Abstract
Reasons for the rising clinical impact of the bacterium Enterococcus faecium include the species’ rapid acquisition of adaptive genetic elements. Here, we focused on the impact of recombination on the evolution of E. faecium. We used the recently developed BratNextGen algorithm to detect recombinant regions in the core genome of 34 E. faecium strains, including three newly sequenced clinical strains. Recombination was found to have a significant impact on the E. faecium genome: of the original 1.2 million positions in the core genome, 0.5 million were predicted to have been affected by recombination in at least one strain. Importantly, strains in one of the two major E. faecium clades (clade B), which contains most of the E. faecium human gut commensals, formed the most important reservoir for donating foreign DNA to the second major E. faecium clade (clade A), which contains most of the clinical isolates. Also, several genomic regions were found to mainly recombine in specific hospital-associated E. faecium strains. One of these regions (the epa-like locus) likely encodes the biosynthesis of cell wall polysaccharides. These findings suggest a crucial role for recombination in the emergence of E. faecium as a successful hospital-associated pathogen.
Keywords: BratNextGen, comparative genomics, phylogenomics, whole-genome sequencing, nosocomial pathogen, antibiotic resistance
Introduction
Enterococci are Gram-positive bacteria that commonly inhabit the gastrointestinal-tract of healthy humans and other animals. However, since the 1980s, the clinical impact of enterococci has continuously increased and they now rank as one of the leading causes of nosocomial infections of the blood-stream, urinary tract, surgical wounds, and other sites (Arias and Murray 2012; Gilmore et al. 2013). In the past, enterococcal hospital-acquired infections (HAIs) were predominantly caused by E. faecalis, but since the 1990s, HAIs have been increasingly associated with E. faecium. Nowadays, E. faecium HAIs are almost as common as HAIs caused by E. faecalis (Hidron et al. 2008).
An important cause for the rise of E. faecium as a nosocomial pathogen has been the species’ capacity to easily acquire novel adaptive traits, including genetic elements encoding antibiotic resistance determinants. For example, nosocomial infections caused by ampicillin-resistant E. faecium were first reported in the United States in the 1980s, after which they spread rapidly around the world (Galloway-Peña et al. 2009). Currently, ampicillin resistance is reported in more than 80% of clinical E. faecium isolates worldwide (Hidron et al. 2008; Willems and van Schaik 2009). Other genetic elements that have been acquired by E. faecium include esp and other genes located on the esp pathogenicity island ICEEfm1, the putative hyaluronidase gene hyl, and several genes encoding surface proteins (Rice et al. 2003; Vankerckhoven et al. 2004; Coque et al. 2005; Klare et al. 2005; Camargo et al. 2006; Hendrickx et al. 2007; van Schaik et al. 2010), a putative phosphotransferase system that contributes to intestinal colonization during antibiotic treatment (Zhang et al. 2013), and specific insertion elements, especially IS16 (Leavis et al. 2007). The cumulative acquisition of these, but probably many more, genetic elements by E. faecium has resulted in the evolutionary development of a genetically distinct E. faecium subpopulation with a progressively increasing fitness in hospitalized patients (Willems et al. 2011). This subpopulation was initially designated lineage C1 (Homan et al. 2002) and later renamed to clonal complex 17 (CC17) on the basis of multilocus sequence typing (MLST) data analysis (Willems et al. 2005, 2011). Phylogenomic studies, using only a limited number of E. faecium strains, strengthened these previous findings as they revealed an ancient phylogenetic split in the E. faecium population, essentially dividing animal and human clinical isolates into one clade (clade A) and human commensal isolates into another clade (clade B) (Galloway-Peña et al. 2012; Palmer et al. 2012). However, in a recent study using Bayesian-based population genetic modeling of an MLST data set of more than 1,700 isolates, Willems et al. (2012) found that nosocomial E. faecium strains clustered into three, instead of one, subgroups, suggesting different evolutionary trajectories for modern clinical isolates.
Pan-genome analyses of E. faecium indicated that E. faecium has an open genome which means that the total available gene pool within this species is essentially unlimited (van Schaik et al. 2010). This also suggests that the E. faecium genome is highly plastic and few barriers exist for the acquisition of foreign genetic elements. Evidence for high levels of recombination was found in recent comparative genomics analyses which noted the existence of hybrid E. faecium strains, of which the genomes were an amalgam of clade A and clade B backgrounds (Galloway-Peña et al. 2012; Palmer et al. 2012). Similarly, MLST data analysis showed that certain E. faecium subpopulations displayed high levels of admixture, indicating that up to 14% of the MLST gene sequences originated from other E. faecium subpopulations (Willems et al. 2012). Furthermore, the majority of gene tree topologies of individual MLST genes were incongruent (Willems et al. 2005), which together with the above suggest that the impact of recombination on the population structure and evolution of E. faecium may be considerable. However, recombination in E. faecium has so far only been studied using either a limited set of strains or only few genes (i.e., seven core genes in the case of MLST data). Recent sequencing efforts have significantly increased the number of E. faecium whole-genome sequences (WGS) available in the public domain. These WGS provide an excellent opportunity to study recombination on a high-resolution whole-genome level. Here, we use the Bayesian Recombination Tracker (BratNextGen) algorithm (Marttinen et al. 2008, 2012; Castillo-Ramírez et al. 2012; McNally et al. 2013) to study the impact of recombination in E. faecium using a set of 34 WGS, including those of three clinical strains that were sequenced for the purpose of this study. We show that most of the recombination detected in clade A originated from a clade B type background and that purging recombinant signals from a genome-wide alignment prior to phylogenetic tree-building widened the ancient split between clade A and B. Moreover, we found that contemporary clinical isolates share specific recombination signals pointing to recombination-driven adaptations to selection pressures associated with hospitalization (e.g., antibiotic treatment).
Materials and Methods
Sequence Information and Genome Sequencing
WGS were retrieved from GenBank (ftp://ftp.ncbi.nih.gov/genomes/Bacteria, last accessed August 3, 2013 and ftp://ftp.ncbi.nih.gov/genomes/Bacteria_DRAFT, last accessed August 3, 2013). Enterococcus faecium WGS were downloaded on May 2012 and included 31 available genomes: 1 complete and 30 draft genomes. Available WGS of 66 other enterococci were downloaded in July 2012. Besides using publicly available WGS, three E. faecium isolates were newly sequenced for the purpose of this study. These included strains E155, E525, and E1165, which all belong to MLST ST17 (Homan et al. 2002; Willems et al. 2005) and have been isolated from hospitalized patients in the United States (Chicago), Australia (Melbourne), and Italy (Genoa), respectively (table 1). Genomic DNA was isolated from cell pellets of these strains using the Wizard Genomic DNA Purification kit (Promega, Leiden, The Netherlands) according to the manufacturer’s instructions. The strains were sequenced using Illumina GAII sequencing technology generating 50-bp paired-end reads from a library with an average insert size of 250 bp (Baseclear B.V., Leiden, The Netherlands). After confirming the quality of the raw Illumina reads with FastQC v0.10.1 (Simon Andrews, Babraham Bioinformatics, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/, last accessed August 3, 2013), reads were assembled de novo using Velvet v1.2.01 (Zerbino and Birney 2008). For each Velvet assembly, a range of different k-mer (hash) lengths (15–49 bp) was empirically tested to gain an optimal N50. In addition, Velvet was run with the options “-exp_cov” (expected k-mer coverage) and “-cov_cutoff” (coverage cutoff). The expected k-mer coverage (Ck) was calculated using [Ck = C × (L − k + 1)/L], where L and k are the read and k-mer lengths (in bp), respectively. C is the estimated sequence coverage given by [C = L × N/S], where N is the number of reads and S is the estimated size of the genome. For the three genome assemblies, an estimated E. faecium genome size (S) of 2.75 Mb was used. The coverage cutoff was set at 0.25 × Ck as suggested by the Velvet developers (Zerbino DR, personal communication).
