Abstract
Rpn11 is a proteasome-associated deubiquitinating enzyme that is essential for viability. Recent genetic studies showed that Rpn11 is functionally linked to Rpn10, a major multiubiquitin chain binding receptor in the proteasome. Mutations in Rpn11 and Rpn10 can reduce the level and/or stability of proteasomes, indicating that both proteins influence its structural integrity. To characterize the properties of Rpn11, we examined its interactions with other subunits in the 19S regulatory particle and detected strong binding to Rpn3. Two previously described rpn3 mutants are sensitive to protein translation inhibitors and an amino acid analog. These mutants also display a mitochondrial defect. The abundance of intact proteasomes was significantly reduced in rpn3 mutants, as revealed by strongly reduced binding between 20S catalytic with 19S regulatory particles. Proteasome interaction with the shuttle factor Rad23 was similarly reduced. Consequently, higher levels of multiUb proteins were associated with Rad23, and proteolytic substrates were stabilized. The availability of Rpn11 is important for maintaining adequate levels of intact proteasomes, as its depletion caused growth and proteolytic defects in Rpn3. These studies suggest that Rpn11 is stabilized following its incorporation into proteasomes. The instability of Rpn11 and the defects of rpn3 mutants are apparently caused by a failure to recruit Rpn11 into mature proteasomes.
Keywords: proteasome, Rpn3, ubiquitin, proteolysis, Rad23
Introduction
Removal of a substrate-linked multiubiquitin chain is required for successful protein degradation and is achieved by proteasome-associated deubiquitinating enzymes such as Rpn11, Ubp6, and Uch37.1–4 A previously described mutant (rpn11-1) harbors a frame-shift mutation in the carboxy terminus that replaces the 31 terminal amino acids with 9 incorrect residues.5,6 This mutant confers viability at 23 °C but not at 37 °C. Incubation at the nonpermissive temperature resulted in proteasome instability and was accompanied by the accumulation of multiubiquitinated (multiUb) proteins and stabilization of well-characterized proteolytic substrates.7 Rpn11 is functionally linked to Rpn10, a proteasome receptor that binds multiUb proteins.8 Loss of Rpn10 (rpn10Δ) can reduce proteasome stability,9 and an rpn11-1 rpn10Δ double mutant is lethal.8 To further examine the functional interactions between Rpn11 and other subunits in the 19S regulatory particle, we tested its interactions with several 19S lid subunits using a coupled in vitro transcription/translation assay.8 These studies revealed a strong interaction with Rpn3, a subunit in the 19S regulatory particle.
Rpn3 represents one of several non-ATPase subunits in the 19S regulatory particle and is present in the lid subcomplex. Rpn3 is poorly characterized, and no specific biochemical functions have been ascribed to this protein. Rpn3 and several other subunits in the regulatory particle contain a conserved motif that facilitates protein–protein interactions within multisubunit complexes, such as the proteasome, COP9, and initiation factor-310,11 (termed the PCI domain). Two temperature-sensitive mutants (rpn3-4 and rpn3-7) were previously identified and found to harbor temperature-sensitive growth defects. Moreover, both mutants showed proteolytic defects that included accumulation of multiUb proteins and stabilization of reporter substrates.12 These and other reports observed an M-phase defect in rpn3 mutants that is related to deficient turnover of regulatory proteins that control growth-phase-specific transitions.12,13
We determined that the phenotypes associated with rpn3-4 and rpn3-7 closely resemble the defects previously observed in rpn11-1. The defects of rpn11-1 are primarily due to the instability of the rpn11-1 protein. We found that Rpn11 is highly unstable in rpn3-4 and rpn3-7 mutants, suggesting that its depletion in rpn3 mutants generates defects previously observed in rpn11-1. We determined that Rpn3 and the mutant proteins (rpn3-4 and rpn3-7) formed equivalent interactions with purified Rpn11. However, Rpn11 was unstable in rpn3 mutants and was poorly assembled into proteasomes. We conclude that Rpn11 interaction with Rpn3 is not the sole determinant for its successful incorporation into mature proteasomes. Rpn11-1 displays growth and proteolytic defects that arise from proteasome instability and the reduced levels of intact proteasomes. We found a close correspondence between the rapid turnover of Rpn11, its absence in purified proteasomes, and proteasome assembly defects in rpn3-4 and rpn3-7. A straightforward interpretation of these results is that Rpn3 performs an important role in recruiting Rpn11 to the proteasome to promote proteasome assembly and function.
Results
Strong interaction between Rpn11 and Rpn3
Rpn11 was expressed in Escherichia coli as a fusion to glutathione S-transferase (GST) and purified following incubation of protein extracts with glutathione Sepharose. DNA constructs expressing proteasome subunits from the T7 promoter were added to an in vitro coupled transcription/translation reaction to generate 35S-labeled proteins, as described previously.8 The 35S-labeled Rpn proteasome subunits were incubated with the immobilized GST-Rpn11 protein. Unbound proteins were removed by washes in binding buffer, and the bound proteins were detected by autoradiography (Fig. 1a). We observed a strong interaction with Rpn3 (lane 6). A slight shift in the position of Rpn3 is due to its close co-migration with GST-Rpn11 (compare lanes 5 and 6). GST-Rpn11 showed weak binding to Rpn1 (lane 2) and Rpn7 (lane 12). No detectable interactions were seen with either Rpn2 or Rpn6 (lanes 4 and 10). The lack of interaction with Rpn5 (lane 8) is not definitive because this proteasome subunit was poorly labeled (lane 7). To determine if the defects of rpn11-1 were caused by an inability of rpn11-1 protein to bind Rpn3, we compared 35S-labeled Rpn3 interactions with recombinant GST-Rpn11 and GST-rpn11-1 mutant proteins (Fig. 1b). Both GST-Rpn11 and GST-rpn11-1 could bind 35S-labeled Rpn3 (upper panel, lanes 2 and 3), demonstrating that carboxy-terminal residues in Rpn11 are not required for its interaction with Rpn3. Nonspecific interaction of 35S-labeled Rpn3 with the control GST beads was not observed (lane 4).
Fig. 1.
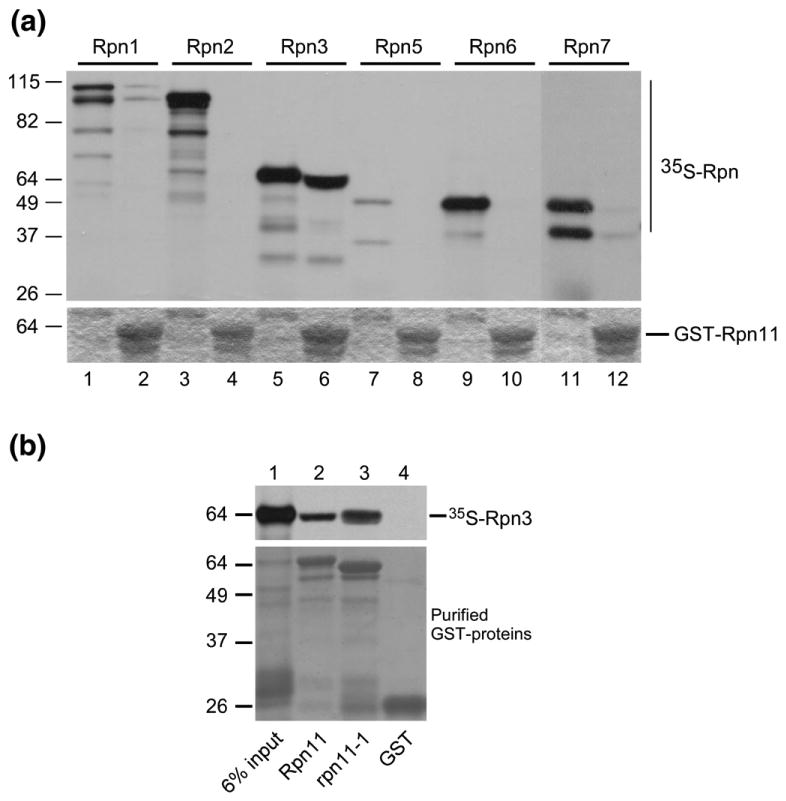
Direct interaction between Rpn11 and Rpn3 proteasome subunits. (a) GST-Rpn11 was expressed in E. coli and purified on glutathione Sepharose matrix. Plasmids expressing proteasome subunits Rpn1-7 were used as templates in an in vitro transcription/translation coupled reaction using [35S]methionine. The radiolabeled proteins were incubated with immobilized GST-Rpn11. The upper panel represents an autoradiogram showing the quality of the 35S-labeled proteins and their binding to GST-Rpn11. Odd-numbered lanes correspond to a control sample of each 35S-labeled Rpn protein that was not applied to the matrix. Even-numbered lanes show the result of the binding reaction. The lower panel shows the expression of GST-Rpn11 in even-numbered lanes. The slight shift in the position of 35S-labeled Rpn3 in lane 6 is caused by its co-migration with high levels of GST-Rpn11. (b) The interaction between 35S-labeled Rpn3 and a mutant form of Rpn11 (rpn11-1) was tested. Nonspecific interaction between 35S-labeled Rpn3 and control GST-bound beads was not observed (lane 4).
