Summary
Fanconi anemia (FA) is a disorder associated with a failure in DNA repair. FANCM (defective in FA complementation group M) and its partner FAAP24 target other FA proteins to sites of DNA damage. FANCM-FAAP24 is related to XPF/MUS81 endonucleases but lacks endonucleolytic activity. We report a structure of an FANCM C-terminal fragment (FANCMCTD) bound to FAAP24 and DNA. This S-shaped structure reveals the FANCM (HhH)2 domain is buried, whereas the FAAP24 (HhH)2 domain engages DNA. We identify a second DNA contact and a metal center within the FANCM pseudo-nuclease domain and demonstrate that mutations in either region impair double-stranded DNA binding in vitro and FANCM-FAAP24 function in vivo. We show the FANCM translocase domain lies in proximity to FANCMCTD by electron microscopy and that binding fork DNA structures stimulate its ATPase activity. This suggests a tracking model for FANCM-FAAP24 until an encounter with a stalled replication fork triggers ATPase-mediated fork remodeling.
Highlights
-
•
FANCM-FAAP24 contains a large head and a curved hook tail
-
•
ATPase activity of FANCM translocase domain is stimulated by forked DNA structures
-
•
An S-shaped FANCM-FAAP24 fragment binds DNA through FAAP24 hairpins
-
•
Both FANCM PND and FAAP24 hairpins are required for FANCM function in vivo
The FANCM-FAAP24 complex anchors the Fanconi anemia core complex onto chromatin and activates DNA repair. Coulthard et al. describe the FANCM-FAAP24 architecture and characterize multiple DNA-binding sites. Forked DNA structures stimulate FANCM ATPase activity, which is important for stalled replication fork remodelling.
Introduction
Fanconi anemia (FA) is a genetically heterogenous disorder characterized by progressive bone marrow failure, elevated hematologic cancer risk, and cellular hypersensitivity to DNA interstrand crosslinking agents (Deans and West, 2011). Sixteen different genes (FANCA-FANCQ) are causative in FA and the gene products participate in the repair of DNA interstrand crosslinks and other lesions that block replication fork progression. Eight of the FA proteins assemble in the FA core complex that associates with chromatin and leads to the mono-ubiquitination of FANCD2 and FANCI (Whitby, 2010). FANCM is the anchor for this core complex in chromatin, and it heterodimerizes with FAAP24 to activate the DNA damage response and promote repair (Ciccia et al., 2007, Kim et al., 2008). Additionally, FANCM facilitates activation of the ATR-mediated DNA damage checkpoint response, defective in Seckel syndrome (Collis et al., 2008, Huang et al., 2010), and damage-mediated targeting of the BLM helicase, defective in Bloom syndrome (Deans and West, 2009). It can thus be thought of as a sensor molecule involved in the activation of several repair and signaling pathways involved in human disease. Full-length FANCM can act as an ATP-dependent branch point translocase that promotes replication fork regression (Gari et al., 2008a, Gari et al., 2008b). The ATPase activity of FANCM is located within the amino-terminal DEAH helicase-like domain, responsible for translocase and branch migration activities (Gari et al., 2008b, Meetei et al., 2005). This ATPase activity is generally found to be dispensable for core complex targeting and FANCD2 ubiquitination but is required for replication fork stability and efficient checkpoint response (Blackford et al., 2012, Collis et al., 2008, Huang et al., 2010).
FANCM is a member of the XPF/MUS81 family of eukaryal heterodimeric endonucleases, many of which are involved in interstrand crosslink repair (Ciccia et al., 2008). These endonucleases are part of a broader nuclease superfamily bearing a PD-(D/E)-X-K catalytic motif that typically uses two-metal-ion catalysis (Steczkiewicz et al., 2012, Yang, 2008). The motif is embedded within a nuclease domain that precedes a tandem helix-loop-helix (HhH) motif at the C-terminal extremity of XPF/MUS81 endonucleases (Nishino et al., 2003). Human FANCM has instead a CD-D-X-R motif and residues G1823 and R1866 that replace the equivalent human XPF residues D676 and K716, known to be essential for XPF endonuclease activity (Enzlin and Schärer, 2002). This has led to the suggestion that FANCM has no intrinsic nucleolytic activity consistent with biochemical evidence (Meetei et al., 2005). While the XPF/MUS81 catalytic motif in FANCM is degenerate, the overall structure of the XPF nuclease domain is retained and it can therefore be described as a pseudo-nuclease domain (PND; Figure 1A).
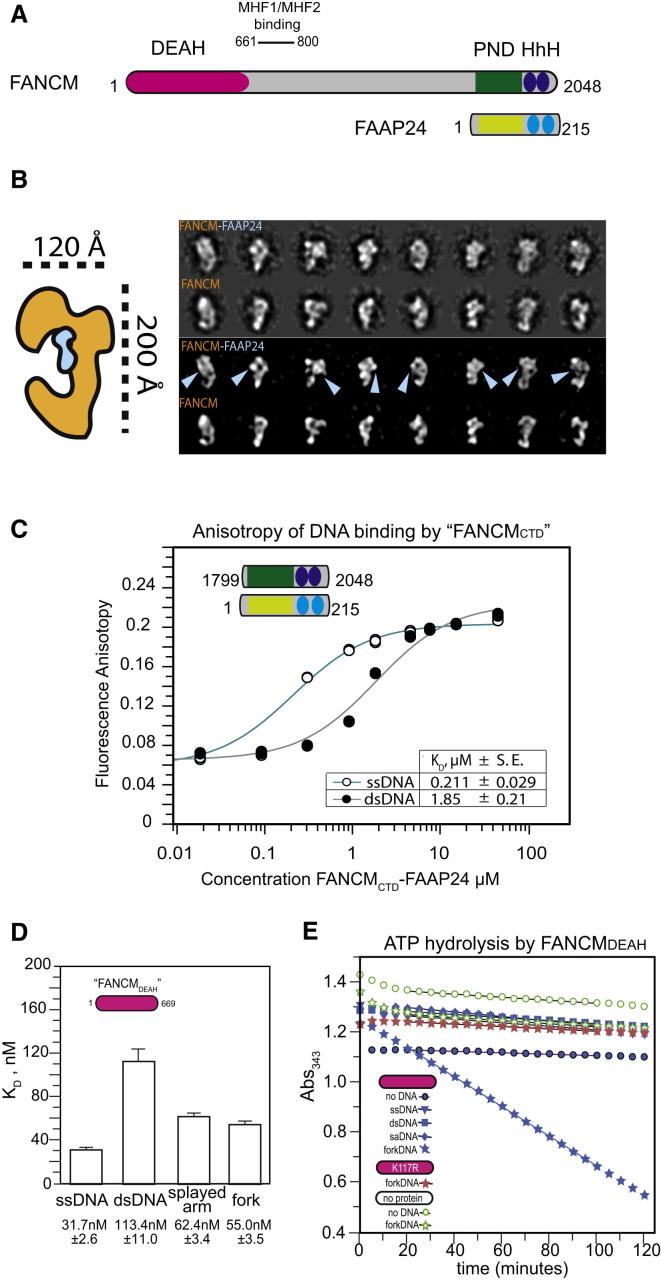
Figure 1.
FANCM-FAAP24 Complex Depicted with Electron Microscopy
(A) Schematic of FANCM-FAAP24 domain structure.
(B) Schematic showing the reconstruction of FANCM (orange) and FAAP24 (blue). Reference-free class averages containing characteristic views of FANCM-FAAP24 particles (raw 1, 3), or of FANCM alone (raw 2, 4). Increased contrast is used in rows 3 and 4 to aid particle comparison. Cyan arrows point at the additional density, which we assign to FAAP24.
(C) Fluorescence polarization assay measurement of FANCMCTD-FAAP24 affinity for ssDNA and dsDNA. Measurements for (C) and (D) were performed in triplicate on at least three independent protein preparations. Kd values were calculated for each experiment, averaged, and an SEM was calculated.
(D) Fluoresence polarization assay measurement of the FANCM1–669 DNA translocase domain’s affinity for different branched and unbranched DNA.
