Abstract
We report here detection of novel intracellular clathrin-coated structures revealed by continuous high-speed imaging of cells expressing green fluorescent protein fusion proteins. These structures, which we operationally term ‘gyrating clathrin’ (G-clathrin), are characterized by localized but extremely rapid movement, leading to the hypothesis that they are coated buds on waving membrane tubules. G-clathrin structures have structurally and functionally distinct features. They lack detectable adaptor proteins AP-1 and AP-2 but contain GGA1 [Golgi-localized, γ-ear-containing, Arf (ADP-ribosylation factor)-binding protein] as well as the cation-dependent mannose-6-phosphate receptor. While they accumulate internalized transferrin (Tf), they do not contain detectable levels of cargos targeted for the late endosome/lysosome pathway such as EGF and dextran. Pulse-chase studies indicate that Tf appears in G-clathrin structures in the cell periphery after sorting endosomes (SEs), but before filling of the perinuclear endocytic recycling compartment. Furthermore, the inhibitors LY294002 and wortmannin, which inhibit direct recycling of Tf from SEs to the plasma membrane, also block its appearance in G-clathrin. These observations suggest that peripheral G-clathrin contributes to rapid recycling, a kinetically defined compartment that has largely eluded structural identification. More generally, the rapid continuous live cell imaging reported here reveals new aspects of membrane trafficking.
Keywords: coated membrane, endocytosis, endosome, GGA, recycling
Clathrin-mediated membrane trafficking is involved in many aspects of cargo and information transmission between the plasma membrane (PM) and intracellular compartments. The structure and function of clathrin-coated membranes in eukaryotic cells have been the focus of many studies, and the involvement of more than 30 proteins in coated pit function has emerged (reviewed in 1,2). However, an understanding of the nature of their individual functions as well as the timing, extent and consequences of their interactions remains unclear in most cases.
In addition to PM coated pits and clathrin in the trans-Golgi network (TGN) region, clathrin-coated structures also occur on both vesicular and tubular portions of endosomal membranes. Pinpointing the exact functions of endosomal clathrin-coated membranes has been challenging, but they have been demonstrated to function in multivesicular body formation and targeting to the late endosomal/lysosomal degradative pathway, in recycling, as well as in retrograde transport of cargo from the endocytic pathway to the TGN (3–7). In situ, endosomal structures have been reported to be relatively stationary or to show microtubule-dependent vectorial movements (≈0.5–1 μm/second) consistent with delivery of cargo to more centripetal organelles within the cell including late endosomes, lysosomes and the TGN (8,9).
We report here the detection of another group of clathrin coat structures in the periphery of cells, using fast imaging of live cells expressing green fluorescent protein (GFP)–clathrin. We characterize their movements, which are much faster than that previously reported for clathrin yet highly localized at the same time, leading to the inference that they reflect a distinctive gyrating or waving movement of coated buds along or on the ends of membrane tubules. Furthermore, we demonstrate that these structures, which we operationally term ‘gyrating clathrin’ (G-clathrin), contain cargo and coat components involved in recycling. It is likely that these structures, which are distinct from peripheral early endosomal vacuolar regions containing early endosomal antigen 1 (EEA1), may comprise part of an endosomal recycling compartment that has been defined kinetically but not previously discretely visualized.
Results
Rapidly moving ‘G-clathrin’ structures
Previously we have reported that high-level exogenous expression of phosphatidylinositide 3-kinase C2α (PI3K-C2α) can induce the proliferation of numerous punctate clathrin structures in the cytoplasm of cells that move extremely rapidly but in a very restricted area, and that these movements were both energy dependent and microtubule mediated (10). We hypothesized that exogenous PI3K-C2α expression might grossly exaggerate the normal occurrence of these structures. Here we asked whether fast, localized GFP–clathrin movement could be detected in the absence of exogenous PI3K-C2α expression, and which may have previously eluded detection at the comparatively long exposure times generally used for live cell microscopy. Accordingly, we investigated more closely the behavior of GFP–clathrin signals in cells in the absence of any other exogenous protein. Using 0.5 to 1 second exposure times generally employed (Video S1), the slight jiggling behavior of relatively stationary coated pits was readily observed (11,12). Interestingly, even under these image acquisition conditions, a sense of rapid clathrin movements could be appreciated as a barely perceptible blur (Video S1, boxed region). However, continuous imaging with short exposure times (≤50 milliseconds) allowed the rapid clathrin behavior in the same field to be more readily appreciated (Video S2, boxed region). These movements, which were both energy and temperature dependent (Figure S1A and B), were rapid but localized, and gave the sensation of a gyrating motion about a relatively fixed locus (Figure 1A). Accordingly, we refer to them here operationally as ‘gyrating clathrin’ or G-clathrin.
Figure 1. G-clathrin structures display localized waving movement.
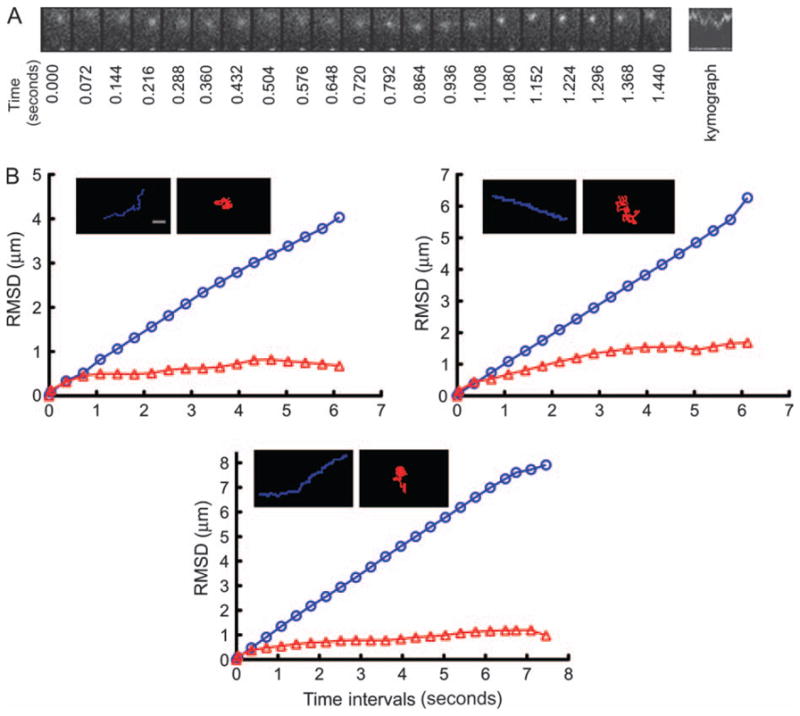
A) GFP–clathrin-transfected COS-1 cells were imaged at 36-millisecond intervals. A motile clathrin spot displaying waving movement is shown in the montage of frames captured at the indicated time-points, and in the corresponding kymograph of distance versus time. The movement is characterized by frequent directionality transitions accompanied by intensity changes, likely because of z-axis movements. Region size is 3 × 2 μm. B) Trajectories and root mean square displacements (RMSD) of clathrin spots during a 5 to 10 second tracking period. Three examples of spots exhibiting either linear vectorial motion (blue) with large RMSD, or gyrating motion (red) with small RMSD, are presented. Bar, 1 μm.
Observation of other cell lines transiently expressing GFP–clathrin using short exposure times demonstrated that G-clathrin structures occur widely. In addition to COS-1 cells, they are readily evident in HEK293, A431, HepG2 and Du145 human prostate cancer cells, and to a lesser but still detectable extent in HeLa and NIH 3T3 cells (data not shown). G-clathrin could be detected by focusing on the bottom PM, although they appear sharper when the focal plane is raised ≈0.4 μm, suggesting that they exist directly above the PM. Quantitative analysis of image stacks captured from the latter focal plane indicated that G-clathrin comprises about one-third of the detectable clathrin structures in this region [29 ± 3% (SEM)], most of which probably reflect coated endosomal membranes (5,12). The structures appear to move in and out of the focal plane as suggested by the intensity and shape changes of individual spots (Video S2, boxed region).