Table 1.
Whole-Genome Sequenced E. faecium Strains Available in July 2012
| Strain | Sourcea | Country | Year | Reported Resistancesb and Virulence Genes | MLSTc | GenBank BioProject Codes |
|---|---|---|---|---|---|---|
| E155 | Clinical isolate (faeces)d | USA | 1995 | AMP, VAN; esp+ | 17 | PRJNA192879 |
| E525 | Clinical isolate (wound) | AUS | 1998 | AMP, VAN; esp+ | 17 | PRJNA192893 |
| E1165 | Clinical isolate (wound) | ITA | 1997 | AMP; esp+ | 17 | PRJNA192894 |
| Aus0004 | Clinical isolate (blood) | AUS | 1998 | VAN; esp+ | 17 | PRJNA87025 |
| C68 | Clinical isolate (faeces)d | USA | 1996 | AMP, VAN; esp+ hyl+ | 16 | PRJNA40855 |
| TX0082 | Clinical isolate (blood) | USA | 1999 | AMP, VAN | 17 | PRJNA61137 |
| E1162 | Clinical isolate (blood) | FRA | 1997 | AMP; esp+ | 17 | PRJNA47013 |
| TX0133A | Clinical isolate (blood) | USA | 2006 | VAN; esp+ | 17 | PRJNA61139 |
| TX0133a01 | Clinical isolate (blood) | USA | 2006 | VAN; esp+ hyl+ | 17 | PRJNA61143 |
| TX0133a04 | Clinical isolate (blood) | USA | 2006 | esp+ | 17 | PRJNA61129 |
| TX0133B | Clinical isolate (blood) | USA | 2006 | esp+ | 17 | PRJNA61141 |
| TX0133C | Clinical isolate (blood) | USA | 2006 | VAN; esp+ | 17 | PRJNA61131 |
| 1,231,410 | Clinical isolate (skin and soft tissue) | USA | 2005 | AMP, VAN; esp+ hyl+ | 17 | PRJNA55719 |
| DO (TX16) | Clinical isolate (blood) | USA | 1992 | AMP; hyl+ | 18 | PRJNA54089 |
| 1,231,502 | Clinical isolate (blood) | USA | 2005 | AMP, VAN; esp+ hyl+ | 203 | PRJNA55713 |
| U0317 | Clinical isolate (urine) | NLD | 2005 | AMP; esp+ hyl+ | 78 | PRJNA47349 |
| 1,230,933 | Clinical isolate (wound) | USA | 2005 | AMP, VAN; esp+ hyl+ | 18 | PRJNA55701 |
| 1,231,408 | Clinical isolate (blood) | USA | 2005 | AMP | 582 | PRJNA55721 |
| E4453 | Commensal isolate (dog faeces) | NLD | 2008 | AMP | 192 | PRJNA179597 |
| E1071 | Commensal isolate (faeces)d | NLD | 2000 | VAN | 32 | PRJNA47015 |
| E4452 | Commensal isolate (dog faeces) | NLD | 2008 | AMP | 266 | PRJNA179613 |
| E1679 | Clinical isolate (vascular catheter) | BRA | 1998 | AMP, VAN; esp+ | 114 | PRJNA47347 |
| E1636 | Clinical isolate (blood) | NLD | 1961 | AMP | 106 | PRJNA47345 |
| D344SRF | Spontaneous mutant of strain D344R | USA | ? | — | 25 | PRJNA46237 |
| TC 6 | Transconjugante | USA | ? | hyl+ | 25 | PRJNA41101 |
| E1039 | Commensal isolate (faeces) | NLD | 1998 | — | 42 | PRJNA47011 |
| 1,231,501 | Clinical isolate (blood) | USA | 2005 | — | 52 | PRJNA55715 |
| LCT-EF90 | Derived from E. faecium type strainf | SWI | ? | — | 76 | PRJNA141665 |
| E980 | Commensal isolate (faeces) | NLD | 1998 | — | 94 | PRJNA47017 |
| Com15 | Commensal isolate (faeces) | USA | 2007 | — | 583 | PRJNA55725 |
| 1,141,733 | Clinical isolate (blood) | USA | 2005 | — | 52 | PRJNA55717 |
| PC4.1 | Commensal isolate (faeces) | AUS | 2008 | — | 720 | PRJNA46979 |
| Com12 | Commensal isolate (faeces) | USA | 2006 | — | 107 | PRJNA55723 |
| TX1330 | Commensal isolate (faeces) | USA | 1994 | — | 107 | PRJNA55481 |
aAll isolates were of human origin unless stated otherwise.
bWe only list reported ampicillin (AMP) and vancomycin (VAN) resistances here.
cMLST data were extracted from the MLST website (Imperial College, London; http://efaecium.mlst.net/, last accessed August 3, 2013) or were determined using MLST v1.6 (Larsen et al. 2012).
dIsolates collected from the faeces of hospitalized patients. Strains E155 and C68 are regarded as clinical isolates because they were representative clones of hospital outbreaks. Strain C68 is described in Carias et al. (1998).
eTransconjugant of mating between strains C68 and D344SRF.
fThis strain was derived from the E. faecium type strain and was cultured at 15 and 37 °C for more than 4 weeks prior to DNA isolation and sequencing. The E. faecium type strain was first deposited to culture collections in 1946, but the impact of this extensive subculturing of this derivative has not been characterized.
Functional Annotation and Core Genome Prediction
The Prokaryotic Annotation pipeline of ISGA (default settings) (Hemmerich et al. 2010) was used to annotate the three newly sequenced E. faecium genomes, as well as one of the available draft genomes (LCT-EF90) for which no annotation was available from GenBank. Annotated proteins were assigned to Clusters of Orthologous Groups (COG) (Tatusov et al. 2001) as described previously (Snel et al. 2002). Protein domains were predicted using SMART v7.0 (Letunic et al. 2012) and Pfam v26.0 (Finn et al. 2010). To determine groups of orthologous proteins, the 97,972 annotated proteins present in the 34 E. faecium strains were used as input for an all-versus-all sequence similarity search using BLASTP v2.2.24 (Altschul et al. 1997) (default settings, except for: -F “m S”; -e 1.0E−05; -z 97,972 [i.e., the total number of proteins]). From the BLAST output, groups of orthologous proteins were predicted using OrthoMCL v2.0.2 (Li et al. 2003). Orthologous groups with exactly one representative protein from each of the 34 input strains were considered to be part of the E. faecium core genome.