The sensitivity of rpn3 mutants to translation inhibitors is suppressed by RPN3
Exponential-phase yeast cells were adjusted to an absorbance of OD600=1, and serial 10-fold dilutions were spotted on agar medium containing paromomycin, hygromycin B, or canavanine (an arginine analog that can cause protein mis-folding). The control strain [wild type (WT)] showed normal growth at 23 °C and 37 °C and grew efficiently on medium containing the drugs (Fig. 2, top row). As expected, rpn3-4 and rpn3-7 showed a severe temperature-sensitive growth defect (37 °C)12 and were also unable to grow efficiently in medium containing the translation inhibitors paromomycin, hygromycin B, and canavanine. These defects were more severe in rpn3-7. Expression of Rpn3 from a plasmid restored WT resistance to these drugs, and growth at 37 °C (see lower two rows), demonstrating that the defects were linked to the rpn3 mutations.
Fig. 2.
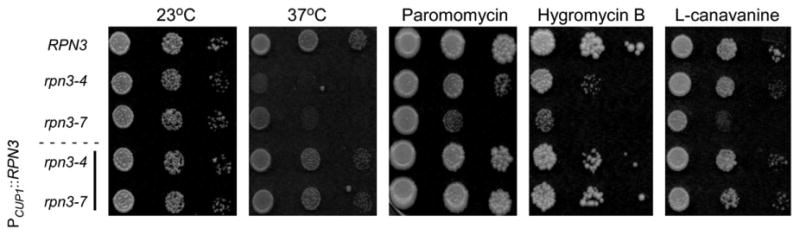
Sensitivity of rpn3 mutants to drugs that cause protein damage is suppressed by Rpn3. RPN3, rpn3-4, rpn3-7, and the mutants expressing high-copy Rpn3 were spotted on agar medium and incubated at either 23 °C or 37 °C (two left panels). Serial dilutions (10-fold) of the same cultures were also spotted on medium containing paromomycin, hygromycin B, and canavanine. Growth was examined after 3–5 days. Drug sensitivity was examined at 23 °C in medium containing 100 μM CuSO4 to induce expression of Rpn3.
rpn3 mutants have a mitochondrial defect
Rinaldi et al. reported a mitochondrial defect in rpn11-1,6,14 which implicated a role for the proteasome in controlling transitions between fission and fusion, which is believed to promote mitochondrial regeneration. Based on the unexpected similarity of the defects of rpn3 and rpn11-1 mutants (as described further below), we expressed Mt-GFP in rpn3 to track the dynamic changes in mitochondrial morphology in living cells.15 Mitochondrial morphology was examined by fluorescence microscopy, and significant defects were detected in both rpn3-4 and rpn3-7 at 37 °C (Fig. 3a). As expected, WT (RPN3) cells showed the normal pattern of extended tubules and a few punctuate bodies. In contrast, very high levels of large aggregates were seen in both rpn3-4 and rpn3-7 at 37 °C (lower left panels). Nomarski (differential interference contrast) imaging showed that rpn3 mutant cells are also larger than RPN3 WT cells. Collectively, these defects resemble the pattern previously seen in rpn11-1, which are also reproduced here (Fig. 3b). To further validate these findings, we examined several control strains, including yeast mutants with well-characterized defects in mitochondrial fission and fusion. An fzo1Δ mutant showed few tubules but high levels of small green fluorescent protein (GFP)-staining bodies, consistent with excessive levels of mitochondrial fission (Fig. 3c). In contrast, examination of dnm1Δ showed dramatic accumulation of large mitochondrial aggregates, as expected for excessive fusion. There is evidence that the turnover of Fzo1 by the proteasome plays a key role in regulating fusion/fission transitions.16,17 These findings demonstrate that altered proteasome function in rpn3 and rpn11 mutants hinders mitochondrial biogenesis and function.17
Fig. 3.
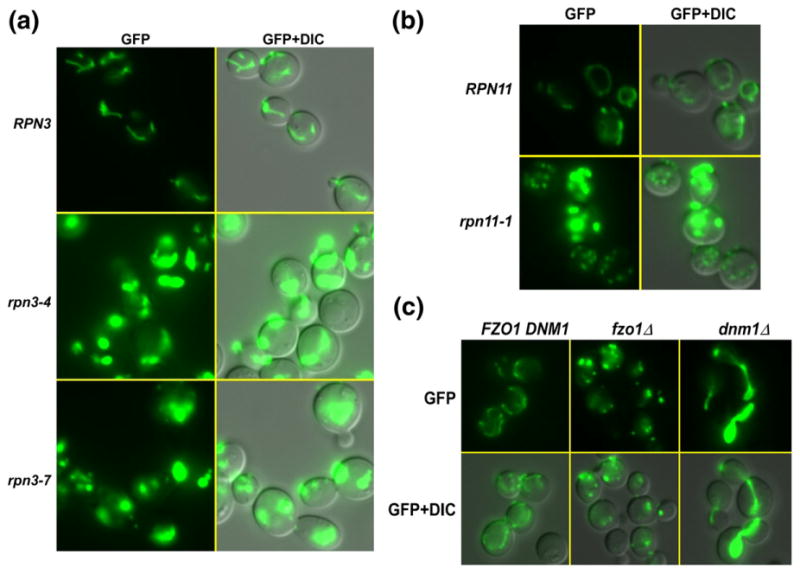
rpn3 mutants show a defect in mitochondrial fission/fusion transitions. (a) A plasmid expressing Mt-GFP was transformed into RPN3, rpn3-4, and rpn3-7. Actively growing yeast cultures were pre-grown at 23 °C and then transferred to 37 °C for 4 h. The cells were examined by fluorescence microscopy. GFP fluorescence showed clear evidence for long tubules in RPN3 cells, which represent mitochondria. The right column shows a merged image of GFP fluorescence and differential interference contrast (DIC) to show the morphology of the cells. Mt-GFP localization in rpn3-4 and rpn3-7 was markedly different from that of the WT. Two striking effects—dramatically elevated levels and a conspicuous lack of tubules—are evident in these images. Most cells contained a single large aggregate. (b) A previously described mitochondrial defect in rpn11-1 mutant is reproduced here. In contrast to rpn3 mutants, rpn11-1 showed the presence of both large aggregates (excessive fusion) and small punctuate fluorescent bodies (excessive fission). (c) To further confirm these results, we examined well-characterized mutants that affect mitochondrial fusion and fission. Mt-GFP was expressed in fzo1Δ and dnm1Δ, and as expected, elevated levels of fission (fzo1Δ) and fission (dnm1Δ) were detected.
Proteasome assembly and function are altered in rpn3 mutants
Whole-cell extracts were prepared from actively growing cells, and an equal amount of protein lysate was separated in native polyacrylamide gels.8 The gels were incubated in buffer containing the chymotryptic substrate LLVY-AMC, and proteasome peptidase activity was measured by detecting in situ fluorescence. Intact proteasomes are indicated by species containing a catalytic particle (CP) that is bound to either one (RP1CP) or two (RP2CP) regulatory particles (arrowheads indicate RP2CP and RP1CP). We found that rpn3-4 and rpn3-7 showed reduced levels of intact 26S proteasomes at both 23 °C and 37 °C, as noted by the lower levels of fluorescence coincident with these high-molecular-weight electrophoretic bands (Fig. 4a). The absence of the RP1CP/RP2CP complexes is more striking in rpn3-7 at both 23 °C and 37 °C (right panel, lanes 8 and 11). Expression of the WT Rpn3 protein in both rpn3 mutants restored RP1CP and RP2CP levels (Fig. 4a, lanes 3 and 6 and lanes 9 and 12). These results confirm that mutations in RPN3 cause a proteasome assembly defect. Intriguingly, high-level expression of Rpn3 appeared to modestly increase peptidase activity of intact proteasomes at 37 °C in rpn3-4 (compare lane 4 to lane 6) and rpn3-7 (compare lane 10 to lane 12), suggesting that it might perform a rate-limiting function. Further evidence that proteasomes are improperly assembled or unstable in the rpn3 mutants was confirmed by measuring peptidase activity in the presence of 0.05% SDS. Low concentration of detergent can stimulate the hydrolytic activity of the free 20S CP (Fig. 4b). While the overall proteasome peptidase activity in the absence of SDS was lower in both rpn3-4 and rpn3-7 at 23 °C (Fig. 4a; lanes 2 and 8) and severely reduced at 37 °C (lanes 5 and 11), addition of SDS significantly increased peptidase activity. The SDS-stimulated activity is generated predominantly by the free 20S particles (Fig. 4b). In rpn3-4, SDS caused a marked increase in 20S activity at 37 °C (compare lanes 4 and 5). Expression of WT Rpn3 restored normal levels of 20S CP activity (lane 6), whereas peptidase activity associated with RP1CP and RP2CP increased. Similarly, 20S CP peptidase activity was stimulated by SDS in rpn3-7 (compare lanes 10 and 11) but reduced following expression of WT Rpn3 (compare lanes 5 and 6 and lanes 11 and 12). The high level of free 20S CP in rpn3-4 and rpn3-7 is consistent with an assembly (or stability) defect. Significantly, exogenous expression of Rpn3 in the rpn3 mutants restored 20S incorporation into intact proteasomes (RP1CP and RP2CP) (Fig. 4b; 37 °C; lanes 6 and 12).
Fig. 4.