To learn more about these structures, cells expressing GFP–clathrin were imaged for extended periods (≈10 seconds) with 36-millisecond exposures. We then analyzed all GFP–clathrin objects in multiple fields that could be continuously and reliably tracked for ≥5 seconds and grouped spots based on their speed and ‘distance index’, the latter reflecting maximal separation displacement achieved during a time sequence compared with total accumulated distance moved. By these parameters, the localized waving movements of G-clathrin structures were readily obvious in having very low values for distance index despite high speeds (Table 1; Figure S2), and were quantitatively distinct from clathrin spots that moved in a predominantly vectorial manner. The latter covered more distance (higher distance index), and likely reflect clathrin-coated endosomes or transport carriers derived from the TGN that have been observed previously (12–15).
Table 1.
G-clathrin structures have unique mobility characteristicsa
| Average speed (μm/second ± SD) | Distance index
|
||
|---|---|---|---|
| Range (%) | Average ± SD (%) | ||
| G-clathrin | 3.73 ± 0.53 | 5.23–12.77 | 8.00 ± 1.78 |
| Vectorial clathrin | 1.43 ± 0.66 | 25.73–85.73 | 63.34 ± 19.14 |
| Stationary clathrin | <0.01 | N/A | N/A |
N/A, not applicable.
GFP–clathrin was imaged in live cells for 10 seconds using 36-millisecond exposures. All moving spots that persisted continuously as distinct structures for more than 5 seconds (56 spots from four cells) were tracked using the Metamorph ‘track points’ function, and the average speed, distance index and root mean square displacement were calculated in Excel. Stationary spots (173 in these images collected at the PM focal plane) had average speed values ≤0.01 μm/second. Distance index: the greatest separation distance achieved by a spot in a time stack divided by its total accumulated distance moved.
The G-clathrin was also readily distinguished from the many clathrin spots in the cell periphery that are essentially stationary, showing only very small, irregular movements and correspondingly small average speeds (Table 1). These structures are PM coated pits or relatively stationary endosomes, and some of their small apparent movement may actually be because of minor shape changes or intensity shifts. As seen in the three representative examples presented in Figure 1B, the unique mobility characteristics of G-clathrin were also apparent in actual spot traces and in root mean square displacement analyses of spot movement traces (16). Here, vectorial motion yielded linear plots of displacement versus interval time, whereas the localized mobility of G-clathrin was highlighted by much smaller and limiting displacement values.
G-clathrin structures are associated with GGA1
To begin to obtain compositional and functional information about G-clathrin structures, we asked whether they might also contain clathrin adaptors such as adaptor proteins AP-1 or AP-2 (17). The G-clathrin structures are clearly not PM coated pits (see above), and as expected did not contain detectable AP-2 as determined by examining cells expressing both mCherry–clathrin and GFP–AP-2 (Figure S3). AP-1 spots are also readily detectable in the peripheral cytoplasm of cells as well as in the Golgi region (13,18). Many of these are stationary, while some move vectorially; however, none of these was observed to be coincident with G-clathrin (Video S3).
Far more extensive co-localization of G-clathrin was observed with GGA, a clathrin adaptor whose role in the TGN has been intensively studied but which has also been reported to occur on peripheral clathrin structures (19,20). In cells expressing yellow fluorescent protein (YFP)-GGA1, in addition to a strong perinuclear Golgi localization, we found that at least two populations of GGA1 signal could be discerned in the periphery (Video S4; Figure 2). First, a substantial amount of YFP-GGA1 signal was essentially stationary on the timescale of our experiments (≈1 second) and all of these structures also contained mCherry–clathrin (Figure 2) as previously reported (13), although not all clathrin spots contained GGA1. More interestingly, highly dynamic YFP-GGA1 structures were also observed, comprising about half of the total GGA1 spots detected in the cell periphery in these cells [48.6 ± 4.9% (SEM)]. Simultaneous high-speed imaging of YFP-GGA1 and mCherry–clathrin (Figure 2, kymograph; Figure S4, Video S4) revealed that virtually all the motile G-clathrin structures also contained YFP-GGA1 [97 ± 1% (SEM)]. This enabled us to use this distinctively motile GGA1 signal as a marker for the G-clathrin structures (denoted G-GGA/clathrin), as it gave a clearer pattern by virtue of lacking the background of stationary coated pits, which did not contain GGA1 (Figure 2; 19).
Figure 2. G-clathrin structures contain GGA1.
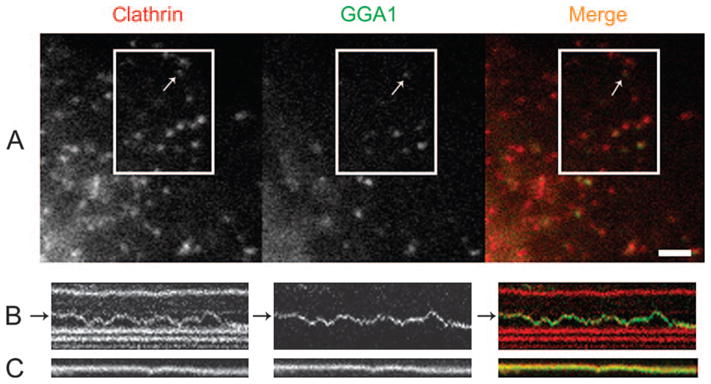
A) YFP-GGA1 signals extensively co-localized with mCherry–clathrin; arrow indicates a G-GGA/clathrin structure. B) Kymographs from several spots in the boxed region reveal the identical behavior over time of GGA1 and clathrin in moving (panel B) and stationary (panel C) GGA1–clathrin structures (also see Video S4); kymograph total duration 6.5 seconds. Bar, 2 μm.
One of the major ligands for GGA adaptors in cells is the cation-dependent mannose-6-phosphate receptor (CD-MPR), which is involved in sorting of lysosomal enzymes in the TGN and throughout the endocytic pathway (reviewed in 21). When both mCherry–clathrin and GFP-CD-MPR were expressed in COS-1 cells and imaged using rapid acquisition, we could readily detect CD-MPR in G-clathrin (Video S5), demonstrating that both adaptor and receptor are in these structures.
Are G-clathrin structures components of the endocytic pathway?
To determine whether G-GGA/clathrin structures accumulate internalized cargos, we incubated cells with Alexa568-transferrin (Tf) for 60 min and then fixed the cells. Under these conditions, internalized Tf was observed in most peripheral YFP-GGA1 structures (Figure 3A), consistent with their being components of the endocytic pathway. Interestingly, simultaneous localization of EEA1 showed only limited coincidence with GGA1, suggesting that most peripheral sorting endosomes (SEs) lack GGA1 coats in these cells (22).
Figure 3. G-GGA/clathrin structures contain endocytosed Tf.
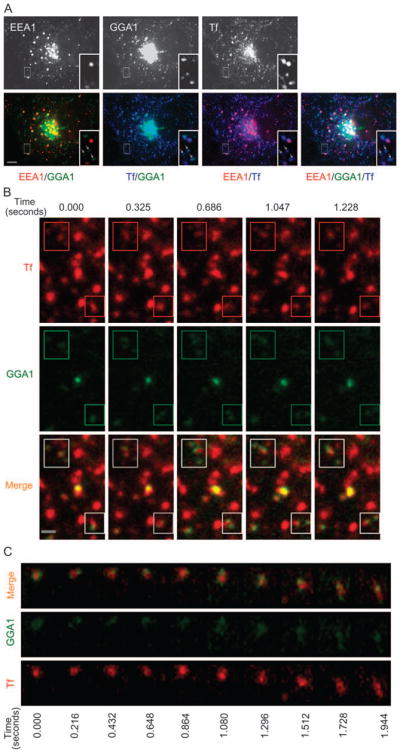
A) COS-1 cells expressing YFP-GGA1 were incubated with Alexa568 Tf for 1 hour at 37°C, then fixed and immunostained with anti-EEA1 antibody. Almost all the peripheral GGA1 signals overlap with Tf signal (arrows), but the majority of peripheral GGA1 structures do not contain EEA1. Inset shows an expanded view of boxed region. Bar, 10 μm. B) COS-1 cells expressing YFP-GGA1 were incubated with Alexa568 Tf for 15 min at 37°C and images of both GGA1 and Tf were simultaneously acquired using 36-millisecond exposures (Video S6). Rapid movements of G-GGA/clathrin structures along with Tf (boxed regions) are presented in a series of frames from Video S6. Bar, 1 μm. C) G-GGA buds decorate a rapidly moving Tf structure. Panel shows a montage of a series of frames from the boxed region (4 × 3.5 μm) of Video S7.