Detection of Recombination
The BratNextGen algorithm (Marttinen et al. 2008, 2012) was used to detect recombination events in E. faecium. A concatenated multiple sequence alignment of the E. faecium core genome was used as input for this algorithm. This concatenated alignment was built as follows: for each orthologous group, the corresponding nucleotide sequences were extracted and aligned using Muscle v3.7 (Edgar 2004), after which gaps were stripped from the alignment using trimAl v1.2 (Capella-Gutiérrez et al. 2009). Stripped alignments were concatenated in the order of orthologous group numbers as assigned by OrthoMCL. Because OrthoMCL assigns orthologous group numbers on a “first-come, first-served” basis, orthologous genes ended up in the concatenated alignment in the order in which they were read by OrthoMCL, thus following the order in which they are located on the genomic contigs and scaffolds. As the first genomic contigs that entered OrthoMCL were those of strain 1,141,733, the alignment followed the exact gene order in the contigs of this strain. Analysis of the alignment showed that the gene order used also exactly matched that of 11 other strains and nearly exactly matched that of the remaining 22 strains (a median of only 3 synteny breaks was observed for these 22 strains with a maximum of only 10 and 11 synteny breaks for strains Aus0004 and 1,231,408, respectively). Thus, the concatenated alignment of orthologous genes accurately reflects the synteny of the average E. faecium genome. The estimation of recombination was performed with the default BratNextGen settings as in (Marttinen et al. 2012) starting out with a partition of the genome alignment into 5 kb blocks and using 20 iterations of the estimation algorithm, which was assessed to be sufficient since changes in the hidden Markov model parameters were already negligible over the last 30% of the iterations. The alpha parameter was estimated using the default options in BratNextGen and the resulting value was 4.96. Significance of a recombining region was determined as in (Marttinen et al. 2012) using a permutation test with 100 permutations executed in parallel on a cluster computer (threshold of 5% was used to conclude significance for each region).
Phylogeny and the Identification of Recombinant Sequence Origins
Phylogenetic trees were built using RAxML v7.2.8 (Stamatakis 2006) under the GTRCAT model. To assess the effect of recombination on the evolution of E. faecium, two phylogenetic trees were built: one using the aligned SNPs from the concatenated core genome alignment, and one using the same information, but filtered for significant recombination signals as determined by BratNextGen. Confidence was inferred by running 1,000 bootstrap replicates under the GTRCAT model. The phylogenetic origin of the identified recombinant sequences was assessed as follows: first, for each recombination event, the corresponding sequence alignment was extracted from the larger E. faecium core genome alignment and a separate phylogenetic subtree was built. Recombinant sequences were classified as belonging to the same recombination event when they had received exactly the same recombination start and end positions by BratNextGen. To prevent obscuring the individual subtrees with signals from other recombination events, all sequences that partially or completely belonged to another recombination event were removed from the alignment prior to building the tree (note that recombination events do not overlap within one sequence). Second, the resulting trees were midpoint rooted using the Phangorn package v1.6-5 (http://cran.r-project.org/web/packages/phangorn/index.html, last accessed August 3, 2013) in R (R Development Core Team, http://www.R-project.org, last accessed August 3, 2013), after which a decision scheme was followed using the topology of the first major split closest to the root (supplementary fig. S1, Supplementary Material online). In the whole-genome recombination-filtered tree, E. faecium was separated into the two clades A and B (see also Results and Discussion). This knowledge was taken into account in the decision scheme, such that recombinant sequences were predicted to have originated from either an (ancestral) clade A or clade B strain. Alternatively, when the recombinant sequences clustered separately from both clades (i.e., as an outgroup), the sequence was predicted to have originated from a species other than E. faecium. Because BratNextGen uses a conservative approach for predicting recombinant sequences to avoid false positives (Marttinen et al. 2012), it is likely that not all recombination events were detected. Therefore, undetected overlapping recombination events could still obscure individual subtrees and blur the separation into clades A and B. To take this into account, a maximum of two supposedly nonrecombinant sequences were deleted from the tree if needed to get a “pure” clade A/B separation, thereby keeping a minimum of three clade A and three clade B strains in the tree. If the tree could not be purified in this way, the phylogenetic source of the recombinant sequence was considered to be either E. faecium or inconclusive (supplementary fig. S1, Supplementary Material online).
Analysis of Recombinant Sequences Originating from Outside E. faecium
In cases where recombinant sequences were predicted to originate from outside E. faecium, an attempt was made to identify the potential donor species. For this, recombinant sequences were split up into their corresponding gene sequences (partial gene sequences <30 bp were not considered). Recombinant gene sequences were then screened against GenBank’s nucleotide sequence database (nt) (ftp://ftp.ncbi.nlm.nih.gov/blast/db/, last accessed August 3, 2013; date of download August 2012) and against the assemblies of 66 non-E. faecium enterococci (discussed earlier) using BLASTN v2.2.24 (default settings, except for: -F F). Target sequences were recovered using an E-value cutoff of 1.0E-10 and a query sequence coverage of ≥75%. Recovered sequences were aligned with the corresponding E. faecium gene sequences, after which midpoint rooted individual gene trees were built using RAxML. Trees were purified as described above if necessary and were inspected manually to assign potential donor species.
Database Submission
The whole genome sequence data of E. faecium strains E155, E525, and E1165 have been deposited at DDBJ/EMBL/GenBank under the accessions AUWX00000000, AUWV00000000, and AUWW00000000, respectively.
Results and Discussion
Enterococcus faecium Phylogeny Based on the Core Genome
To study the effects of recombination on the genome evolution of E. faecium, we used the genome sequence information of 34 E. faecium strains, including 31 publicly available strains and 3 strains that were newly sequenced for the purpose of this study. The newly sequenced strains were E155, E525, and E1165 and were all modern clinical isolates, originating from hospitalized patients in the United States, Australia, and Italy, respectively, and belonging to ST17, which is an important node in the hospital-associated subpopulation of E. faecium (Homan et al. 2002; Willems et al. 2005, 2011, 2012) (table 1). De novo assembly of the sequence reads resulted in an average genome size of 2.94 Mb (±0.14 Mb) and a mean scaffold N50 of 43.4 kb (±2.9 kb). The number of scaffolds of size ≥200 bp ranged from 185 to 242 (supplementary table S1, Supplementary Material online). Annotation of the new genomes using the ISGA pipeline resulted in an average set of 2,971 (±170) protein-encoding genes per genome.
