Abstract
The mammalian sperm nucleus is characterized by unique properties that are important for fertilization. Sperm DNA retains only small numbers of histones in distinct positions and the majority of the genome is protamine associated, which allows for extreme condensation and protection of the genetic material. Furthermore, sperm nuclei display a highly ordered architecture that is characterized by a centrally located chromocenter comprising the pericentromeric chromosome regions and peripherally positioned telomeres. Establishment of this unique and well conserved nuclear organization during spermiogenesis is not well understood.
Utilizing fluorescence in situ hybridization (FISH), we show that a large fraction of the histone associated sperm genome is repetitive in nature, while a smaller fraction is associated with unique DNA sequences. Coordinated activity of Poly(ADP-ribose) (PAR) polymerase (PARP) and topoisomerase II beta (TOP2B) has been shown to facilitate DNA relaxation and histone to protamine transition during spermatid condensation, and altered PAR metabolism is associated with an increase in sperm histone content. Combining FISH with 3D laser scanning microscopy technology, we further show that altered PAR metabolism by genetic or pharmacological intervention leads to a disturbance of the overall sperm nuclear architecture with a lower degree of organization and condensation of the chromocenters formed by chromosomal pericentromeric heterochromatin.
Keywords: sperm, spermatogenesis, nuclear architecture, chromatin, poly(ADP-) ribosylation, PARG, PARP1
Introduction
The structure of sperm chromatin is very different from that of somatic cells (Braun 2001, Rousseaux et al. 2004). Sperm function requires extreme compaction of the paternal genome to ensure its protection from environmental impact during transport. Mammalian sperm chromatin mainly consists of DNA associated with protamines in a nearly paracrystalline complex crosslinked by disulfide bonds. The transition from nucleosomal to a highly condensed chromatin structure takes place during the haploid phase of spermatogenesis, termed spermiogenesis, when most histones are first replaced by transition proteins TP1 and TP2, which are then replaced by protamines (Kimmins and Sassone-Corsi 2005, Sassone-Corsi 2002, Lewis et al. 2003, Zhao et al. 2004). Only a very small portion of the sperm genome retains a histone-based, nucleosomal form similar to chromatin organization in the archetypal somatic cell, and sperm DNA is packed into the most highly condensed, naturally occurring chromatin state in mammals. The chromatin remodeling process allows for extremely dense DNA packaging into toroid-shaped DNA loops (Balhorn 2007) which are, together with the remaining histone associated DNA regions, folded into a sperm-specific and highly organized nuclear structure supported by the proteinaceous nuclear matrix (Shaman et al. 2007, Ward and Coffey 1991). The global sperm nuclear architecture is likewise different from somatic cells. It is characterized by a centrally located chromocenter, which consists of clustered pericentromeric chromosome segments (Zalensky et al. 1993), and peripherally localized telomeric regions with pairwise intrachromosomal clustering of the telomeres (Ward and Zalensky 1996, Zalensky et al. 1995). Moreover, chromosomes preferentially occupy defined spatial positions within the sperm nucleus (Mudrak et al. 2012, Zalenskaya and Zalensky 2004, Meyer-Ficca et al. 1998). The intricate organization of the condensed sperm nucleus has been proposed to provide a potential, largely unexplored form of epigenetic information layered on top of the genetic information contained in the paternal chromosome complement (Zalensky and Zalenskaya 2007, Yamauchi et al. 2011)
Completion of chromatin condensation is required for functional sperm development (Agarwal and Said 2003, Spano et al. 2005). In mature mouse sperm, less than 5% of the genome remains associated with histones, whereas in humans this fraction varies from ~10% to ~15% (Carrell et al. 2008, Gusse et al. 1986). In humans, residual histone retention in excess of >25% is associated with infertility, i.e., completing the nucleoprotein exchange is essential to form a functional sperm.
During nucleoprotein exchange in spermiogenesis, the DNA structure changes from a supercoiled nucleosomal form to a non-supercoiled, relaxed state. The dramatic transition in DNA topology is required for condensation and shaping of the nucleus, and requires controlled DNA strand breaks concomitantly with exchange of histones for transition proteins in spermatid steps 9–11 (Smith and Haaf 1998, Marcon and Boissonneault 2004, McPherson and Longo 1993, Risley et al. 1986, Meyer-Ficca et al. 2005a). The transient DNA strand breaks are, at least in part, formed by topoisomerase II beta (TOP2B) (Leduc et al. 2008, Laberge and Boissonneault 2005) and activate the enzymes poly(ADP-ribose) polymerase 1 and 2 (PARP1, PARP2) (Meyer-Ficca et al. 2011). These proteins are among the first proteins to detect and bind to DNA strand breaks, which activates their catalytic domains. Upon activation, PARPs cleave NAD+ into nicotinamide and ADP-ribose, which becomes polymerized in the synthesis of long chains of poly(ADP-ribose) (PAR) as a posttranslational modification of target proteins such as histones and, importantly, of PARP itself. PARPs are inactivated by auto-poly(ADP-ribosyl)ation and dissociate from the DNA break. The high electronegative charge enables PAR to compete with DNA for binding to core histones, histone H1, and other proteins associated with DNA, which affords local chromatin decondensation. Because PAR glycohydrolase (PARG) degrades PAR (to ADPR-ribose monomers) shortly after PAR formation, PARP-mediated chromatin decondensation is extremely rapid and transient ((Brochu et al. 1994, Desnoyers et al. 1995, Meyer et al. 2004). Removal of PAR by PARG allows PARP to bind again to remaining breaks and become activated with ensuing auto-modification, which results in repeated cycles of PAR synthesis and degradation that are linked to massive NAD+ consumption. PAR turnover catalyzed by PARPs and PARG is therefore classically viewed as a system providing “histone shuttling” to facilitate local removal of core histones and H1 linker proteins from DNA (Althaus et al. 1993), thereby opening chromatin for remodeling. PAR degradation is strongly compromised in Parg(110)−/− mice (Cortes et al. 2004) and hence PARP1 and 2 remain auto-modified and inactivated longer, which disrupts PARP’s ability to execute rapid auto-modification cycles.
We previously reported that the metabolism of PAR is essential for proper execution of nuclear condensation in spermiogenesis and interfering with it causes poor chromatin condensation with abnormal retention of core histones and histone H1 like linker proteins H1t and HILS1 in mature sperm concomitant with impaired embryonic survival (Meyer-Ficca et al. 2009, Meyer-Ficca et al. 2011). In the present study, we used mouse models of altered PAR metabolism to identify regions of the sperm genome affected by normal and excessive nucleosome retention and to determine effects of elevated sperm histone content on nuclear architecture.
The results demonstrate that the majority of normally histone-associated sperm DNA localizes to distinct nuclear regions in the sperm, i.e. the nuclear periphery and the centrally located sperm chromocenter, pointing towards a structural relevance of retained histones in overall nuclear organization. A large portion of the sperm chromatin that remains nucleosomal is represented by repetitive DNA sequences. Moreover, the data show that formation and condensation of the sperm chromocenter was affected in sperm from animals with impaired PARsylation. The findings presented here indicate that an aberrantly increased histone content in sperm not only affects gene coding regions, which may have epigenetic consequences to the resulting embryo that have yet to be determined, but importantly also impinges upon the overall sperm nuclear architecture with potentially adverse effects on sperm function.
Materials and Methods
Mouse Models
Parp1 gene disrupted mice (Parp1tm1Zqw (Wang et al. 1995) (Parp1−/−), Parg gene disrupted mice (Parg(110)−/−) (Cortes et al. 2004), Parp1 and Parg gene disrupted mice (Parp1−/−/ Parg(110)−/−), as well as wild-type control animals (129S6/SvEvTac) were maintained and used according to the guidelines of the University of Pennsylvania Institutional Animal Care and Use Committee.
Alternative splicing of the Parg gene leads to expression of several PARG isoforms (Meyer-Ficca et al. 2004, Meyer-Ficca et al. 2005b). Parg(110)−/− mice have a targeted deletion of exons 2 and 3 in the Parg gene, so that the gene knock out ablates expression of three large PARG protein isoforms of 110 kDa, 102 kDa, and 98 kDa, but allows for expression of two smaller ones of 63 kDa (ubiquitous) and 58 kDa (mitochondrial), resulting in a 75% reduction of cellular PARG activity (Cortes et al. 2004). Parp1−/− gene disrupted mice are deficient in PARP1 expression, but express PARP2, which has overlapping functions with PARP1. Both enzymes are highly expressed during spermatogenesis (Meyer-Ficca et al. 2009, Dantzer et al. 2006) and simultaneous deletion of both Parp1 and Parp2 genes leads to embryonic lethality (Menissier de Murcia et al. 2003), preventing studies with completely abolished PARP1 and PARP2 activity. Wild-type 129SVE males (129S6/SvEvTac) used in the PARP inhibitor studies were purchased from Taconic Inc. (Hudson, NY).
Mean litter size information was calculated using breeding data collected over a total time frame of six years from the following number of breeding cages (generally containing one male and two females) per genotype: wild type, 19; Parg(110)−/−, 29; Parp1−/−, 24; Parp1−/−Parg(110)−/− double knock out, 18, Parg(110)+/−, 7. For each breeding cage, the average litter size over a breeding time frame of 6 month was determined. The mean values of those average litter sizes were determined for each genotype, and significance analyzes were performed using ANOVA followed by Bonferroni’s Multiple Comparison Correction in GraphPad Prism 5.04.
Embryo / Oocyte collection
In order to compare the fertilization potential of sperm from Parg(110)−/− mice with wild-type sperm, four individual Parg(110)−/− males, and five wild-type control males were naturally mated with at least two virgin wild-type females per test male. One day after a copulation plug was observed (1.5dpc), the female animals were sacrificed, and embryos / oocytes were isolated from the fallopian tubes. Embryo development was determined by microscopic analysis, and successful fertilization was concluded when embryos had undergone cell cleavage to the 2-cell or 4-cell state. Fertilization rates were calculated, and statistical significance was calculated using Fisher exact test in Prism version 5.04 (GraphPad).
PARP inhibitor studies
Wild-type 129SvEv male mice were each treated with PJ34 ([N-(6-Oxo-5,6-dihydro-phenanthridin-2-yl)-N,N-dimethylacetamide / HCl] (Abdelkarim et al. 2001, Mabley et al. 2001, Fonfria et al. 2004, Gambi et al. 2008), (Axxora, San Diego, CA) at a concentration of 10 mg/kg bodyweight (bw) (PJ34 group), or with saline (control group) by daily i.p. injections beginning on days 21 post partum (pp) until collection of sperm on day 80 postpartum. In addition, the PARP inhibitor ABT-888 (Penning et al. 2009) was used in a manner comparable to PJ34 to show validity of the drug approach (Figure S1), and subsequent sperm isolation, slide preparation, chromomycin A3 staining and major satellite fluorescence in situ hybridization was performed as described.