We then used simultaneous dual color imaging of YFP-GGA1 and Alexa568-Tf to directly visualize Tf cargos in G-GGA/clathrin structures in live cells. This revealed that after 15 min incubation, virtually all the dynamic G-GGA/clathrin structures (90 ± 2% SEM) were associated with Tf (Video S6; Figure 3B). Interestingly, in some cases, G-GGA/clathrin decorated only part of the Tf-labeled membrane (Video S7), appearing to have a bipolar distribution. Here, the presence of one or more distinct YFP-GGA1 buds on the same structure could best be appreciated by observing the rapid, coordinated movement of the GGA1 and Tf signals over time (Video S7; Figure 3C).
To determine more precisely what part of the endocytic pathway these Tf-containing G-GGA/clathrin structures comprised, we performed pulse/chase uptake experiments with Alexa568-Tf at 23°C to slow internalization and recycling in cells expressing YFP-GGA1. After a 1.5 min pulse, washout and about 2 min of chase, rapid imaging of cells indicated that very few of the peripheral structures had become loaded with Tf. However, upon chasing for 10 min, many G-GGA/clathrin structures contained Tf (Video S8), which diminished after additional chasing in the presence of unlabeled Tf (data not shown).
These experiments provided a direct but qualitative indication of the time–course of Tf trafficking into the G-GGA/clathrin compartment in live cells, and to get more quantitative information, we used cells treated identically at 23°C but then fixed and stained for EEA1 to mark SEs. At the earliest times after a pulse of Alexa568-Tf uptake and immediate washout and fixation, some EEA1-positive SEs containing Tf can be detected (Figure 4A). By 2 min after Tf washout, most SEs now contained Tf (Figure 4B), consistent with previous observations of the uptake of Tf, although very few of the peripheral GGA1 structures had become loaded with Tf. However, by 10 min of chase, about 90% of the peripheral GGA1 spots were now Tf positive (Figure 4C), consistent with the live cell imaging data (Video S8). Quantification of multiple experiments similar to that shown in Figure 4 demonstrated that the increasing degree of overlap of Tf signal with peripheral GGA1 structures, many of which must actually be G-clathrin (≈50%, see above), clearly demonstrated that the G-GGA/clathrin compartment loads after the SE, and that the signals become largely coincident by 10 min of chase (Figure 4D). In contrast, under these conditions, the extent of labeling of the perinuclear endocytic recycling compartment with Tf is not substantial until well after 10 min (23, and data not shown). Together, these results indicate that the G-GGA/clathrin compartment is downstream of the SE, but clearly upstream or coincident with the perinuclear endocytic recycling compartment.
Figure 4. G-GGA/clathrin are post-sorting endosomal structures.
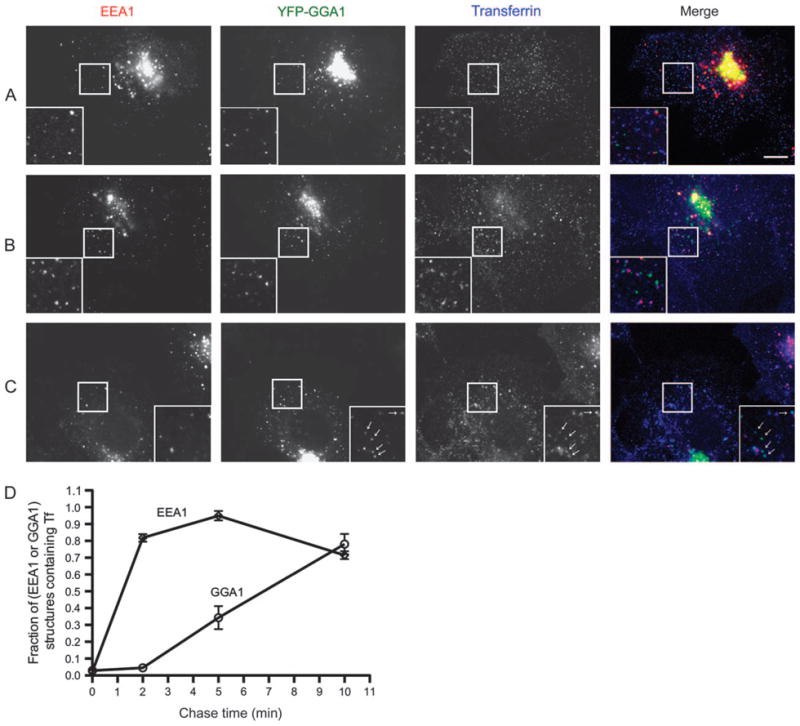
Cells transfected with YFP-GGA1 were pulse-labeled with Alexa647 Tf at 23°C for 1.5 min, chased for 0, 2 or 10 min, fixed and stained with EEA1 antibody. Inset shows an expanded view of boxed region. A) At start of chase, Tf labels only a few small peripheral EEA1-positive endosomes, and Tf is not detectable in GGA1 structures. Bar, 10 μm in all panels. B) After 2 min chase, almost all the EEA1 endosomes are labeled by Tf, while GGA1 structures still lack Tf. C) After 10 min chase, almost all the GGA1 structures contain Tf (arrows). D) Quantitative analysis of appearance of Tf in GGA1- and peripheral EEA1-positive structures (mean ± SEM). Ten regions from 10 cells at each time-point were analyzed.
We then asked whether G-GGA/clathrin structures discriminate cargos destined for the late endosomal and lysosomal compartments from those targeted to the recycling pathway. First, cells expressing YFP-GGA1 were incubated overnight in Texas Red-tagged dextran (Mr ≈ 70 kDa) to saturate the fluid phase uptake pathway leading to late endosomes/lysosomes. Upon live cell imaging, no evidence for Texas Red-dextran in the G-GGA/clathrin structures was detected (data not shown). Second, we examined the distribution of EGF, a ligand primarily sorted to late endosomes and lysosomes (2). Cells were labeled with Texas Red-EGF (100 ng/mL) for 1, 4 or 10 min at 37°C, washed briefly and observed after chasing for 1–30 min. Again, Texas Red-tagged EGF was absent from G-GGA/clathrin structures although it was detectable in stationary or vectorially moving structures bearing GGA coats (Video S9).
As fluid phase markers do not accumulate in recycling compartments (24,25), the absence of internalized dextran, as well as of EGF targeted to late endosomes/lysosomes, suggested that G-GGA/clathrin structures might function in recycling pathways. Accordingly, we examined peripheral GGA1 structures for Rab11, a protein that is known to be involved in recycling pathways in mammalian cells (26,27). We found that mCherry–Rab11 often moves coordinately with G-clathrin and, like Tf, labels tubules whose end(s) are decorated by GGA1 (Figure 5) or clathrin (Video S10, arrows). Collectively, the presence of Rab11 and of recycling cargo and receptors [CD-MPR (Video S5) and Tf receptors] are consistent with the G-GGA/clathrin structures participating in a recycling pathway from the SE.
Figure 5. G-GGA decorates Rab11-containing structures.
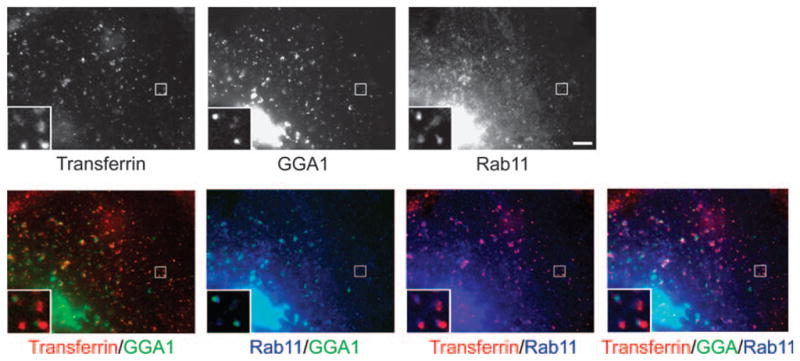
COS-1 cells expressing YFP-GGA1 and cyan fluorescent protein –Rab11 were incubated with Alexa568 Tf for 15 min at 37°C. GGA1 partially co-localizes with Rab11-positive membranes, sometimes decorating the ends of these membranes. Inset shows an expanded view of boxed region. Bar, 5 μm.