(E) Stimulation of FANCM1–669 ATPase activity by forked DNA structures.
FANCM is constitutively associated with several partner proteins including FAAP24, the MHF histone-fold complex, and HCLK2 (Ciccia et al., 2007, Collis et al., 2008, Yan et al., 2010). The structurally related FAAP24 partner binds the C-terminal region of FANCM containing the pseudo-nuclease and dual helix-loop-helix (HhH) domains. MHF binds to FANCM residues 661–800, while HCLK2 binds to both the FANCM translocase and C-terminal portion. FAAP24 itself has a divergent nuclease-like domain (NLD) that precedes a tandem HhH domain like FANCM. As with other XPF/MUS81 family members, heterodimerization through the C-terminal region may contribute to protein stability and DNA interaction (Chang et al., 2008). The structure of a monomeric FAAP24 HhH domain in the absence of DNA was recently defined, which together with in vitro data, suggested a role in DNA-interaction (Wienk et al., 2013).
Structural analysis of FANCM has focused on the interaction of residues 661–800 with the MHF1/2 histone-like complex (Tao et al., 2012). This portion of FANCM adopts a double “V” shape structure when bound to the MHF1/2 complex to generate a double-stranded DNA (dsDNA) binding site. Other regions of FANCM have not yet been characterized structurally, in particular the amino-terminal FANCM translocase domain or the C-terminal part of FANCM. Here, we describe the structure and biochemical properties of a C-terminal fragment of FANCM containing the PND and (HhH)2 domains bound to full length FAAP24 (referred to as FANCMCTD-FAAP24) and show that it can bind DNA. We identify multiple points of contact between FANCMCTD-FAAP24 and dsDNA, suggesting a DNA tracking role for FANCMCTD-FAAP24. Using EM and biochemical data, we show that the FANCM translocase lies in proximity to FANCMCTD-FAAP24 within the full-length complex and binds DNA. Surprisingly, we found that the FANCM translocase ATPase activity is only stimulated by fork DNA structures, leading us to propose a model for FANCM-FAAP24 tracking and translocase activation upon engagement of a stalled replication fork.
Results
Single Particle Analysis of the Complete FANCM-FAAP24 Complex
To define the overall shape of the FANCM-FAAP24 complex, we imaged full-length FANCM protein alone, and bound to FAAP24 (Figure 1B). The isolated FANCM particle is 200 Å × 120Å and contains a concave, large head and an elongated, curved tail, resembling a hook. When bound to FAAP24, FANCM appears to maintain its overall structure, allowing us to directly compare matching views of the monomeric and heterodimeric particles. Additional electron density, which we assign to FAAP24, fills the FANCM concave head, indicating that FAAP24 and FANCMCTD occupy the central core in this dimeric assembly. Our observations imply that the FANCMCTD and FANCM N-terminal translocase domain are likely to be arranged in proximity, rather than mapping on the two distal poles of the particle, and may therefore cooperate in the recognition of DNA damage.
DNA-Binding Properties of FANCMCTD-FAAP24 and FANCM1–669
To define how FANCM-FAAP24 engages dsDNA structures, we used fluorescence anisotropy to characterize the DNA-binding properties of a truncated C-terminal fragment of FANCM (FANCMCTD, residues 1,799–2,048) containing the pseudo-nuclease and (HhH)2 domains bound to FAAP24 (Figure 1C) and the FANCM DNA translocase domain (residues 1–669). This reductionist approach was necessary given the relatively small amounts of full-length recombinant FANCM-FAAP24 that could be purified (Figure 1D). Purified FANCMCTD-FAAP24 complex bound to both single-stranded DNA (ssDNA) and dsDNA with dissociation constants (Kd) of 0.21 and 1.85 μM, respectively. These affinities are comparable to values reported for the XPF-ERCC1 endonuclease under identical conditions (Tripsianes et al., 2005). The FANCM translocase domain bound more tightly to ssDNA than the FANCMCTD-FAAP24 complex with a Kd of 30 nM. It also bound well to branched DNA structures such as a splayed arm (63.4 nM) and a replication fork (55 nM; Figure 1D). These data show that FANCM-FAAP24 has multiple discrete DNA-binding sites present at either end of FANCM in addition to the site previously described for the MHF1/2-binding region.
ATPase Activity of FANCM1–669
Having established that FANCM1-669 is able to bind DNA, we characterized the ATPase activity of this protein fragment (Figure 1E). Previous studies have reported that the translocase domain of FANCM shows either spontaneous ATPase activity or an ATPase activity that is stimulated by ssDNA and dsDNA (Mosedale et al., 2005, Xue et al., 2008). However, in our hands, in the absence of DNA, FANCM1–669 showed no detectable ATPase activity. Furthermore, the addition of a variety of DNA structures that are bound tightly by FANCM1–669 were unable to stimulate ATPase activity. The only exception was static fork-structured DNA, which induced a dramatic increase in ATP consumption, corresponding to an ATPase rate of ∼16 molecules of ATP per minute per molecule of FANCM1–669. This stimulation by fork DNA was not observed with a mutant protein containing a point mutation in the Walker A motif (FANCM1–669-K117R). The ability of FANCM1–669 to be activated by fork-structured DNAs is reminiscent of Pyrococcus furiosus XPF, which has a 4- to 12-fold higher rate of ATP hydrolysis with branched duplex DNAs than linear duplex DNA (Komori et al., 2004). Our data therefore suggest that FANCM1–669 is a DNA-stimulated ATPase that is highly specific for fork-structured DNA.
Structure of the FANCMCTD-FAAP24-dsDNA Complex
To understand the basis for DNA-binding and recognition by FANCMCTD-FAAP24, the complex was crystallized with an 11 base-pair DNA duplex. The crystals diffracted to 2.15 Å and the structure was determined using a single mercury derivative (Table 1; Figure S1A available online). The structure revealed that the FANCMCTD-FAAP24 heterodimer has an S-shaped conformation and uses the FAAP24 (HhH)2 domain to bind both DNA strands across a minor groove of duplex DNA (Figure 2A). The FAAP24 (HhH)2 domain has a footprint spanning five base pairs of DNA similar to that observed for the Aeropyrum pernix (HhH)2 domain (Newman et al., 2005) and in both cases, the DNA is only slightly distorted from a classical B-DNA form (Figure 2B). The structure consists of two parts: the FANCMCTD PND bound to the NLD of FAAP24 (defined hereafter as the PND/NLD heterodimer) connected by flexible linkers to a heterodimer of (HhH)2 domains from FANCM and FAAP24 (defined hereafter as [HhH]2 dimer). The PND/NLD dimer and the (HhH)2 dimer have nonintersecting approximate 2-fold axes giving the complex an overall asymmetric shape. The PND/NLD dimer and the (HhH)2 dimer have independent dimerization interfaces (Figures S2 and S3) and are coupled through an interface between the FANCM (HhH)2 domain and the FANCM PND (Figure 2A). This interface provides rigidity to the complex, which is supported by an analysis of translation-liberation-screw (TLS) parameters (Figures S1B and S1C). The FANCM PND and (HhH)2 domains are therefore flanked by the equivalent FAAP24 domains.
Table 1.
Data Collection and Refinement Statistics
| Native | EMTS | |
|---|---|---|
| Data Collection | ||
| Space group | P41212 | P41212 |
| Cell dimensions | ||
| a, b, c (Å) | 125.1, 125.1, 74.8 | 124.4, 124.4, 75.2 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Wavelength (Å) | 1.5418 | 1.5418 |
| Resolution (Å) | 2.15 | 3.2 |
| Rmerge | 0.058 (0.371) | 0.153 (0.354) |
| I/σI | 11.4 (2.1) | 4.9 (2.2) |
| Completeness (%) | 100 (100) | 99.8 (100) |
| Redundancy | 10.0 (9.9) | 24.2 (24.7) |
| Refinement | ||
| Resolution (Å) | 25.433–2.15 | |
| No. reflections | 30,696 | |
| Rwork/Rfreea | 0.207/0.253 | |
| No. atoms | ||
| Protein | 3,079 | |
| DNA | 418 | |
| Ligand/ion | 4 | |
| Water | 100 | |
| B-factors | ||
| Protein | 30.373 | |
| DNA | 68.645 | |
| Ligand/ion | 29.935 | |
| Water | 32.54 | |
| Rmsd | ||
| Bond lengths (Å) | 0.014 | |
| Bond angles (°) | 1.449 | |
Values in parentheses are for highest-resolution shell. EMTS, ethylmercury thiosalicylate.