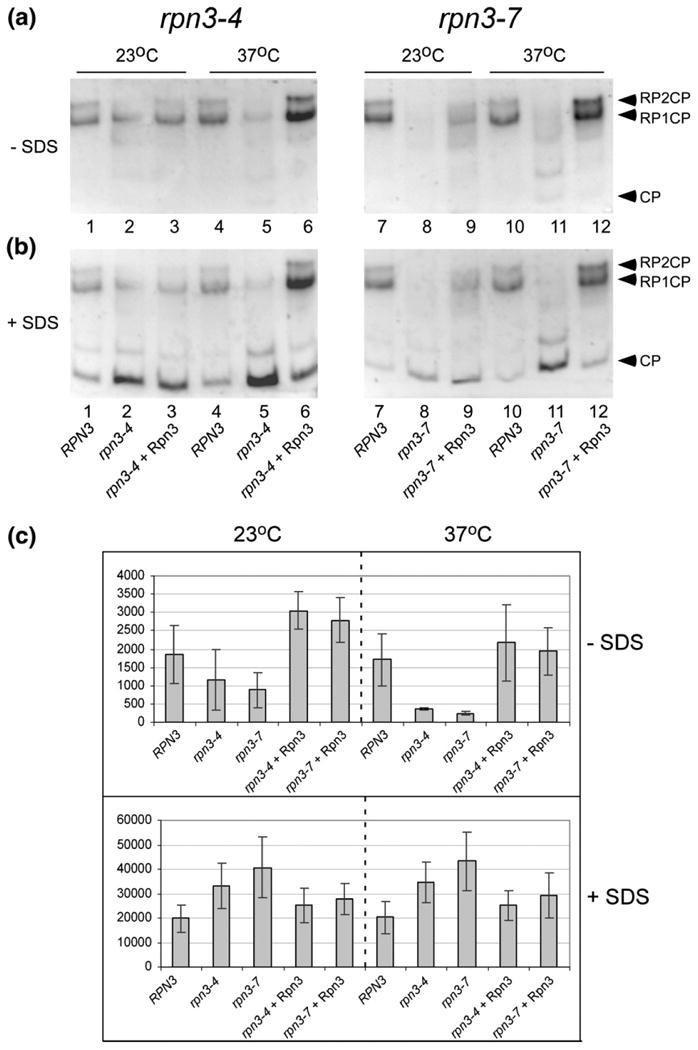
Proteasome assembly and activity are altered in rpn3 mutants. (a) Protein extracts were prepared from RPN3, rpn3-4, and rpn3-7, and an equal amount of protein was separated in a native polyacrylamide gel. After electrophoresis, the gels were immersed in buffer containing the chymotryptic substrate LLVY-AMC, and in situ fluorescence was measured. (b) The same gels were subsequently incubated in buffer that contained 0.05% SDS, and fluorescence was reexamined. The positions of free 20S CP and intact proteasomes comprising either one (RP1CP) or two (RP2CP) regulatory particles bound to CP are indicated on the right side of the panel. (c) Epoxomicin-sensitive proteasome peptidase activity, as well as its stimulation by SDS, was measured and plotted.
Based on the in-gel analysis (Fig. 4a and b), we sought quantitative data to define the changes in proteasome activity in rpn3 mutants. By comparing peptidase activity in the presence and absence of SDS, we could gauge the level of proteasome assembly in the rpn3 mutants. LLVY-AMC hydrolysis was measured using equal amounts of protein extract from RPN3 and rpn3 mutants grown at either 23 °C or 37 °C, and the data were plotted (Fig. 4c). At 23 °C, both rpn3-4 and rpn3-7 showed reduced activity. Expression of Rpn3 restored the higher levels of steady-state peptidase activity in both rpn3 mutants. At 37 °C, peptidase activity was significantly reduced in the rpn3 mutants in the absence of detergent. However, WT levels of activity were restored by exogenous expression of Rpn3. Strikingly, addition of 0.05% SDS yielded higher overall activity in both mutants (lower panel), indicating higher levels of free catalytic (20S) particles (consistent with the in-gel analysis). Normal levels of peptidase activity were restored by expression of the WT (Rpn3) protein in rpn3-4 and rpn3-7. Moreover, the resulting decrease in SDS-stimulated activity (upon endogenous expression of Rpn3) indicated that the free CP was incorporated into intact proteasomes.
Proteolytic substrates are stabilized in rpn3 mutants
The in-gel studies (Fig. 4) uncovered a defect in proteasome assembly and/or stability in rpn3-4 and rpn3-7. To examine the effect of diminished proteasome function in rpn3, we measured the abundance of well-characterized proteolytic substrates. A plasmid expressing Ub-Pro-βgal18,19 was transformed into RPN3, rpn3-4, and rpn3-7, and protein extracts were prepared from yeast cells grown at 23 °C and 37 °C. An immunoblot was incubated with antibodies against ubiquitin and βgal to assess the stability of Ub-Pro-βgal (Fig. 5a). A WT strain expressing a control substrate (Met-βgal) that is not degraded by the ubiquitin/proteasome system was also examined (lane 1). In agreement with previous studies,12 we detected high levels of multiUb proteins in rpn3-7 at 23 °C (lane 4), indicating reduced substrate turnover at the permissive temperature. The accumulation of multiUb proteins increased further upon incubation of rpn3-4 and rpn3-7 at 37 °C (lanes 7 and 8). The filter was subsequently treated with antibodies against βgal, and a small increase in Ub-Pro-βgal levels was detected in rpn3 mutants at 23 °C (Fig. 5a, lower panel, compare lane 2 to lanes 3 and 4). At 37 °C, Ub-Pro-βgal was degraded in RPN3 (lane 6), whereas it was stabilized in rpn3-4 and rpn3-7 (lanes 7 and 8). The abundance of the stable Met-βgal protein was unaffected (lanes 1 and 5).
Fig. 5.
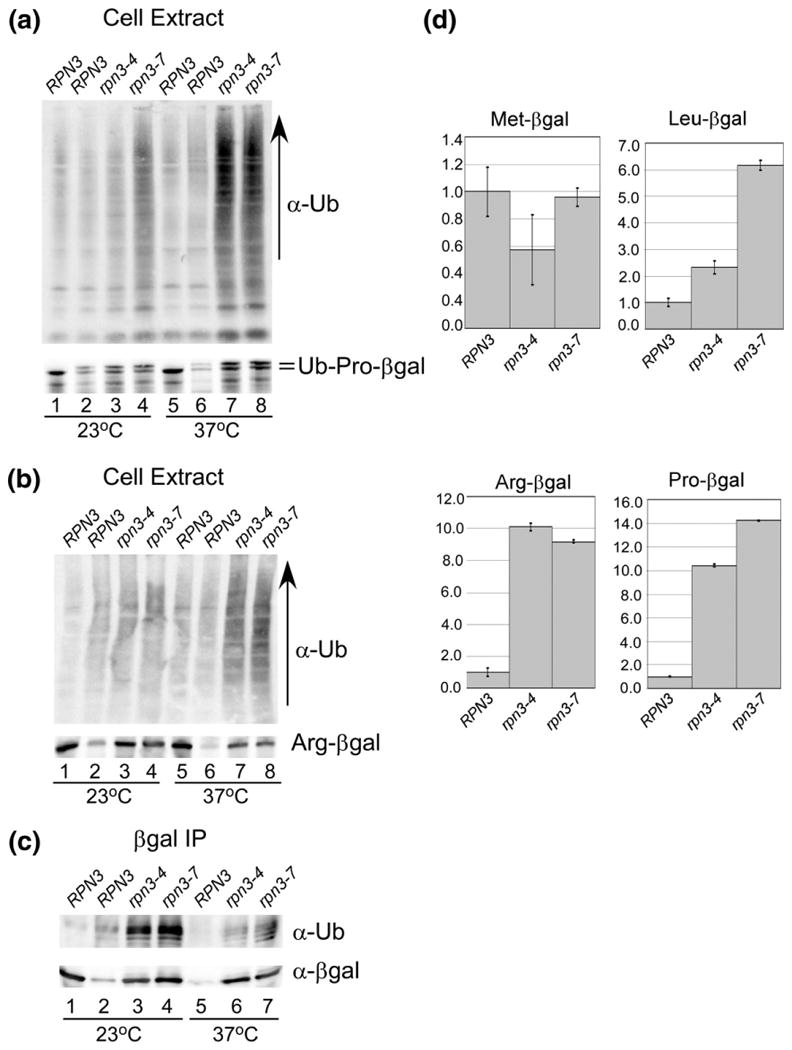
Defective proteolysis in rpn3 mutants. (a) Protein extracts were prepared from RPN3, rpn3-4, and rpn3-7 expressing Ub-Pro-βgal. RPN3 expressing a control protein (Met-βgal) is characterized in lane 1. An equal amount of protein extract was resolved by SDS/PAGE and examined by immunoblotting using antibodies against ubiquitin and βgal. (b) Similar to (a), protein extracts expressing Arg-βgal were characterized. RPN3 expressing a control protein (Met-βgal) is characterized in lane 1. An equal amount of protein extract was resolved by SDS/PAGE and examined by immunoblotting using antibodies against ubiquitin and βgal. (c) The extracts examined in (b) were incubated with βgal antibody to immunoprecipitate Arg-βgal, and the immunoprecipitated proteins were examined with antibodies against ubiquitin (upper panel) and βgal (lower panel). (d) Substrates Leu-βgal, Arg-βgal, and Ub-Pro-βgal were expressed in RPN3, rpn3-4, and rpn3-7, and β-galactosidase activity was measured in triplicate. The data were quantified and standardized to the expression of the protein in the wildtype strain, and plotted.
To more clearly establish if increased levels of a specific substrate could be detected, we expressed Arg-βgal in RPN3, rpn3-4, and rpn3-7 (which, unlike Ub-Pro-βgal, does not retain an amino-terminal ubiquitin moiety). Protein extracts were characterized as described above, and higher levels of multiUb proteins were detected in rpn3-4 and rpn3-7 at 37 °C (Fig. 5b). Incubation with anti-βgal antibodies showed clear evidence for accumulation of Arg-βgal in rpn3-4 and rpn3-7 but not in RPN3 (Fig. 5b, lower panel). The level of stable Met-βgal is shown as a control (lanes 1 and 5). Protein extracts were incubated with anti-βgal antibodies to immunoprecipitate Arg-βgal (Fig. 5c). The filter was incubated with antibodies against ubiquitin, and high levels were detected in rpn3-4 and rpn3-7. Evidence for multiple ubiquitin conjugates can also be detected (see lanes 6 and 7). As expected, the ubiquitin blot showed minimal reaction against Met-βgal (lane 1), although it was efficiently immunoprecipitated (lane 1).