Having demonstrated that Tf accumulates in SEs before reaching G-GGA/clathrin structures, we exploited the observation that incubation of cells with cargo at 16°C allows uptake into SEs, but further transport is inhibited (28). First, we confirmed these observations in our system by incubating cells with Alexa568-Tf for up to 1 hour at 16°C, and then either fixed and stained for EEA1 to reveal SEs, or imaged directly to evaluate loading of G-GGA/clathrin with Alexa568-Tf. Under these conditions, appearance of internalized Tf in G-GGA/clathrin structures was barely detectable, with only 8.9 ± 3.2% (SEM) of G-GGA/clathrin spots containing Tf after 1 hour incubation at 16°C. In contrast and as expected, EEA1-positive SEs were strongly labeled under these conditions (data not shown, and reference 28,29). Furthermore, upon washing the cells and raising the temperature to 37°C for 5 min, endocytosed Tf was now readily detected in the G-GGA/clathrin structures (84 ± 5% SEM), consistent with earlier observations that the temperature block is readily reversible (29), and with our observations that the G-GGA/clathrin structures lie downstream of the SE (Figure 4D).
To determine more directly whether G-GGA/clathrin structures participate in recycling from the SE, we used LY294002 and wortmannin. These reagents, which inhibit PI3Ks by disparate chemical mechanisms (30), have been shown to inhibit recycling of endocytosed receptors and cargo to the PM (31–33). To enhance accumulation of Tf in SEs, we examined cells incubated at 16°C (as above) in the presence or absence of LY294002. We then used flow cytometry to quantify the extent of Tf recycling upon releasing the temperature block by chasing at 37°C. First, our observations (Figure 6A) are consistent with earlier reports (33) in demonstrating an inhibition in rapid recycling from SEs in cells incubated with LY294002 (≈80% inhibition at 5–10 min), with lesser effects on recycling at longer times likely corresponding to transport through the kinetically slower perinuclear recycling compartment. Similar results were observed in cells treated with wortmannin (data not shown). Importantly, live cell imaging of cells that had internalized fluorescent Tf under these conditions revealed that LY294002 treatment did not appear to detectably affect the appearance or behavior of G-GGA/clathrin, but rather greatly inhibited its loading with Tf on warm-up and chase for 5 min at 37°C (Figure 6B; Video S11). Quantification of these results revealed that LY294002 treatment resulted in almost 85% inhibition of Tf appearance in G-GGA/clathrin under these conditions (Figure 6C). This correlates closely with the approximately 80% inhibition in rapid recycling at this time-point measured by flow cytometry. Together, these results strongly suggest that G-GGA/clathrin structures function in rapid recycling from SEs to the PM.
Figure 6. LY294002 inhibits rapid recycling in COS-1 cells.
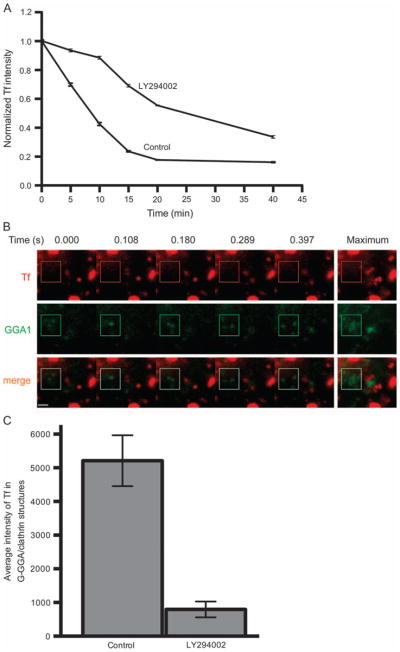
A) COS-1 cells were incubated with 20 μg/ml Alexa568-Tf for 1 hour at 16°C, then chased in 37°C medium supplemented with unlabeled Tf for indicated times, all in the presence or absence of LY294002. Intracellular Tf levels were measured by flow cytometry. Rapid Tf recycling in the initial 5 to 10 min chase was markedly inhibited by LY294002. B) COS-1 cells transiently expressing YFP-GGA1 were treated as in (A), and images were acquired after 5 min chase (Video S11). A series of still images and the maximal image from a time stack of 50 continuous frames indicated that LY294002 significantly inhibited the loading of Tf into G-GGA/clathrin structures. In contrast, control cells were not affected (data not shown, but compare Figure 4D and Video S6). Boxed region includes several G-GGA structures. Bar, 1 μm. C) Quantitative analysis indicated about 85% inhibition of Tf loading of G-GGA structures after 5 min chase in the presence of LY294002.
Discussion
The rapid, gyrating G-GGA/clathrin movements that we have identified and analyzed here have not, to the best of our knowledge, been reported previously. Key to their clear visualization and study has been the use of continuous image acquisition with very short exposure times, generally ≤50 milliseconds. G-GGA/clathrin structures occur widely and to differing extents in all cell lines studied. Many are located in close proximity to SEs in the cell periphery, although others that were not further characterized here can be detected further toward the cell interior.
Clathrin-coated structures on internal membranes in the cell periphery have been identified in a number of settings. For example, clathrin coats on vesicular portions of early endosomes, often displaying distinctive bilayer coats at the ultrastructural level, have been observed (4–7), but these must be distinct from G-GGA/clathrin as the latter does not co-localize with SE markers. Coated tubular regions of early endosomes have also been described (3), often containing AP-1 as well (34), and some of these structures have been implicated in lysosomal targeting and retrograde transport of cargo from the endocytic pathway to the TGN (35,36). G-GGA/clathrin may comprise a subset of these structures although not those containing AP-1, which we observe to either be stationary or exhibit vectorial motion distinct from the whirling G-GGA/clathrin movement described here. Finally, most clathrin structures observed by total internal reflection fluorescence microscopy are stationary coated pits, but a very small fraction have been reported to exhibit linear lateral movement in the field of view (37). Their vectorial movement spatially confined to the evanescent field directly abutting the PM is inconsistent with the waving motion of the structures generally more distal to the membrane reported here.
Virtually all the peripheral G-clathrin structures also contain expressed GGA1. The majority of GGA is in the TGN region and functions in sorting of late endosomal cargos (22). However, peripheral GGA structures have also been identified (19,20,38), and the subset containing AP-1 (39), distinct from G-clathrin which lacks this adaptor, have been shown to function in transport between the TGN and peripheral endosomes (13,14). In light of our findings of G-GGA/clathrin’s involvement in receptor recycling at the SE, it is interesting that a role for GGA-dependent sorting of MPR in the perinuclear endocytic recycling compartment has recently been reported (40). Poor viability of cells with silenced GGA expression, as reported earlier (22), has hampered our attempts to further dissect the role of GGA in G-clathrin function.
In this study, we show that the G-clathrin structures become loaded with internalized Tf and identify them as early recycling structures based upon several criteria. First, kinetically they are loaded after the EEA1-marked SE has acquired substantial Tf but before the perinuclear endocytic recycling compartment has acquired detectable cargo. Second, in addition to Tf, G-clathrin also contains recycling receptors including the CD-MPR, and is associated with rab11, a marker for the recycling pathway (41), but it lacks detectable levels of cargos that are delivered to the late endosomal/lysosomal compartment and are largely spared from recycling, for example, EGF and dextran. Kinetic studies have shown that recycling from SEs can occur rapidly either directly back to the PM, or more slowly by way of a perinuclear endocytic recycling compartment (42,43). In this context, our finding that inhibitors of PI3K that inhibit rapid recycling (33) also greatly decrease or block the appearance of recycling Tf in G-clathrin structures strongly suggests that the latter contribute or are responsible for rapid recycling from the SE. These structures are thus excellent candidates for the previously unidentified physical compartment whose recycling properties have been appreciated from kinetic studies. The extent, dynamics and regulation of physical connection between G-clathrin and SEs remains to be established, but we speculate that G-GGA/clathrin may comprise part of a tubular endosomal network (3,44) responsible for the complex sorting functions of endosomes and the recycling compartment. Further work at the ultrastructural level will be needed to resolve this important question, as well as the nature of other adaptors and regulators that traverse this compartment.