A total of 5% of the data were set aside to compute Rfree.
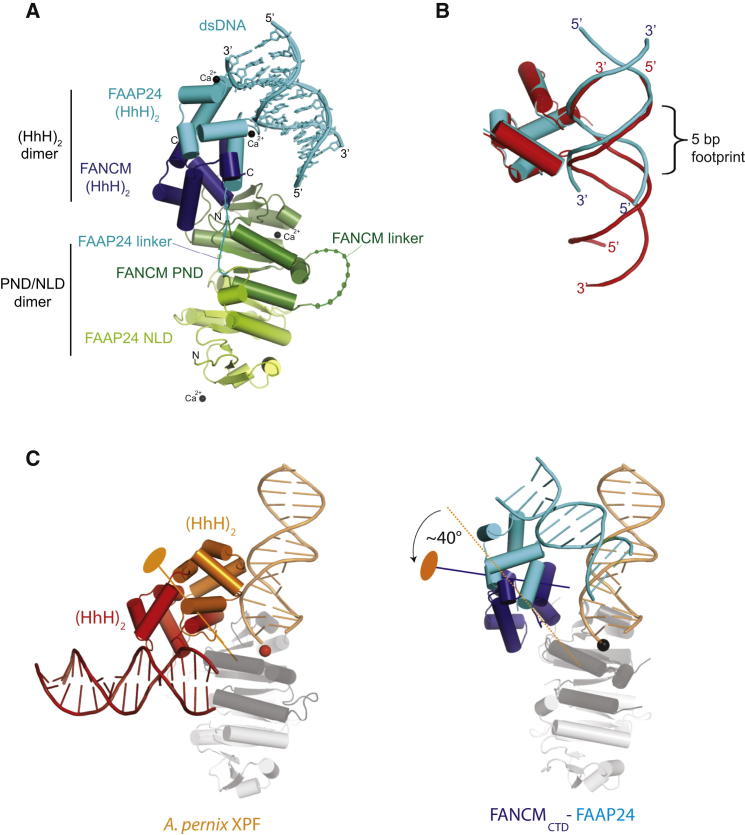
Figure 2.
Crystal Structure of an S-Shaped FANCMCTD-FAAP24 Complex Bound to dsDNA
(A) Schematic representation of the asymmetric unit. Calcium ions are shown as black spheres. Linked colored spheres show number and approximate location of residues for the linker connecting the FANCM-PND to its (HhH)2 and the FAAP24 NLD to its (HhH)2. Both linkers and one loop that had no electron density in the crystal structure are presumably disordered.
(B) DNA footprint on dsDNA for A. pernix XPF (red) and FAAP24 (cyan) (HhH)2 domains.
(C) Rotation of the (HhH)2 domains relative to the superposed ND/PND/NLD-domains from the A. pernix XPF and FANCMCTD-FAAP24 structures. Approximate location of the (HhH)2 dimer axis is shown in blue for FANCM-FAAP24, orange for A. pernix XPF.
See also Figure S2.
An important consequence of the FANCM PND-(HhH)2 domain interaction is that the relative angle between the PND/NLD dimer and the (HhH)2 dimer is distinct from other XPF family structures, thus altering the trajectory of the dsDNA relative to the FANCM PND metal center (as discussed later). The angle differs by at least 40° from that observed in the homodimeric XPF from A. pernix (Newman et al., 2005; Figure 2C) and by an even larger angle from the human MUS81-EME1 complex (not shown; Chang et al., 2008). Superposition of the A. pernix (HhH)2 homodimer onto the FANCMCTD-FAAP24 (HhH)2 heterodimer shows an almost identical 45° angle between the (HhH)2 dimer molecular 2-fold axis (or approximate 2-fold for FANCMCTD-FAAP24) and principle axis of the DNA duplex (Newman et al., 2005; Figure 2C). We conclude that FANCMCTD-FAAP24 provides a rigid platform that sets up a unique trajectory for engaging dsDNA, distinct from other XPF/MUS81 structures.
The FANCM (HhH)2 Hairpins Are Buried against the FANCM Pseudo-Nuclease Domain
The FANCM intramolecular coupling interface described above buries the FANCM (HhH)2 domain hairpins completely. The functional role of the FANCM-(HhH)2 domain is thus quite distinct from that observed for FAAP24-(HhH)2. Unlike those found in FAAP24 and the HhH domains of many other proteins, the FANCM hairpins lack the conserved PGhG motif (where h is any hydrophobic residue; Figure S2), suggesting it is unlikely to bind dsDNA even if it were accessible. Indeed, the intramolecular interactions between the pseudo-nuclease and the (HhH)2 domains of FANCM observed in our structure give the structure its distinctive S-shape (Figure 2A). I1985 and T1986 from the first FANCM-HhH are completely buried in the FANCM pseudo-nuclease:(HhH)2 interface, contributing to the 559.7 Å2 buried surface area at the interface. These residues are equivalent to V172 and K173 of the first FAAP24 HhH that directly contacts dsDNA (discussed later).
There is good conservation of FANCM hairpin residues at the PND:(HhH)2 interface but only within the FANCM family. The structure of an equivalent fragment from A. pernix XPF also shows a nuclease-(HhH)2 domain interface comprising a similar hydrophobic patch on the nuclease domain but quite different contact residues from the (HhH)2 domain (Figure 2C; Figure S2; Newman et al., 2005). This difference is attributable to the different relative position of the (HhH)2 domain to the XPF nuclease domain. A similar hydrophobic patch is also seen in the nuclease domain of the MUS81 structure but in this case the patch is shielded by a linker helix between the MUS81 nuclease and (HhH)2 domains (Chang et al., 2008). This comparison shows the considerable variation in the relative orientations of PND/nuclease domains to the (HhH)2 domains, which for active endonucleases may be crucial for proper guiding of substrates into the active site. The appreciable PND domain-(HhH)2 domain interface in FANCM ensures a rigid platform for engaging dsDNA structures.
FAAP24 Hairpin Residues Are Functionally Required
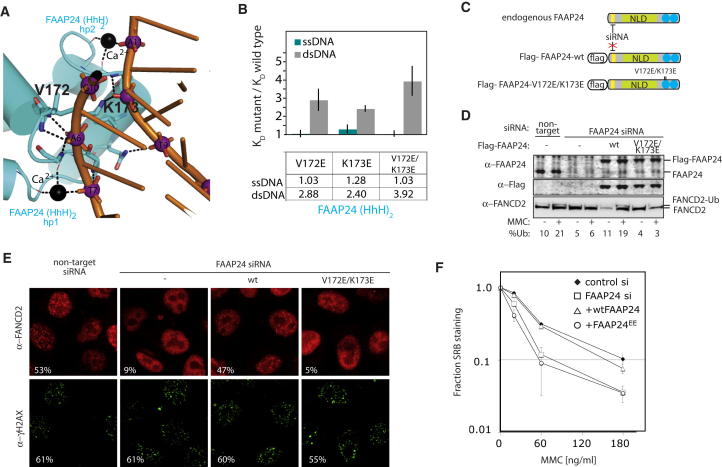
The FANCMCTD-FAAP24-dsDNA structure reveals how the FAAP24 (HhH)2 domain uses four main-chain amide groups from each hairpin motif to engage the phosphodiester backbone of each DNA strand (Figure 3A). This interaction allows the (HhH)2 domain to bridge across the minor groove of the duplex near its blunt end. The side chains of V172 and K173 from the first FAAP24 hairpin also make important DNA contacts. The V172 side chain packs against a DNA ribose close to the first hairpin, whereas K173 side chain contacts the phosphodiester bridge bound by the second FAAP24 hairpin. Two metal ions are also bound to backbone carbonyl groups and directly contact the DNA phosphate moieties. To validate the functional importance of the two FAAP24 contact residues in solution, we mutated V172 and K173 to glutamate. Using the fluorescence polarization assay, both mutants were found to have a 2- to 3-fold weaker affinity for dsDNA compared to a wild-type FANCMCTD-FAAP24 control but had little effect on ssDNA binding (Figure 3B). A double V172E/K173E mutation in the hairpin regions had an additive effect, resulting in an approximate 4-fold decrease in affinity specific for dsDNA. These data indicate that FAAP24 hairpins are involved in crosslinking both strands of a dsDNA duplex in vitro.