To confirm that rpn3-4 and rpn3-7 harbored a defect in the general turnover of proteolytic substrates, we quantified degradation by the N-end rule targeting system (Arg-βgal and Leu-βgal),18 and the ubiquitin fusion degradation system20 (Ub-Pro-βgal). β-Galactosidase activity was measured in triplicate using two independent preparations of protein extract (Fig. 5d). Enzymatic activity was compared to the level detected in RPN3 (which was arbitrarily set to a value of 1). High βgal activity was measured in rpn3-4 and rpn3-7 for all three proteolytic substrates, indicating that overall substrate turnover is affected. Moreover, the significant accumulation of multiUb proteins in rpn3 mutants (Fig. 5a and b) is also consistent with a global effect of substrate turnover.
Reduced levels of Rpn11 in rpn3 mutants
The 20S subunit Pup1 was expressed at physiological levels with a carboxy-terminal hemagglutinin (HA) epitope (Pup1-HA). Protein extracts were prepared from exponential-phase yeast cells, resolved by SDS/PAGE, and characterized by immunoblotting (Fig. 6). Protein extracts prepared from RPN3, rpn3-4, and rpn3-7 grown at 23 °C showed similar levels of proteasome subunits and multiUb proteins (Fig. 6a, lanes 1–3). Expression of Rpn3 from a plasmid in rpn3-4 and rpn3-7 had no effect on the expression of proteasome subunits or multiUb proteins at 23 °C (lanes 4 and 5). Examination of extracts prepared from cells grown at 37 °C showed higher levels of multiUb proteins in both rpn3 mutants (compare lane 6 to lanes 7 and 8). Immunoblotting studies showed that the expression of Rpn10, Rpt1, and Pup1 proteasome subunits was not affected in rpn3-4 and rpn3-7 at 23 °C or 37 °C. However, the expression of Rpn11 was strongly reduced in both mutants at 37 °C (lanes 7 and 8). (Rpn12 levels were also moderately reduced at 37 °C in both mutants). Expression of Rpn3 from a plasmid (bottom panel; lanes 4 and 5 and lanes 9 and 10) restored the levels of Rpn11 to WT levels.
Fig. 6.
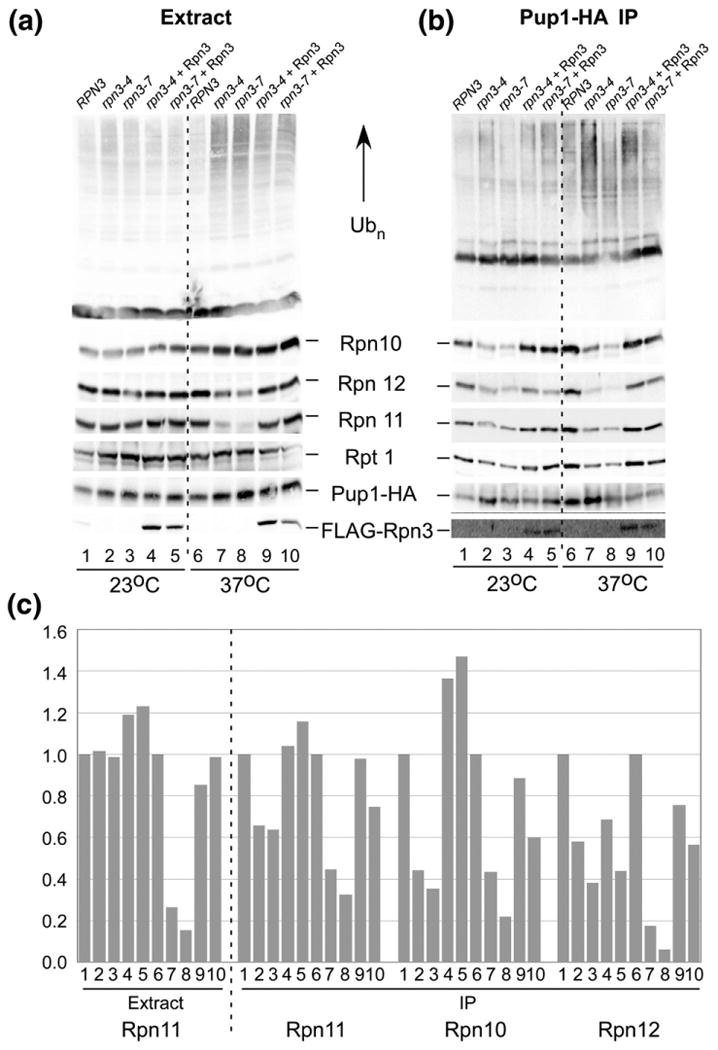
Proteasome instability in rpn3 mutants. (a) Protein extracts were prepared from RPN3, rpn3-4, and rpn3-7 containing Pup1-HA expressed at physiological levels. Immunoblots were incubated with antibodies to detect the indicated proteasome subunits. (b) Protein extracts were incubated with HA agarose, and proteasome subunits purified with Pup1-HA were detected. (c) Proteasome subunits associated with Pup1-HA in (b) were quantified. [Lane numbers in (c) represent samples detected in the corresponding lanes in (a) and (b).]
The co-purification of the 19S regulatory particle with Pup1-HA provides a useful way to examine proteasome assembly and stability. Immunoblotting showed that reduced levels of Rpn10, Rpn12, Rpn11, and Rpt1 were co-purified with Pup1-HA from extracts prepared from rpn3 mutant cells grown at 37 °C (Fig. 6b, lanes 7 and 8). Based on the specific interaction between Rpn3 and Rpn11 (Fig. 1), we speculate that the depletion of Rpn11 in rpn3-4 and rpn3-7 cells (Fig. 1) might be linked to defective proteasome assembly at 37 °C (Fig. 6b), consistent with studies described in Fig. 4. Whereas the abundance of proteasome subunits was unchanged in total extracts (Fig. 6a), with the exception of Rpn11 (and Rpn12), the decrease in the level of multiple 19S subunits in association with Pup1-HA is consistent with the loss of intact 26S proteasomes. Surprisingly, high levels of multiUb proteins were detected in proteasomes isolated from rpn3-4 and rpn3-7 at 37 °C. This finding suggests that proteasomes that are assembled in rpn3 mutants may be unable to effectively clear multiUb substrates. The proteasome assembly defect in rpn3 mutants was suppressed by FLAG-Rpn3 (Fig. 6b, lanes 9 and 10). The binding data were quantified by densitometry and plotted (Fig. 6c). The level of Rpn11 in RPN3, rpn3-4, and rpn3-7 in extracts is shown on the left, and its reduced interaction with the proteasome at 37 °C is apparent (lanes 7 and 8). A corresponding reduction in other proteasome subunits (Rpn10 and Rpn12) in association with immunopurified proteasomes (using Pup1-HA) confirmed that rpn3 mutants have a proteasome assembly/stability defect.
Shuttle factor interaction with the proteasome is reduced in rpn3 mutants
Rad23 is the archetype of a shuttle factor that can transport multiUb substrates to the proteasome.21,22 Multiple shuttle factors are expressed in yeast,21,23–25 and their interaction with the proteasome is reduced in rpn11-17 due to the instability of the proteasome. Because Rpn11 is unstable in rpn3 mutants, we investigated if the resulting proteasome assembly defect would lead to reduced interaction with shuttle factors. FLAG-Rad23 was expressed in RPN3, rpn3-4, and rpn3-7 at 23 °C and then transferred to 37 °C for 3 h. Equal amounts of protein extracts were prepared and applied to FLAG agarose. The bound proteins were separated by SDS/PAGE to determine if Rpn11 was incorporated into the proteasome (Fig. 7a). Immunoblotting showed that multiUb proteins were efficiently co-purified with FLAG-Rad23 from all three strains at 23 °C (lanes 1–3). However, this interaction was noticeably reduced in rpn3-7 at 37 °C (lane 6). We did not detect an appreciable decrease in Rad23 interaction with multiUb chains in rpn3-4 at 37 °C (lane 5).
Fig. 7.
