Abstract
Protein synthesis requires several GTPase factors, including Elongation Factor Tu (EF-Tu), which delivers aminoacyl-tRNAs to the ribosome. In order to understand how the ribosome triggers GTP hydrolysis in translational GTPases, we have determined the crystal structure of EF-Tu and aminoacyl-tRNA bound to the ribosome with a GTP analog, to 3.2 Å resolution. EF-Tu is in its active conformation, the Switch I loop is ordered, and the catalytic histidine is coordinating the nucleophilic water in position for in-line attack on the γ-phosphate of GTP. This activated conformation is due to a critical and conserved interaction of the histidine with A2662 of the Sarcin-Ricin Loop of the 23S rRNA. The structure suggests a universal mechanism for GTPase activation and hydrolysis in translational GTPases on the ribosome.
In all stages of protein synthesis the ribosome requires exogenous protein factors, including several GTP hydrolyzing enzymes known as GTPases. These proteins are essential and highly conserved, and include Elongation Factor Tu (EF-Tu), which delivers aminoacyl-tRNA to the ribosome as part of a ternary complex (TC) with GTP, as well as Elongation Factor G and Initiation Factor 2 (1). The architecture of the GTP binding domain is similar in all translational GTPases from bacteria to higher eukaryotes, and hydrolysis is accompanied by conformational changes in the conserved switch I and II regions (2). Furthermore, translational GTPases bind to the same region of the ribosome in all species (3,4). The conserved binding site and structural similarities suggest that there is a common mechanism by which the ribosome activates GTP hydrolysis in these factors. However, despite over 40 years of research, this mechanism has remained elusive.
The catalysis of GTP hydrolysis in translational GTPases requires an invariant histidine (His84 in EF-Tu) that acts as a general base, abstracting a proton from a water molecule, for in-line attack on the γ-phosphate of GTP (5,6,7). The intrinsic GTPase activity of translational GTPases is low (8) and increases radically upon productive binding to the ribosome (9, 10). For EF-Tu, efficient GTPase activation requires the binding of an aminoacyl-tRNA to its cognate mRNA codon. Activation could be accomplished either indirectly, i.e. proper ribosome binding induces an active conformation of the G-factor, and/or more directly, with the ribosome playing an active role in catalysis. The ribosomal components that compose the GTPase binding site and therefore may be important for hydrolysis include the Sarcin-Ricin Loop (SRL) of the 23S rRNA (3, 11), the L11 protein and rRNA (12), and the protein L12(13).
Premature GTP hydrolysis in EF-Tu is thought to be prevented by a “hydrophobic gate,” consisting of residues Val20 of the P-loop and Ile60 of Switch I, which restricts access of His84 to the catalytic water (6). Proposals of how the hydrophobic gate is “opened” include that His84 would simply “push” through the gate when activated (6), or that a disordering (14, 15) or radical rearrangement of Switch I removes this steric block (16-18). However, these hypotheses were made based on structures of EF-Tu in isolation, or studies of the post-hydrolysis, kirromycin-stalled complex on the ribosome. A conclusive understanding of how GTPase activation occurs on the ribosome requires the structure of a complex of EF-Tu prior to GTP hydrolysis.
Here we report the crystal structure of EF-Tu bound to the 70S ribosome along with Trp-tRNATrp, and the antibiotic paromomycin, stalled in the activated conformation by the GTP analog β-γ-methyleneguanosine 5′-triphosphate (GDPCP), at 3.2 Å resolution. The structure reveals the GTPase-competent form of EF-Tu and suggests a universal mechanism for GTP hydrolysis on the ribosome.