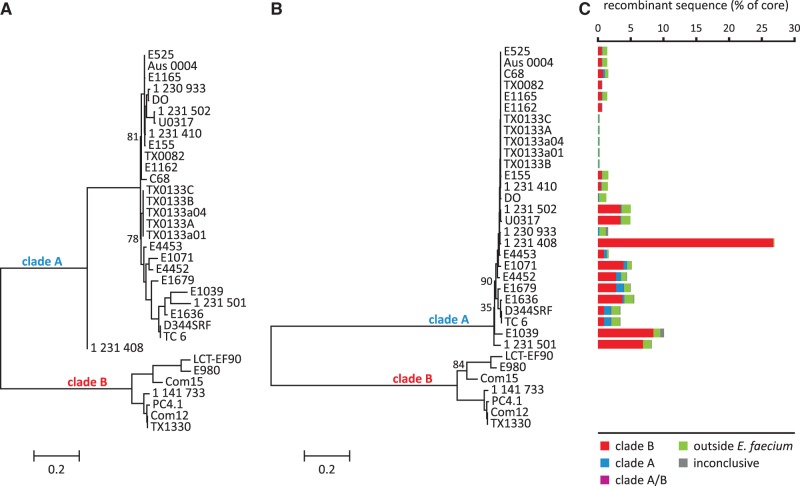
A total of 97,927 protein sequences were extracted from the 34 E. faecium genomes and used as input for assessing the core genome. This resulted in a set of 5,075 orthologous groups, of which 1,512 were considered to belong to the core genome as they contained exactly one protein representative from each of the 34 input strains. After stripping the individual core gene alignments for gaps, all alignments were concatenated to generate a whole-genome core alignment of 1,223,137 bp. From this alignment, we extracted 85,488 SNPs, which were used for the construction of a phylogenetic tree. The topology of the resulting tree (fig. 1A and supplementary fig. S2, Supplementary Material online, for details on bootstrap support) was in agreement with what has been observed before by others who used an E. faecium core genome extracted from 8 to 29 isolates to build a phylogenetic tree (Galloway-Peña et al. 2012; Lam et al. 2012; Palmer et al. 2012; Willems et al. 2012). The tree displayed a deep phylogenetic split between two major E. faecium clades. These clades were previously observed by Palmer et al. (2012), who designated them clades A and B, and by Galloway-Peña et al. (2012), who designated them clades HA (hospital-associated) and CA (commensal/community-associated), respectively. Clade A contained most of the clinical and hospital-associated isolates as well as animal isolates, whereas clade B consisted mainly of human commensal isolates. The clinical isolate 1,231,408 clustered consistently (100% bootstrap support) with clade A, albeit relatively distantly from all other clade A strains. This is also in line with previously built phylogenetic trees and with the observation that 1,231,408 is a hybrid strain that appears to have received a large part of its genome from a clade B background (Palmer et al. 2012) (discussed later). As expected on the basis of their ST (table 1), the newly sequenced clinical isolates clustered closely together with other modern clinical isolates of the same ST, as well as with strain C68 (ST16), which is a single locus variant of ST17 (Homan et al. 2002).
Fig. 1.—
Impact of recombination on E. faecium phylogeny and quantification and determination of origins of recombinant sequences. (A) Midpoint rooted phylogenetic tree (RAxML) built from an alignment of 85,488 variable positions in the E. faecium core genome. No recombination filtering was applied. (B) Midpoint rooted phylogenetic tree (RAxML) build from an alignment of 46,823 nonrecombinant variable positions in the E. faecium core genome. Numbers near branches indicate percentage bootstrap support: only numbers <95% are indicated and only for main branches (for all bootstrap supports, see supplementary fig. S2, Supplementary Material online). Branch lengths correspond to the scale bar, in units of changes/nucleotide position. (C) Predicted levels and origin of recombinant sequences per strain.
Enterococcus faecium Phylogeny Based on a Recombination-Filtered Core Genome
To estimate the effects of recombination on the evolution of E. faecium, we used the algorithm BratNextGen (Marttinen et al. 2008, 2012). Running this algorithm on the E. faecium core genome alignment resulted in the prediction of 1.2 Mb of recombinant sequence, divided over 383 recombination events (supplementary fig. S3, Supplementary Material online). Of the original 1,223,137 positions in the core genome alignment, 534,961 (44%) were predicted to be affected by recombination in at least one strain. On the SNP level, this rate was comparable with 38,665 of the original 85,488 positions (45%) identified as being recombinant. The remaining 46,823 nonrecombinant SNPs were used to build a recombination-free phylogenetic tree. As shown in figure 1B (supplementary fig. S2, Supplementary Material online, for details on bootstrap support), the topology of this tree was largely similar to that of the unfiltered tree presented in figure 1A in the sense that the ancient split between clade A and B was still observed after purging for recombination. Interestingly, BratNextGen did not detect any recombination signals in clade B. This does not necessarily mean that clade B strains are completely unaffected by recombination. BratNextGen uses a conservative approach for predicting recombinant sequences to avoid false positives (Marttinen et al. 2012). Consequently, true recombination signals may go undetected (i.e., false negatives) when there is low statistical power. The reasons for the absence of detectable recombination in clade B may be that 1) clade B strains were underrepresented in the available WGS data set used here, and 2) the diversity within clade B was relatively large as compared to the diversity within clade A (fig. 1A). Because BratNextGen has higher statistical power to detect significant recombination events when they are shared across strains, these factors (i.e., limited sample of strains and high diversity between them) may have hampered the detection of recombinant sequence segments within clade B. Indeed, we noticed that, when using our decision scheme for predicting recombinant sequence origins (supplementary fig. S1, Supplementary Material online), tree purifications were made 74 times corresponding to the removal of 103 sequences. This observation may point to a recombinant nature for the corresponding sequences although other reasons for sequence divergence (i.e., mutation) cannot be ruled out. Of the 103 removed sequences, 60 belonged to clade B strains, mostly to strains LCT-EF90 (33 cases) and E980 (23 cases), thus potentially indicating the presence of recombinant sequences in clade B. Nevertheless, the fact that BratNextGen detected no recombination signals at all in clade B at least strongly suggests that, in general, clade A is more prone to recombination than clade B. Consequently, the recombination-filtering procedure effectively meant a purification of clade A only. As became evident by visually comparing the trees in figure 1A and B, the purification resulted in a strongly increased cohesiveness of the phylogenetic tree for clade A. To quantitatively support this observation, we extracted all strain-to-strain pairwise branch-length distances from both trees and compared the average distance within clade A and B and between both clades, pre- and post-recombination-filtering. Potential over-sampling of lineages was taken into account by including only one strain (the one furthest from the root) for branches where all pairwise strain distances were less than 5.0E−04 in the purified tree (e.g., strain TX0133a01 was picked as a representative for all 5 TX0133 strains). This analysis showed that upon recombination-filtering, the average distance within clade A significantly decreased (from 0.1 to 0.03; P < 0.01; Wilcoxon signed rank test), whereas the average distance between clade A and B significantly increased (from 1.4 to 2.0; P < 0.01). The average distance within clade B (0.2) remained the same. In summary, recombination-filtering widened the ancient clade A/B split by bringing the clade A strains closer together.