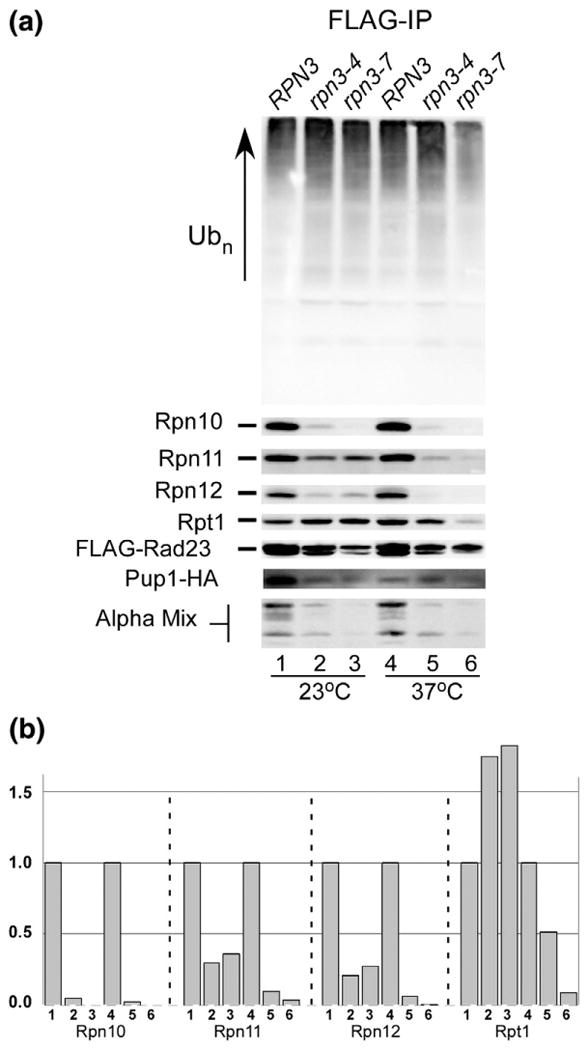
The Rad23 shuttle factor fails to precipitate intact proteasomes from rpn3 mutants. (a) RPN3, rpn3-4, and rpn3-7 were transformed with a high-copy plasmid expressing FLAG-Rad23. An equal amount of protein extracts was prepared from cultures grown at either 23 °C or 37 °C and incubated with anti-FLAG Sepharose. The bound proteins were released in SDS-containing gel electrophoresis buffer and separated by SDS/PAGE. Immunoblots were incubated with antibodies to detect the proteasome subunits indicated on the left. (b) Protein levels detected in (a) were quantified by densitometry and plotted. [The lanes in (b) correspond to the same lanes in (a).]
Rad23 binds the Rpn1 subunit that is present in the base of the 19S regulatory particle, which also contains six AAA-class Rpt ATPases,9 and the Rpn2 and Rpn13 subunits. FLAG-Rad23 was immunoprecipitated, and the co-purification of Rpn11 was found to be strongly reduced at 23 °C in both rpn3 mutants (lanes 2 and 3) and was almost undetectable at 37 °C (lanes 5 and 6). Similarly, much lower amounts of Rpn10 and Rpn12 were co-purified with FLAG-Rad23 from the rpn3 mutants. In contrast, at 23 °C, Rpt1 that is present in the base was efficiently co-purified with FLAG-Rad23 from all three strains. This finding is consistent with the known interaction between Rad23 and Rpn1 in the base and suggests that Rad23 might initially bind an intermediate complex that contains the base, but not lid, or the core particles. This idea is illustrated in lane 5, where Rpt1 is efficiently purified with FLAG-Rad23, whereas reduced levels of lid subunits (Rpn10, Rpn11, and Rpn12) and 20S core subunits (alpha mix and Pup1-HA) were detected. The binding data were quantified by densitometry, and the failure of FLAG-Rad23 to co-precipitate 19S subunits is shown (Fig. 7b). The co-purification of both 19S and 20S subunits with FLAG-Rad23 confirms its interaction with intact proteasomes in WT cells. However, these interactions are strongly diminished in rpn3-4 and rpn3-7.
Rpn11 cannot overcome the defects of rpn3 mutants
Previous studies showed that the proteolytic defects of rpn11-1 are caused by a proteasome assembly or stability defect.7 Similarly, we show here that depletion of Rpn11 in rpn3 is associated with defective proteasome assembly. To determine if Rpn11 instability contributed directly to the biochemical and growth defects of rpn3-4 and rpn3-7, we examined the in vivo stability of native Rpn11. RPN3, rpn3-4, and rpn3-7 strains were transferred from the permissive temperature (23 °C) to 37 °C, and aliquots were withdrawn at 60-min intervals (Fig. 8a). Protein lysates were prepared, and equal amounts were resolved by SDS/PAGE and examined by immunoblotting. Incubation with anti-Rpn11 antibodies showed constant levels of Rpn11 in RPN3 WT cells. However, incubation at 37 °C resulted in rapid depletion of Rpn11 in both rpn3-4 and rpn3-7. The same filter was probed with antibodies against Rpn10, and no significant change in abundance was observed in rpn3 mutants. To examine the effect of overexpressing Rpn11, we transformed RPN3, rpn3-4, and rpn3-7 with a high-copy plasmid expressing FLAG-Rpn11. The cells were transferred from 23 °C to 37 °C (Fig. 8b), and the abundance of native Rpn11 and FLAG-Rpn11 was determined by immunoblotting. The filter was also incubated with antibodies against the 19S regulatory subunits Rpn10, Rpn12, and Rpt1. High levels of FLAG-Rpn11 were detected in all strains after a brief 3-h incubation at 37 °C, and expression was significantly higher than that of native Rpn11 (lanes 1 and 5 lack FLAG-Rpn11). Rpn12 levels were slightly reduced at 37 °C. Protein extracts were also incubated with anti-HA antibodies to immunoprecipitate Pup1-HA and the proteasome (Fig. 8c). Lower levels of FLAG-Rpn11 were isolated with Pup1-HA from both rpn3-4 and rpn3-7 at 23 °C (lanes 3 and 4). Virtually no 19S subunits were co-purified with Pup1-HA at 37 °C, confirming significant proteasome instability (lanes 7 and 8).
Fig. 8.
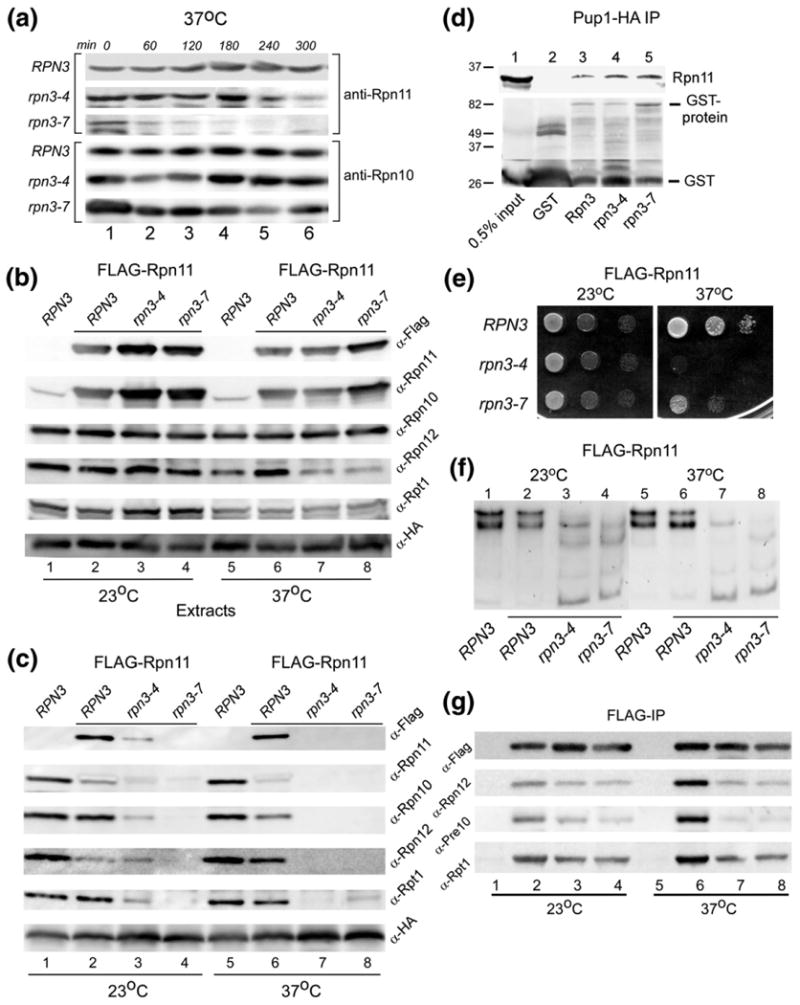
WT Rpn11 is destabilized in rpn3 mutants. (a) The stability of native Rpn11 was determined in RPN3, rpn3-4, and rpn3-7. Yeast cells were pre-grown at 23 °C and then transferred to 37 °C. Aliquots were withdrawn at the intervals shown, and the level of Rpn11 was determined by immunoblotting using anti-Rpn11 antibodies. The same filter was subsequently probed with antibodies against Rpn10 (lower panel). (b) RPN3, rpn3-4, and rpn3-7 were transformed with a high-copy plasmid expressing FLAG-Rpn11. Cultures were grown at 23 °C and then transferred to 37 °C for 3 h. An equal amount of total protein extract was examined by immunoblotting using antibodies indicated on the right. Lanes 1 and 5 represent control WT strain, lacking FLAG-Rpn11 overexpression. The level of both native Rpn11 and FLAG-Rpn11 was estimated by comparing the reaction with both anti-FLAG and anti-Rpn11 antibodies (compare lanes 1 and 2). (c) The extracts described in (a) were incubated with anti-HA antibody to immunoprecipitate (targeting Pup1-HA in the 20S core particle). The immunoprecipitates were resolved by SDS/PAGE and characterized using antibodies indicated on the right. (d) GST-Rpn3, GST-rpn3-4, and GST-rpn3-7 were bound to glutathione Sepharose, and their interaction with recombinant His6-Rpn11 was determined by immunoblotting. A control lane containing only GST is also shown (lane 2). (e) Yeast cells overexpressing FLAG-Rpn11 were spotted on agar medium (using serial 10-fold dilutions), and suppression of the rpn3 growth defect at 37 °C was investigated. (f) The protein extracts described in (b) were separated in a native polyacrylamide gel, and in situ hydrolysis of proteasome substrate LLVY-AMC was determined. (g) The protein extracts described in (b) were incubated with FLAG agarose to immunoprecipitate FLAG-Rpn11. The co-purification of 19S and 20S subunits was determined by immunoblotting.