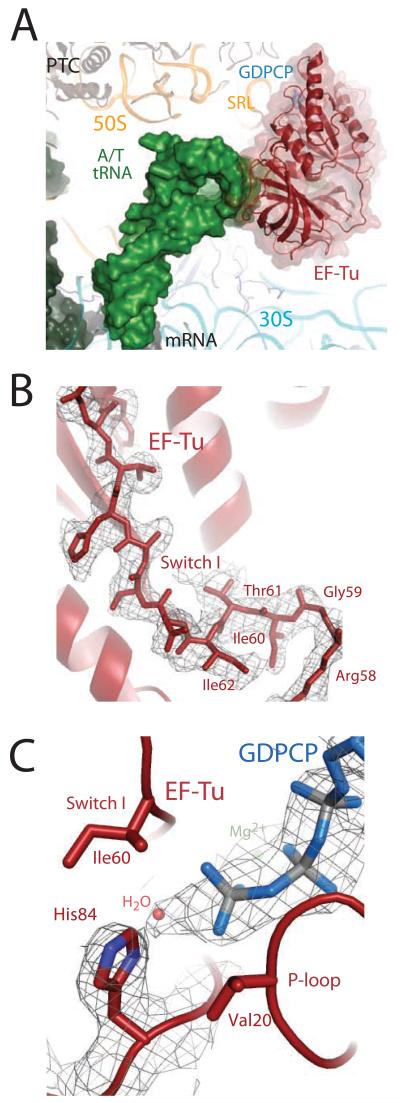
The overall conformations of the ribosome, EF-Tu, and the aminoacyl-tRNA are as previously reported (15) (Fig. 1a), and all the conformational changes predicted to communicate codon recognition to EF-Tu are observed, with the exception that the Switch I loop is ordered in the GTPase center (Fig. 1b). Additionally, the catalytic histidine is activated and coordinating a water for inline attack on the γ-phosphate of the GTP analog (Fig. 1c).
Figure 1.
The ternary complex bound to the ribosome in the activated state. A) EF-Tu (red) and aminoacyl-tRNA (green) bound to the 70S ribosome, stabilized by GDPCP (blue). B) Unbiased Fo-Fc electron density for GDPCP, the water molecule (orange), and His84 positioned in a catalytically active conformation. E.coli number is used throughout the text. C) Unbiased Fo-Fc electron density displayed for the Switch I loop.
As well as providing insight into the mechanism of GTP hydrolysis of all translational GTPases, this structure represents a key, missing state in the decoding pathway. High-resolution crystal structures have previously been determined for the isolated TC (19) and the post-hydrolysis state stalled on the ribosome by kirromycin (15). In conjunction with these two structures, this structure of the activated GTP state bound to the ribosome provides insight into the chemical details of the complete hydrolysis pathway of EF-Tu.
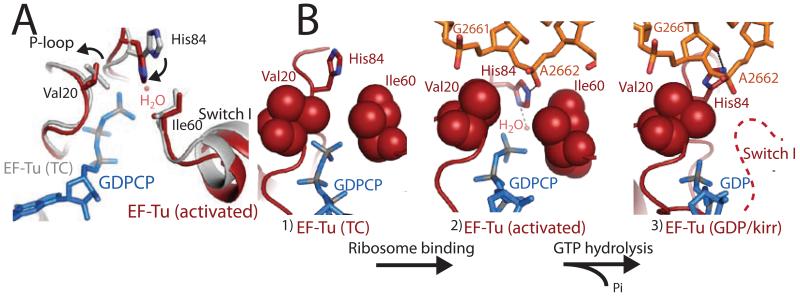
Prior to ribosome binding, the TC contains an unbent tRNA, and Switch I, Switch II, and the P-loop are ordered around the GTP molecule in the active site. However, the catalytic His84 (Switch II) is rotated away from GTP in an inactive conformation (19). Ribosome binding and codon recognition induces the bent A/T tRNA (20), as well as conformational changes in the tRNA, 30S ribosomal subunit, and EF-Tu essential for triggering GTP hydrolysis (15). Productive binding of the TC to the ribosome results in a shift of the G-domain of EF-Tu by up to ~7Å, to avoid a steric clash between Switch I and the SRL of the 23S rRNA. Ribosome binding also causes a distortion in the 3′ end of the aminoacyl-tRNA between residues 72-75 as reported (15), which releases contacts between the tRNA backbone and the Switch I loop (fig s1). However, in contrast to our previous prediction, this alone does not disorder Switch I, which is largely unchanged from its conformation in the isolated TC (Fig 2a) (19).
Figure 2.
The active site of EF-Tu during decoding. A) Superposition of the GTPase centers from the isolated TC (grey)(19) and the TC-GDPCP-ribosome (red). Small movements are observed for hydrophobic gate residues Val20 and Ile60. B) GTPase activation allows the phosphate of A2662 of the SRL (orange) to position His84 into the active site. After GTP hydrolysis (15) and Pi release Switch I becomes disordered (dashed-line) and His84 rotates away from GTP.