Figure 3.
FAAP24 (HhH)2 Domain Makes Functionally Important Contacts with dsDNA
(A) Detailed view of FAAP24 (HhH)2 domain interaction with dsDNA through minor groove phosphates.
(B) Targeted mutations in a FAAP24 hairpin affect the dsDNA equilibrium dissociation constants relative to a wild-type control in the fluorescence polarization assay (average of three experiments; error bars show SEM). Values shown are the quantification of the histogram. Measurements were performed in triplicate on at least three independent preparations. The ratio of equilibrium dissociation constants of the mutant protein relative to wild-type was calculated for each experiment, averaged, and an SEM was calculated.
(C) Scheme of the complementation system. Endogenous FAAP24 is knocked down by transfection of STEALTH RNAi but the exogenous Flag-FAAP24 contains an siRNA-resistant wobble mutation (yellow).
(D) Western blotting of FAAP24 and FANCD2. FANCD2 ubiquitinated and nonubiquitinated forms are indicated. %Ubi is the quantified ratio of ubiquitinated to nonubiquitinated FANCD2 forms.
(E) Indirect immunofluorescence of cells treated with 40 ng/ml mitomycin C for 7 hr as described in the Experimental Procedures. The percentage of nuclei with > 5 foci scored from 300 nuclei counted is shown in the inset for each example.
(F) Cell survival after treatment with mitomycin C was measured using the SRB assay. Points represent the average of two experiments, each performed in quadruplicate, ± average SE.
To assess the in vivo requirement for functional FAAP24 hairpins in this role, we expressed the V172E/K173E mutant using a 293 cell-based complementation system (Figure 3C). FAAP24 was previously shown to be required for activation of the FA DNA repair pathway in response to the interstrand crosslinking drug mitomycin C (Ciccia et al., 2007). As previously observed, siRNA-mediated knockdown of FAAP24 resulted in reduced FANCD2 ubiquitination and foci formation and increased sensitivity to mitomycin C (Figures 3D–3F). While expression of siRNA-resistant wtFAAP24 protein could rescue these phenotypes, FAAP24-V172E/K173E mutant protein could not. These results provide cellular evidence that direct DNA binding by FAAP24 is required for the activation of the FA pathway, and consequently the cellular response to interstrand crosslink (ICL) damage.
The FANCM Pseudo-Nuclease Domain Has a Metal Center and Makes DNA Contacts Required for FANCD2 Ubiquitination
The FANCM PND domain structure exhibits the characteristic αβββαβ-fold topology of PD-(D/E)-X-K nucleases and is closely related to the catalytically active nuclease domain of A. pernix XPF (55.83% identity, root-mean-square deviation [rmsd] of 1.322 Å over 116 Cα atoms; Newman et al., 2005, Nishino et al., 2003). Within the central cleft of the FANCM PND domain, we observed a single metal ion, distinguished by coordination geometry and ligand distances (Figure S4). This metal ion, most likely a calcium ion derived from the crystallization buffer, is coordinated by acidic side chains of D1854 and E1864, as well as the backbone carbonyl of R1865 (Figure 4A). The calcium ion superposes almost exactly with the metal center found in XPF and one of the metal ions present in type II restriction enzymes (Figure S4; Newman et al., 2005, Nishino et al., 2003). In FANCM, the metal-binding D1854 side chain also hydrogen-bonds to the H1825 side chain and a water molecule. Residues in spatial proximity to the metal site are well placed to contact the DNA phosphodiester backbone. These include residues such as E1870 and R1866, supported by comparisons with the type II restriction enzymes bound to their cognate dsDNA.
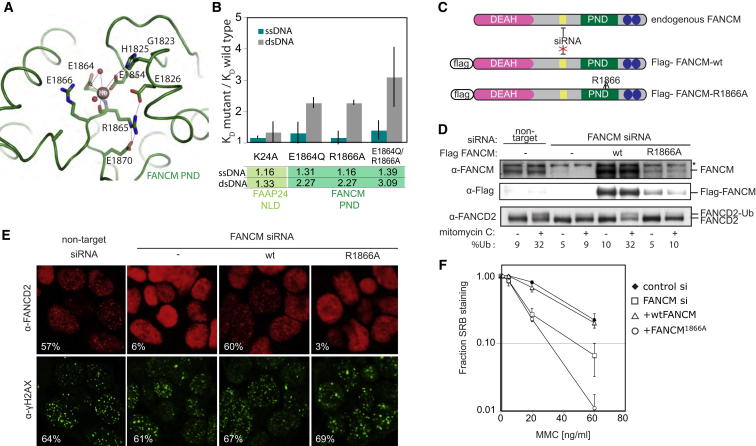
Figure 4.
A Divalent Metal Site within the FANCMCTD Pseudo-Nuclease Domain Is Essential for FANCM Function
(A) Schematic of selected side chains close to the divalent metal binding site within FANCM PND.
(B) Mutations in the metal-binding cleft, but not the equivalent cleft of FAAP24, increase the equilibrium dissociation constants for dsDNA relative to the wild-type protein. Values shown are the quantification of the histogram. Measurements were performed in triplicate on at least three independent preparations. The ratio of equilibrium dissociation constants of the mutant protein relative to wild-type was calculated for each experiment, averaged, and an SEM was calculated.
(C) Scheme of the complementation system. Endogenous FANCM is knocked down by the stable siRNA but the exogenous Flag-FANCM contains an siRNA-resistant wobble mutation (yellow).
(D) Western blot analysis of FANCM and FANCD2. Asterisk denotes nonspecifc band detected with FANCM antibody. FANCD2 ubiquitinated and nonubiquitinated forms are indicated. %Ubi is the quantified ratio of ubiquitinated to nonubiquitinated FANCD2 forms.
(E) Indirect immunofluorescence of cells treated with 40 ng/ml mitomycin C for 7 hr as described in the Experimental Procedures. The percentage of nuclei with > 5 FANCD2 or γH2AX foci scored from 300 nuclei counted is shown in the inset for each example.
(F) Cell survival after treatment with mitomycin C was measured using the SRB assay. Points represent the average of three experiments, each performed in quadruplicate, ± SE.
See also Figure S4.
Within the crystal lattice, access to the FANCM PND metal center is blocked by the packing of a symmetry-related FAAP24 molecule, precluding binding of dsDNA. We therefore tested in solution whether the FANCM PND metal-binding cleft is involved in binding dsDNA, by characterizing the effect of two mutations (E1864Q and R1866A) on DNA binding by the FANCMCTD-FAAP24 complex (Figure 4B). E1864Q was designed to disrupt the metal binding site, and R1866 is a nearby residue that could interact with DNA bound in the cleft. Both mutations selectively reduced binding of dsDNA by more than 2-fold relative to the wild-type protein but did not affect binding to ssDNA (Figure 4B). An equivalent mutation within the central cleft of FAAP24, K24A, did not alter the affinity of the complex for dsDNA (Figure 4B). This suggests that FAAP24, like ERCC1 (the heterodimeric partner of XPF), does not bind to DNA using this region (Tripsianes et al., 2007). We also mutated of R1860, located adjacent to a symmetry-related dsDNA, to alanine, which perturbed both ssDNA and dsDNA binding (data not shown).