Comparison of the two trees showed that most of the strains that clustered tightly together in the unfiltered tree also clustered tightly together upon recombination-filtering. The most notable exceptions were 1) the separation of strains E1039 and 1,231,501 from each other and from the larger cluster containing for instance strain E1679, 2) the shift of the 5 × TX0133 branch to a position within the ST17 cluster of modern clinical isolates, and 3) the shift of the branch 1,231,502/U0317 to a position just outside the ST17 cluster (fig. 1 and supplementary fig. S2 [Supplementary Material online] for details). These last two modifications resulted in a robust branch consisting only of modern clinical isolates of ST17 (and 16) and with ST78 and ST203 isolates together forming a distinct lineage. Another major change that occurred upon recombination-filtering was the displacement of strain 1,231,408 from a “hybrid” position in-between clade A and B toward a position branching deeper inside clade A and tightly clustering with strain 1,230,933, which clustered with strain DO in the unfiltered tree. The strong shift of strain 1,231,408 toward clade A and away from clade B fits with the previously observed hybrid nature of this strain (Palmer et al. 2012). However, a similar post-recombination-filtering displacement of strain 1,231,408 was not observed in a previous study (Qin et al. 2012), in which the authors used PHI (Bruen et al. 2006) to detect and remove recombination signals prior to phylogenetic tree building. In their resulting tree, strain 1,231,408 still occupied a “hybrid” position similar to the one observed in the unfiltered tree presented here (fig. 1A).
Quantities of Recombination in Clade A
Recombination signals were detected in each of the 27 clade A strains included in this study. However, the amount of detected recombinant DNA as a percentage of the 1.2 Mb core genome varied widely over the different clade A strains (fig. 1C). When correcting for potential over-sampling of genetic lineages, the average amount of recombination detected in clade A was 4.4% of the core genome. The strain with the highest amount of recombination was strain 1,231,408 with as much as 26.9% of its core genome predicted to be recombinant. Other strains with a relatively high load of recombination were E1039 (10.1%) and 1,231,501 (8.2%). At the other end of the spectrum were strains TX0133a01, E1162, and TX0082 with only 0.3%, 0.7%, and 0.7% of their core genome predicted to be of foreign origin, respectively (fig. 1C). Interestingly, the modern clinical isolates of ST16 and ST17 had a significantly lower amount of recombination (average of 1.1%) than the other strains in clade A (6.0%) (P < 0.01; Mann–Whitney U test). This finding appears to be in agreement with a previous observation made by Willems et al. (2012), who applied a genetic admixture analysis on MLST sequence data from a large collection of E. faecium strains and found that hospital isolates displayed lower levels of recombination than isolates from nonhospitalized persons and pigs. This may point to genetic and/or ecological isolation of E. faecium hospital isolates and fits with the previously proposed hypothesis that novel, highly invasive, E. faecium strains may arise through horizontal gene transfer, but once they have adopted their new lifestyle, they become genetically isolated from other bacteria and become less prone to recombination (Willems et al. 2012).
The Recombinant DNA in Clade A Mainly Had a Clade B Origin
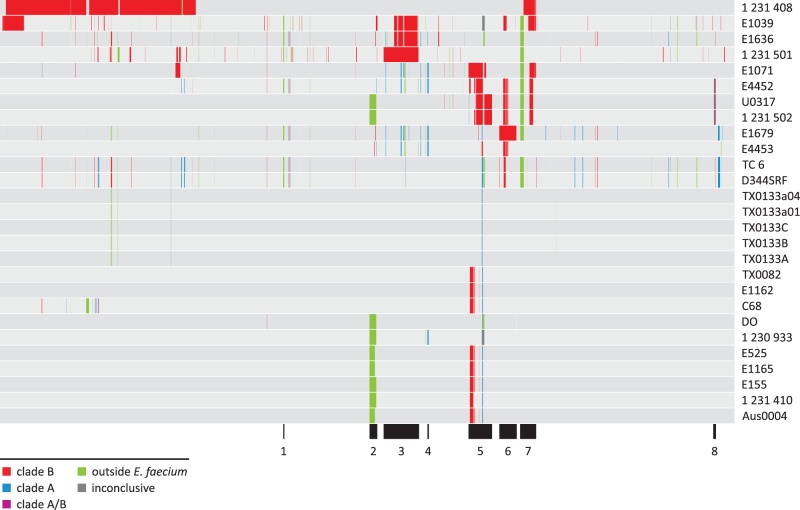
Interestingly, the vast majority of the E. faecium recombinant sequence was predicted to have originated from a clade B type E. faecium background (fig. 1C). More specifically, we found that of the 1.2 Mb of recombinant sequence, 71% had a predicted clade B type source, followed by 19.6% with a predicted source from outside E. faecium and 7.6% with a predicted clade A type origin. For only 0.1% of the recombinant sequence, it was unclear whether it originated from a clade A or clade B type background. Also, for only 1.7% of the recombinant sequence, the phylogenetic source was inconclusive according to the decision scheme that we employed (supplementary fig. S1, Supplementary Material online). These results indicated that clade B E. faecium strains have formed, and possibly still form, an important source for donating core genomic DNA to clade A E. faecium strains. Especially strains 1,231,408, E1039, and 1,231,501 were found to have received a large amount of core genomic DNA from a clade B type source, totalling 327, 103 and 84 kb, respectively. Also with respect to the total amount of foreign DNA detected in their core genomes, the amount of DNA with a potential clade B type source in these strains was high at 99%, 84%, and 84%, respectively. In figure 2, we plotted the detected recombinant sequences and their potential sources onto the core genome sequence of the completed strain Aus0004, as determined in this study. The results presented in figure 2 expand on previous results of Palmer et al. (2012), who studied genome mosaicism in five clade A and three clade B E. faecium strains. For each gene in these eight strains they determined whether it had either a clade A- or clade B-type signature. Mosaicism was most evident for two clade A strains they analyzed, namely strains 1,231,408 and 1,231,501. Here, when using the currently much larger data set of 34 E. faecium genomes, we found that strains 1,231,408 and 1,231,501 still ranked among the top strains with respect to the amount of recombinant sequence coming from a clade B type background. In accordance with the observations made by Palmer et al. (2012), we found that these recombinant sequences clustered into a few long genomic stretches taking up around 27% and 7.8% of the 1,231,408 and 1,231,501 core genomes, respectively (fig. 2).
Fig. 2.—
Distribution of recombinant sequences over the Enterococcus faecium core genome. Recombinant sequences were plotted onto the E. faecium Aus0004 core genome, as determined in this study, starting with the first core gene (efau0004_00001, dnaA) on the left and proceeding to the last core gene (efau0004_02867) on the right. Black segments below the recombinant regions indicate recombination hotspots.