Sperm Isolation and Slide Preparation
Sperm from closely age-matched individuals were isolated by carefully slicing the cauda epididymidis of freshly dissected epididymides several times with scissors and subsequently allowing sperm to swim out in phosphate buffered saline (PBS) (Invitrogen, Carlsbad, CA). Small aliquots of the sperm suspensions were distributed on glass slides, air-dried and stored at −20°C prior to analysis, the remaining sperm were purified of somatic cell contaminations and used in the micrococcal nuclease digestions as described below. Mitotic chromosome spreads from primary murine fibroblasts and embryonic stem cells were prepared according to the procedure described for rat cells (Meyer-Ficca et al. 1998).
Isolation and labeling of micrococcal nuclease digestion sensitive sperm chromatin fractions (MNDS)
Sperm samples were obtained and used to generate separate MNDS fractions from three individual wild-type and from three individual Parg(110)−/− mice as follows. Sperm suspensions were treated with somatic lysis buffer as described (Hammoud et al. 2009). After absence of somatic cells was verified by microscopy, micrococcal nuclease-sensitive fractions were isolated according to a robust and well established method (Arpanahi et al. 2009, Nazarov et al. 2008, Zalenskaya et al. 2000) with minor modifications. Briefly, after somatic cell lysis, sperm were washed twice in PBS (Mediatech, Inc., Manassas, VA; MT21–031-CV) containing 1mM PMSF (Sigma). They were resuspended in PBS containing 1mM PMSF (Sigma) and 10 mM DTT (Roche, Indianapolis, IN) and incubated at 37°C. Calcium, magnesium and micrococcal nuclease (MNase; New England Biolabs, Ipswich, MA) were added to a final concentration of 2 mM, 2 mM, and 0.25U, respectively. The digestion was stopped by addition of EDTA (Sigma) to a final concentration of 10 mM. The supernatant containing the micrococcal nuclease-sensitive soluble fraction was separated from the pellet containing the micrococcal nuclease insoluble fraction by centrifugation (5 min at 1500 x g). The soluble chromatin was precipitated with ethanol, pelleted and purified by gel electrophoresis on 2% agarose gels according to (Zalenskaya et al. 2000). Equal amounts of gel-purified fragments obtained for each individual animal from the identical genotypes (either wild type or Parg(110)−/−) were combined and subsequently digoxigenin-labeled using the amplification steps of the Whole Genome Amplification kit WGA3 (Sigma) supplemented with digoxigenin-labeled dUTP (Roche) as described (Krylov et al. 2010).
Fluorescence in situ hybridization (FISH)
Probes
Cy3-conjugated mouse pan-centromeric probes (Cambio, Cambridge, UK) were used for detecting the pericentromeric located mouse major satellite DNA (GenBank EF028077.1). For marking of telomeres, 5'-Biotin-labeled oligonucleotides complementary to mammalian telomeric sequences (5'-(CCC TAA)6-3' and 5'-(TTA GGG)6-3' (IDT, Coralville, IA) were diluted into a 100ng each / µl stock solution in 1× TE. The telomere probe mixture was diluted to 1ng/ µl in hybridization buffer (10% dextrane sulfate, 30% formamide, deionized, 10% phosphate buffer (160mM Na2HPO4, 60 mM NaH2PO4, pH7.4) (all chemicals from Sigma, St. Louis, MO), 1 µg/µl E. coli DNA (Invitrogen, Carlsbad, CA)) prior to hybridization. Pan-centromeric probe mixture was either mixed 1:1 (vol:vol) with telomere probe mixture (“2-color FISH”) or used alone (“1-color-FISH” for quantitative analyses). For hybridization of the sperm MNDS fractions, digoxigenin labeled MNDS DNA probe consisting of pooled fractions from either three wild type animals or three nine Parg(110)−/− animals was diluted to 10ng/ul in hybridization buffer either with 1.5 µg/µl E. coli DNA to visualize all MNDS signals, or in the presence of 1.5 µg/µl murine Cot DNA (Invitrogen), to quench signals from repetitive DNA elements contained in the MNDS fraction and to visualize the MNDS fraction complementary to unique gene sequences.
Hybridization procedure
FISH on sperm and murine metaphase preparations were performed as previously described (Meyer-Ficca et al. 1998, Scherthan 2009). For sperm in situ hybridizations, the following additional steps were included prior to the hybridization procedure: Sperm slides were incubated in pretreatment solution (4× SSC (Invitrogen), 0.5% Tween 20, 20% Formamide (chemicals from Sigma, St. Louis, MO)0 for 10 min at 95°C, washed briefly in water, and treated with 1M NaSCN for 45min at 73°C. Slides were then denatured and in situ hybridization was performed for 72 h. Nuclear DNA was counterstained with DAPI (Sigma) or SYTOX Green (Invitrogen) and slides were mounted with Vectashield (Vector Laboratories, Burlingame, CA). For volume measurement analyses, slides detected with pan-centromeric probe only were used. Three independent replicates of the hybridization experiments were performed.
Microscopy and Image Analysis
Standard fluorescence microscopy was performed using a Nikon TE2000-U, equipped with monochrome CCD camera (Photometrics Nikon Cool snap) connected to a computer work station using ImagePro Plus version 5.1 software (Media Cybernetics) for quantification and documentation. Qualitative confocal microscopy of two-color FISH samples was performed on a Leica DM 6000 CFS upright microscope equipped with a TCS-SP5 tunable confocal and multiphoton and a Leica SP5-II FLIM inverted microscope, utilizing a HCX PL APO CS 63x oil lens (N.A. = 1.4) system (Leica Microsystems, Wetzlar, Germany), equipped for FITC and Cy3 analysis. DAPI visualization was achieved by multi-photon excitation at 720nm produced by a Chameleon Ultra II Ti:sapphire pulse laser (Coherent, Santa Clara, CA, USA ). Quantitative confocal microscopy of one-color FISH samples was performed on a Leica TCS SP5 system utilizing a HCX PL APO CS 63.0x water lens (N.A. = 1.2). LAS-AF software (Leica) was used to collect Z-stacks of images on both microscopes. Z-stacks of images were processed and analyzed using Volocity® 3D Image analysis software, Version 5.5 (PerkinElmer). In 1-color FISH staining, red signals, representing signals from pan-centromeric probe labeling the pericentromeric heterochromatin, and green signals, representing total nuclear DNA, were detected and measured. Volume measurements were exported into Excel 2003 (Microsoft) for manual sorting.
Measurements resulting from overlapping cells were excluded from the analyses. For each individual cell, the number of individual red signals representing pericentromeric heterochromatin was determined and the ratio of total red object volume to total green object volume representing total nuclear DNA calculated. To validate the automated three-dimensional method utilizing the Volocity program, a comparable analysis was performed by an independent researcher on two-dimensional (2D) images obtained by conventional epifluorescence microscopy of the same samples on a Nikon TE2000-U microscope equipped with a monochrome CCD camera (Photometrics Coolsnap) using the ImageProTM Plus version 5,1.2 software (Media Cybernetics) for documentation. Results obtained with this method were less precise but comparable (data not shown). Two-dimensional images of two-color FISH samples were obtained by compression of z-stacks using the Volocity software.
Statistical Analysis
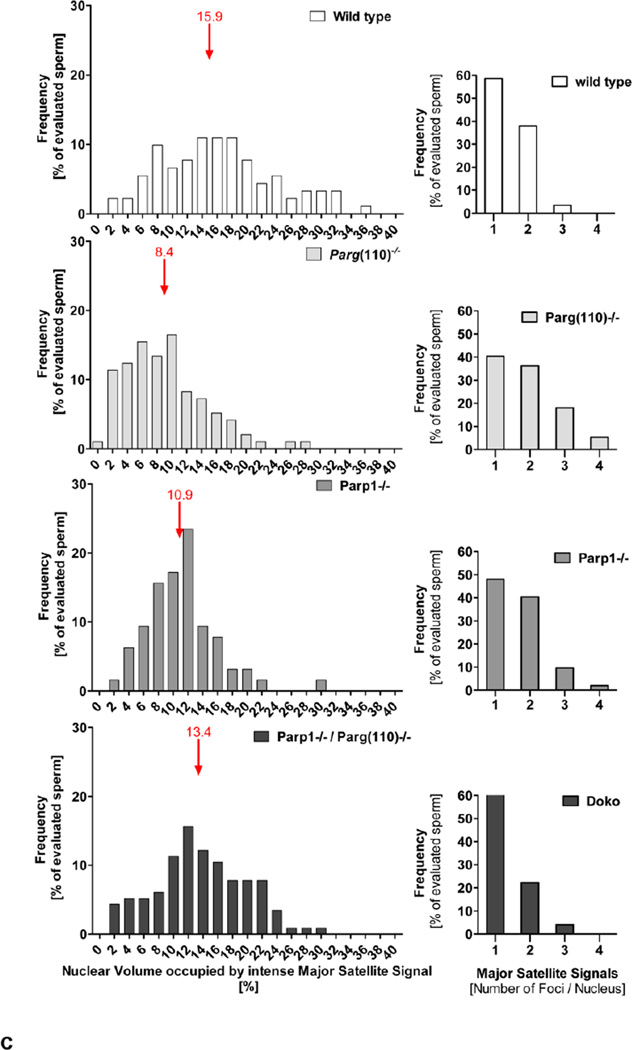
Volume ratios were collected for 158–284 sperm per individual for each of the genotypes and 58–84 sperm per individual from the PARP inhibitor treated and control males, with 3 technical replicates of 2 biological replicates analyzed for each group. Statistical analysis was performed using 1-way-ANOVA with Bonferroni multiple comparison adjustments (for different genotypes), and Mann-Whitney tests with Bonferroni multiple comparison adjustments (PARP inhibitor treated and control samples). Box plots indicate the interquartile range; whiskers indicate the 5–95% percentile (Fig. 4b and Fig. 6b). Frequency distribution analyses of Nuclear Volumes used a bin width of 2% of total nuclear volume (X-axis), and plotted percentage of evaluated sperm (Y-axes) (Fig. 4c and Fig. 6c). Median values are indicated in red. Significance was classified as follows: *, p<0.05–0.01; **, p <0.01, ***, p<0.001, ****, p<0.0001. Percentage of evaluated sperm (Y-axes) was plotted with respect to the number of individual major satellite foci observed (X-axis) ) (Fig. 4c and Fig. 6c).