Although Switch I remains ordered in the active site, the catalytic histidine is in its activated conformation (Fig 2). We see no evidence that large rearrangements are required for catalysis (fig s1)(15-18). Instead, the “open” state is reached by a 1.2 Å shift in the side chain of Ile60, an ~80° rotation of the side chain of Val20, and a small movement of the backbone of residues 19-22 away from the GTP analog (Fig 2). The conformation of Switch I is similar to that modeled for the eukaryotic ortholog of EF-G bound to GDPNP on the ribosome (21).
These small changes in the active site would subtly increase the space between Switch I and the P-loop, possibly facilitating GTP hydrolysis. However, our analysis suggests that even the ‘closed’ hydrophobic gate (19) could accommodate the activated conformation of His84 and would not preclude binding of a water in its catalytic position, though a water is not observed. Instead, in order to prevent GTP hydrolysis prior to codon recognition, the activated conformation of His84 must be unfavorable in solution. Indeed, the adjacent residues, Gly83 and Pro82 are over 98% conserved, and mutational data suggest the flexibility of these residues may be important (19, 22).
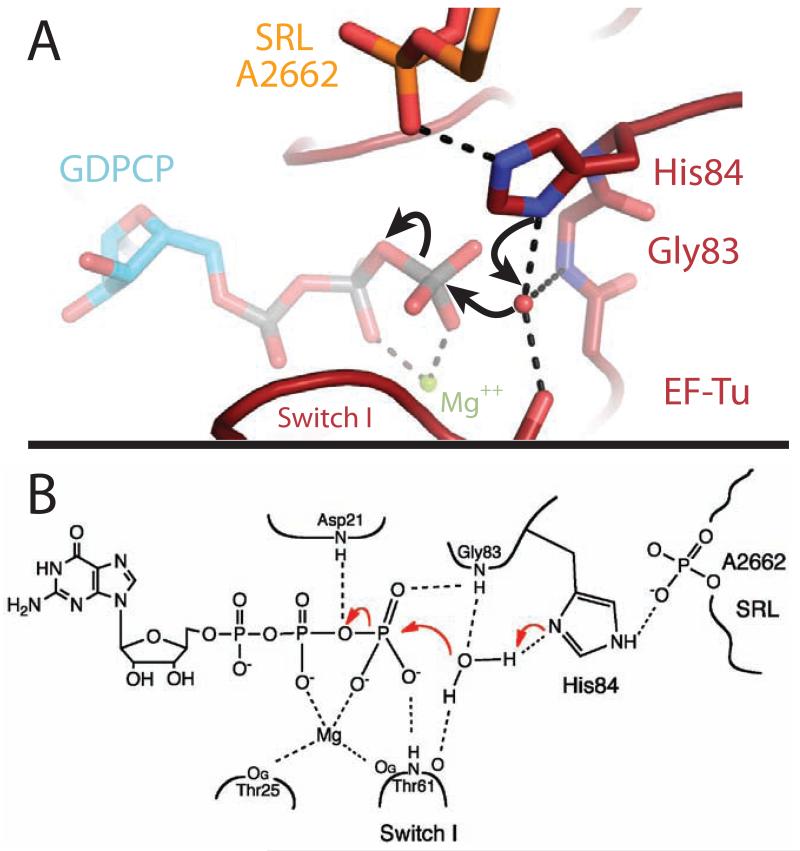
GTPase activation by the ribosome therefore does not involve an opening of the hydrophobic gate, but rather the specific positioning of His84 into the active site. In this activated structure of EF-Tu, the phosphate of residue A2662 of the SRL orders His84 in its catalytic conformation, which then acts as a general base to allow the nucleophilic water molecule to attack the γ-phosphate of GTP (Fig. 2, 3). When positioning His84, the phosphate of A2662 could also conceivably participate in a charge-relay system that facilitates the deprotonation of the water by His84, playing an analogous role to the catalytic aspartic acid in serine proteases (23). However the existence and contribution of such a mechanism would need to be assessed by biochemical experiments. The activation does not require movements within the SRL, as its overall conformation is similar to that in previous 70S ribosome structures (15, 24). This critical role of the Sarcin-Ricin Loop answers the long-standing question of why the SRL is important for binding and GTPase activation by the ribosome. The toxin α-sarcin acts by cleaving the 23S rRNA between residues G2661 and A2662(11), exactly the phosphate group activating His84. It is therefore clear why cleaving this bond, or introducing mutations at nearby sites in the SRL (25), leads to defects in His84 positioning and GTP hydrolysis (11,26). α-sarcin is active against both bacterial and eukaryotic ribosomes and causes similar defects in the function of their respective translational GTPases (11,26). We therefore predict that the positioning of the catalytic histidine into the active site by A2662 of the SRL is the universal mechanism for GTPase activation on the ribosome. EF-G has additional interactions between its domain 3 and A2660 of the SRL, which explains why depurination of A2660 by the toxin ricin affects EF-G, but not EF-Tu (26).