The observed dynamics of G-clathrin structures are consistent with waving movements of fluorescently tagged coated buds along or on the ends of membrane tubules. It is intriguing to consider what might be the functional advantages of this extremely rapid, localized movement. For example, it might promote fission of the tubule, enhance its contact with surrounding transport vesicles or even promote diffusion and segregation of receptors along the tubule membrane.
An important aspect of this report is that it reveals intracellular membrane dynamics on a timescale not previously appreciated. G-GGA/clathrin structures have been the focus of our interest here, but similar rapid movements can be seen in the absence of expression of any exogenous protein in cells that have endocytosed fluorescent Tf (Video S12). Thus, live cell imaging at this sampling rate reveals a novel arena of membrane trafficking, and it seems reasonable to expect that these kinds of movements play widespread and critical roles in intracellular transport.
Materials and Methods
Constructs
Preparation of fluorescent protein fusions with clathrin LCa has been described (10,11). YFP-GGA1 (45) and GFP-CD-MPR (45) were generous gifts of Dr Juan Bonifacino (NIH). mCherry–Rab11a was generated by cloning full length Rab11 (46) from EGFP-C2-Rab11a (a generous gift from Dr Marci A. Scidmore, Cornell University) into the XhoI and EcoRI sites of mCherry-C1. YFP–AP-1γ and GFP–AP-2α were constructed as described (47).
Antibodies and reagents
Alexa568/633/647-Tf, Texas Red-EGF and Texas Red-dextran (molecular weight ≈ 70 kDa) were purchased from Molecular Probes, Inc. Anti-EEA1 antibody was from BD Biosciences. LY294002, wortmannin and dimethyl sulfoxide (DMSO) were purchased from Sigma.
Fluorescence microscopy and analysis
For immunostaining, transiently transfected COS-1 cells were fixed with 3.7% formaldehyde at room temperature for 10 min and subsequent immunostaining was performed as described previously (11).
For live cell imaging with transfected COS-1 cells in glass bottom culture dishes (MatTek), the complete DMEM medium was replaced with HEPES-supplemented F12 medium and images were acquired at indicated time intervals. For Tf pulse-chase experiments at 23°C, cells were first incubated with 20 μg/ml fluorescently labeled Tf for 1.5 min, rapidly washed, chased for indicated times and either fixed immediately or loaded on the microscope for imaging. Texas Red-EGF (100 ng/ml) or Texas Red-dextran (1 mg/ml) was used to challenge the cells for 1–10 min or 12 hour, respectively, at 37°C, washed briefly and observed. Samples were imaged on Zeiss Axiovert S100 TV widefield microscope system (63×/1.4 NA objective) thermostatted with a Harvard TC202-A controller and PDMI-2 sample holder, as well as a fabricated objective thermostat fitted to a Lauda water bath. Images were captured on a Cascade 650 camera (Photometrics, Inc.) controlled by Metamorph software (Universal Imaging, Inc.). Unless otherwise indicated, live cell imaging was performed with continuous streaming using the specified exposure time. Z-stack control and acquisition used a piezo-driven objective with an E-662 LVPZT-Amplifier/Servo controller. Simultaneous imaging of two fluorescent signals was accomplished using a Dual-view system (Roper Scientific, Inc.).
For quantitative analyses in all experiments, between 100 and 120 spots in 10–20 randomly selected regions of interest (ROIs) in the cell periphery, each comprising 50–70 pixels square (≈ 5.5–7.7 μm square), were analyzed. For quantification of overlap in fixed cells (e.g. Tf with EEA1 or GGA1 structures), all the EEA1/GGA1 structures in the ROI were accurately outlined and the region was transferred to the corresponding Tf image to determine coincidence. For quantification of overlap of G-clathrin with GGA1, Metamorph ‘stack arithmetic/maximum’ was applied to the time stack to find moving structures, identified by a characteristic blurred, often rosetted, area. Accurate outlines were drawn around these areas, the regions transferred to GGA images and overlap quantified. For quantification of overlap of G-GGA with Tf, the G-GGA structures were similarly identified and transferred to Tf images. Tf intensities in these regions ≥20% above the nearby background were considered positive, and the co-localization of GGA1 and Tf was further confirmed by visually testing for coordinated movement in the color-combined stack. For quantitative analysis of the fraction of G-clathrin structures near the PM, GFP–clathrin expressing interphase cells were first located and in-focus images of the basal PM were collected. The objective was then driven upward 0.40 μm and 200 image frames were streamed at 36 millisecond/frame. In these time stacks and for quantification of the proportion of G-GGA in total GGA1 structures, Metamorph ‘stack arithmetic/minimum’ was used to find stationary spots, ‘stack arithmetic/maximum’ to reveal vectorially motile spots, and nonlinear motile spot were counted manually, confirmed by the maximal image rosettes and visualization through the time stack. Spots persisting in less than three continuous frames were excluded.
Flow cytometry and quantification of Tf uptake and recycling
COS-1 cells were incubated at 16°C with 20 μg/ml Tf conjugated with Alexa Fluor 633 (Invitrogen) in the presence of 100 μM LY294002 in 0.2% DMSO (or 0.2% DMSO for control, both present throughout all loading, washing and chase buffers) in HEPES-buffered (pH 7.2) serum-free F12 medium for 1 hour. Bound Tf was removed from the cell surface by washing at 16°C twice with F12 medium (pH 7.2), a brief acid wash in 50 mM sodium MES [2-(4-morpholino)-ethane sulfonic acid], 150 mM NaCl, pH 5.0, followed by an additional wash at pH 7.2. Subsequently cells were rapidly warmed in a water bath and chase initiated by rinsing and incubating with F12 medium containing 2 mg/ml unlabeled Tf for the indicated times at 37°C. Incubation was stopped by washing cells with F12 medium, chilling on ice and analyzed using a FACSCalibur flow cytometer (Becton Dickinson). Data were collected from 5000 events, and the mean fluorescent intensity was determined for each time-point. Data were normalized against the mean fluorescent intensity of 0 min chase of control cells. Error bars represent the SEM from triplicate samples. Cell viability was confirmed by staining with trypan blue.
For live cell imaging of the effects of LY294002 treatment on Tf transport, the procedure was identical to that in flow cytometry except that Tf conjugated with Alexa Fluor 568 was used with COS-1 cells expressing YFP-GGA1. Both Tf and GGA1 signals were simultaneously imaged with 36-millisecond exposures for 50 continuous frames after 5 min chase at 37°C. To measure the amount of Tf in G-GGA/clathrin structures, G-GGA/clathrin structures were identified by review of the time stacks, and for each structure, the brightest frame, indicating the best focal plane in the time stack, was determined. An outline was drawn around the G-GGA/clathrin structure, transferred to the corresponding Tf image in the time stack, and the average Tf intensity in the outline was quantified. This was background corrected by the average intensity of several nearby comparable regions devoid of Tf. Tf intensities measured in this way were obtained in 22 G-GGA/clathrin spots from three cells, and the corresponding mean value was determined.
Supplementary Material
Figure S1: G-clathrin movements are energy and temperature dependent. In these maximal projection images of time series, the G-clathrin movements in control cells generated a ‘rosetted’ blurred image at 23°C (A and B, left panels, arrows), reflecting their characteristic localized, waving movement. In contrast, in the presence of energy inhibitors (50 mM 2-deoxyglucose and 25 mM sodium azide for 20 min; A, right panel) or at 10°C (B, right panel) the maximal projection shows much smaller spots, indicating that the movements are greatly inhibited. Images of the same fields were acquired at 36-millisecond intervals before and after treatments. Bar: 3 μm (A), 1 μm (B).