We speculated that the strong interaction between Rpn11 and Rpn3 (Fig. 1) might confer a stabilizing effect on Rpn11. Specifically, the rapid turnover of Rpn11 in rpn3 mutants might have been caused by defective interaction with rpn3-4 and rpn3-7 proteins. To test this idea, we immobilized GST-Rpn3, GST-rpn3-4, and GST-rpn3-7 on glutathione Sepharose. His6-tagged Rpn11 was expressed in E. coli, and total bacterial lysates were incubated with the immobilized GST-Rpn3/rpn3 proteins (Fig. 8d). Immunoblotting confirmed that Rpn11 interacted with Rpn3 (lane 3) and also formed efficient interactions with both rpn3 mutant proteins (lanes 4 and 5). No interaction was seen with the control GST beads (lane 2). Although these findings indicate that purified Rpn11 can bind mutant rpn3 proteins, it is conceivable that these interactions are impaired in vivo.
We investigated if high-level expression of FLAG-Rpn11 would suppress the temperature-sensitive growth defect of rpn3 mutants. Serial 10-fold dilutions of yeast cultures were spotted on agar medium, and growth was examined after 2 days of incubation (Fig. 8e). FLAG-Rpn11 overexpression conferred partial growth at 37 °C in rpn3-7, but not in rpn3-4. However, this modest level of growth recovery was not sufficient to appreciably affect the proteasome assembly defect of either rpn3 mutant (Fig. 8f). In-gel peptidase activity measurement showed that RPN3 expressing FLAG-Rpn11 (lane 2) had equivalent proteasome assembly as an untransformed cell (lane 1). Proteasome assembly in rpn3-4 and rpn3-7 (lanes 3 and 4 and lanes 7 and 8) was not noticeably improved by expressing FLAG-Rpn11 (compare to untransformed cells in Fig. 4a).
Proteasomes purified using antibodies against a 20S subunit (Pup1-HA; Fig. 8c above) showed significantly reduced levels of Rpn11 and other 19S subunits in rpn3 mutants. In a reciprocal experiment, FLAG-Rpn11 was immunoprecipitated, and its interaction with 19S and 20S subunits was determined by immunoblotting (Fig. 8g). We found that FLAG-Rpn11 interaction with a base subunit (Rpt1) was efficient in rpn3 mutants at 23 °C but was reduced at 37 °C (lanes 7 and 8). Interaction with Rpn12 (a lid subunit) and Pre10 (a 20S core subunit) was strongly reduced at 37 °C. There were no nonspecific reactions in extracts that did not express FLAG-Rpn11 (lanes 1 and 5). We note that the shuttle factor Rad23 also formed a preferred interaction with the base subcomplex in rpn3 mutants (Fig. 7a).
Discussion
The atomic structure of the 19S regulatory particle, as well as the identification of interacting partners, has resisted detailed understanding due to its dynamic nature. However, genetic and proteomic studies have yielded a wealth of information that has functionally or biochemically linked specific subunits to other components in the proteasome. In this regard, previous studies uncovered weak interactions between Rpn11 and specific proteasome subunits, including Rpn8 and Rpn10.26 In an effort to further characterize Rpn11, we conducted direct protein–protein binding studies and confirmed a weak association with Rpn1. An unexpectedly strong interaction with Rpn3 was detected (Fig. 1). Rpn3 is a poorly characterized proteasome subunit, and no biochemical function has been described. However, genetic studies revealed a role in cell cycle progression and proteolysis.12
rpn3 and rpn11-1 mutants share a number of phenotypic traits that are linked to proteasome instability. We and others reported that mutations in RPN11 can have distinct effects on proteasome assembly and function.3,7,27 Proteasome are destabilized in the well-characterized rpn11-1 mutant, which results in reduced proteolysis and lethality at high temperature.5 In contrast, a catalytically inactive mutant (Rpn11AXA) that does not affect proteasome assembly3 appears to trap proteasomes in nonfunctional complexes that are bound to high levels of multiUb substrates.7 The growth and proteolytic defects of rpn3 and rpn11-1 are caused by proteolytic deficiencies.12 We speculate that these defects may arise from a failure of Rpn11 to be properly incorporated into proteasomes. The phenotypes of rpn11-1 and rpn3 mutations cannot be attributed entirely to the interaction between Rpn11 and Rpn3 proteins. For instance, the efficient binding between rpn3 mutant proteins and Rpn11 does not overcome the rpn3 deficiencies. Similarly, a robust rpn11-1–Rpn3 interaction does not overcome the proteolytic and growth defects of rpn11-1. Our biochemical and genetic studies8 suggest that Rpn3 might play an important role in promoting Rpn11 incorporation into proteasomes. The presence of native Rpn3 in the proteasome might create a structural environment that is favorable for Rpn11/proteasome association.
An intriguing defect in rpn11-1 is the imbalance in mitochondrial fission/fusion interconversion,14 which is thought to facilitate mitochondrial regeneration. High levels of mitochondrial aggregates were detected in rpn11-1, intimating high levels of fusion. Similarly, we observed dramatically elevated mitochondrial fusion in both rpn3-4 and rpn3-7. While the cause of this effect is unknown, we note that the stability of a key regulatory protein (Fzo1) that guides mitochondrial fission/fusion transitions is controlled by the ubiquitin/proteasome system.16 One interpretation of these results is that a failure to properly control Fzo1 stability might alter the cellular capacity to promote mitochondrial fusion. This finding underscores a plausible mechanistic link between proteasome activity and mitochondrial fusion. Whereas proteasome instability is the proximate effect of Rpn11 depletion (in rpn11-1 and rpn3), a secondary consequence is a failure to regulate mitochondria morphogenesis and function.
The significant interaction with Rpn11 led us to determine if Rpn3 contributed to any of the defects that were previously reported for the rpn11-1 mutant. In the present studies, we uncovered an unexpected role for Rpn3 in recruiting Rpn11 into proteasomes. Both rpn3-4 and rpn3-7 contained significantly reduced levels of Rpn11, suggesting that the free form is unstable. However, these two mutant alleles display distinct sensitivities to drugs and differential proteasome instability. Intriguingly, these properties correspond to the differential stability of Rpn11 in rpn3-4 and rpn3-7. Specifically, the more rapid turnover of Rpn11 in rpn3-7 is associated with more severe defects. The defects of rpn3 are caused by the mutant rpn3 proteins, since expression of WT Rpn3 fully suppressed all the defects. We recognize that the proteolytic defects of rpn3 mutants are not unexpected because Rpn3 is an essential proteasome subunit. However, its role in promoting Rpn11/proteasome association is the first biochemical effect reported for Rpn3. The association of other subunits in the 19S regulatory particle with the purified 20S core particle is also noticeably diminished in rpn3.
Previous studies showed that proteasomes in rpn11-1 are unstable (or poorly assembled).7 The temperature-sensitive growth defect of rpn11-1 was closely associated with reduced levels of intact proteasomes. The rpn11-1 protein contains a frame-shift mutation in the carboxy terminus, resulting in the substitution of 31 residues with incorrect amino acids. It was possible that the defects of rpn11-1 were caused by the presence of nonnative carboxy-terminal residues. However, we dismissed this possibility by expressing a mutant that lacked the entire carboxy-terminal region (implicated in the frameshift) and found that rpn111–277 displayed the same properties as rpn11-1. These results demonstrated that the loss of native carboxy-terminal residues and not the presence of nonnative residues exerted the effects observed in rpn11-1. We also note that both Rpn11 and rpn11-1 could interact with Rpn10, a multiubiquitin chain binding receptor in the proteasome. Whereas both single mutants cause proteasome assembly defects, a double mutant (rpn11-1 rpn10Δ) is inviable. Expression of just the carboxy-terminal residues of Rpn11 (GST-CRpn11, residues 267–306) partially suppressed the defects of rpn11-1, but not those of rpn11-1 rpn10Δ or rpn3 mutants (data not shown). These residues are functionally important, as they can suppress the defects of rpn11-1 in trans, but they are insufficient for overcoming the defects contributed by the loss of Rpn3 (rpn3) or Rpn10 (rpn11-1 rpn10).
In the present work, we show that the abundance of Rpn11 significantly influences proteasome stability. While it is conceivable that depletion of any proteasome subunit might cause this effect, we note that the role of Rpn3 to selectively stabilize Rpn11 is a unique effect. We also show that the loss of Rpn11 in rpn3 mutants mimics many of the proteasome assembly and growth defects that were previously observed in rpn11-1, which expresses native Rpn3. However, Rpn11 and Rpn3 are not functionally redundant, as overexpression of Rpn3 did not overcome the defects of rpn11-1 (data not shown). Additionally, overexpression of Rpn11 in rpn3 mutants did not suppress the temperature-sensitive or proteolytic defects of rpn3-4 and rpn3-7 (Fig. 8 and data not shown). This latter finding was surprising, since we anticipated that providing higher levels of Rpn11 would overcome the reduced levels of native Rpn11 in rpn3 mutants. High levels of FLAG-Rpn11 were detected in rpn3 mutants (Fig. 8a), although it failed to support proteasome assembly (Fig. 8b). The failure of overexpressed Rpn11 to suppress the defect of rpn3 is possibly due to inefficient recruitment into proteasomes, which could cause turnover of the free form of the protein.