To explore the role of metal binding in FANCM activation of DNA repair, we tested the R1866A mutant for its ability to complement a stable knockdown of endogenous FANCM (Figure 4C; Deans and West, 2009). The siRNA resistant mutant protein could not rescue defective FANCD2 ubiquitination, reduced FANCD2 foci formation or increased mitomycin C sensitivity of cells depleted of endogenous FANCM (Figures 4D–4F). This is despite this mutant having only 2- to 3-fold reduction in in vitro DNA binding assays (Figure 4B) and the fact that all cell lines tested had similar levels of mitomycin C-induced damage as measured by γH2AX foci formation. Mutation of the R1866 equivalent within human XPF eliminates its nuclease activity (Enzlin and Schärer, 2002). Since human FANCM is inactive as an endonuclease, we assume the phenotype of the R1866A is due to reduced ability of FANCM to bind DNA and target the FA core complex to DNA damage. These data confirm that the residues within the PND cleft of FANCM are crucial for the in vivo function of FANCM despite the apparent lack of endonucleolytic activity.
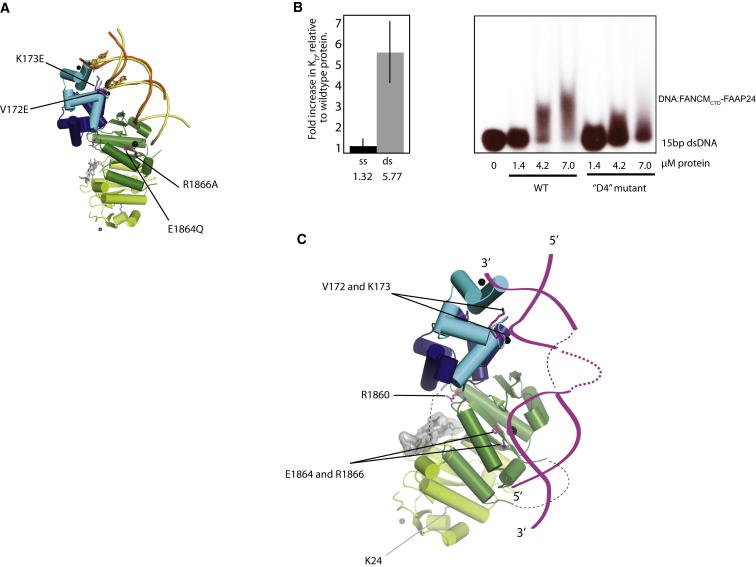
The presence of two discrete DNA-binding sites within the C-terminal portion of FANCM (one within the PND and a second via the FAAP24 hairpins) suggests they engage the same DNA structure or structures containing duplex region(s), such as stalled replication forks (Figure 5B). To assess the impact of combining mutations from both FANCM-PND and FAAP24 (HhH)2 toward dsDNA, we prepared a quadruple mutation (D4) combining all four mutations. This resulted in a 6-fold reduction in dsDNA binding by FANCMCTD-FAAP24 (Figure 5B), whereas ssDNA binding was only decreased by 1.3-fold. This finding was confirmed by electromobility shift experiments where the D4 mutant showed a reduced level of duplex DNA shift compared with ssDNA (Figure 5C). These data suggest both regions do indeed contribute to binding the same dsDNA. We then considered the trajectory of the linear DNA bound to FAAP24 hairpins relative to the FANCM PND cleft. Extending the 11 base-pair duplex bound by the FAAP24 (HhH)2 domain to 15–16 base pairs as a linear B-DNA does not bring the duplex close enough to R1866 to interact with the metal center directly (Figure 5A), suggesting the DNA has to be distorted to contact the FANCM PND (Figure 5C). Whether such a distortion is driven by the FANCM ATPase activity or by encountering damaged DNA structures requires further experiments and is outside the scope of this study.
Figure 5.
Bipartite Duplex Recognition within FANCMCTD-FAAP24 Requires Distortion of dsDNA
(A) Extending the 11 base pair dsDNA (yellow) compared to the experimentally observed dsDNA (orange) glances past the metal center of FANCM PND domain.
(B) Effect of combining all four DNA-binding mutations (D4) in FANCMCTD-FAAP24 on the equilibrium dissociation constant for ss- and dsDNA normalized against wild-type protein (left). D4 refers to FANCM:E1864Q/R1866A-FAAP24:V172E/K173E. Right, electrophoretic mobility shift assay showing wild-type and D4 mutant binding to ss and dsDNA. Measurements were performed in triplicate on at least three independent preparations. The ratio of equilibrium dissociation constants of the mutant protein relative to wild-type was calculated for each experiment, averaged, and an SEM was calculated.
(C) Distortion of dsDNA is required to proceed from the FAAP24 hairpins into the FANCM PND metal-binding cleft to permit simultaneous engagement of both DNA-binding regions of FANCMCTD-FAAP24.
Selected residues referred to in the text are shown as sticks including K24 from FAAP24 and R1860 near a dsDNA symmetry contact (grey surface).
Discussion
Understanding the function of the FA core complex in the repair of DNA damage requires a molecular understanding of how the complex is recruited to DNA by FANCM and its associated partner proteins. We have shown that the FAAP24 subunit, despite having some FANCM-independent roles (Wang et al., 2013) plays two important roles in the function of the much larger FANCM subunit. First, it is necessary for organization of the FANCM pseudo-nuclease and (HhH)2 domains into a rigid S-shaped structure. Second, it is also involved in a direct DNA targeting role for FANCM similar to that proposed for ERCC1 within the XPF-ERCC1 endonuclease complex from nuclear magnetic resonance titration of labeled DNA (Tripsianes et al., 2007). Our observation that the FAAP24 (HhH)2 domain engages both DNA strands of a duplex is at odds with a recent solution study on an isolated monomeric FAAP24 (HhH)2 domain that engaged only a single strand (Wienk et al., 2013). Large structural differences (rmsd of 2.3 Å over 51 Cα) between our structure and the isolated FAAP24 (HhH)2 domain of Wienk et al. (2013) are likely to account for the different DNA-binding preferences (Figure S5).
In this crystallographic study we have shown FANCMCTD-FAAP24 forms a heterodimer through independent dimerization of both the (HhH)2 and PND/NLD domains (Figure S3). This is similar to interfaces seen in MUS81-EME1, archaeal XPF (HhH)2, and ERCC1(HhH)2 structures and suggests a common mode of dimerization in members of the XPF/MUS81 family (de Laat et al., 1998, Tsodikov et al., 2005). It then raises the question: why is FANCM:FAAP24 not a nuclease? This is especially pertinent as we observe that FANCM PND retains a single metal-ion site directly equivalent to site A of many type II restriction enzymes and other PD-D/E-X-K nucleases (Yang, 2008). Metal site B, crucial for catalysis in related nucleases, is not found in our structure but is also known to be notoriously difficult to trap even in the presence of substrate (Yang, 2008). Both metal sites (A and B) are often bridged by a single acidic side chain (PD-[D/E]-X-K in type II restriction enzymes), equivalent to FANCM D1854 (Pingoud et al., 2005). Simple electrostatic calculations using the program ABPS (Baker et al., 2001) suggest that D1854 has a very low pKa, similar to the two metal ion-coordinating D52 side chain of A. pernix XPF. Therefore, it still remains a possibility that FANCM could bind a second B-site metal ion through D1854. The impact of the three FANCM-specific residues (G1823, H1825, and R1866) that cluster around the acidic side chains D1854 and E1864 may influence and stabilize the low pKa values for these residues. However, no detectable endonuclease activity has been observed for human FANCM-FAAP24. Moreover, substituting the GD-E-X-K catalytic motif of human XPF-ERCC1 by GD-E-X-R (to mimic the FANCM substitution) has a significant negative impact on XPF-ERCC1 activity, also consistent with a FANCM-FAAP24 role as a pseudo-nuclease (M.B., unpublished data). Comparisons with other FANCM orthologs indicate that only hominid FANCM orthologs have substitutions deviating from the PD-(D/E)-X-K nuclease consensus, suggesting that the loss of its nuclease activity in hominids is a quite recent event (Figure S4). So the inferences made here for the human FANCM PND metal-binding site may not hold for FANCM orthologs in lower vertebrates. The absence of human FANCM PND endonuclease activity may permit the retention of the domain for targeting without cutting branched DNA molecules.