Figure S2: Grouping of motile clathrin structures. Moving clathrin spots, comprising all the distinct structures that could be tracked for more than 5 seconds (56 spots from four cells) were tracked and quantified as indicated in Table 1. The velocity (green spots, right ordinate) and distance index (blue bars, abscissa) data for each spot are presented in scatter plot and column, respectively, sorted according to decreasing distance index (a distinctive measure of overall movements; see Table 1). Two distinct groups are distinguished by these parameters: one group comprising vectorially moving clathrin spots (left) displays consistently higher distance index and lower velocity values. The other group (right) shows higher velocity but strikingly lower and uniform distance index values, reflecting the pronounced localized yet rapid motion apparent visually as gyrating or G-clathrin.
Figure S3: AP-2 is absent from G-clathrin structures. mCherry–clathrin and GFP–AP-2 were simultaneously imaged at 50-millisecond intervals. A G-clathrin structure lacking detectable AP-2 signal but with characteristic rapid movement (evident in animation) is noted by the arrow. Bar, 0.5 μm.
Figure S4: GGA1 is present in G-clathrin structures. mCherry–clathrin and YFP-GGA1 were simultaneously imaged using 36-millisecond exposures. A series of frames taken at indicated time-points shows a G-clathrin spot (in the lower part) containing GGA1 moving up and down very quickly, creating a curved line in the kymograph. In contrast, a static clathrin spot (likely coated pit) lacking GGA1 signal creates a straight line in kymograph. Bar, 1 μm.
Video S1: Rapid clathrin movements are imperceptible using long exposures. COS-1 cells expressing GFP-clathrin were imaged using 1 second exposures. Only a barely perceptible blur (box) due to rapid clathrin movements is observed. 40 frames at 1 second/frame, playback at 5 frames/second (5X real-time). Bar, 2 μm.
Video S10: Coordinated movements of G-clathrin and Rab11. Arrows point to regions of G-clathrin (YFP–clathrin) movement with associated mCherry–Rab11. 50 milliseconds/frame, 50 frames total, playback at 20 frames/second (real time). Bar, 1 μm.
Video S11: Tf loading of G-GGA structures is inhibited by LY294002. COS-1 cells expressing YFP-GGA1 were incubated with 20 μg/ml Alexa568-Tf for 1 hour at 16°C, then chased in 37°C medium supplemented with unlabeled Tf for 5 min, all in the presence of 100 μM LY294002, and imaged immediately using 36-millisecond exposures for 50 frames. Tf appearance in G-GGA/clathrin structures (boxed region) is markedly inhibited. Playback at 27 frames/second (real time). Bar, 1 μm.
Video S12: Rapid gyrating movement occurs in nontransfected cells. COS-1 cells were incubated with 20 μg/ml Alexa568-Tf for 15 min at 37°C, washed and images were acquired using 36-millisecond exposures for 100 frames. The boxed region indicates one example of fast gyrating movement. Playback at 27 frames/second (real time). Bar, 1 μm.
Video S2: Rapid clathrin movements revealed using rapid imaging. The same field of COS-1 cells as in Video S1 was imaged using streaming 36-millisecond exposures. Rapid clathrin movements are evident (box) with high speed imaging. 40 frames at 36 milliseconds/frame, playback at 27 frames/second (real-time). Bar, 2 μm. It may be useful for the reader to play back this and other supplemental videos at slower and faster speeds (see ‘A/V control’ in Quicktime).
Video S3: AP-1 is excluded from G-clathrin structures. Essentially no co-localization is observed between mCherry tagged G-clathrin and YFP-tagged AP-1. The boxed region shows an example of G-clathrin structures lacking AP-1. 36 milliseconds/frame, 50 frames total, playback at 27 frames/second (real time). Bar, 2 μm.
Video S4: G-clathrin structures also contain GGA1. Both stationary and highly mobile (near arrows) YFP-tagged GGA1 (middle panel, green) structures also contain mCherry–clathrin (left panel, red). 36 milliseconds/frame, 50 frames total, playback at 27 frames/second (real time). Playback of this and subsequent videos at half-speed may be advantageous. Bar, 1 μm.
Video S5: CD-MPR is present in G-clathrin. COS-1 cells co-expressing mCherry–clathrin and GFP-CD-MPR were imaged using 40-millisecond exposures. Most G-clathrin structures (e.g. boxed region) contain CD-MPR. 50 frames total, playback at 25 frames/second (real time). Bar, 1 μm.
Video S6: Endocytosed Tf appears in G-GGA/clathrin structures. After a 15 min incubation (37°C) with Alexa568-Tf, the majority of G-GGA/clathrin structures (YFP-GGA1) contain Tf (boxed regions). 36 milliseconds/frame, 50 frames total, playback at 27 frames/second (real time). Bar, 1 μm.
Video S7: Moving Tf tubules with GGA1 buds. YFP-GGA1 expressing COS-1 cells were incubated with Alexa568-Tf for 15 min (37°C) and imaged at 36 milliseconds/frame. 140 frames total, playback at 27 frames/second (real time). Boxed region, 4 × 3.5 μm.
Video S8: Endocytosed Tf gradually moves into G-GGA/clathrin structures during a 10 min chase. YFP-GGA1 expressing COS-1 cells were incubated with Alexa568-Tf for 1.5 min at 23°C, washed, chased and the same field was imaged at 2 or 10 min. Very few G-GGA/clathrin structures (green) contain Tf (red) at the earlier time, but virtually all do after 10 min (examples near arrows). 36 milliseconds/frame, 40 frames total, playback at 27 frames/second (real time). Bar, 1 μm.
Video S9: Internalized EGF is absent from G-GGA/clathrin structures. COS-1 cells expressing YFP-GGA1 were incubated with Texas Red-EGF for 10 min (37°C), washed and chased for 10 min, then simultaneously imaged for both EGF (red) and GGA1 (green). Although EGF is present in some GGA1 structures that are stationary or show slow, vectorial movement, essentially none of the G-GGA structures contain EGF. 36 milliseconds/frame, 50 frames total, playback at 27 frames/second (real time). Bar, 3 μm.
Acknowledgments
We are grateful to Drs Juan Bonifacino (NIH) and Marci A. Scidmore (Cornell) for gifts of reagents, I. Gaidarov and Jianke Zhang for helpful discussion, and the KCC Bioimaging and Flow Cytometry (Matthew Farabaugh) Facilities for assistance. This work was supported by NIH grant GM-49217 (J. H. K.).