The Rad23 shuttle factor can bind multiUb proteins through two ubiquitin-associated domains28 and the proteasome by an amino-terminal ubiquitin-like domain.22 Since Rad23 is thought to promote the delivery of ubiquitinated proteins to the proteasome,21 it is likely that free Rad23 does not form this interaction in vivo. The failure of Rad23 to be co-purified with the 20S complex in rpn3 mutants suggests that all or most components of the 19S particle are dissociated from the catalytic core particle in rpn3 (at the nonpermissive temperature). Intriguingly, FLAG-Rad23 retained the ability to co-precipitate Rpt1 in rpn3-4 at 23 °C and 37 °C. Rad23 binds the Rpn1 subunit,29 which is present in the base subcomplex of the proteasome and mediates interaction with both the lid subcomplex and the 20S core particle. We found that FLAG-Rad23 successfully co-precipitated Rpt1 that is present in the base subcomplex. However, FLAG-Rad23 co-purified low levels of the core particle (20S) from both rpn3-4 and rpn3-7 at 37 °C. It is unclear why Rad23 interaction with multiUb proteins is reduced at 37 °C in rpn3-7, although the overall levels of multiUb proteins were similar to that seen in RPN3. Further study will be required to determine if rpn3 mutants affect Rad23 function.
In vitro binding studies showed that rpn3 mutant proteins could bind Rpn11. However, the effect of this interaction on Rpn11 stability in vivo is uncertain. The loss of Rpn11 in total cell extracts coincides with its depletion in the proteasome and the loss of intact proteasomes (Fig. 6). This is evident by the reduced levels of Rpn11 in extracts, after transfer of rpn3 cells from 23 °C to 37 °C, and its progressive loss in proteasomes. Moreover, expression of Rpn3 in these mutants restored Rpn11 levels in the proteasome (Fig. 6). It is presently not known if Rpn11 is degraded by the proteasome itself or by other cellular proteases.
The rpn3 proteins showed allele-specific sensitivity to translation inhibitors and an amino acid analog (Fig. 2). These properties were closely coupled to distinguishable biochemical defects associated with rpn3-4 and rpn3-7 mutants. Specifically, the additional growth and proteolytic defects of rpn3-7 were coupled to a more rapid turnover of Rpn11 and reduced levels of intact proteasomes. Consequently, proteasomes in rpn3-7 showed reduced activity and weaker interaction with multiUb proteins and shuttle factors. Although the underlying mechanism of Rpn3 action is not known, our results show that the phenotypic defects associated with rpn3-4 and rpn3-7 are closely coupled to Rpn11 instability in these mutants. Overexpression of Rpn3 suppressed the defects of rpn3-4 and rpn3-7 by overcoming the proteasome assembly defect. However, elevated levels of multiUb proteins remained stably associated with proteasomes. One interpretation of this result is that expression of Rpn3 in rpn3-4 and rapn3-7 results in the accumulation of both WT and mutant proteins (Rpn3/rpn3-4 and Rpn3/rpn3-7). Since both WT and mutant proteins can bind the proteasome (data not shown), an intermediate effect is not unexpected. A previous report characterized proteasome integrity by size-exclusion chromatography and found that it was unaffected in rpn3 mutants.12 In contrast, we show here using functional (in-gel analysis) and biochemical approaches (immunoprecipitation of proteasomes) that the association of the 19S regulatory particle with the 20S particle is significantly reduced in rpn3 mutants. One possible explanation for this discrepancy is that size-exclusion chromatography will not necessarily distinguish functionally competent complexes from other assembly intermediates (or dead end complexes) that are similar in size to native proteasomes. Taken together, our genetic and biochemical studies suggest that, by stabilizing Rpn11, Rpn3 performs an essential role in promoting proteasome integrity and function.
Materials and Methods
Reagents
Proteasome inhibitor (epoxomicin) and substrate (Suc-Leu-Leu-Val-Tyr-AMC), as well as antibodies against the His6 epitope, were purchased from Boston Biochem. Anti-FLAG antibodies and GST-affinity beads were purchased from Sigma Chemical Co. and GE Healthcare, respectively. Polyclonal antibodies against Rpn11, Rpt1, and Rad23 were generated by Pocono Rabbit Farm and Laboratory, Inc. (Canadensis, PA, USA) using recombinant proteins. Anti-Rpn10 and anti-Rpn12 antibodies were kindly provided by D. Skowyra (St. Louis University). Monoclonal antibodies against β-galactosidase were purchased from Promega (Madison, WI). Immunodetection was achieved using an enhanced chemiluminescence reagent from PerkinElmer and captured using a Kodak GelLogic 1500 Imager and quantified using Kodak software. Fluorescence released by the hydrolysis of LLVY-AMC was measured using a Tecan Infinite F200 plate reader.
Yeast strains and plasmids
Integrative rpn3-4 and rpn3-7 plasmids were kindly provided by Dr. Eric Bailly.3 DNA was digested by SacI and transformed into JD47-13c (Table 1). JD47-13c strain and plasmid DNA for generating an integrative derivative of Pup1-HA were provided by Dr. Jurgen Dohmen (University of Cologne). DNA of integrative of PUP1-HA was digested with BstXI and transformed into RPN3, rpn3-4, and rpn3-4 strains. Mt-GFP plasmid DNA was kindly provided by Dr. B. Westermann (Table 2). GST fusions of Rpn3, rpn3-4, and rpn3-7 were generated by amplifying the full-length gene, encoding amino acids 1– 523 from WT, rpn3-4, and rpn3-7 genomic DNA, and cloned into pGEX-4T-3 using BamHI and EcoRI restriction sites. FLAG-tagged Rpn3, rpn3-4, and rpn3-7 were cloned into YEplac vector as EcoRI and XbaI fragments and expressed with an N-terminal FLAG epitope from a PCUP1 promotor. His6-Rpn11 and FLAG-Rad23 were previously described.10 Expression from the PCUP1 promoter was induced by the addition of 100 μM CuSO4 to the medium.
Table 1.
Yeast strains
| Strain name | Genotype | Source |
|---|---|---|
| KMY576 | MATa his3Δ200 trp1Δ 63 lys2-801 ura3-52 leu2-2,112 | JD47-13C |
| LCY1908 | KMY576 rpn3-4∷TRP1 | This study |
| LCY1909 | KMY576 rpn3-7∷TRP1 | This study |
| KMY1241 | KMY576 PUP1-HA∷URA3 | JD139 |
| NTY51 | LCY1908 PUP1-HA∷URA3 | This study |
| NTY52 | LCY1909 PUP1-HA∷URA3 | This study |
| NTY60 | NTY51 PCUP1-FLAG-RPN3 | This study |
| NTY61 | NTY52 PCUP1-FLAG-RPN3 | This study |
| KKJY47 | KMY1241 PCUP1-FLAG-RAD23 | This study |
| KKJY48 | NTY51 PCUP1-FLAG-RAD23 | This study |
| KKJY49 | NTY52 PCUP1-FLAG-RAD23 | This study |
| KKJY28 | KMY576 Leu-β-galactosidase | This study |
| KKJY29 | KMY576 Met-β-galactosidase | This study |
| KKJY30 | KMY576 Ub-Pro-β-galactosidase | This study |
| KKJY31 | KMY576 Arg-β-galactosidase | This study |
| KKJY32 | LCY1908 Leu-β-galactosidase | This study |
| KKJY33 | LCY1908 Met-β-galactosidase | This study |
| KKJY34 | LCY1908 Ub-Pro-β-galactosidase | This study |
| KKJY35 | LCY1908 Arg-β-galactosidase | This study |
| KKJY36 | LCY1909 Leu-β-galactosidase | This study |
| KKJY37 | LCY1909 Met-β-galactosidase | This study |
| KKJY38 | LCY1909 Ub-Pro-β-galactosidase | This study |
| KKJY39 | LCY1909 Arg-β-galactosidase | This study |
| LCY1725 | KMY576 Mt-GFP | This study |
| LCY 2371 | LCY1908 Mt-GFP | This study |
| LCY 2372 | LCY1909 Mt-GFP | This study |
Table 2.
Plasmids
| Plasmids | Description | Origin |
|---|---|---|
| LEP547 | yIP204-rpn3-4 | E. Bailly and S. Reed |
| LEP548 | yIP204-rpn3-7 | E. Bailly and S. Reed |
| pJD508 | yIPlac211-Pup1-HA | J. Dohman |
| LEP542 | pGEX-GST-Rpn3 | This study |
| KKJEP1 | pGEX-GST-rpn3-4 | This study |
| KKJEP2 | pGEX-GST-rpn3-7 | This study |
| LEP522 | pGEX-GST-Rpn11 | A. Chandra |
| LEP523 | pGEX-GST-rpn11-1 | A. Chandra |
| LEP601 | pET28a-His6-Rpn11 | A. Chandra |
| LEP52 | PCUP1–FLAG-Rad23∷LEU2 | L. Chen |
| HEP76 | PCUP1–FLAG-Rpn3∷TRP1 | This study |
| LEP529 | pXY211-Mt-GFP | B. Westermann |
In vitro translation reaction and binding assay
In vitro35S-labeled protein transcription/translation reactions were performed as described previously.8
Interaction of recombinant Rpn3/rpn3 and Rpn11
GST-Rpn3, GST-rpn3-4, and GST-rpn3-7 were expressed in E. coli and purified on GST beads. An equal amount of bacterial lysates containing recombinant His6-Rpn11 was applied to GST proteins. The binding reaction was incubated for 4 h at 4 °C. Unbound proteins were removed by several washes in binding buffer, and the adsorbed proteins were released in SDS-containing sample buffer and resolved in a 10% SDS polyacrylamide gel. The separated proteins were transferred to nitrocellulose and characterized by immunoblotting.