Another significant difference between our structure and that of MUS81-EME1 and XPF-ERCC1 lies in the FANCM pseudo-nuclease-(HhH)2 interface. The interface results in a rigid FANCM-FAAP24 complex and directs the duplex DNA bound by the FAAP24 (HhH)2 domains toward the FANCM metal binding site. This interface is not conserved within the XPF family, and the MUS81-EME1 and A. pernix XPF crystal structures have a different orientation of the two domains. The relative orientation of the pseudo-nuclease and (HhH)2 domains has functional consequences for FANCM-FAAP24. Instead of directly entering the FANCM pseudo-catalytic center as proposed for the A. pernix homolog (Newman et al., 2005), longer B-DNA duplexes would glance past the PND cleft, needing an appreciable deformation to engage proximal residues to the metal ion center (Figure 5).
DNA targeting is important when considering how FANCM activates the FA core complex. Taken together, our data suggest that although the FANCMCTD-FAAP24 module binds to DNA with a much weaker affinity than the translocase domain of FANCM, this is a critical interaction in vitro and in vivo. The overall ability of the CTD of FANCM to interact with DNA is of course greatly enhanced by the fact that the N-terminal domain provides constitutive association with chromatin. This association comes from a stable complex with the MHF1/2 histone-fold dimer as well as intrinsic DNA binding in the DEAH domain (Xue et al., 2008, Yang, 2008). A similar situation is observed for the archaeal protein HEF, where cooperativity of DNA binding is seen between an N-terminal DEAH translocase domain and a C-terminal XPF-family nuclease domain (Nishino et al., 2005). For HEF, this cooperativity enhances the nuclease activity of an N-terminal fragment in trans by a separately expressed C terminus (Komori et al., 2004). We therefore suggest that the DNA binding affinities of FAAP24 and both the FANCM DNA translocase and C-terminal domains are required for the initial recognition of the complex DNA structure caused by a stalled replication at the ICL (Figure 6). This substrate is bound by the FANCM-translocase, FANCM pseudo-nuclease, and FAAP24-(HhH)2 domains in a cooperative fashion. As the ATPase activity of the translocase domain appears not to be required for DNA targeting (Xue et al., 2008), facilitated diffusion may be sufficient for substrate searching. This searching may be assisted by the metal ions bound to the FAAP24 (HhH)2 domain and the FANCM PND (our modeling studies suggest the DNA would need to be distorted to do this). At a stalled replication fork, the FANCM ATPase activity is stimulated and initiates repair (Figure 6). Our electron microscopy data show that the N- and C-terminal domains may lie in proximity and therefore a subsequent encounter with a branched DNA molecule could trigger a conformational change affecting the central region of the FANCM protein. This large intervening region contains domains required for core complex interaction and interaction with the Bloom’s complex (Deans and West, 2009), which may be exposed to promote recruitment of these complexes upon such a conformational change. The model proposed herein provides insights into the structural aspects of FANCM-FAAP24 DNA-binding crucial for targeting of the FA core complex to sites of interstrand crosslinks stalled/damaged replication forks.
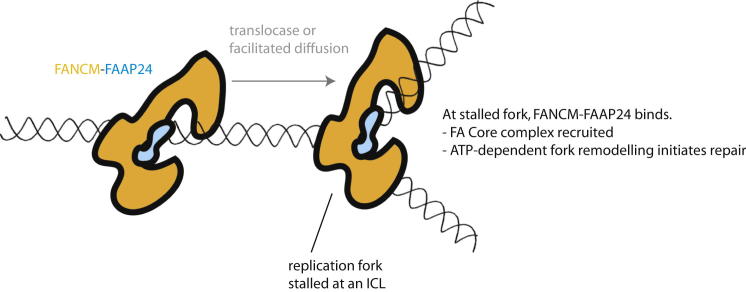
Figure 6.
Model for FANCM-FAAP24 Tracking and Stimulation of ATPase Activity by Fork Encounter
A cartoon representation of FANCM-FAAP24 is shown tracking along dsDNA until an encounter with a stalled replication fork. At this point, the fork stimulates FANCM ATPase activity, resulting in fork remodeling and initiation of DNA repair.
Experimental Procedures
Protein Preparation
FANCM1799–2948-FAAP24 (FANCMCTD–FAAP24) was expressed and purified as described previously (Ciccia et al., 2007). Mutants were produced by Quikchange (Stratagene) using site-directed mutagenic primers (Sigma, UK) to amplify a mutant form of the original plasmid in a PCR reaction, and confirmed by sequence analysis. These were expressed and purified identically to the wild-type protein in Escherichia coli. FANCM1–669 (spanning the FANCM DNA translocase domain) was expressed as a GST-fusion protein in E. coli using standard glutathione-resin affinity tag protocols. Further purification was performed with a heparin affinity column that purified FANCM translocase to homogeneity.
For purification of full-length FANCM protein, the FANCM cDNA was cloned into MCS1 of pFBDM with an in frame N-terminal Flag-tag and C-terminal 8His-tag (Berger et al., 2004). For dual expression of FANCM and FAAP24, untagged FAAP24 was cloned into MCS2 of the same construct. Baculovirus were generated using the Multibac system (Berger et al., 2004). Sf9 cells (2 l at 1 × 106/ml) were infected with baculovirus at a multiplicity of infection of 2 and grown at 27°C for 72 hr. Cells were harvested, washed twice with PBS, and resuspended in buffer K50 (50 mM potassium phophate, pH 7.5, 50 mM KCl, 10% glycerol, 1 mM dithiothreitol (DTT), 1 mM EDTA, and complete protease inhibitors [Sigma]). Cells were incubated for 30 min on ice followed by 40 strokes in a dounce (pestle A), with the addition of further KCl to a 400 mM final concentration and 40 more strokes in dounce (pestle B). Lysates were centrifuged at 30,000 × g twice for 30 min and cleared supernatants applied to a 1 ml Flag affinity resin column (Sigma M2). The column was washed with buffer K400 (as per K50 but 400 mM KCl) and an intermediate wash of K400 + 1 mM ATP + 3 mM MgCl2 (this is to remove HSP70 contaminant) and then eluted with buffer N (50 mM potassium phosphate, pH 7.5, 150 mM KCl, 10 mM imidazole, 10% glycerol) containing 125 μg/ml 3× Flag peptide (Sigma). Fractions containing FANCM were then incubated with 250 μl of Nickel-NTA resin (QIAGEN) washed in buffer N and then eluted in buffer N+300 mM imidazole. Peak fractions were pooled, desalted against buffer N, and loaded onto a 1 ml heparin column (GE). Protein was eluted in buffer N+1 mM DTT with a gradient of 150 mM to 1.5 M KCl. FANCM-containing fractions eluted at around 350 mM KCl and were snap-frozen and stored in aliquots at −70°C.
Single Particle Electron Microscopy
Full-length FANCM or FANCM-FAAP24 complexes were deposited onto continuous carbon grids and stained with a 2% uranyl acetate solution. Electron micrographs were collected using a Tecnai G2 Spirit (FEI) operating at 120 keV. Samples were imaged at a nominal magnification of 30,000× (3.5 Å/pixel at the specimen level). Data were acquired using an electron dose of 35 electrons/Å2 on a 2k × 2k GATAN Ultrascan 1000 camera. Image stacks were prepared using the Eman2 image processing package (Tang et al., 2007). Reference-free two-dimensional class averages were calculated using routine multivariate statistical analysis/multireference alignment protocols in IMAGIC (van Heel et al., 1996).