Footnotes
Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Dell’Angelica EC. Clathrin-binding proteins: got a motif? Join the network! Trends Cell Biol. 2001;11:315–318. doi: 10.1016/s0962-8924(01)02043-8. [DOI] [PubMed] [Google Scholar]
- 2.Benmerah A, Lamaze C. Clathrin-coated pits: vive la difference? Traffic. 2007;8:970–982. doi: 10.1111/j.1600-0854.2007.00585.x. [DOI] [PubMed] [Google Scholar]
- 3.Stoorvogel W, Oorschot V, Geuze HJ. A novel class of clathrin-coated vesicles budding from endosomes. J Cell Biol. 1996;132:21–33. doi: 10.1083/jcb.132.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raiborg C, Bache KG, Mehlum A, Stang E, Stenmark H. Hrs recruits clathrin to early endosomes. EMBO J. 2001;20:5008–5021. doi: 10.1093/emboj/20.17.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sachse M, Urbe S, Oorschot V, Strous GJ, Klumperman J. Bilayered clathrin coats on endosomal vacuoles are involved in protein sorting toward lysosomes. Mol Biol Cell. 2002;13:1313–1328. doi: 10.1091/mbc.01-10-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raiborg C, Wesche J, Malerod L, Stenmark H. Flat clathrin coats on endosomes mediate degradative protein sorting by scaffolding Hrs in dynamic microdomains. J Cell Sci. 2006;119:2414–2424. doi: 10.1242/jcs.02978. [DOI] [PubMed] [Google Scholar]
- 7.Myromslien FD, Grovdal LM, Raiborg C, Stenmark H, Madshus IH, Stang E. Both clathrin-positive and -negative coats are involved in endosomal sorting of the EGF receptor. Exp Cell Res. 2006;312:3036–3048. doi: 10.1016/j.yexcr.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen E, Severin F, Backer JM, Hyman AA, Zerial M. Rab5 regulates motility of early endosomes on microtubules. Nat Cell Biol. 1999;1:376–382. doi: 10.1038/14075. [DOI] [PubMed] [Google Scholar]
- 9.Bananis E, Murray JW, Stockert RJ, Satir P, Wolkoff AW. Regulation of early endocytic vesicle motility and fission in a reconstituted system. J Cell Sci. 2003;116:2749–2761. doi: 10.1242/jcs.00478. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, Gaidarov I, Keen JH. Phosphoinositide 3-kinase C2alpha links clathrin to microtubule-dependent movement. J Biol Chem. 2007;282:1249–1256. doi: 10.1074/jbc.M606998200. [DOI] [PubMed] [Google Scholar]
- 11.Gaidarov I, Santini F, Warren RA, Keen JH. Spatial control of coated-pit dynamics in living cells. Nat Cell Biol. 1999;1:1–7. doi: 10.1038/8971. [DOI] [PubMed] [Google Scholar]
- 12.Keyel PA, Watkins SC, Traub LM. Endocytic adaptor molecules reveal an endosomal population of clathrin by total internal reflection fluorescence microscopy. J Biol Chem. 2004;279:13190–13204. doi: 10.1074/jbc.M312717200. [DOI] [PubMed] [Google Scholar]
- 13.Puertollano R, van der Wel NN, Greene LE, Eisenberg E, Peters PJ, Bonifacino JS. Morphology and dynamics of clathrin/GGA1-coated carriers budding from the trans-Golgi network. Mol Biol Cell. 2003;14:1545–1557. doi: 10.1091/mbc.02-07-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polishchuk RS, San Pietro E, Di Pentima A, Tete S, Bonifacino JS. Ultrastructure of long-range transport carriers moving from the trans Golgi network to peripheral endosomes. Traffic. 2006;7:1092–1103. doi: 10.1111/j.1600-0854.2006.00453.x. [DOI] [PubMed] [Google Scholar]
- 15.Rappoport JZ, Taha BW, Lemeer S, Benmerah A, Simon SM. The AP-2 complex is excluded from the dynamic population of plasma membrane-associated clathrin. J Biol Chem. 2003;278:47357–47360. doi: 10.1074/jbc.C300390200. [DOI] [PubMed] [Google Scholar]
- 16.Kusumi A, Sako Y, Yamamoto M. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys J. 1993;65:2021–2040. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brett TJ, Traub LM. Molecular structures of coat and coat-associated proteins: function follows form. Curr Opin Cell Biol. 2006;18:395–406. doi: 10.1016/j.ceb.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 18.Huang F, Nesterov A, Carter RE, Sorkin A. Trafficking of yellow-fluorescent-protein-tagged mu1 subunit of clathrin adaptor AP-1 complex in living cells. Traffic. 2001;2:345–357. doi: 10.1034/j.1600-0854.2001.25020506.x. [DOI] [PubMed] [Google Scholar]
- 19.Boman AL, Zhang C, Zhu X, Kahn RA. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol Biol Cell. 2000;11:1241–1255. doi: 10.1091/mbc.11.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dell’Angelica EC, Puertollano R, Mullins C, Aguilar RC, Vargas JD, Hartnell LM, Bonifacino JS. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J Cell Biol. 2000;149:81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonifacino JS. The GGA proteins: adaptors on the move. Nat Rev Mol Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh P, Griffith J, Geuze HJ, Kornfeld S. Mammalian GGAs act together to sort mannose 6-phosphate receptors. J Cell Biol. 2003;163:755–766. doi: 10.1083/jcb.200308038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- 24.Baravalle G, Schober D, Huber M, Bayer N, Murphy RF, Fuchs R. Transferrin recycling and dextran transport to lysosomes is differentially affected by bafilomycin, nocodazole, and low temperature. Cell Tissue Res. 2005;320:99–113. doi: 10.1007/s00441-004-1060-x. [DOI] [PubMed] [Google Scholar]
- 25.Luzio JP, Pryor PR, Gray SR, Gratian MJ, Piper RC, Bright NA. Membrane traffic to and from lysosomes. Biochem Soc Symp. 2005;72:77–86. doi: 10.1042/bss0720077. [DOI] [PubMed] [Google Scholar]
- 26.Brown MS, Anderson RG, Goldstein JL. Recycling receptors: the round-trip itinerary of migrant membrane proteins. Cell. 1983;32:663–667. doi: 10.1016/0092-8674(83)90052-1. [DOI] [PubMed] [Google Scholar]
- 27.Sonnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multi-color imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149:901–914. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren M, Xu G, Zeng J, De Lemos-Chiarandini C, Adesnik M, Sabatini DD. Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc Natl Acad Sci U S A. 1998;95:6187–6192. doi: 10.1073/pnas.95.11.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Dam EM, Stoorvogel W. Dynamin-dependent transferrin receptor recycling by endosome-derived clathrin-coated vesicles. Mol Biol Cell. 2002;13:169–182. doi: 10.1091/mbc.01-07-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 31.Kurashima K, Szabo EZ, Lukacs G, Orlowski J, Grinstein S. Endosomal recycling of the Na+/H+ exchanger NHE3 isoform is regulated by the phosphatidylinositol 3-kinase pathway. J Biol Chem. 1998;273:20828–20836. doi: 10.1074/jbc.273.33.20828. [DOI] [PubMed] [Google Scholar]
- 32.Martys JL, Wjasow C, Gangi DM, Kielian MC, McGraw TE, Backer JM. Wortmannin-sensitive trafficking pathways in Chinese hamster ovary cells. Differential effects on endocytosis and lysosomal sorting. J Biol Chem. 1996;271:10953–10962. doi: 10.1074/jbc.271.18.10953. [DOI] [PubMed] [Google Scholar]
- 33.van Dam EM, Ten Broeke T, Jansen K, Spijkers P, Stoorvogel W. Endocytosed transferrin receptors recycle via distinct dynamin and phosphatidylinositol 3-kinase-dependent pathways. J Biol Chem. 2002;277:48876–48883. doi: 10.1074/jbc.M206271200. [DOI] [PubMed] [Google Scholar]
- 34.Futter CE, Gibson A, Allchin EH, Maxwell S, Ruddock LJ, Odorizzi G, Domingo D, Trowbridge IS, Hopkins CR. In polarized MDCK cells basolateral vesicles arise from clathrin-gamma-adaptin-coated domains on endosomal tubules. J Cell Biol. 1998;141:611–623. doi: 10.1083/jcb.141.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer C, Zizioli D, Lausmann S, Eskelinen EL, Hamann J, Saftig P, von Figura K, Schu P. mu1A-adaptin-deficient mice: lethality, loss of AP-1 binding and rerouting of mannose 6-phosphate receptors. EMBO J. 2000;19:2193–2203. doi: 10.1093/emboj/19.10.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theos AC, Tenza D, Martina JA, Hurbain I, Peden AA, Sviderskaya EV, Stewart A, Robinson MS, Bennett DC, Cutler DF, Bonifacino JS, Marks MS, Raposo G. Functions of adaptor protein (AP)-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol Biol Cell. 2005;16:5356–5372. doi: 10.1091/mbc.E05-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rappoport JZ, Taha BW, Simon SM. Movement of plasma-membrane-associated clathrin spots along the microtubule cytoskeleton. Traffic. 2003;4:460–467. doi: 10.1034/j.1600-0854.2003.00100.x. [DOI] [PubMed] [Google Scholar]
- 38.Puertollano R, Bonifacino JS. Interactions of GGA3 with the ubiquitin sorting machinery. Nat Cell Biol. 2004;6:244–251. doi: 10.1038/ncb1106. [DOI] [PubMed] [Google Scholar]
- 39.Doray B, Ghosh P, Griffith J, Geuze HJ, Kornfeld S. Cooperation of GGAs and AP-1 in packaging MPRs at the trans-Golgi network. Science. 2002;297:1700–1703. doi: 10.1126/science.1075327. [DOI] [PubMed] [Google Scholar]
- 40.Tortorella LL, Schapiro FB, Maxfield FR. Role of an acidic cluster/dileucine motif in cation-independent mannose 6-phosphate receptor traffic. Traffic. 2007;8:402–413. doi: 10.1111/j.1600-0854.2007.00541.x. [DOI] [PubMed] [Google Scholar]
- 41.Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao M, Maxfield FR. Characterization of rapid membrane internalization and recycling. J Biol Chem. 2000;275:15279–15286. doi: 10.1074/jbc.275.20.15279. [DOI] [PubMed] [Google Scholar]
- 43.Hopkins CR, Trowbridge IS. Internalization and processing of transferrin and the transferrin receptor in human carcinoma A431 cells. J Cell Biol. 1983;97:508–521. doi: 10.1083/jcb.97.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol. 2006;7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- 45.Puertollano R, Aguilar RC, Gorshkova I, Crouch RJ, Bonifacino JS. Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science. 2001;292:1712–1716. doi: 10.1126/science.1060750. [DOI] [PubMed] [Google Scholar]
- 46.Rzomp KA, Scholtes LD, Briggs BJ, Whittaker GR, Scidmore MA. Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect Immun. 2003;71:5855–5870. doi: 10.1128/IAI.71.10.5855-5870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X, Zhao X, Puertollano R, Bonifacino JS, Eisenberg E, Greene LE. Adaptor and clathrin exchange at the plasma membrane and trans-Golgi network. Mol Biol Cell. 2003;14:516–528. doi: 10.1091/mbc.E02-06-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: G-clathrin movements are energy and temperature dependent. In these maximal projection images of time series, the G-clathrin movements in control cells generated a ‘rosetted’ blurred image at 23°C (A and B, left panels, arrows), reflecting their characteristic localized, waving movement. In contrast, in the presence of energy inhibitors (50 mM 2-deoxyglucose and 25 mM sodium azide for 20 min; A, right panel) or at 10°C (B, right panel) the maximal projection shows much smaller spots, indicating that the movements are greatly inhibited. Images of the same fields were acquired at 36-millisecond intervals before and after treatments. Bar: 3 μm (A), 1 μm (B).