Recombinant DNA with a Predicted Source Outside E. faecium
As mentioned earlier, almost 20% of the recombinant core genomic DNA in E. faecium was predicted to come from a source outside E. faecium. To find potential donor species for these recombinant segments, we extracted the corresponding gene sequences from each recombinant segment and screened them against a database of enterococcal genome sequences (Materials and Methods) and GenBank’s nt database. Similar sequences were extracted and used, together with the corresponding recombinant and nonrecombinant E. faecium gene sequences, to build phylogenetic trees (Materials and Methods). The resulting trees were manually inspected to find close homologs of (i.e., potential donor species for) the recombinant E. faecium sequences. In cases where one recombination event covered multiple genes, all the resulting gene trees had to be in agreement with each other concerning potential donor species. If not, for instance because one of the recombinant genes did not yield BLAST hits outside E. faecium, the donor species was deemed unknown. Unfortunately, we did not find potential donor species for the majority (84 out of 111) of the corresponding recombination events. This was due to the fact that for these 84 events, no similar sequences were detected for at least one of the recombinant genes, other than E. faecium sequences themselves. The most likely explanation for obtaining so few BLAST hits may be that current microbial sequence databases simply do not (yet) contain sequences from the actual donor species or from species closely related to these donors. Still, for the remaining 27 recombination events (24%), we did find more specific evidence concerning the nature of the potential donor species. For instance, for 25 out of the 27 recombination events, the recombinant gene sequences clustered in between E. faecium and other enterococcal species or clustered together with multiple different enterococcal species other than E. faecium, suggesting that these recombinant sequences have originated from the genus Enterococcus (results not shown). Finally, for two recombination events, we found even more specific evidence about the nature of the potential donor species. One of these events was found in strain C68 and involved genes efxg_00252–efxg_00259. Each of these genes clustered together with a gene from E. hirae ATCC 9790 (ehr_06405–ehr_06440, respectively), suggesting that E. hirae or a closely related species has been the responsible recombination donor for these genes. Similarly, we found that gene efme1071_1769 in strain E1071 had an E. faecalis signature. These observations indicate that there is also detectable recombination between E. faecium and other species in the genus Enterococcus thereby significantly enlarging the potential reservoir for genetic diversity in E. faecium.
Functional Analysis of Recombinant Genes
To describe the functional impact of recombination on the E. faecium genome, we analyzed whether certain functional gene categories were more affected by recombination than others. To this aim, we assigned COGs to each orthologous protein group in the E. faecium genome. We then compared the composition of the COG functional categories found in the recombinant regions versus those found in the recombination-free regions. After correcting for potential over-sampling, we found that 1,832 proteins were affected by recombination compared with 40,502 recombination-free proteins, corresponding to 2,086 and 45,654 assigned COG functional categories, respectively. As shown in supplementary figure S4A, Supplementary Material online, a linear regression analysis between the COG category compositions of these two data sets showed a strong linear correlation (R2 = 0.87), suggesting that in general most functional groups are equally prone to recombination in the E. faecium core genome. Still, when we used Grubb’s test for outliers using a 99% confidence level, we found that COG categories C (energy production and conversion) and S (function unknown) were identified as outliers, potentially being relatively more (category C) and less (category S) prone to recombination in the E. faecium core genome (supplementary fig. S4A, Supplementary Material online). Further analysis of the categories C and S showed that their skewed representation in recombinant DNA was not due to specific COGs that were over- or underrepresented, but was caused by a proportional change of all COGs in the entire category (results not shown). By a similar approach, we also evaluated whether specific functional categories originated from specific recombination donors. Supplementary figure S4B–D, Supplementary Material online, shows the COG distributions for the recombinant genes originating from one of the three major donors of recombinant DNA (clade A, clade B, and outside E. faecium) compared with the COG distributions for the recombinant genes originating from all donors, except the one of interest. This analysis revealed that recombinant genes belonging to COG category G (carbohydrate transport and metabolism) mainly originated from clade A. Two individual COGs from category G that were overrepresented in recombinant DNA from clade A, were COG2723 and COG1486, which both encode putative beta-glucosidases. Together these two COGs made up 62% of all recombinant COGs of category G originating from clade A, whereas they only made up 9% of all recombinant COGs of this category originating from other sources. Other functions that originated mostly from clade A included a putative sugar uptake permease (COG4975), a dihydroxyacetone kinase (COG2376), and an ABC type polysaccharide transporter (COG1134) (results not shown). These results suggest that genes involved in sugar uptake and metabolism are extensively transferred between clade A E. faecium strains. This is of interest because genes involved in these processes may be important for successful colonization of the gut during antibiotic treatment, as has been demonstrated recently (Zhang et al. 2013).
Functional Analysis of Recombination Hotpsots
To locate the position of conserved major recombinant regions, we searched for recombination events that were detected in at least two strains (correcting for potential over-sampling) with a total summed length of at least 5 kb. This revealed a set of 18 recombination events (listed in supplementary table S2, Supplementary Material online), which merged into eight regions of overlapping and closely flanking recombination events (fig. 2). Interestingly, six of these eight regions (regions 2–7), including the four largest ones with a length ranging from 26 to 59 kb, were located in the third quadrant of the Aus0004 core genome. These data indicate a nonrandom distribution of recombination events over the E. faecium core genome and suggest the existence of a hotspot for recombination in E. faecium, which, when translating to the Aus0004 genome, roughly lies between open reading frames efau004_01354 and efau004_02178. Besides identifying recombination events that were conserved across multiple isolates, we searched for sites in the E. faecium core genome with clear traces of recombination-driven gene acquisition. For this, we screened for genomic regions of at least five consecutive accessory genes that were flanked on both sides by the same recombination event. The accessory genes and the flanking recombinant core genes had to be located on one single contig or scaffold. A total of 17 recombination events met these criteria, several of which were clearly associated with mobile genetic elements, such as phages, transposons, recombinases, and integrases (supplementary table S2, Supplementary Material online). Of the 17 events, five were found in strain 1,231,408, corresponding to the first quadrant of the Aus0004 core genome. Still, 9 of the remaining 12 events were found within the recombination hotspot described earlier, which further indicates that this region on the E. faecium genome is relatively prone to recombination.
Recombination of an Ampicillin Resistance Determinant
As mentioned earlier, we found that the clinical E. faecium strains of ST16/17 displayed relatively low levels of recombination. Considering this, it is of interest to note that 6 of the 18 conserved recombination events listed earlier were associated with ST16/17 strains, indicating that the recombination-driven diversity of these genomic regions may reflect adaptation to selection pressures associated with hospitalization. Four of the corresponding recombination events (events 8–11, supplementary table S2, Supplementary Material online) were located within region 5 (fig. 2). Among these were events 8 and 9, which were predicted by BratNextGen to be independent recombination events, but which covered exactly the same set of core genes. Together, these two events were found in all ST16/17 strains analyzed, except TX0133. Other events that partly overlapped with events 8 and 9 were found in strains E4452 and E1071 (results not shown). One of the genes within the affected region was ddcP (efau004_01934 in Aus0004), which has previously been identified as a membrane-associated d-alanyl-d-alanine carboxypeptidase that confers resistance to ampicillin and lysozyme in E. faecium E1162 (Zhang et al. 2012). Recombinant ddcP was predicted to stem from an ancestral clade B type E. faecium. On the protein level, three positions gave recombinant DdcP a clear clade B type signature, including an Ile-to-Val at position 101, an Asp-to-Ala at position 298 and an Asp-to-Glu at position 392 (supplementary fig. S5, Supplementary Material online). Although these amino acid variations do not by themselves fully explain DdcP-mediated ampicillin resistance in E. faecium (e.g., strain E1071 and all clade B strains are ampicillin sensitive), they do suggest a scenario in which clinical strains, like E1162, have recombined their ddcP gene with a clade B type ddcP gene, which in combination with other factors like mutations in pbp5, has resulted in clinical strains becoming more prone to developing ampicillin resistance.