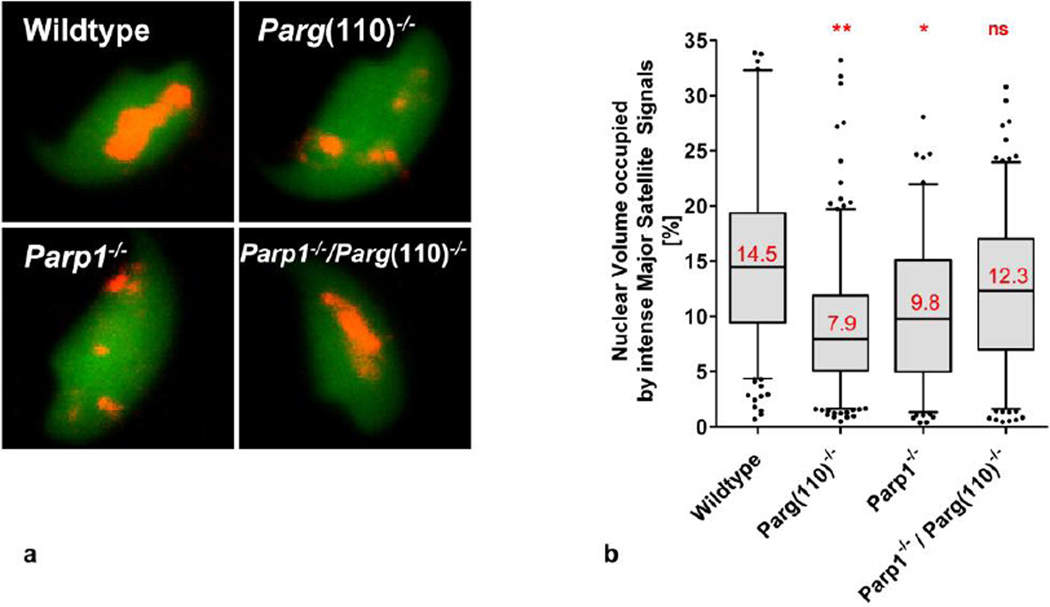
Fig. 4.
Quantitative analysis of abnormal chromocenter condensation in mice with perturbed PAR metabolism. a. Three-dimensional reconstructions of laser scan microscopy z-stack images of individual sperm were used to quantify the percentage of nuclear volume occupied by intensely stained major satellite DNA, i.e. the highly condensed portion of the heterochromatin in the chromocenter that yielded signals above the imaging threshold, as well as the individual number of intense major satellite signals per sperm nucleus. b. Statistical analysis of quantification results demonstrated that sperm from Parp1−/− and Parg(110)−/− animals had a significantly lower degree of condensation of their pericentromeric heterochromatin (n= 252 (wild type), 284 (Parg(110)−/−), 154 (Parp1−/−) and 230 (Parp1−/−Parg(110)−/− double knock out)) individual sperm were analyzed in 3 independent hybridization experiments, significance was determined on the means obtained in 3 individual hybridization experiments, and calculated by 1-way ANOVA with Bonferroni correction for multiple measurements; mean values are indicated in red). c. Graphs on the left side of the panel show frequency distributions of nuclear volumes occupied by intense major satellite signals, i.e. pericentromeric heterochromatin that is condensed above signal detection limit (bin width = 2, position and value of median binned value is indicated by red arrows). The right hand side of the panel illustrates the corresponding frequency distribution of numbers of individual Major Satellite FISH signal per sperm nucleus (n= 52–99 sperm per genotype; the analysis was repeated in three independent experiments; one-way ANOVA followed by Dunnett’s Multiple Comparison Test showed significant differences between results from wild type compared to Parp1−/− or Parg(110)−/−, but not to Parp1−/− / Parg(110)−/− double knock out
Fig. 6.
Quantitative analysis of chromocenter condensation after pharmacological PARP inhibition. Statistical analysis (b) after signal quantification and three dimensional volume analysis on laser scan microscopy images (a) reveals a significant reduction of the median nuclear volume occupied by intense major satellite DNA signals and indicates a lower general degree of chromocenter condensation (I; n=59–87 individually imaged sperm per treatment group and experiment, in 3 independent hybridization experiments, significance was determined on values obtained in 3 individual hybridization experiments, and calculated by students t-test followed by Bonferroni correction for multiple measurements; mean values are indicated in red). c. Frequency distribution of nuclear volume of major satellite FISH signals condensed above signal detection limit per sperm (left panels; bin width = 2, median binned value is indicated by red arrows) (n= 58–87 sperm were analyzed per treatment group; and the analysis was repeated in three independent experiments. Mann Whitney tests indicate significant differences between treatment groups, ****, p<0.0001). Right panel shows the numbers of individual FISH foci visible for major satellite DNA probe in sperm from control and PARP inhibitor (PJ34) treated animals
Chromomycin A3 (CMA3) staining assays
CMA3 assays were performed as described previously (Meyer-Ficca et al. 2011, Bianchi et al. 1996, Singleton et al. 2007). Standard fluorescence microscopy was performed as described above. Fluorescence microphotographs of sperm smear slides stained with DAPI and CMA3 were taken and automated analysis of large numbers of pictures was performed by first measuring nuclear area as defined by DAPI staining, and subsequently quantifying CMA3 fluorescence signal intensity (‘mean signal intensity”) within those areas to eliminate background. CMA3 fluorescence intensity was recorded for each nucleus (n>200 per animal, 2 animals per group). For the table of CMA3 stained sperm, representative monochrome images from in the quantitative analyses are shown.
Statistical analysis of fluorescence intensity values was done using Prism 5 (GraphPad Software, Inc). Box plots indicate the interquartile range; whiskers indicate the 5–95% percentile (Fig. 1i and j). Median values are indicated in red. Significance was tested calculated using Mann-Whitney test followed by Bonferroni correction, and significance classified as follows: *, p<0.05–0.01; **, p <0.01, ***, p<0.001, ****, p<0.0001.
Fig. 1.
Poor sperm chromatin quality in Parg(110)−/− animals or after pharmacological PARP inhibition is characterized by increased CMA3 staining. Representative images of CMA3 and DAPI stained sperm from wild type animals (a, b), Parg(110)−/− animals (c, d), saline control animals (e, f) and PARP inhibitor treated animals (g, h) are shown. Quantitative analyses of CMA3 staining intensity for wild type and Parg(110)−/− sperm (i) and for sperm from long-term saline or PJ34 treated animals are summarized in box plots (j). Box, interquartile range; whiskers, 5–95% percentile. Median values and statistical significance are indicated in red. ***, p<0.001
Results
Perturbation of PAR metabolism compromises completeness of sperm chromatin compaction
Alterations of PAR metabolism either due to reduced PARG activity in the Parg(110)−/− animals (Cortes et al. 2004), or due to pharmacological PARP inhibition using long-term treatments with PJ34 cause poor sperm chromatin condensation characterized by increased CMA3 staining intensity. Figure 1 shows examples of CMA3 stained sperm from Parg(110)−/− animals (Fig. 1a–d) and the respective quantitative analysis (Fig.1, I),. Figure 1e–h and Fig. 1j demonstrates that a comparable effect was observed in sperm from long-term PARP inhibitor treated animals. We have shown previously that this increased CMA3 staining intensity correlates with an increased amount of histones retained in sperm and that the additional deletion of the Parp1 gene in Parp1−/−Parg(110)−/− double knock out sperm in part reverses this chromatin compaction defect (Meyer-Ficca et al. 2011).
A large portion of histone associated DNA is of repetitive nature and is localized in distinct structures of the sperm nucleus
To gain insight into the nature of the DNA that remains predominantly histone associated in sperm, we used partial micrococcal nuclease digestion of purified sperm of wild type or Parg(110)−/− mice to selectively isolate nucleosomal DNA fractions (MNDS DNA). These MNDS DNA fractions were then labeled as probes for fluorescence in situ hybridizations (FISH) to visualize the sperm nuclear regions that these DNA fragments originate from. Hybridization of these probes to wild type sperm in situ demonstrated a preferential localization of the MNDS fractions to the nuclear periphery and the sperm chromocenter. The signals colocalized with murine alpha satellite DNA, a pericentromeric heterochromatin sequence (Fig. 2a–j, arrows), and outlined the nuclear periphery, a preferred location of subtelomeric and telomeric heterochromatin regions in sperm (Fig. 2a–j, arrowheads). The DNA halo that surrounds the sperm nucleus represents chromatin loops extruded from the nucleus due to the swelling step, which is a necessary part of the sperm FISH procedure. It was also recognized by this probe. No obvious general difference was observed between sperm MNDS signals from either wild type or Parg(110)−/− animals.
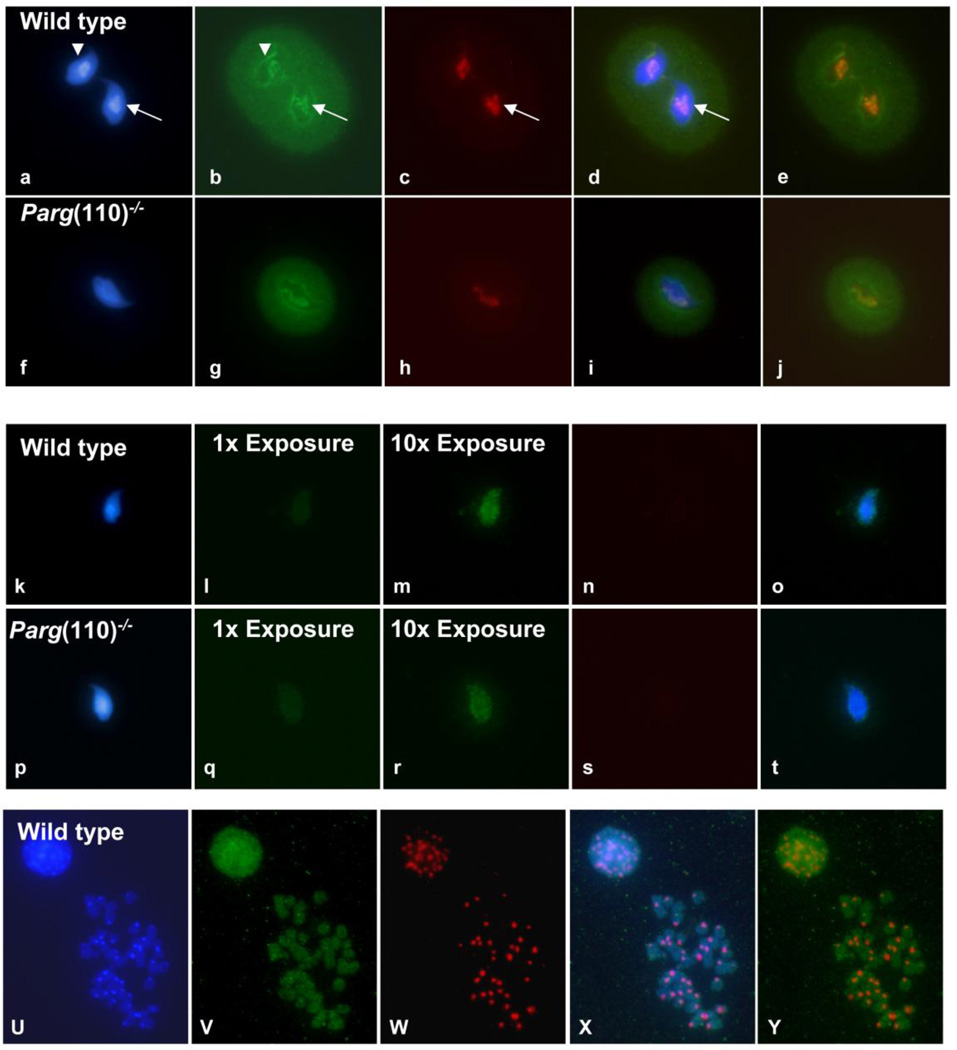
Fig. 2.