Figure S2: Grouping of motile clathrin structures. Moving clathrin spots, comprising all the distinct structures that could be tracked for more than 5 seconds (56 spots from four cells) were tracked and quantified as indicated in Table 1. The velocity (green spots, right ordinate) and distance index (blue bars, abscissa) data for each spot are presented in scatter plot and column, respectively, sorted according to decreasing distance index (a distinctive measure of overall movements; see Table 1). Two distinct groups are distinguished by these parameters: one group comprising vectorially moving clathrin spots (left) displays consistently higher distance index and lower velocity values. The other group (right) shows higher velocity but strikingly lower and uniform distance index values, reflecting the pronounced localized yet rapid motion apparent visually as gyrating or G-clathrin.
Figure S3: AP-2 is absent from G-clathrin structures. mCherry–clathrin and GFP–AP-2 were simultaneously imaged at 50-millisecond intervals. A G-clathrin structure lacking detectable AP-2 signal but with characteristic rapid movement (evident in animation) is noted by the arrow. Bar, 0.5 μm.
Figure S4: GGA1 is present in G-clathrin structures. mCherry–clathrin and YFP-GGA1 were simultaneously imaged using 36-millisecond exposures. A series of frames taken at indicated time-points shows a G-clathrin spot (in the lower part) containing GGA1 moving up and down very quickly, creating a curved line in the kymograph. In contrast, a static clathrin spot (likely coated pit) lacking GGA1 signal creates a straight line in kymograph. Bar, 1 μm.
Video S1: Rapid clathrin movements are imperceptible using long exposures. COS-1 cells expressing GFP-clathrin were imaged using 1 second exposures. Only a barely perceptible blur (box) due to rapid clathrin movements is observed. 40 frames at 1 second/frame, playback at 5 frames/second (5X real-time). Bar, 2 μm.
Video S10: Coordinated movements of G-clathrin and Rab11. Arrows point to regions of G-clathrin (YFP–clathrin) movement with associated mCherry–Rab11. 50 milliseconds/frame, 50 frames total, playback at 20 frames/second (real time). Bar, 1 μm.
Video S11: Tf loading of G-GGA structures is inhibited by LY294002. COS-1 cells expressing YFP-GGA1 were incubated with 20 μg/ml Alexa568-Tf for 1 hour at 16°C, then chased in 37°C medium supplemented with unlabeled Tf for 5 min, all in the presence of 100 μM LY294002, and imaged immediately using 36-millisecond exposures for 50 frames. Tf appearance in G-GGA/clathrin structures (boxed region) is markedly inhibited. Playback at 27 frames/second (real time). Bar, 1 μm.
Video S12: Rapid gyrating movement occurs in nontransfected cells. COS-1 cells were incubated with 20 μg/ml Alexa568-Tf for 15 min at 37°C, washed and images were acquired using 36-millisecond exposures for 100 frames. The boxed region indicates one example of fast gyrating movement. Playback at 27 frames/second (real time). Bar, 1 μm.
Video S2: Rapid clathrin movements revealed using rapid imaging. The same field of COS-1 cells as in Video S1 was imaged using streaming 36-millisecond exposures. Rapid clathrin movements are evident (box) with high speed imaging. 40 frames at 36 milliseconds/frame, playback at 27 frames/second (real-time). Bar, 2 μm. It may be useful for the reader to play back this and other supplemental videos at slower and faster speeds (see ‘A/V control’ in Quicktime).
Video S3: AP-1 is excluded from G-clathrin structures. Essentially no co-localization is observed between mCherry tagged G-clathrin and YFP-tagged AP-1. The boxed region shows an example of G-clathrin structures lacking AP-1. 36 milliseconds/frame, 50 frames total, playback at 27 frames/second (real time). Bar, 2 μm.
Video S4: G-clathrin structures also contain GGA1. Both stationary and highly mobile (near arrows) YFP-tagged GGA1 (middle panel, green) structures also contain mCherry–clathrin (left panel, red). 36 milliseconds/frame, 50 frames total, playback at 27 frames/second (real time). Playback of this and subsequent videos at half-speed may be advantageous. Bar, 1 μm.
Video S5: CD-MPR is present in G-clathrin. COS-1 cells co-expressing mCherry–clathrin and GFP-CD-MPR were imaged using 40-millisecond exposures. Most G-clathrin structures (e.g. boxed region) contain CD-MPR. 50 frames total, playback at 25 frames/second (real time). Bar, 1 μm.
Video S6: Endocytosed Tf appears in G-GGA/clathrin structures. After a 15 min incubation (37°C) with Alexa568-Tf, the majority of G-GGA/clathrin structures (YFP-GGA1) contain Tf (boxed regions). 36 milliseconds/frame, 50 frames total, playback at 27 frames/second (real time). Bar, 1 μm.
Video S7: Moving Tf tubules with GGA1 buds. YFP-GGA1 expressing COS-1 cells were incubated with Alexa568-Tf for 15 min (37°C) and imaged at 36 milliseconds/frame. 140 frames total, playback at 27 frames/second (real time). Boxed region, 4 × 3.5 μm.
Video S8: Endocytosed Tf gradually moves into G-GGA/clathrin structures during a 10 min chase. YFP-GGA1 expressing COS-1 cells were incubated with Alexa568-Tf for 1.5 min at 23°C, washed, chased and the same field was imaged at 2 or 10 min. Very few G-GGA/clathrin structures (green) contain Tf (red) at the earlier time, but virtually all do after 10 min (examples near arrows). 36 milliseconds/frame, 40 frames total, playback at 27 frames/second (real time). Bar, 1 μm.
Video S9: Internalized EGF is absent from G-GGA/clathrin structures. COS-1 cells expressing YFP-GGA1 were incubated with Texas Red-EGF for 10 min (37°C), washed and chased for 10 min, then simultaneously imaged for both EGF (red) and GGA1 (green). Although EGF is present in some GGA1 structures that are stationary or show slow, vectorial movement, essentially none of the G-GGA structures contain EGF. 36 milliseconds/frame, 50 frames total, playback at 27 frames/second (real time). Bar, 3 μm.