Crystallization and Data Processing
FANCMCTD-FAAP24 was co-crystallized with an 11 base pair dsDNA substrate. High performance liquid chromatography-purified oligo GATGATGCTGA was annealed to its complementary strand (Jena Bioscience, Germany) and then purified by fast protein liquid chromatography with a mono Q ion exchange column (GE Healthcare). The duplex DNA was mixed in a 3:1 molar ratio with purified FANCMCTD-FAAP24 and crystallized at 4°C with the sitting drop vapor diffusion method. The crystallization condition was 18% PEG 3350, 0.18 M calcium acetate, and 10 mM barium or strontium chloride. Native and derivative X-ray diffraction data sets were collected on a home source at 100 K. Crystals were cryo-protected by transfer into a final solution of 20% PEG 3350, 20% PEG 400, and 0.2 M calcium acetate. Both data sets were processed in Mosflm and scaled using Scala (Evans, 1993). Experimental phases were obtained to 3.5 Å using single anomalous derivative phasing in the direct methods package BnP (Weeks et al., 2002; Table 1) and improved by Resolve (Terwilliger, 2000). A model was built using Coot (Emsley and Cowtan, 2004) and refined using Refmac5 (Murshudov et al., 1997) and rebuilding in Coot (Table 1), including TLS toward the end of refinement. The FANCMCTD structure contains residues T1818–L2035, lacking a loop in the PND domain (E1903–F1912) and part of the FANCM PND-(HhH)2 linker, which were not modeled. The FAAP24 chain spans residues V11–P214, excepting seven residues from the interdomain linker. All residues in the refined structure fell within the allowed dihedral angles (4.2%) or most favored (95.8%) of the Ramachandran plot (Lovell et al., 2003).
DNA Affinity Measurements by Fluorescence Anisotropy
The fluorescence polarization-based DNA-binding assays were performed as described previously (Ciccia et al., 2007). The DNA was fluorescently tagged with a 5′ 6-FAM label (Sigma UK) and the anisotropy in the polarization of light emitted by this probe was recorded at equilibrium. This anisotropy is dependent on rotational tumbling time of the molecule: when the DNA is bound by a protein, the emitted light is more anisotropic. Anisotropy measurements were performed after a 1 hr incubation time on a TECAN SAFIRE-2 at a range of protein concentrations (0 to ∼100 μM) and the resulting binding curves were fitted to a standard equation (Equation 1) using the nonlinear regression software GraFit (Erithacus Software):
| (Equation 1) |
where A is anisotropy, Af is anisotropy of free ligand, Ab is anisotropy at saturation, [L] is concentration of protein, and Kd is the apparent dissociation constant.
Assays were performed with 50 nM DNA substrate in a final buffer composition of 20 mM NaCl; 25 mM Tris, pH 8; 2 mM MnCl2, 0.1 mg/ml BSA, and 4 mM 2-mercaptoethanol. Oligos were purchased PAGE-purified from SIGMA-GENOSYS UK and are listed below.
5′-6FAM-CCGGTGGCCAGCGCTCGGCGT(20) (O-1)
5′-A(20)CGCCGAGCGCTGGCCACCGG (O-2)
O-1 was used as the ssDNA and also annealed with O-2 to form the dsDNA substrates; both are 40 nucleotides long and identical to those used to characterize ERCC1-XPF (Tsodikov et al., 2005). Measurements were performed in triplicate on at least three independent preparations. The ratio of equilibrium dissociation constants of the mutant protein relative to wild-type was calculated for each experiment, averaged, and an SEM was calculated.
The competition assay was performed under similar conditions as the DNA binding assay, with a reaction condition containing 20 mM NaCl, 25 mM Tris (pH 8), 2 mM MnCl2, 0.1 mg/ml BSA, 2.5 μM 6-FAM dsDNA, 1.75 μM FANCMCTD-FAAP24, and a range of unlabeled DNA concentrations (0–12.5 μM). Each DNA concentration was performed in duplicate for both ssDNA and dsDNA. As a control, the experiment was also performed with buffer replacing the FANCMCTD-FAAP24. Anisotropy measurements were performed after a 1 hr incubation time on a TECAN SAFIRE-2.
shRNAmir Experiments
A clone with stable knockdown of FANCM was generated in 293-FLPin cells (Invitrogen) by transfection with Expression Arrest human shRNAmir clone V2HS_203688 (Open Biosystems, Huntsville, AL) and selection with 4 μg/ml puromycin. A clone transfected with Expression Arrest nonsilencing shRNAmir was used as a control. The sequence 5′-CGTTCTACTCCAAGAGTTAA-3′ of FANCM was changed to 5′-AGGTCGACACCCCGGGTAA-3′ by Phusion site-directed mutagenesis (NEB) of pDONR-FANCM to create wild-type FANCM resistant to the shRNAmir. Further site-directed mutagenesis was used to create the R1866A mutant in this vector. FANCM or FANCM-R1866A were transferred to pFLAG-FLP-In by LR Gateway recombination (Invitrogen), and integrated by FLP-mediated recombination into the single genomic FRT site of 293-FLPin cells as per manufacturers recommendations (Invitrogen). Pools were selected in 200 μg/ml Hygromycin B and expression confirmed by western blotting using α-Flag. siRNA knockdown of endogenous FANCM was maintained in control FLP-recombined cells (not shown). For FAAP24 siRNA knockdown experiments, Stealth RNAi (Invitrogen) with the oligo sequence 5′-GCUCAUUUUCGAGGAUGGCUUGACA-3′ was used. We mutated the target sequence in FAAP24 from 5′-ATTTTCGAGGATGGCTTGACA-3′ to 5′-ATCTTTGAAGACGGTCTCACT-3′ and generated stable 293-FLPin cell lines as described above. Cells were transfected with 100 nM oligo or control sequence using Lipofectamine 2000 (Invitrogen) and assayed after 48–72 hr.
Analysis of Complemented Cell Lines
Sulforhodamine B assays were performed essentially as described (Skehan et al., 1990) using 3,000 cells/well exposed to mitomycin C for 72 hr before fixation. Western blotting was performed on extracts from cells treated with 40 ng/ml mitomycin C for 16 hr. Membranes were blocked in Tris-buffered saline + 0.05% tween-20 + 5% nonfat dry milk powder and then probed with α-FANCM (M3823, Fanconi Anemia Research Fund, 1:1,000), α-FANCD2 (NB-100-182, Novus, 1:2,000), α-Flag (M2, Sigma, 1:5,000), or α-FAAP24 (SWE54, Ciccia et al., 2007, 1:1,000). Respective HRP-coupled secondary antibodies were detected using ECL Plus (GE Healthcare). The extent of knockdown of FANCM or FAAP24 was quantitatively measured using IR680/IR800CW-labeled secondary antibodies (Li-Cor Biosciences) detected using an Odyssey scanner and accompanying software (Li-Cor Biosciences).
Immunofluorescence Staining
Cells were grown in eight-well chamber slides (BD) coated with 2.5 μg/ml polyethylenimine (Sigma). Cells were treated for 7 hr with 40 ng/ml mitomycin C, fixed in 3.7% paraformaldehyde for 20 min at room temperature, followed by 1 min in ice-cold methanol. Fixed samples were then permeabilized in 0.3% (v/v) Triton X-100, blocked for 1 hr in PBS + 2% BSA + 0.05% Tween 20 and incubated with primary antibodies (1:500 rabbit α-FANCD2 [Abcam] and 1:1,000 mouse α-γH2A.X [Upstate]) overnight at 4°C. Finally, samples were incubated with secondary antibodies (1:1,000 Alexa Fluor 488 donkey-α-mouse or Alexa Fluor 555 donkey-α-rabbit) for 1 hr at room temperature. Images were collected using a Zeiss LSM 510 Meta confocal system coupled to an Axioplan 2 microscope at 100× magnification.
Acknowledgments
This work was supported by core funding from Cancer Research UK, the National Health and Medical Research Council, Australia (#1031104), the Victorian Government’s OIS Program, the Fanconi Anemia Research Fund, the ERC, Swiss Bridge, and the Louis-Jeantet Foundation. A.D. is a National Breast Cancer Foundation Fellow (Australia). The FANCM and FAAP24 cDNAs were a gift from Alberto Ciccia.