Histone associated sperm chromatin is localized at distinct sperm nuclear structures and appears to be enriched in repetitive sequences. (a–j) MNDS fractions were obtained from sperm samples of three individual wild-type and from three Parg(110)−/− animals, and equal amounts of MNDS DNA fragments obtained from all animals of the identical genotype were combined to generate the wild-type and the Parg(110)−/− MNDS fraction, respectively. The pooled wild-type and the pooled Parg(110)−/− MNDS fractions were labeled, and used together with mouse major satellite DNA as probes for FISH experiments onto wild-type mouse sperm only. MNDS fractions were detected as green signals (b, g) and major satellite DNA as red signals (major satellite; c,h), respectively. MNDS signals were prominent along the nuclear periphery (white arrowheads), and a strong colocalization of green MNDS signals with the major satellite positive (red) chromocenter were observed (white arrows). d and i display three color merges of a–c and f–h; e and j displays a merge of red and green only. Suppression of FISH signals from repetitive sequences by addition of an excess of mouse Cot DNA abolished the major satellite signal as expected (n, s), and dramatically diminished the MNDS signals (l, q). At 10 fold overexposure compared to the exposure times used in b, g, l and q, we observed a punctuate pattern of MNDS signals distributed over the sperm nucleus, representing single copy sequences present in the MNDS fraction (m, r, two color merge in o, t). FISH of MNDS fractions isolated from wild type sperm on mitotic metaphase chromosomes yielded homogenous signals covering all chromosome regions (u, DAPI stained DNA; v, green MNDS FISH signals; w, red major satellite signals; x, three color merge; y, merge of green and red signals.) Comparable staining patterns were observed for MNDS probes isolated from Parg(110)−/− (not shown), indicating that the increased histone content in the Parg(110)−/− model does not disturb the general histone placement in sperm.
These results indicate that the MNase digestion procedure appropriately extracted nucleosomal DNA located in the sperm chromocenter as the innermost regions of the sperm nucleus. The intense staining of chromocenter and nuclear periphery confirms that histones are preferentially retained in repetitive, heterochromatic regions of the sperm nucleus. Furthermore the data indicate that the excessive histone retention observed in Parg(110)−/− sperm does not cause a large scale alteration of the general histone localization pattern in sperm, i.e. the preference of sperm histones to be retained at repetitive regions of the chromocenter and the nuclear periphery, but increases the total amount of histones retained within the entire nucleus.
A relatively small portion of the histone associated sperm DNA contains unique genomic sequences
To determine the repetitive DNA portion in the MNDS fraction, hybridizations were repeated in the presence of a 150-fold excess of unlabelled mouse Cot DNA to quench signals generated by repetitive DNA (Fig. 2k–t). Cot DNA is a mixture of DNA fragments containing repetitive sequences. Those sequences are very abundant in the genome, and are enriched for during the Cot preparation procedure by their rapid reannealing rate after heat denaturation. Signals of the murine alpha satellite DNA labeling pericentromeric heterochromatin were completely abolished (Fig. 2n–s), demonstrating effectiveness of the competitive suppression of repetitive sequences by Cot DNA. The fluorescence intensity of the MNDS probe was dramatically reduced by the Cot DNA quenching of repetitive DNA signals, but images taken at a 10fold longer exposure reveal specific punctuate MNDS signals (Fig. 2m and r, merge with nuclear signals in o and t). These punctuate / granular signals covered most of the sperm nucleus, but had a dull or uniform appearance in the region of the chromocenter (Fig.2k and m, arrow). FISH of MNDS fractions on mitotic chromosome spreads yielded a relatively homogenous staining pattern without clear preference for distinct chromosomal regions (Fig. 2u–y).
Taken together these experiments demonstrate that a large portion of sperm histones are associated with repetitive genome portions that occupy very distinct nuclear regions in the sperm nucleus, while a smaller portion is also associated with unique sequences across the genome. The absence of a large increase in fluorescence in the presence of Cot DNA indicates that excessively retained histones in mice with perturbed PAR metabolism are not preferentially associated with non-repetitive sequences.
Impaired formation of a continuous heterochromatic chromocenter in CMA3 positive sperm from Parg(110)−/− animals
To address if abnormalities in sperm histone content result in any changes to the sperm nuclear architecture, we performed two color FISH using a telomeric probe (green) and a mouse major satellite DNA probe (red) to detect telomeric and pericentric heterochromatin, respectively, in sperm from Parg(110)−/− mice, Parp1−/− and Parg and Parp double gene disrupted mice (Parp1−/−/ Parg(110)−/−), and respective wild-type control animals (Fig. 3a–p). The expected localization of probes at the pericentromeric and telomeric chromosome regions was confirmed by FISH on metaphase spreads (Fig. 3q), demonstrating specificity of the probes used. The major satellite probes were visible as bright red signals near the pericentric heterochromatin region of most chromosomes (red signals in Fig. 3q, white arrow). Mouse chromosomes are acrocentric, so that each chromosome has signals for one set of telomeres signals (green) in close proximity to the pericentric chromatin (i.e. the major satellite signal) (white arrow in Fig. 3q), while the other set is at the distal end of the chromosome (arrowhead in Fig. 3q).
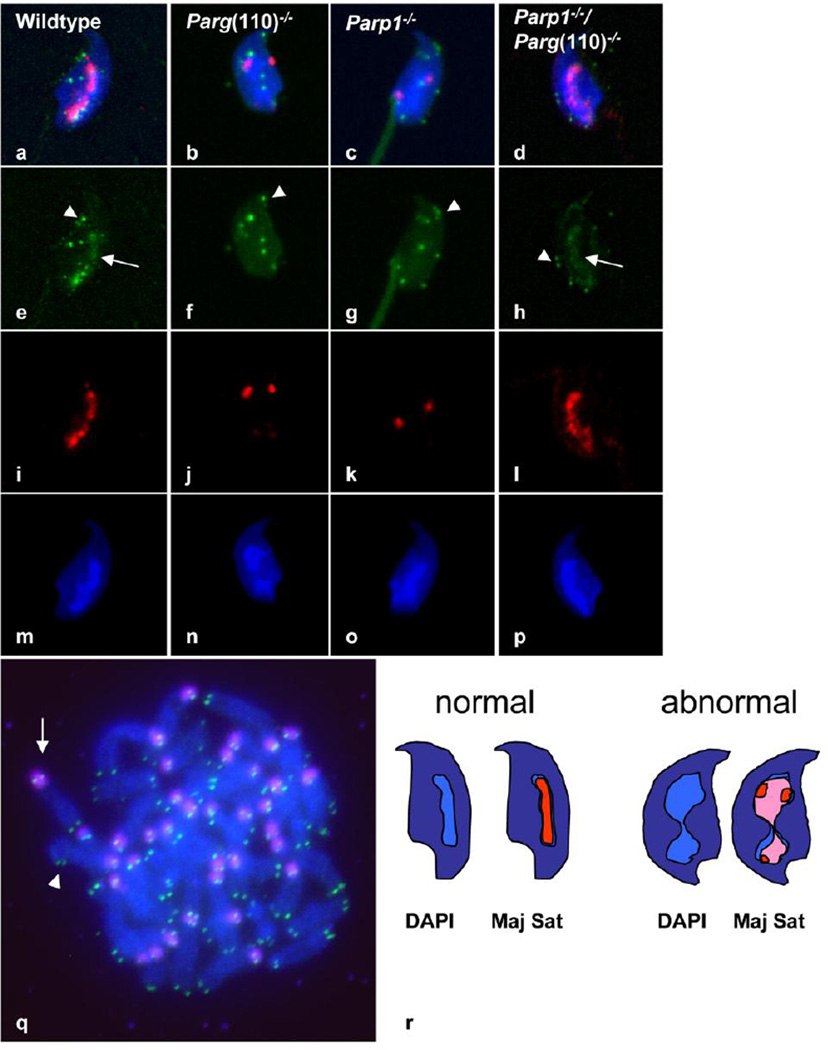
Fig. 3.
