DOI] [PubMed] [Google Scholar]
- 25.Heimpel H, Schwarz K, Ebnother M, Goede JS, Heydrich D, Kamp T, et al. Congenital dyserythropoietic anemia type I (CDA I): molecular genetics, clinical appearance, and prognosis based on long-term observation. Blood. 2006;107(1):334–40 [DOI] [PubMed] [Google Scholar]
- 26.Tamary H, Dgany O, Proust A, Krasnov T, Avidan N, Eidelitz-Markus T, et al. Clinical and olecular variability in congenital dyserythropoietic anaemia type I. Br J Haematol. 2005;130(4):628–34 [DOI] [PubMed] [Google Scholar]
- 27.Pop R, Shearstone JR, Shen Q, Liu Y, Hallstrom K, Koulnis M, et al. A key commitment step in erythropoiesis is synchronized with the cell cycle clock through mutual inhibition between PU.1 and S-phase progression. PLoS Biol. 2010; 8(9):e1000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bracker TU, Giebel B, Spanholtz J, Sorg UR, Klein-Hitpass L, Moritz T, et al. Stringent regulation of DNA repair during human hematopoietic differentiation: a gene expression and functional analysis. Stem Cells. 2006;24(3):722–30 [DOI] [PubMed] [Google Scholar]