R.C. performed all crystallographic and biochemical experiments with the FANCM NTD and CTD. A.D. purified full-length FANCM:FAAP24 and performed the complementation and knockdown experiments. M.B. characterized the D4 mutant by EMSA assay. The EM work was carried out by P.S.; both P.S. and A.C. analyzed and interpreted the EM data. The results were interpreted by R.C., N.Q.M., and A.D. All authors contributed to writing the paper.
Published: August 8, 2013
Footnotes
Supplemental Information includes five figures and can be found with this article online at http://dx.doi.org/10.1016/j.str.2013.07.006.
Accession Numbers
The Protein Data Bank accession number for the coordinates for the FANCMCTD:FAAP24 structure reported in this paper is 4BXO.
Supplemental Information
References
- Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger I., Fitzgerald D.J., Richmond T.J. Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 2004;22:1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- Blackford A.N., Schwab R.A., Nieminuszczy J., Deans A.J., West S.C., Niedzwiedz W. The DNA translocase activity of FANCM protects stalled replication forks. Hum. Mol. Genet. 2012;21:2005–2016. doi: 10.1093/hmg/dds013. [DOI] [PubMed] [Google Scholar]
- Chang J.H., Kim J.J., Choi J.M., Lee J.H., Cho Y. Crystal structure of the Mus81-Eme1 complex. Genes Dev. 2008;22:1093–1106. doi: 10.1101/gad.1618708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., Ling C., Coulthard R., Yan Z., Xue Y., Meetei A.R., Laghmani H., Joenje H., McDonald N., de Winter J.P. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Ciccia A., McDonald N., West S.C. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu. Rev. Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- Collis S.J., Ciccia A., Deans A.J., Horejsí Z., Martin J.S., Maslen S.L., Skehel J.M., Elledge S.J., West S.C., Boulton S.J. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol. Cell. 2008;32:313–324. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- de Laat W.L., Sijbers A.M., Odijk H., Jaspers N.G., Hoeijmakers J.H. Mapping of interaction domains between human repair proteins ERCC1 and XPF. Nucleic Acids Res. 1998;26:4146–4152. doi: 10.1093/nar/26.18.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans A.J., West S.C. FANCM connects the genome instability disorders Bloom’s Syndrome and Fanconi Anemia. Mol. Cell. 2009;36:943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Deans A.J., West S.C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Enzlin J.H., Schärer O.D. The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. EMBO J. 2002;21:2045–2053. doi: 10.1093/emboj/21.8.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, P.R. (1993). Data reduction. Proceedings of CCP4 Study Weekend on Data Collection & Processing, 114–122.
- Gari K., Décaillet C., Delannoy M., Wu L., Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc. Natl. Acad. Sci. USA. 2008;105:16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K., Décaillet C., Stasiak A.Z., Stasiak A., Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- Huang M., Kim J.M., Shiotani B., Yang K., Zou L., D’Andrea A.D. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol. Cell. 2010;39:259–268. doi: 10.1016/j.molcel.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Kee Y., Gurtan A., D’Andrea A.D. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood. 2008;111:5215–5222. doi: 10.1182/blood-2007-09-113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori K., Hidaka M., Horiuchi T., Fujikane R., Shinagawa H., Ishino Y. Cooperation of the N-terminal Helicase and C-terminal endonuclease activities of Archaeal Hef protein in processing stalled replication forks. J. Biol. Chem. 2004;279:53175–53185. doi: 10.1074/jbc.M409243200. [DOI] [PubMed] [Google Scholar]
- Lovell S.C., Davis I.W., Arendall W.B., 3rd, de Bakker P.I.W., Word J.M., Prisant M.G., Richardson J.S., Richardson D.C. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Meetei A.R., Medhurst A.L., Ling C., Xue Y., Singh T.R., Bier P., Steltenpool J., Stone S., Dokal I., Mathew C.G. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosedale G., Niedzwiedz W., Alpi A., Perrina F., Pereira-Leal J.B., Johnson M., Langevin F., Pace P., Patel K.J. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat. Struct. Mol. Biol. 2005;12:763–771. doi: 10.1038/nsmb981. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Newman M., Murray-Rust J., Lally J., Rudolf J., Fadden A., Knowles P.P., White M.F., McDonald N.Q. Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition. EMBO J. 2005;24:895–905. doi: 10.1038/sj.emboj.7600581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Komori K., Ishino Y., Morikawa K. X-ray and biochemical anatomy of an archaeal XPF/Rad1/Mus81 family nuclease: similarity between its endonuclease domain and restriction enzymes. Structure. 2003;11:445–457. doi: 10.1016/s0969-2126(03)00046-7. [DOI] [PubMed] [Google Scholar]
- Nishino T., Komori K., Tsuchiya D., Ishino Y., Morikawa K. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure. 2005;13:143–153. doi: 10.1016/j.str.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Pingoud A., Fuxreiter M., Pingoud V., Wende W. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehan P., Storeng R., Scudiero D., Monks A., McMahon J., Vistica D., Warren J.T., Bokesch H., Kenney S., Boyd M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- Steczkiewicz K., Muszewska A., Knizewski L., Rychlewski L., Ginalski K. Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res. 2012;40:7016–7045. doi: 10.1093/nar/gks382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Tao Y., Jin C., Li X., Qi S., Chu L., Niu L., Yao X., Teng M. The structure of the FANCM-MHF complex reveals physical features for functional assembly. Nat. Commun. 2012;3:782. doi: 10.1038/ncomms1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripsianes K., Folkers G., Ab E., Das D., Odijk H., Jaspers N.G., Hoeijmakers J.H., Kaptein R., Boelens R. The structure of the human ERCC1/XPF interaction domains reveals a complementary role for the two proteins in nucleotide excision repair. Structure. 2005;13:1849–1858. doi: 10.1016/j.str.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Tripsianes K., Folkers G.E., Zheng C., Das D., Grinstead J.S., Kaptein R., Boelens R. Analysis of the XPA and ssDNA-binding surfaces on the central domain of human ERCC1 reveals evidence for subfunctionalization. Nucleic Acids Res. 2007;35:5789–5798. doi: 10.1093/nar/gkm503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsodikov O.V., Enzlin J.H., Schärer O.D., Ellenberger T. Crystal structure and DNA binding functions of ERCC1, a subunit of the DNA structure-specific endonuclease XPF-ERCC1. Proc. Natl. Acad. Sci. USA. 2005;102:11236–11241. doi: 10.1073/pnas.0504341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M., Harauz G., Orlova E.V., Schmidt R., Schatz M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Wang Y., Leung J.W., Jiang Y., Lowery M.G., Do H., Vasquez K.M., Chen J., Wang W., Li L. FANCM and FAAP24 maintain genome stability via cooperative as well as unique functions. Mol. Cell. 2013;49:997–1009. doi: 10.1016/j.molcel.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks C.M., Blessing R.H., Miller R., Mungee R., Potter S.A., Rappleye J., Smith G.D., Xu H., Furey W. Towards automated protein structure determination: BnP, the SnB-PHASES interface. Z. Kristallogr. 2002;217:686–693. [Google Scholar]
- Whitby M.C. The FANCM family of DNA helicases/translocases. DNA Repair (Amst.) 2010;9:224–236. doi: 10.1016/j.dnarep.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Wienk H., Slootweg J.C., Speerstra S., Kaptein R., Boelens R., Folkers G.E. The Fanconi anemia associated protein FAAP24 uses two substrate specific binding surfaces for DNA recognition. Nucleic Acids Res. 2013;41:6739–6749. doi: 10.1093/nar/gkt354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Li Y., Guo R., Ling C., Wang W. FANCM of the Fanconi anemia core complex is required for both monoubiquitination and DNA repair. Hum. Mol. Genet. 2008;17:1641–1652. doi: 10.1093/hmg/ddn054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Delannoy M., Ling C., Daee D., Osman F., Muniandy P.A., Shen X., Oostra A.B., Du H., Steltenpool J. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol. Cell. 2010;37:865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. An equivalent metal ion in one- and two-metal-ion catalysis. Nat. Struct. Mol. Biol. 2008;15:1228–1231. doi: 10.1038/nsmb.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.