Chromocenter condensation is altered in mice with perturbed PAR metabolism. a–q. Typical images obtained by in situ hybridization on sperm obtained from wild type (a, e, I, m) and Parg(110)−/− (b, f, j, n), Parp1−/− (c, g, k, o) and Parp1−/−/ Parg(110)−/− (d, h, l, p) animals with probes that detect the mouse major satellite as markers for pericentromeric chromatin (red) and telomeric repeat sequences (green). Image of a hybridized metaphase spread of mouse chromosomes confirms correct localization of signals for the major satellite (red, arrow in q) and telomeric probes (green, arrowhead in q). Merged images (a–d) and individual images are shown for telomeres signals (e–h), major satellite signals (i–l) and the corresponding DAPI images (m–p) of the respective genotypes. Signals representing telomeric sequences partially colocalized with the chromocenter (e.g. arrow in e and h), and were partially visible near the nuclear periphery in all genotypes (e.g. arrowheads e–h), with no obvious difference between the individual sperm genotypes. Signals representing major satellite DNA colocalized with the DAPI intense chromocenter in the wild type and the Parp1−/−/Parg(110)−/− sperm (I, m and a for wild type; l, p and d for Parp1−/−/Parg(110)−/−). In Parp1−/− and Parg(110)−/− sperm the in situ hybridization signals were disperse and more widespread over the nucleus, and thus yielded weaker signals, as all imaging used identical signal intensity thresholds. r, interpretative illustration visualizing how a lower degree of chromocenter condensation (light blue streak in center of dark blue sperm nucleus in scheme of normal sperm at left) and clustering might be reflected in a lower nuclear volume and an increase in individual number of FISH signals obtained for major satellite DNA (red). The lower degree of chromocenter condensation (hourglass shaped light blue structure) causes a “dilution” of FISH fluorescence signals (pink), so that for a larger portion of the chromocenter the FISH signals remain below the laser scan microscopy detection threshold. The detected “red” signals are smaller in area and appear dispersed
In qualitative laser scan microscopy of sperm preparations, the signals obtained for the major satellite DNA in wild type sperm nuclei typically appeared as continuous or almost continuous centrally located streak (red signals in Fig. 3a and i) that was partially colocalized with the centrally located sperm chromocenter. The telomere signals partially overlapped with the major satellite signals (arrow) (green signals in Fig. 3a and e), and were partially located to the nuclear periphery and adjacent to the position of the nuclear envelope (arrowheads). These observations are consistent with earlier observations of telomeric DNA being peripherally arranged in sperm (Zalensky et al. 1995, Mudrak et al. 2012, Meyer-Ficca et al. 1998), reviewed in (Zalenskaya and Zalensky 2002). The partial colocalization of telomeric signals with the chromocenter, which is composed of pericentromeric heterochromatin, is in line with the murine acrocentric chromosome organization. Telomere signals in sperm from all analyzed knock out genotypes appeared similar to the usual representation seen for wild type (Fig. 3a–d and e–h). Signals for the major satellite probes in wild-type sperm formed a relatively continuous intense streak of signals colocalized with the DAPI-intensive chromocenter. In contrast, Parg(110)−/− and Parp1−/− sperm were marked by distinct bright but dispersed fluorescent signals that did not tend to form a contiguous signal. Overall, they were more diffuse and hence of lower intensity, indicating a disruption of normal centromere clustering. The Parg(110)−/− / Parp1−/− double knock out sperm resembled the wild-type signal morphology consistent with the previous notion that ablation of both, PARP1 and PARG expression provides at least a partial rescue of the individual knock out phenotypes (Meyer-Ficca et al. 2011).
These observations prompted us to further investigate a possible centromeric clustering defect. We analyzed the volume of pericentromeric DNA by quantitative measurement of the individual signals comprising each cluster to determine the total volume occupied by pericentromeric DNA in each single sperm nucleus.
As the amount of major satellite DNA present per sperm genome is expected to be constant in all genotypes, we interpreted the decreased signal intensity with increased signal dispersion as a sign of a lower degree of chromatin condensation and centromere clustering in the chromocenter. We hypothesized that this dispersion causes the same number of fluorescent probe molecules to be distributed over a larger nuclear volume, and consequently a larger portion of the signals will remain below the detection threshold which had to remain constant to allow for comparison of quantitative signals (Principle is illustrated in Fig. 3r). We used three-dimensional reconstruction of laser scan z-stack images obtained from 2-color FISH samples (Fig. 4a) to quantify differences. The volume of whole sperm nuclei was measured, and the percentage of nuclear volume occupied by highly condensed major satellite DNA presenting as bright red signals was calculated. Statistical analysis of values obtained in three independent experiments indicated a significant reduction of the nuclear volume occupied by intense major satellite signals, which represent highly condensed pericentromeric heterochromatin, in Parg(110)−/− and Parp1−/− nuclei compared to wild type sperm (Fig. 4b). The observed shift to a lower degree of chromocenter condensation and simultaneous increase in number of individual signals is also visible in the frequency distribution graphs (Fig. 4c).
Parg(110)−/− mice have highly significantly reduced litter sizes with a mean litter size of 4.38 (±0.23 SEM) pups/litter compared to wild type (6.3 ±0.14), Parp1−/− (5.2 ±0.3) and Parp1−/−Parg(110)−/− double knock out (5.65 ±0.48) and Parg(110)+/− animals (6.7 ±0.4), (significance was determined by ANOVA analysis followed by Bonferroni Multiple Comparison Test, P-value < 0.05). The reduction of litter size in Parp1−/− and Parp1−/−Parg(110)−/− double knock out animals did not reach significance after Bonferroni correction.
To determine if the litter size reduction in Parg(110)−/− animals can be attributed to a male factor, i.e. problems with sperm functionality, age-matched wild type or Parg(110)−/− males were mated to wild type females, and the number of successfully fertilized oocytes that could be retrieved 1.5 days after copulation was determined. At this time point, most of the zygotes sired by wild type males had undergone successful cleavage (39/45; retrieved from matings of 4 males and 9 females), while matings between with Parg(110)−/− males and wild type females resulted in a significantly reduced cleavage rate (27/59; obtained from matings of 5 males and 11 females; p<0.0001 using Fisher’s exact test) (table 1), indicating a reduced fertilization potential of the Parg(110)−/− sperm.
Table 1.
Cleavage rates observed in embryos sired by Parg(110)−/− sperm and wild-type sperm.
| Embryo development 36 h after in vivo mating | ||||
|---|---|---|---|---|
| Genotype of ♂ | Uncleaved | Cleaved | p-value (Fisher’s Test) |
|
| Wild type | 6 | 39 | n = 45 embryos / oocytes, from 4 ♂ and 9 ♀ |
P<0.0001 |
| Parg(110)−/− | 32 | 27 | n = 59 embryos / oocytes, from 5 ♂ and 11 ♀ |
|
Pharmacological PARP inhibition impairs sperm chromocenter condensation
To confirm that alteration of the PAR metabolism interferes with normal chromocenter formation or pericentric heterochromatin condensation, or both, a non-genetic mouse model of PARP inhibition was added to the investigations. In this model, genetically identical, inbred siblings were continuously treated with PJ34, a potent PARP inhibitor, beginning at puberty up to the time of tissue collection. As predicted, PARP inhibitor treatment resulted in a decrease of the volume occupied by bright major satellite FISH signals in sperm from PJ34-treated wild type animals relative to the total nuclear volume, compared to saline-treated controls.
While telomere signals occupied similar positions as those seen in untreated control sperm (Fig. 5a and c and Fig. 5b and d), the major satellite DNA signals were again more likely to occupy dispersed individual positions than one continuous streak characteristic for wild type sperm (Fig. 5a and e, and Fig. 5b and f; and right panels in Fig 6c). Comparable results were also obtained after treating animals with ABT-888, a different and novel PARP inhibitor (Fig. S1).
Fig. 5.
Pharmacological PARP inhibition changes heterochromatin condensation in the sperm chromocenter, similar to the genetic mouse models. (a – h) In situ hybridization was performed on sperm obtained from long-term PARP inhibitor treated and control wild type animals with probes detecting the mouse major satellite (red; e, f) and telomeric repeat sequences (green; c, d). No obvious difference was observed for telomeric signal distribution between the treatments (c, d). Signals representing the major satellite DNA in control treated animals mostly appeared as relatively continuous string of signals (e), while in PARP inhibitor treated sperm the signals often appeared more dispersed and weaker (f)
The quantitative analysis of 2-color FISH samples (Fig. 6 a) showed that intensely fluorescent major satellite DNA signals characteristic for the condensed chromocenter occupied a significantly smaller portion of the nuclear volume (Fig. 6b), similar to the previous observation in Parg(110)−/− animals. The frequency distribution shows that a high percentage of individual sperm in the analyzed population was affected by this phenomenon (Fig 6c).
Discussion
It is becoming increasingly clear that perturbation of the well-orchestrated nuclear reorganization in spermiogenesis may have adverse, even detrimental effects on offspring. Abnormal sperm nuclear structure and poor chromatin quality have been shown to be of direct clinical relevance due to their implications of causing infertility and recurrent pregnancy loss (D'Occhio et al. 2007, Dada et al. 2012, Jenkins and Carrell 2012). A number of genes that are required for correct morphogenesis of the sperm nucleus have already been be identified through studies of fertility defects in knock out mouse models (Zhao et al. 2004, Shirley et al. 2004, Zhao et al. 2001), but our current knowledge base is by far still not able to explain, treat or prevent sperm chromatin defects associated with male factor infertility. As outlined above, mature sperm retain only a small fraction of core histones bound to DNA encoding specific genes involved in embryo development, raising the possibility that these retained histones regulate gene expression after fertilization (Hammoud et al. 2009, Arpanahi et al. 2009, Gatewood et al. 1987, Gardiner-Garden et al. 1998, Kramer and Krawetz 1996, Brykczynska et al. 2010). Abnormal histone retention could therefore perturb epigenetic mechanisms involving regulatory histone marks and ensuing abnormal de- and re-methylation of DNA in the zygote (Vavouri and Lehner 2011), thereby contributing to the observed frequent embryonic failure in Parg(110)−/− mice. However, when studying the epigenetic influence of sperm histones, it has been widely overlooked that earlier studies have demonstrated that heterochromatic genome regions, which are known to be enriched in repetitive sequences, also remain preferentially nucleosomal in sperm nuclei (Zalenskaya et al. 2000, Pittoggi et al. 1999) and most investigations have been focused on histones retained in gene coding chromatin domains. One of the reasons for this is that microarray tiling arrays and high throughput sequencing selectively exclude the analysis of repetitive DNA for practical reasons. They rely on suppression of repetitive sequences using either Cot DNA (microarrays) or mapping of sequences to assembled genome builds. In the case of repetitive sequences, the latter remains to be computationally challenging (Treangen and Salzberg 2011), and sequence information of repetitive regions, e.g. in pericentromeric heterochromatin genomes is still simply not available, precluding correct mapping of most highly repetitive DNA.
Nucleosomal organization may provide additional, perhaps structural functions and thus may provide additional layers of epigenetic information contained in sperm. Besides histone versus protamine association at the chromatin level, distinct matrix attachment of defined regions and the sperm specific spatial chromosome arrangement likely constitute an important epigenetic “user manual” at the more global nuclear level that may be crucial for proper genome unpacking and utilization in the zygote and thus successful embryonic development (Zalensky and Zalenskaya 2007, Miller et al. 2010, Ward 2010).