Figure 3.
Chemical mechanism of GTP hydrolysis. A) Highly conserved His84 acts as a general base to activate the catalytic water molecule, which is positioned by interactions with Thr61, Gly83 and His84. B) Chemical drawing depicting the interactions between EF-Tu and GTP that stabilize the β and γ phosphates, allowing GTP hydrolysis to occur (see also fig s3).
Ribosome association alone is not sufficient for A2662 to activate His84. Rather, all of the conformational changes that occur upon cognate codon recognition in the ribosome (codon-anticodon monitoring, domain closure), the tRNA (A/T state, 3′ end distortion), and EF-Tu (ß-loop interaction with 30S shoulder, G domain shift)(15) are essential for properly positioning the GTPase center of EF-Tu for activation by the SRL. The energy required to induce these movements is balanced against the energy derived from binding of a cognate tRNA, including that from interactions of the 16S rRNA residues A1492, A1493, and G530 with the minor groove of the codon-anticodon helix (27). Even subtle changes in the position of the G-domain relative to the SRL would cause defects in GTPase activation by preventing A2662 from properly placing His84 into the active site.
The activation of GTP hydrolysis in EF-Tu by the SRL is similar to the regulation of cellular GTPases by their partner proteins. Unlike the classic cases of Rho and Ras, in which their GTPase Activator Proteins (GAP) directly donate catalytic residues (28, 29), other G proteins are activated by “Regulator of G-protein Signaling” (RGS) proteins, which instead stabilize the active conformation of the GTPase(30). By analogy, the SRL acts as an RGS-type GAP for translational GTPases.
All components required for GTPase activation should be present in this catalytically active structure. Notably, we see no evidence that ribosomal protein L12, predicted to be important for hydrolysis, is interacting with EF-Tu, though such an interaction would be compatible with the crystal packing. Indeed, it is sterically impossible for L12 or any other protein to interact with GTP, consistent with data suggesting that L12 does not function directly in catalysis (13, 31, 32). Furthermore, deletion of the eukaryotic orthologs of L12 from yeast ribosomes is not lethal (33), indicating that L12 may not be critical for achieving GTP hydrolysis.
Following hydrolysis, Pi is released from EF-Tu. The γ-phosphate of GTP makes several contacts with Switch I through residue Thr62 (Fig 3, s3), and release of Pi could destabilize and perhaps even require movement of Switch I. This and the loss of interactions with the 3′ end of the tRNA backbone (fig s1), leads to the previously observed disordering of Switch I (15, 18). Furthermore, comparison of the kirromycin- and the GDPCP-stalled complexes shows a rotation of ~4° of the G-domain relative to domains 2 and 3 (sfig 4). There is a corresponding movement in the acceptor stem of the tRNA, in order to maintain interactions with Switch II(15), which could explain the subtle difference in fluorescence signal between the activated and kirromycin stalled states(34).
This structure of 70S-tRNA-EF-Tu-GDPCP provides an atomic resolution model of a translational GTPase in its activated state. Codon recognition leads to a series of conformational changes in the 30S subunit, tRNA, and EF-Tu that position EF-Tu for GTP hydrolysis. GTPase activation does not require a large opening of the “hydrophobic gate,” but instead the positioning of the catalytic histidine into the active site by the SRL residue A2662. The high level of sequence and structural conservation between all translational GTPases suggests that while each factor recognizes a distinct ribosomal state, each must bind in such a way that the SRL interacts with its catalytic histidine. Therefore GTPase activation by the SRL is the universal mechanism for triggering GTP hydrolysis on the ribosome across all kingdoms of life.
Supplementary Material
Acknowledgments
We thank R. Green for reagents and D. de Sanctis at ESRF for facilitating data collection. This work was supported by the Medical Research Council UK, the Wellcome Trust, the Agouron Institute and the Louis-Jeantet Foundation. RMV is supported by a Gates-Cambridge scholarship, TMS by HFSP and Emmanuel College. The structures are available at the Protein Data Bank with accession codes 2xqd and 2xqe.