Recombination of the epa-Like Locus Is Reminiscent of Pneumococcal “Capsule-Switching”
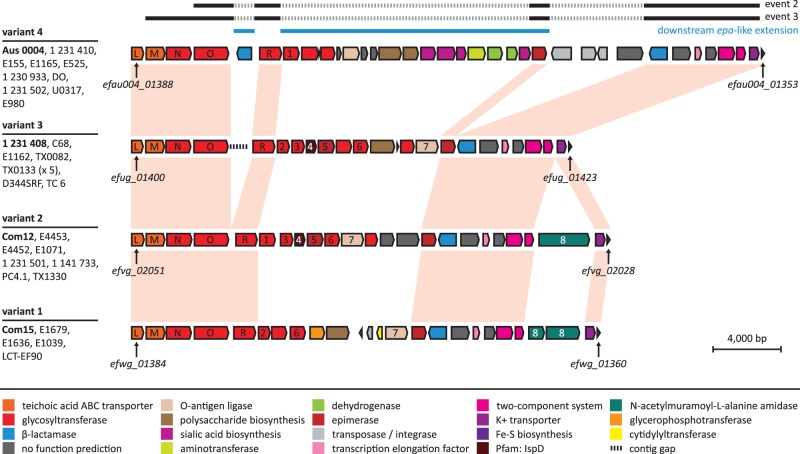
The two remaining recombination events that were associated with ST16/17 E. faecium strains were both found in region 2 (fig. 2) and were predicted to have originated from an unidentified source outside E. faecium. Both events covered and flanked almost the same set of core and accessory genes, respectively, namely efau004_01354 to efau004_1385 in strain Aus0004 and its orthologs in strains E1165 and E525 (event 2, supplementary table S2, Supplementary Material online) and a somewhat larger region extending toward the orthologs of efau004_01387 in strains 1,231,410, E155, 1,230,933, DO, 1,231,502, and U0317 (event 3). In Aus0004, the region between efau004_01354 to efau004_1387 mostly consists of accessory genes, including two ISL3-family transposases and a retroviral integrase, further pointing toward a history of lateral gene transfer for this complete genomic region. The region involved has previously been described as a downstream extension of the E. faecium epa-like locus (Palmer et al. 2012; Qin et al. 2012), a gene cluster that is putatively involved in the biosynthesis of antigenic cell wall polysaccharide, based on homology with the E. faecalis epa locus (Xu et al. 1998; Teng et al. 2009). The extended region of the epa-like locus (corresponding to efau004_01365–efau004_01382, plus efau004_01384 in Aus0004) is known to be highly variable across E. faecium and has been divided into four locus variants (Qin et al. 2012). Analysis of the current set of 34 E. faecium strains showed that the same division into four locus variants still holds. Figure 3 shows representatives of these four locus variants as well as the regions affected by recombination. Notably, all the epa-like loci affected by recombination events 2 and 3 belonged to locus variant four. Only one epa-like locus of this type, the one present in the clade B strain E980, did not contain recombination signals according to BratNextGen. This does not necessarily mean that this locus has not recombined in this strain, but may suggest that the potential recombination event was not recent enough, so that any strong recombination signals may have been neutralized over time. Besides recombination events 2 and 3, additional independent events were detected within the same genomic region of strains E1165, E4453, E1679, E4452, E1636, and E1039, but these events covered only small segments within epaMNO, never extending beyond three consecutive core genes (results not shown). Nevertheless, the observation that multiple independent recombination events were detected in the downstream epa-like extension, suggests that this region of the E. faecium genome is prone to recombination. As shown in figure 3, the variable epa-like region that was flanked and thus potentially affected by events 2 and 3 contained a predicted class C beta-lactamase gene (efau004_01384 in Aus0004) and sialic acid biosynthesis genes neuABCD (efau004_01366 and efau004_01370–efau004_01372) specific for locus variant four. Sialic acid decoration of the bacterial cell surface may be a form of molecular mimicry that affects detection by the host’s immune system (Vimr and Lichtensteiger 2002). Interestingly, events 2 and 3 both extended beyond the epa-like locus into a core genomic region containing yet another class C beta-lactamase gene (efau004_01360). A recombinant cell surface biosynthesis cluster flanked by two beta-lactamase genes has also been described in Streptococcus pneumoniae. In this species, serotype variants have arisen through recombinational replacements within and around this so-called cps locus (Trzciński et al. 2004; Brueggemann et al. 2007; Croucher et al. 2011; Golubchik et al. 2012; Marttinen et al. 2012). Our observations suggest that recombination may have a role in capsule variation in E. faecium.
Fig. 3.—
Enterococcus faecium epa-like locus variants and recombination signals detected in locus variant four. Gene clusters of the four epa-like locus variants are displayed for representative strains Aus0004 (variant four), 1,231,408 (variant three), Com12 (variant two), and Com15 (variant one). The occurrence of the four variants across the other strains is indicated to the left. Core genomic genes are connected by pink shades. Other orthologous and paralogous genes are indicated by numbers. Part of the conserved upstream epa-like locus (epaL–epaO and epaR) is indicated by L, M, N, O, and R. The downstream epa-like extension is indicated by blue solid lines above the illustration of locus variant four. Locus variant four was predicted to be affected by large core genomic recombination events 2 and 3 (indicated by solid black lines above the locus variant four illustration) that potentially spanned multiple accessory genes (indicated by dashed gray lines). Genes are color-coded according to functional category. Drawings are to scale.
Conclusions
In this study, we have analyzed the impact of recombination on the whole-genome level in E. faecium. We show that filtering for recombination prior to phylogenetic tree building had a purifying effect on the ancient E. faecium clade A/B split. Closer inspection of the origin of the recombinant sequences indicated that this purifying effect was mainly caused by the fact that clade A strains have received foreign DNA mainly from clade B type strains. However, no recombination signals were detected in clade B strains themselves, suggesting that the transfer of DNA within the E. faecium population is mainly a unidirectional event: from clade B to clade A, and not vice versa. However, it is important to note that the number of clade B strains in our data set was relatively low, which may have hampered the detection of recombination signals within this clade. The inclusion of more clade B strains in future recombination detection analyses will identify whether the apparent lack of recombination within clade B is genuine or is merely a result of a lack of resolution. Still, the results presented here strongly indicate that clade A is more prone to recombination than clade B. Given the observation that recombination seems to act as a coalescent force for clades A and B, it is of interest to note that, within clade A, contemporary clinical isolates of ST16/17 contained relatively low levels of recombination. This fits with previous hypotheses (Willems et al. 2012; McNally et al. 2013) that novel, highly successful, clinical strains may arise through gene acquisition, but become genetically isolated and less prone to recombination once they have adopted their new life-style. Still, several recombination events were associated with contemporary clinical isolates. These events involved the ampicillin resistance determinant ddcP and the putative cell wall polysaccharide biosynthesis epa-like locus. These findings point to important recombination-driven mechanisms that facilitate adaptation of E. faecium to selection pressures associated with hospitalization.
Supplementary Material
Supplementary figures S1–S5 and tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org).