In addition to the variety of known defects of spermatogenesis caused by genetic mutation, environmental effects due to exposure to adverse situations and substances are of increasing interest, particularly in the light of a growing awareness of connections between pollution and sperm defects (Sanchez-Pena et al. 2004, Rubes et al. 2005), as well as the unexplained and ongoing deterioration of semen quality observed in several developed countries (Rolland et al. 2013, Almagor et al. 2003). Recent evidence suggests that exposure of adult men to environmental or medical toxicants is able to disrupt the paternal epigenome (Miller et al. 2009, Godmann et al. 2009). PAR modifications (and hence PAR metabolism) are a central locus of regulation for critical biological processes that involve chromatin remodeling (Malanga and Althaus 2005, Althaus et al. 1994, Wacker et al. 2007, Tulin et al. 2003). PAR is generated by the ubiquitous PAR polymerases 1 and 2 (PARP1, PARP2) using NAD+ as a substrate PAR metabolism is emerging as a cellular gauge of environmental stress factors due to (i) its dependence on NAD+ as the substrate, whose synthesis in humans is limited by dietary uptake of vitamin B3, (ii) its cross-talk with sirtuins, which are NAD+-dependent histone deacetylases, (iii) its critical roles in DNA repair, inflammation and ageing, and (iv) the functions of PARP1 as cofactor of NFkappaB-, steroid receptor- and hypoxia-inducible transcription (Meyer-Ficca et al. 2005b, Malanga and Althaus 2005, Kim et al. 2005, Bai and Canto 2012, Hassa and Hottiger 2008, Elser et al. 2008, Altmeyer and Hottiger 2009, Erener et al. 2012). Taken together, PAR metabolism is currently emerging as a gauge that is capable of integrating environmental exposures with appropriate cellular responses and epigenetic modifications (Luo and Kraus 2012, Luo and Kraus 2011).
Thus, PARPs are aptly positioned to mediate certain epigenetic changes in the male germ line in response to the environment. The presence of physiologically active PARP enzyme during male germ cell development has been described for several species (Meyer-Ficca et al. 2005a, Quesada et al. 1996, Corominas and Mezquita 1985, Maymon et al. 2006). Chromatin remodeling steps during spermiogenesis are characterized by the presence of transient, likely topoisomerase II mediated physiological DNA strand breaks that trigger PAR formation and link DNA relaxation to histone removal during nucleoprotein transition to protaminated DNA (Smith and Haaf 1998, Meyer-Ficca et al. 2005a, Leduc et al. 2008). Genetically or pharmacologically impaired PARsylation during mouse spermiogenesis causes increased histone retention and reduced sperm chromatin condensation (Meyer-Ficca et al. 2009, Meyer-Ficca et al. 2011, Quenet et al. 2009).
The defects in sperm from animals treated with PARP inhibitor PJ34 resemble the phenotype observed in the Parg(110)−/− knock out, while the Parp1−/−Parg(110)−/− double knock out sperm appear to have relatively normal sperm nuclear architecture (this study) and sperm chromatin composition (Meyer-Ficca et al. 2011). Two seemingly opposite situations, i.e. reduced PAR levels following direct pharmacological PARP inhibition in the PJ34 treated animals, and increased PAR levels in the Parg(110)−/− genetic model due to significantly reduced enzymatic PAR degrading activity of the residual PARG protein, cause comparable defects, while a combination of both in the Parp1−/−Parg(110)−/− double knock out animals reverse the observed phenotypes. This counterintuitive situation can be explained by the importance of proper metabolic PAR turnover (i.e. the necessity for cells to synthesize PAR and, equally important, to degrade PAR rapidly), which depends on a proper balance of PARP and PARG activity. In the Parg(110)−/− knock out mice, normal PARP activity combined with reduced PARG activity leads to PAR accumulation and traps PARP1 in an automodified (PARsylated) state that is enzymatically inactive. Reduced PARG activity thus indirectly inhibits PARP activity similar to direct pharmacological PARP inhibition using PJ34, explaining the observed comparable phenotypes.
In Parp1−/− knock animals, PARP1 enzyme as the major cellular PARP is absent, but PARP2 serves as a back up enzyme that provides PARP activity albeit at a reduced level. Interestingly, combining the reduced ability to synthesize PAR (in the Parp1−/−) with the reduced ability to degrade PAR (in the Parg(110)−/−) in the double knock out animals (Parp1−/−Parg(110)−/−) appears to restore the required balance of PARP/PARG activity, and prevents the defects observed in PJ34 treated and in Parg(110)−/− animals, indicating that not absolute PAR levels, but rather proper turnover of PAR are the determining factor for proper nuclear organization and chromatin organization. In summary, our data presented here, in combination with previously reported findings of male subfertility in mice with disrupted PAR metabolism (Meyer-Ficca et al. 2011), therefore also suggest that environmental factors can have an impact on sperm nuclear architecture, contributing to an unknown extent to the alteration of epigenetic information in sperm. PARP inhibitors, similar to PJ34 which has been used here to simulate inhibition by naturally occurring endogenous and exogenous substances (Banasik et al. 2012), are now being clinically tested to treat cancer, stroke, heart infarction and chronic inflammation (Ratnam and Low 2007, Curtin 2005, Graziani and Szabo 2005, Phillips et al. 2009, Bedikian et al. 2009), adding some translational relevance to the work presented here.
Supplementary Material
Fig. S1 Verification of pharmacological PARP inhibitor approach using ABT-888. ABT-888 treatment results in poor sperm chromatin quality, as observed by increased sperm CMA3 staining after pharmacological PARP inhibition using ABT-888. Representative images of CMA3 and DAPI stained sperm from saline treated animals (a, b), and PARP inhibitor ABT-888 treated animals (c, d) are shown. Quantitative analyses of CMA3 staining intensity for sperm from long-term saline or ABT-888 treated animals are shown in box plot (e; Box, interquartile range; whiskers, 5–95% percentile. Median values obtained for sperm from n=4 animals per treatment group, and 11–60 sperm per animals. Statistical significance calculated using Mann-Whitney test is indicated in red. **,p<0.01).
PARP inhibition using ABT-888 changes heterochromatin condensation in the sperm chromocenter (similar to observation in Fig. 3–6). (f-k) Representative images of signals obtained by In situ hybridization with mouse major satellite DNA probes on sperm obtained from long-term PARP inhibitor ABT-888 treated and saline treated control wildtype animals. Mouse major satellite signal are shown in j and k (red signals), total nuclear DNA was counter-stained with DAPI (blue, h, i), and merges are visible in f and g. Signals representing the major satellite DNA in control treated animals mostly appeared as relatively continuous string of signals (j, merge with image of DAPI stained image in f), while the signals often appeared more dispersed and weaker in sperm from ABT-888 treated animals (k, merge with DAPI image in g). Frequency distribution of the number of individual FISH foci visible for major satellite DNA probe in sperm from control and PARP inhibitor (ABT-888) treated animals indicates increased incidence of unclustered chromocenter (l).
Acknowledgements
This work was supported by grants from the National Institutes of Health (NIH R01HD048837 and U54HD068157) and the Mari Lowe Center for Comparative Oncology at the University of Pennsylvania. We are grateful to Dr. Lingli Zhang and Dr. Gordon Ruthel (University of Pennsylvania, School of Veterinary Medicine Imaging Facility) for help with confocal microscopy, and to Dr. Larry L Laster and to the University of Pennsylvania’s Center for Clinical Epidemiology and Biostatistics for help with statistical analyses.
References
- Abdelkarim GE, Gertz K, Harms C, Katchanov J, Dirnagl U, Szabo C, Endres M. Protective effects of PJ34, a novel, potent inhibitor of poly(ADP-ribose) polymerase (PARP) in in vitro and in vivo models of stroke. Int J Mol Med. 2001;7:255–260. [PubMed] [Google Scholar]
- Agarwal A, Said TM. Role of sperm chromatin abnormalities and DNA damage in male infertility. Hum Reprod Update. 2003;9:331–345. doi: 10.1093/humupd/dmg027. [DOI] [PubMed] [Google Scholar]
- Almagor M, Ivnitzki I, Yaffe H, Baras M. Changes in semen quality in Jerusalem between 1990 and 2000: a cross-sectional and longitudinal study. Arch Androl. 2003;49:139–144. doi: 10.1080/01485010390129296. [DOI] [PubMed] [Google Scholar]
- Althaus FR, Hofferer L, Kleczkowska HE, Malanga M, Naegeli H, Panzeter P, Realini C. Histone shuttle driven by the automodification cycle of poly(ADP-ribose)polymerase. Environ Mol Mutagen. 1993;22:278–282. doi: 10.1002/em.2850220417. [DOI] [PubMed] [Google Scholar]
- Althaus FR, Hofferer L, Kleczkowska HE, Malanga M, Naegeli H, Panzeter PL, Realini CA. Histone shuttling by poly ADP-ribosylation. Mol Cell Biochem. 1994;138:53–59. doi: 10.1007/BF00928443. [DOI] [PubMed] [Google Scholar]
- Altmeyer M, Hottiger MO. Poly(ADP-ribose) polymerase 1 at the crossroad of metabolic stress and inflammation in aging. Aging (Albany NY) 2009;1:458–469. doi: 10.18632/aging.100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 2009;19:1338–1349. doi: 10.1101/gr.094953.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Canto C. The role of PARP-1 and PARP-2 enzymes in metabolic regulation and disease. Cell Metab. 2012;16:290–295. doi: 10.1016/j.cmet.2012.06.016. [DOI] [PubMed] [Google Scholar]
- Balhorn R. The protamine family of sperm nuclear proteins. Genome Biol. 2007;8:227. doi: 10.1186/gb-2007-8-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banasik M, Stedeford T, Strosznajder RP. Natural inhibitors of poly(ADP-ribose) polymerase-1. Mol Neurobiol. 2012;46:55–63. doi: 10.1007/s12035-012-8257-x. [DOI] [PubMed] [Google Scholar]
- Bedikian AY, Papadopoulos NE, Kim KB, Hwu WJ, Homsi J, Glass MR, Cain S, Rudewicz P, Vernillet L, Hwu P. A phase IB trial of intravenous INO-1001 plus oral temozolomide in subjects with unresectable stage-III or IV melanoma. Cancer Invest. 2009;27:756–763. doi: 10.1080/07357900802709159. [DOI] [PubMed] [Google Scholar]