Footnotes
References
- 1.Margus T, Remm M, Tenson T. BMC Genomics. 2007;8:15. doi: 10.1186/1471-2164-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dever TE, Glynias MJ, Merrick WC. Proc Natl Acad Sci U S A. 1987;84:1814. doi: 10.1073/pnas.84.7.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moazed D, Robertson JM, Noller HF. Nature. 1988;334:362. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 4.Holmberg L, Nygard O. Biochemistry. 1994;33:15159. doi: 10.1021/bi00254a027. [DOI] [PubMed] [Google Scholar]
- 5.Eccleston JF, Webb MR. J Biol Chem. 1982;257:5046. [PubMed] [Google Scholar]
- 6.Berchtold H, et al. Nature. 1993;365:126. doi: 10.1038/365126a0. [DOI] [PubMed] [Google Scholar]
- 7.Kjeldgaard M, Nissen P, Thirup S, Nyborg J. Structure. 1993;1:35. doi: 10.1016/0969-2126(93)90007-4. [DOI] [PubMed] [Google Scholar]
- 8.Jacquet E, Parmeggiani A. Embo J. 1988;7:2861. doi: 10.1002/j.1460-2075.1988.tb03142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pape T, Wintermeyer W, Rodnina MV. Embo J. 1998;17:7490. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mesters JR, Potapov AP, de Graaf JM, Kraal B. J Mol Biol. 1994;242:644. doi: 10.1006/jmbi.1994.1614. [DOI] [PubMed] [Google Scholar]
- 11.Hausner TP, Atmadja J, Nierhaus KH. Biochimie. 1987;69:911. doi: 10.1016/0300-9084(87)90225-2. [DOI] [PubMed] [Google Scholar]
- 12.Bourne HR, Sanders DA, McCormick F. Nature. 1991;349:117. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 13.Mohr D, Wintermeyer W, Rodnina MV. Biochemistry. 2002;41:12520. doi: 10.1021/bi026301y. [DOI] [PubMed] [Google Scholar]
- 14.Vogeley L, Palm GJ, Mesters JR, Hilgenfeld R. J Biol Chem. 2001;276:17149. doi: 10.1074/jbc.M100017200. [DOI] [PubMed] [Google Scholar]
- 15.Schmeing TM, et al. Science. 2009;326:688. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sengupta J, et al. J Mol Biol. 2008;382:179. doi: 10.1016/j.jmb.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villa E, et al. Proc Natl Acad Sci U S A. 2009;106:1063. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuette JC, et al. Embo J. 2009;28:755. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nissen P, et al. Science. 1995;270:1464. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 20.Valle M, et al. Embo J. 2002;21:3557. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor DJ, et al. EMBO J. 2007;26:2421. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knudsen C, Wieden HJ, Rodnina MV. J Biol Chem. 2001;276:22183. doi: 10.1074/jbc.M102186200. [DOI] [PubMed] [Google Scholar]
- 23.Blow DM, Birktoft JJ, Hartley BS. Nature. 1969;221:337. doi: 10.1038/221337a0. [DOI] [PubMed] [Google Scholar]
- 24.Selmer M, et al. Science. 2006;313:1935. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 25.Bilgin N, Ehrenberg M. J Mol Biol. 1994;235:813. doi: 10.1006/jmbi.1994.1041. [DOI] [PubMed] [Google Scholar]
- 26.Rambelli F, et al. Biochem J. 1989;259:307. doi: 10.1042/bj2590307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogle JM, et al. Science. 2001;292:897. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 28.Mittal R, Ahmadian MR, Goody RS, Wittinghofer A. Science. 1996;273:115. doi: 10.1126/science.273.5271.115. [DOI] [PubMed] [Google Scholar]
- 29.Rittinger K, Walker PA, Eccleston JF, Smerdon SJ, Gamblin SJ. Nature. 1997;389:758. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- 30.Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Cell. 1997;89:251. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 31.Diaconu M, et al. Cell. 2005;121:991. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Wieden HJ, Wintermeyer W, Rodnina MV. J Mol Evol. 2001;52:129. doi: 10.1007/s002390010141. [DOI] [PubMed] [Google Scholar]
- 33.Remacha M, et al. Mol Cell Biol. 1995;15:4754. doi: 10.1128/mcb.15.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodnina MV, Fricke R, Wintermeyer W. Biochemistry. 1994;33:12267. doi: 10.1021/bi00206a033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.