Acknowledgments
This work was supported by the European Union Seventh Framework Programme (FP7-HEALTH-2011-single-stage) “Evolution and Transfer of Antibiotic Resistance” (EvoTAR) (grant number 282004 to M.B., W.S., and R.J.W.); the European Research Council (ERC) (grant number 239784 to J.C.); the Academy of Finland (grant number 251170 to J.C.); and the Sigrid Juselius Foundation (grant number 4702281 to J.C.). The Centre for Molecular and Biomolecular Informatics (CMBI, Radboud University Nijmegen, Nijmegen, the Netherlands) is acknowledged for providing computing space. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Literature Cited
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueggemann AB, Pai R, Crook DW, Beall B. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 2007;3:e168. doi: 10.1371/journal.ppat.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo ILBC, Gilmore MS, Darini ALC. Multilocus sequence typing and analysis of putative virulence factors in vancomycin-resistant and vancomycin-sensitive Enterococcus faecium isolates from Brazil. Clin Microbiol Infect. 2006;12:1123–1130. doi: 10.1111/j.1469-0691.2006.01496.x. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinforma. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carias LL, Rudin SD, Donskey CJ, Rice LB. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J Bacteriol. 1998;180:4426–4432. doi: 10.1128/jb.180.17.4426-4434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Ramírez S, et al. Phylogeographic variation in recombination rates within a global clone of methicillin-resistant Staphylococcus aureus. Genome Biol. 2012;13:R126. doi: 10.1186/gb-2012-13-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coque TM, et al. Population structure of Enterococcus faecium causing bacteremia in a Spanish university hospital: setting the scene for a future increase in vancomycin resistance? Antimicrob Agents Chemother. 49. 2005:2693–2700. doi: 10.1128/AAC.49.7.2693-2700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway-Peña J, Roh JH, Latorre M, Qin X, Murray BE. Genomic and SNP analyses demonstrate a distant separation of the hospital and community-associated clades of Enterococcus faecium. PLoS One. 2012;7:e30187. doi: 10.1371/journal.pone.0030187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway-Peña JR, Nallapareddy SR, Arias CA, Eliopoulos GM, Murray BE. Analysis of clonality and antibiotic resistance among early clinical isolates of Enterococcus faecium in the United States. J Infect Dis. 2009;200:1566–1573. doi: 10.1086/644790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore MS, Lebreton F, van Schaik W. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol. 2013;16:10–16. doi: 10.1016/j.mib.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubchik T, et al. Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event. Nat Genet. 2012;44:352–355. doi: 10.1038/ng.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich C, Buechlein A, Podicheti R, Revanna KV, Dong Q. An Ergatis-based prokaryotic genome annotation web server. Bioinforma. 2010;26:1122–1124. doi: 10.1093/bioinformatics/btq090. [DOI] [PubMed] [Google Scholar]
- Hendrickx APA, van Wamel WJB, Posthuma G, Bonten MJM, Willems RJL. Five genes encoding surface-exposed LPXTG proteins are enriched in hospital-adapted Enterococcus faecium clonal complex 17 isolates. J Bacteriol. 2007;189:8321–8332. doi: 10.1128/JB.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidron AI, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- Homan WL, et al. Multilocus sequence typing scheme for Enterococcus faecium. J Clin Microbiol. 2002;40:1963–1971. doi: 10.1128/JCM.40.6.1963-1971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klare I, et al. Spread of ampicillin/vancomycin-resistant Enterococcus faecium of the epidemic-virulent clonal complex-17 carrying the genes esp and hyl in German hospitals. Eur J Clin Microbiol Infect Dis. 2005;24:815–825. doi: 10.1007/s10096-005-0056-0. [DOI] [PubMed] [Google Scholar]
- Lam MMC, et al. Comparative analysis of the first complete Enterococcus faecium genome. J Bacteriol. 2012;194:2334–2341. doi: 10.1128/JB.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen MV, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavis HL, et al. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS Pathog. 2007;3:e7. doi: 10.1371/journal.ppat.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marttinen P, et al. Bayesian modeling of recombination events in bacterial populations. BMC Bioinformatics. 2008;9:421. doi: 10.1186/1471-2105-9-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marttinen P, et al. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res. 2012;40:e6. doi: 10.1093/nar/gkr928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally A, Cheng L, Harris SR, Corander J. The evolutionary path to extraintestinal pathogenic, drug-resistant Escherichia coli is marked by drastic reduction in detectable recombination within the core genome. Genome Biol Evol. 2013;5:699–710. doi: 10.1093/gbe/evt038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, et al. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. MBio. 2012;3:e00318–00311. doi: 10.1128/mBio.00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, et al. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol. 2012;12:135. doi: 10.1186/1471-2180-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LB, et al. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J Infect Dis. 2003;187:508–512. doi: 10.1086/367711. [DOI] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. The identification of functional modules from the genomic association of genes. Proc Natl Acad Sci U S A. 2002;99:5890–5895. doi: 10.1073/pnas.092632599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinforma. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Singh KV, Bourgogne A, Zeng J, Murray BE. Further characterization of the epa gene cluster and Epa polysaccharides of Enterococcus faecalis. Infect Immun. 2009;77:3759–3767. doi: 10.1128/IAI.00149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzciński K, Thompson CM, Lipsitch M. Single-step capsular transformation and acquisition of penicillin resistance in Streptococcus pneumoniae. J Bacteriol. 2004;186:3447–3452. doi: 10.1128/JB.186.11.3447-3452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankerckhoven V, et al. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol. 2004;42:4473–4479. doi: 10.1128/JCM.42.10.4473-4479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schaik W, et al. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics. 2010;11:239. doi: 10.1186/1471-2164-11-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr E, Lichtensteiger C. To sialylate, or not to sialylate: that is the question. Trends Microbiol. 2002;10:254–257. doi: 10.1016/s0966-842x(02)02361-2. [DOI] [PubMed] [Google Scholar]
- Willems RJ, van Schaik W. Transition of Enterococcus faecium from commensal organism to nosocomial pathogen. Future Microbiol. 2009;4:1125–1135. doi: 10.2217/fmb.09.82. [DOI] [PubMed] [Google Scholar]
- Willems RJL, Hanage WP, Bessen DE, Feil EJ. Population biology of Gram-positive pathogens: high-risk clones for dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35:872–900. doi: 10.1111/j.1574-6976.2011.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems RJL, et al. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis. 2005;11:821–828. doi: 10.3201/eid1106.041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems RJL, et al. Restricted gene flow among hospital subpopulations of Enterococcus faecium. MBio. 2012;3:e00151–00112. doi: 10.1128/mBio.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Murray BE, Weinstock GM. A cluster of genes involved in polysaccharide biosynthesis from Enterococcus faecalis OG1RF. Infect Immun. 1998;66:4313–4323. doi: 10.1128/iai.66.9.4313-4323.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, et al. Genome-wide identification of ampicillin resistance determinants in Enterococcus faecium. PLoS Genet. 2012;8:e1002804. doi: 10.1371/journal.pgen.1002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, et al. Identification of a genetic determinant in clinical Enterococcus faecium strains that contributes to intestinal colonization during antibiotic treatment. J Infect Dis. 2013;207:1780–1786. doi: 10.1093/infdis/jit076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.