- Bianchi PG, Manicardi G, Bizzaro D, Campana A, Bianchi U, Sakkas D. Use of the guanine-cytosine (GC) specific fluorochrome, chromomycin A3, as an indicator of poor sperm morphology. J Assist Reprod Genet. 1996;13:246–250. doi: 10.1007/BF02065944. [DOI] [PubMed] [Google Scholar]
- Braun RE. Packaging paternal chromosomes with protamine. Nat Genet. 2001;28:10–12. doi: 10.1038/ng0501-10. [DOI] [PubMed] [Google Scholar]
- Brochu G, Duchaine C, Thibeault L, Lagueux J, Shah GM, Poirier GG. Mode of action of poly(ADP-ribose) glycohydrolase. Biochim Biophys Acta. 1994;1219:342–350. doi: 10.1016/0167-4781(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- Carrell DT, Emery BR, Hammoud S. The aetiology of sperm protamine abnormalities and their potential impact on the sperm epigenome. Int J Androl. 2008;31:537–545. doi: 10.1111/j.1365-2605.2008.00872.x. [DOI] [PubMed] [Google Scholar]
- Corominas M, Mezquita C. Poly(ADP-ribosylation) at successive stages of rooster spermatogenesis. Levels of polymeric ADP-ribose in vivo and poly(ADP-ribose) polymerase activity and turnover of ADP-ribosyl residues in vitro. J Biol Chem. 1985;260:16269–16273. [PubMed] [Google Scholar]
- Cortes U, Tong WM, Coyle DL, Meyer-Ficca ML, Meyer RG, Petrilli V, Herceg Z, Jacobson EL, Jacobson MK, Wang ZQ. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol Cell Biol. 2004;24:7163–7178. doi: 10.1128/MCB.24.16.7163-7178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin NJ. PARP inhibitors for cancer therapy. Expert Rev Mol Med. 2005;7:1–20. doi: 10.1017/S146239940500904X. [DOI] [PubMed] [Google Scholar]
- Dada R, Kumar M, Jesudasan R, Fernandez JL, Gosalvez J, Agarwal A. Epigenetics and its role in male infertility. J Assist Reprod Genet. 2012;29:213–223. doi: 10.1007/s10815-012-9715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer F, Mark M, Quenet D, et al. Poly(ADP-ribose) polymerase-2 contributes to the fidelity of male meiosis I and spermiogenesis. Proc Natl Acad Sci U S A. 2006;103:14854–14859. doi: 10.1073/pnas.0604252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers S, Shah GM, Brochu G, Hoflack JC, Verreault A, Poirier GG. Biochemical properties and function of poly(ADP-ribose) glycohydrolase. Biochimie. 1995;77:433–438. doi: 10.1016/0300-9084(96)88156-9. [DOI] [PubMed] [Google Scholar]
- D'Occhio MJ, Hengstberger KJ, Johnston SD. Biology of sperm chromatin structure and relationship to male fertility and embryonic survival. Anim Reprod Sci. 2007;101:1–17. doi: 10.1016/j.anireprosci.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Elser M, Borsig L, Hassa PO, Erener S, Messner S, Valovka T, Keller S, Gassmann M, Hottiger MO. Poly(ADP-ribose) polymerase 1 promotes tumor cell survival by coactivating hypoxia-inducible factor-1-dependent gene expression. Mol Cancer Res. 2008;6:282–290. doi: 10.1158/1541-7786.MCR-07-0377. [DOI] [PubMed] [Google Scholar]
- Erener S, Hesse M, Kostadinova R, Hottiger MO. Poly(ADP-ribose)polymerase-1 (PARP1) controls adipogenic gene expression and adipocyte function. Mol Endocrinol. 2012;26:79–86. doi: 10.1210/me.2011-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfria E, Marshall IC, Benham CD, Boyfield I, Brown JD, Hill K, Hughes JP, Skaper SD, McNulty S. TRPM2 channel opening in response to oxidative stress is dependent on activation of poly(ADP-ribose) polymerase. Br J Pharmacol. 2004;143:186–192. doi: 10.1038/sj.bjp.0705914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambi N, Tramontano F, Quesada P. Poly(ADPR)polymerase inhibition and apoptosis induction in cDDP-treated human carcinoma cell lines. Biochem Pharmacol. 2008;75:2356–2363. doi: 10.1016/j.bcp.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Ballesteros M, Gordon M, Tam PP. Histone- and protamine-DNA association: conservation of different patterns within the beta-globin domain in human sperm. Mol Cell Biol. 1998;18:3350–3356. doi: 10.1128/mcb.18.6.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatewood JM, Cook GR, Balhorn R, Bradbury EM, Schmid CW. Sequence-specific packaging of DNA in human sperm chromatin. Science. 1987;236:962–964. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- Godmann M, Lambrot R, Kimmins S. The dynamic epigenetic program in male germ cells: Its role in spermatogenesis, testis cancer, and its response to the environment. Microsc Res Tech. 2009;72:603–619. doi: 10.1002/jemt.20715. [DOI] [PubMed] [Google Scholar]
- Graziani G, Szabo C. Clinical perspectives of PARP inhibitors. Pharmacol Res. 2005;52:109–118. doi: 10.1016/j.phrs.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Gusse M, Sautiere P, Belaiche D, Martinage A, Roux C, Dadoune JP, Chevaillier P. Purification and characterization of nuclear basic proteins of human sperm. Biochim Biophys Acta. 1986;884:124–134. doi: 10.1016/0304-4165(86)90235-7. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- Jenkins TG, Carrell DT. The sperm epigenome and potential implications for the developing embryo. Reproduction. 2012;143:727–734. doi: 10.1530/REP-11-0450. [DOI] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: 'PAR-laying' NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- Kimmins S, Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature. 2005;434:583–589. doi: 10.1038/nature03368. [DOI] [PubMed] [Google Scholar]
- Kramer JA, Krawetz SA. Nuclear matrix interactions within the sperm genome. J Biol Chem. 1996;271:11619–11622. doi: 10.1074/jbc.271.20.11619. [DOI] [PubMed] [Google Scholar]
- Krylov V, Kubickova S, Rubes J, Macha J, Tlapakova T, Seifertova E, Sebkova N. Preparation of Xenopus tropicalis whole chromosome painting probes using laser microdissection and reconstruction of X. laevis tetraploid karyotype by Zoo-FISH. Chromosome Res. 2010;18:431–439. doi: 10.1007/s10577-010-9127-x. [DOI] [PubMed] [Google Scholar]
- Laberge RM, Boissonneault G. On the nature and origin of DNA strand breaks in elongating spermatids. Biol Reprod. 2005;73:289–296. doi: 10.1095/biolreprod.104.036939. [DOI] [PubMed] [Google Scholar]
- Leduc F, Maquennehan V, Nkoma GB, Boissonneault G. DNA damage response during chromatin remodeling in elongating spermatids of mice. Biol Reprod. 2008;78:324–332. doi: 10.1095/biolreprod.107.064162. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Abbott DW, Ausio J. A haploid affair: core histone transitions during spermatogenesis. Biochem Cell Biol. 2003;81:131–140. doi: 10.1139/o03-045. [DOI] [PubMed] [Google Scholar]
- Luo X, Kraus WL. A one and a two … expanding roles for poly(ADP-ribose) polymerases in metabolism. Cell Metab. 2011;13:353–355. doi: 10.1016/j.cmet.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabley JG, Jagtap P, Perretti M, et al. Anti-inflammatory effects of a novel, potent inhibitor of poly (ADP-ribose) polymerase. Inflamm Res. 2001;50:561–569. doi: 10.1007/PL00000234. [DOI] [PubMed] [Google Scholar]
- Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- Marcon L, Boissonneault G. Transient DNA strand breaks during mouse and human spermiogenesis new insights in stage specificity and link to chromatin remodeling. Biol Reprod. 2004;70:910–918. doi: 10.1095/biolreprod.103.022541. [DOI] [PubMed] [Google Scholar]
- Maymon BB, Cohen-Armon M, Yavetz H, Yogev L, Lifschitz-Mercer B, Kleiman SE, Botchan A, Hauser R, Paz G. Role of poly(ADP-ribosyl)ation during human spermatogenesis. Fertil Steril. 2006;86:1402–1407. doi: 10.1016/j.fertnstert.2006.03.063. [DOI] [PubMed] [Google Scholar]
- McPherson SM, Longo FJ. Nicking of rat spermatid and spermatozoa DNA: possible involvement of DNA topoisomerase II. Dev Biol. 1993;158:122–130. doi: 10.1006/dbio.1993.1173. [DOI] [PubMed] [Google Scholar]
- Menissier de Murcia J, Ricoul M, Tartier L, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RG, Meyer-Ficca ML, Jacobson EL, Jacobson MK. Enzymes in poly(ADP-ribose) metabolism. In: Burkle A, editor. Poly(ADP-ribosyl)ation. Landes Bioscience. 2004. [Google Scholar]
- Meyer-Ficca M, Muller-Navia J, Scherthan H. Clustering of pericentromeres initiates in step 9 of spermiogenesis of the rat (Rattus norvegicus) and contributes to a well defined genome architecture in the sperm nucleus. J Cell Sci. 1998;111(10):1363–1370. doi: 10.1242/jcs.111.10.1363. [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Scherthan H, Burkle A, Meyer RG. Poly(ADP-ribosyl)ation during chromatin remodeling steps in rat spermiogenesis. Chromosoma. 2005a;114:67–74. doi: 10.1007/s00412-005-0344-6. [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Meyer RG, Jacobson EL, Jacobson MK. Poly(ADP-ribose) polymerases: managing genome stability. Int J Biochem Cell Biol. 2005b;37:920–926. doi: 10.1016/j.biocel.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp Cell Res. 2004;297:521–532. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Lonchar JD, Ihara M, Meistrich ML, Austin CA, Meyer RG. Poly(ADP-ribose) polymerases PARP1 and PARP2 modulate topoisomerase II beta (TOP2B) function during chromatin condensation in mouse spermiogenesis. Biol Reprod. 2011;84:900–909. doi: 10.1095/biolreprod.110.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Lonchar J, Credidio C, Ihara M, Li Y, Wang ZQ, Meyer RG. Disruption of Poly(ADP-Ribose) Homeostasis Affects Spermiogenesis and Sperm Chromatin Integrity in Mice. Biol Reprod. 2009;81:46–55. doi: 10.1095/biolreprod.108.075390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Ihara M, Lonchar JD, Meistrich ML, Austin CA, Min W, Wang ZQ, Meyer RG. Poly(ADP-ribose) Metabolism Is Essential for Proper Nucleoprotein Exchange During Mouse Spermiogenesis. Biol Reprod. 2011;84:218–228. doi: 10.1095/biolreprod.110.087361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D, Brinkworth M, Iles D. Paternal DNA packaging in spermatozoa: more than the sum of its parts? Reproduction. 2009 doi: 10.1530/REP-09-0281. [DOI] [PubMed] [Google Scholar]
- Miller D, Brinkworth M, Iles D. Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction. 2010;139:287–301. doi: 10.1530/REP-09-0281. [DOI] [PubMed] [Google Scholar]