Sensitivity to translation inhibitors and an amino acid analog
Yeast cultures were grown in selective medium and normalized to an absorbance of A600=1.0. Serial 10-fold dilutions were prepared, spotted on yeast extract/peptone/dextrose agar plates, and incubated at either 23 °C or 37°C. Aliquots were also spotted onto medium containing 0.8 mM paromomycin, 0.1 mM hygromycin B, and 0.5 μg/ml L-canavanine.
Native PAGE
A non-denaturing polyacrylamide gel was used to examine proteasome assembly and activity, as described previously.8 Briefly, 50 μg of protein lysate was resolved in a native gel consisting of discontinuous steps of 3%, 4%, and 5% polyacrylamides. The positions of proteasome intermediates were detected by monitoring the in situ hydrolysis of the chymotryptic substrate LLVY-AMC.
Immunoprecipitation and immunoblotting
Yeast cells were suspended in buffer A [50 mM Hepes (pH 7.5), 150 mM NaCl, 5 mM ethylenediaminetetraacetic acid, and 1% Triton X-100] containing a protease inhibitor cocktail (Roche) and lysed with a FastPrep disruptor (FP120-Thermo Savant). Cell debris was removed by centrifugation at 14,000g for 2 min at 4 °C. An equal amount of protein extracts (Bradford; Bio-Rad) was incubated with anti-HA affinity matrix (Roche), FLAG-M2 affinity agarose beads (Sigma Chemical Co.), or protein-A matrix that was bound to anti-β-galactosidase antibody. The reactions were incubated at 4 °C for 4 h with constant mixing. The beads were washed three times with buffer A, and the bound proteins were released by boiling in SDS loading buffer and separated in either 10% or 12% SDS–tricine polyacrylamide gels, transferred to nitrocellulose, and examined by immunoblotting.
Measurement of Suc-LLVY-AMC hydrolysis
An equal amount of protein lysate (5 μg in 50 μl) was premixed with 200 ng of proteasome inhibitor (epoxomicin was prepared in 50% dimethyl sulfoxide) or an equivalent volume of vehicle (50% dimethyl sulfoxide lacking the inhibitor), followed by the addition of proteasome assay buffer [200 μl; 25 mM Hepes (pH 7.5) and 0.5 mM ethylenediaminetetraacetic acid] containing the chymotryptic substrate (40 μM LLVY-AMC), with and without 0.05% SDS. Reactions were incubated at 30 °C for 1 h, and fluorescence was detected using a Tecan Infinite F200 detector. Epoxomicin-sensitive activity was generated with triplicate measurements from two independent experiments, quantified, and plotted.
β-Galactosidase activity assay
Cultures of RPN3, rpn3-4, and rpn3-7 were grown at 37 °C for 4 h, and 1 ml was collected by brief centrifugation. The cells were resuspended in 1 ml Z-buffer [100 mM NaPO4 (pH 7.0), 10 mM KCl, 1 mM MgSO4, and 40 μM β-mercaptoethanol]. One-half milliliter of cell suspension was used to measure OD600. Another one-half milliliter of the suspended cells was combined with 20 μl of 0.1% SDS and lysed. Following the addition of 20 μl chloroform and brief mixing, 10 μl chlorophenol red-β-D-galactopyranoside (20 mg/ml chlorophenol red-β-D-galactopyranoside) was added. The reaction was stopped by addition of 0.5 ml 1 M Na2CO3. Following centrifugation, 0.9 ml of the clear supernatant was transferred to a cuvette, and absorbance was measured (OD574).
Fluorescence microscopy
One milliliter of yeast cells was collected and washed with phosphate-buffered saline. The cells were suspended in phosphate-buffered saline and spotted on Poly-Prep slides (Sigma Chemical Co.). Images were obtained with a Zeiss Imager M1 microscope. All exposures for the GFP signal were set to 45 ms (using Zeiss filter set #38 HE).
Acknowledgments
We thank E. Bailly for rpn3 mutant strains and Rpn3 plasmids, B. Westermann for providing Mt-GFP, J. Dohmen for a plasmid expressing Pup1-HA, and D. Skowyra for antibodies against Rpn10 and Rpn12. These studies were supported by a grant from the National Institutes of Health (CA083875) to K.M.
Abbreviations
- multiUb
multiubiquitinated
- GST
glutathione S-transferase
- GFP
green fluorescent protein
- CP
catalytic particle
- HA
hemagglutinin
- WT
wild type
References
- 1.Guterman A, Glickman MH. Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J Biol Chem. 2004;279:1729–1738. doi: 10.1074/jbc.M307050200. [DOI] [PubMed] [Google Scholar]
- 2.Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006;25:4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 4.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 5.Rinaldi T, Ricci C, Porro D, Bolotin-Fukuhara M, Frontali L. A mutation in a novel yeast proteasomal gene, RPN11/MPR1, produces a cell cycle arrest, overreplication of nuclear and mitochondrial DNA, and an altered mitochondrial morphology. Mol Biol Cell. 1998;9:2917–2931. doi: 10.1091/mbc.9.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinaldi T, Ricordy R, Bolotin-Fukuhara M, Frontali L. Mitochondrial effects of the pleiotropic proteasomal mutation mpr1/rpn11: uncoupling from cell cycle defects in extragenic revertants. Gene. 2002;286:43–51. doi: 10.1016/s0378-1119(01)00799-5. [DOI] [PubMed] [Google Scholar]
- 7.Chandra A, Chen L, Liang H, Madura K. Proteasome assembly influences interaction with ubiquitinated proteins and shuttle factors. J Biol Chem. 2010;285:8330–8339. doi: 10.1074/jbc.M109.076786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandra A, Chen L, Madura K. Synthetic lethality of rpn11-1 rpn10Delta is linked to altered proteasome assembly and activity. Curr Genet. 2010;56:543–557. doi: 10.1007/s00294-010-0321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glickman MH, Rubin DM, Fu H, Larsen CN, Coux O, Wefes I, et al. Functional analysis of the proteasome regulatory particle. Mol Biol Rep. 1999;26:21–28. doi: 10.1023/a:1006928316738. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci. 1998;23:204–205. doi: 10.1016/s0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- 11.Kawamura M, Kominami K, Takeuchi J, Toh-e A. A multicopy suppressor of nin1-1 of the yeast Saccharomyces cerevisiae is a counterpart of the Drosophila melanogaster diphenol oxidase A2 gene, Dox-A2. Mol Gen Genet. 1996;251:146–152. doi: 10.1007/BF02172912. [DOI] [PubMed] [Google Scholar]
- 12.Bailly E, Reed SI. Functional characterization of rpn3 uncovers a distinct 19S proteasomal subunit requirement for ubiquitin-dependent proteolysis of cell cycle regulatory proteins in budding yeast. Mol Cell Biol. 1999;19:6872–6890. doi: 10.1128/mcb.19.10.6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kominami K, Okura N, Kawamura M, DeMartino GN, Slaughter CA, Shimbara N, et al. Yeast counterparts of subunits S5a and p58 (S3) of the human 26S proteasome are encoded by two multicopy suppressors of nin1-1. Mol Biol Cell. 1997;8:171–187. doi: 10.1091/mbc.8.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rinaldi T, Hofmann L, Gambadoro A, Cossard R, Livnat-Levanon N, Glickman MH, et al. Dissection of the carboxyl-terminal domain of the proteasomal subunit Rpn11 in maintenance of mitochondrial structure and function. Mol Biol Cell. 2008;19:1022–1031. doi: 10.1091/mbc.E07-07-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westermann B. Molecular machinery of mitochondrial fusion and fission. J Biol Chem. 2008;283:13501–13505. doi: 10.1074/jbc.R800011200. [DOI] [PubMed] [Google Scholar]
- 16.Cohen MM, Leboucher GP, Livnat-Levanon N, Glickman MH, Weissman AM. Ubiquitin–proteasome-dependent degradation of a mitofusin, a critical regulator of mitochondrial fusion. Mol Biol Cell. 2008;19:2457–2464. doi: 10.1091/mbc.E08-02-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neutzner A, Benard G, Youle RJ, Karbowski M. Role of the ubiquitin conjugation system in the maintenance of mitochondrial homeostasis. Ann N Y Acad Sci. 2008;1147:242–253. doi: 10.1196/annals.1427.012. [DOI] [PubMed] [Google Scholar]
- 18.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 19.Bachmair A, Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989;56:1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 20.Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 23.Rao H, Sastry A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J Biol Chem. 2002;277:11691–11695. doi: 10.1074/jbc.M200245200. [DOI] [PubMed] [Google Scholar]
- 24.Saeki Y, Saitoh A, Toh-e A, Yokosawa H. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem Biophys Res Commun. 2002;293:986–992. doi: 10.1016/S0006-291X(02)00340-6. [DOI] [PubMed] [Google Scholar]
- 25.Zhang D, Chen T, Ziv I, Rosenzweig R, Matiuhin Y, Bronner V, et al. Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol Cell. 2009;36:1018–1033. doi: 10.1016/j.molcel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukunaga K, Kudo T, Toh-e A, Tanaka K, Saeki Y. Dissection of the assembly pathway of the proteasome lid in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2010;396:1048–1053. doi: 10.1016/j.bbrc.2010.05.061. [DOI] [PubMed] [Google Scholar]
- 27.Rinaldi T, Pick E, Gambadoro A, Zilli S, Maytal-Kivity V, Frontali L, Glickman MH. Participation of the proteasomal lid subunit Rpn11 in mitochondrial morphology and function is mapped to a distinct C-terminal domain. Biochem J. 2004;381:275–285. doi: 10.1042/BJ20040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L, Shinde U, Ortolan TG, Madura K. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multiubiquitin chain assembly. EMBO Rep. 2001;2:933–938. doi: 10.1093/embo-reports/kve203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]