- Mudrak OS, Nazarov I B, Jones EL, Zalensky AO. Positioning of chromosomes in human spermatozoa is determined by ordered centromere arrangement. PLoS One. 2012;7:e52944. doi: 10.1371/journal.pone.0052944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarov IB, Shlyakhtenko LS, Lyubchenko YL, Zalenskaya IA, Zalensky AO. Sperm chromatin released by nucleases. Syst Biol Reprod Med. 2008;54:37–46. doi: 10.1080/19396360701876849. [DOI] [PubMed] [Google Scholar]
- Penning TD, Zhu GD, Gandhi VB, et al. Discovery of the Poly(ADP-ribose) polymerase (PARP) inhibitor 2-[(R)-2-methylpyrrolidin-2-yl]-1H-benzimidazole-4-carboxamide (ABT-888) for the treatment of cancer. J Med Chem. 2009;52:514–523. doi: 10.1021/jm801171j. [DOI] [PubMed] [Google Scholar]
- Phillips LR, Hill KD, Majerova E. Liquid Chromatographic Determination of NSC 737664 (ABT-888: an Inhibitor of Poly(ADP-ribose) Polymerase (PARP)) in Plasma and Urine in a Phase 0 Clinical Trial. J Liq Chromatogr Relat Technol. 2009;32:261–272. doi: 10.1080/10826070802603351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittoggi C, Renzi L, Zaccagnini G, Cimini D, Degrassi F, Giordano R, Magnano AR, Lorenzini R, Lavia P, Spadafora C. A fraction of mouse sperm chromatin is organized in nucleosomal hypersensitive domains enriched in retroposon DNA. J Cell Sci. 1999;112(20):3537–3548. doi: 10.1242/jcs.112.20.3537. [DOI] [PubMed] [Google Scholar]
- Quenet D, Mark M, Govin J, van Dorsselear A, Schreiber V, Khochbin S, Dantzer F. Parp2 is required for the differentiation of post-meiotic germ cells: identification of a spermatid-specific complex containing Parp1, Parp2, TP2 and HSPA2. Exp Cell Res. 2009;315:2824–2834. doi: 10.1016/j.yexcr.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Quesada P, Atorino L, Cardone A, Ciarcia G, Farina B. Poly(ADPribosyl)ation system in rat germinal cells at different stages of differentiation. Exp Cell Res. 1996;226:183–190. doi: 10.1006/excr.1996.0217. [DOI] [PubMed] [Google Scholar]
- Ratnam K, Low JA. Current development of clinical inhibitors of poly(ADP-ribose) polymerase in oncology. Clin Cancer Res. 2007;13:1383–1388. doi: 10.1158/1078-0432.CCR-06-2260. [DOI] [PubMed] [Google Scholar]
- Risley MS, Einheber S, Bumcrot DA. Changes in DNA topology during spermatogenesis. Chromosoma. 1986;94:217–227. doi: 10.1007/BF00288496. [DOI] [PubMed] [Google Scholar]
- Rolland M, Le Moal J, Wagner V, Royere D, De Mouzon J. Decline in semen concentration and morphology in a sample of 26 609 men close to general population between 1989 and 2005 in France. Hum Reprod. 2013;28:462–470. doi: 10.1093/humrep/des415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseaux S, Faure AK, Caron C, Lestrat C, Govin J, Hennebicq S, Sele B, Khochbin S. Organizing the sperm nucleus. Gynecol Obstet Fertil. 2004;32:785–791. doi: 10.1016/j.gyobfe.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Rubes J, Selevan SG, Evenson DP, Zudova D, Vozdova M, Zudova Z, Robbins WA, Perreault SD. Episodic air pollution is associated with increased DNA fragmentation in human sperm without other changes in semen quality. Hum Reprod. 2005;20:2776–2783. doi: 10.1093/humrep/dei122. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pena LC, Reyes BE, Lopez-Carrillo L, Recio R, Moran-Martinez J, Cebrian ME, Quintanilla-Vega B. Organophosphorous pesticide exposure alters sperm chromatin structure in Mexican agricultural workers. Toxicol Appl Pharmacol. 2004;196:108–113. doi: 10.1016/j.taap.2003.11.023. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- Scherthan H. Analysis of telomere dynamics in mouse spermatogenesis. Methods Mol Biol. 2009;558:383–399. doi: 10.1007/978-1-60761-103-5_22. [DOI] [PubMed] [Google Scholar]
- Shaman JA, Yamauchi Y, Ward WS. Function of the sperm nuclear matrix. Arch Androl. 2007;53:135–140. doi: 10.1080/01485010701329378. [DOI] [PubMed] [Google Scholar]
- Shirley CR, Hayashi S, Mounsey S, Yanagimachi R, Meistrich ML. Abnormalities and reduced reproductive potential of sperm from Tnp1- and Tnp2-null double mutant mice. Biol Reprod. 2004;71:1220–1229. doi: 10.1095/biolreprod.104.029363. [DOI] [PubMed] [Google Scholar]
- Singleton S, Zalensky A, Doncel GF, Morshedi M, Zalenskaya IA. Testis/sperm-specific histone 2B in the sperm of donors and subfertile patients: variability and relation to chromatin packaging. Hum Reprod. 2007;22:743–750. doi: 10.1093/humrep/del439. [DOI] [PubMed] [Google Scholar]
- Smith A, Haaf T. DNA nicks and increased sensitivity of DNA to fluorescence in situ end labeling during functional spermiogenesis. BioTechniques. 1998;25:496–502. doi: 10.2144/98253rr05. [DOI] [PubMed] [Google Scholar]
- Spano M, Seli E, Bizzaro D, Manicardi GC, Sakkas D. The significance of sperm nuclear DNA strand breaks on reproductive outcome. Curr Opin Obstet Gynecol. 2005;17:255–260. doi: 10.1097/01.gco.0000169102.77504.66. [DOI] [PubMed] [Google Scholar]
- Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 2011;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin A, Chinenov Y, Spradling A. Regulation of chromatin structure and gene activity by poly(ADP-ribose) polymerases. Curr Top Dev Biol. 2003;56:55–83. doi: 10.1016/s0070-2153(03)01007-x. [DOI] [PubMed] [Google Scholar]
- Vavouri T, Lehner B. Chromatin organization in sperm may be the major functional consequence of base composition variation in the human genome. PLoS Genet. 2011;7:e1002036. doi: 10.1371/journal.pgen.1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker DA, Frizzell KM, Zhang T, Kraus WL. Regulation of chromatin structure and chromatin-dependent transcription by poly(ADP-ribose) polymerase-1: possible targets for drug-based therapies. Subcell Biochem. 2007;41:45–69. doi: 10.1007/1-4020-5466-1_3. [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner EF. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- Ward WS. Function of sperm chromatin structural elements in fertilization and development. Mol Hum Reprod. 2010;16:30–36. doi: 10.1093/molehr/gap080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward WS, Coffey DS. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod. 1991;44:569–574. doi: 10.1095/biolreprod44.4.569. [DOI] [PubMed] [Google Scholar]
- Ward WS, Zalensky AO. The unique, complex organization of the transcriptionally silent sperm chromatin. Crit Rev Eukaryot Gene Expr. 1996;6:139–147. doi: 10.1615/critreveukargeneexpr.v6.i2-3.30. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Shaman JA, Ward WS. Non-genetic contributions of the sperm nucleus to embryonic development. Asian J Androl. 2011;13:31–35. doi: 10.1038/aja.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalenskaya IA, Zalensky AO. Telomeres in mammalian male germline cells. Int Rev Cytol. 2002;218:37–67. doi: 10.1016/s0074-7696(02)18011-9. [DOI] [PubMed] [Google Scholar]
- Zalenskaya IA, Zalensky AO. Non-random positioning of chromosomes in human sperm nuclei. Chromosome Res. 2004;12:163–173. doi: 10.1023/b:chro.0000013166.04629.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalenskaya IA, Bradbury EM, Zalensky AO. Chromatin structure of telomere domain in human sperm. Biochem Biophys Res Commun. 2000;279:213–218. doi: 10.1006/bbrc.2000.3917. [DOI] [PubMed] [Google Scholar]
- Zalensky A, Zalenskaya I. Organization of chromosomes in spermatozoa: an additional layer of epigenetic information? Biochem Soc Trans. 2007;35:609–611. doi: 10.1042/BST0350609. [DOI] [PubMed] [Google Scholar]
- Zalensky AO, Breneman JW, Zalenskaya IA, Brinkley BR, Bradbury EM. Organization of centromeres in the decondensed nuclei of mature human sperm. Chromosoma. 1993;102:509–518. doi: 10.1007/BF00368344. [DOI] [PubMed] [Google Scholar]
- Zalensky AO, Allen MJ, Kobayashi A, Zalenskaya IA, Balhorn R, Bradbury EM. Well-defined genome architecture in the human sperm nucleus. Chromosoma. 1995;103:577–590. doi: 10.1007/BF00357684. [DOI] [PubMed] [Google Scholar]
- Zhao M, Shirley CR, Hayashi S, Marcon L, Mohapatra B, Suganuma R, Behringer RR, Boissonneault G, Yanagimachi R, Meistrich ML. Transition nuclear proteins are required for normal chromatin condensation and functional sperm development. Genesis. 2004;38:200–213. doi: 10.1002/gene.20019. [DOI] [PubMed] [Google Scholar]
- Zhao M, Shirley CR, Yu YE, et al. Targeted disruption of the transition protein 2 gene affects sperm chromatin structure and reduces fertility in mice. Mol Cell Biol. 2001;21:7243–7255. doi: 10.1128/MCB.21.21.7243-7255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Verification of pharmacological PARP inhibitor approach using ABT-888. ABT-888 treatment results in poor sperm chromatin quality, as observed by increased sperm CMA3 staining after pharmacological PARP inhibition using ABT-888. Representative images of CMA3 and DAPI stained sperm from saline treated animals (a, b), and PARP inhibitor ABT-888 treated animals (c, d) are shown. Quantitative analyses of CMA3 staining intensity for sperm from long-term saline or ABT-888 treated animals are shown in box plot (e; Box, interquartile range; whiskers, 5–95% percentile. Median values obtained for sperm from n=4 animals per treatment group, and 11–60 sperm per animals. Statistical significance calculated using Mann-Whitney test is indicated in red. **,p<0.01).
PARP inhibition using ABT-888 changes heterochromatin condensation in the sperm chromocenter (similar to observation in Fig. 3–6). (f-k) Representative images of signals obtained by In situ hybridization with mouse major satellite DNA probes on sperm obtained from long-term PARP inhibitor ABT-888 treated and saline treated control wildtype animals. Mouse major satellite signal are shown in j and k (red signals), total nuclear DNA was counter-stained with DAPI (blue, h, i), and merges are visible in f and g. Signals representing the major satellite DNA in control treated animals mostly appeared as relatively continuous string of signals (j, merge with image of DAPI stained image in f), while the signals often appeared more dispersed and weaker in sperm from ABT-888 treated animals (k, merge with DAPI image in g). Frequency distribution of the number of individual FISH foci visible for major satellite DNA probe in sperm from control and PARP inhibitor (ABT-888) treated animals indicates increased incidence of unclustered chromocenter (l).