Abstract
The LIM homeobox 2 transcription factor Lhx2 is known to control crucial aspects of neural development in various species. However, its function in human neural development is still elusive. Here, we demonstrate that LHX2 plays a critical role in human neural differentiation, using human embryonic stem cells (hESCs) as a model. In hESC-derived neural progenitors (hESC-NPs), LHX2 was found to be expressed before PAX6, and co-expressed with early neural markers. Conditional ectopic expression of LHX2 promoted neural differentiation, whereas disruption of LHX2 expression in hESCs significantly impaired neural differentiation. Furthermore, we have demonstrated that LHX2 regulates neural differentiation at two levels: first, it promotes expression of PAX6 by binding to its active enhancers, and second, it attenuates BMP and WNT signaling by promoting expression of the BMP and WNT antagonist Cerberus 1 gene (CER1), to inhibit non-neural differentiation. These findings indicate that LHX2 regulates the transcription of downstream intrinsic and extrinsic molecules that are essential for early neural differentiation in human.
INTRODUCTION
The neuroepithelium is derived from epiblasts and is the origin of the nervous system. Its formation is based on the interplay between intrinsic transcription factors and extrinsic signaling (1). Experiments in Xenopus and rodents suggest that extrinsic signaling molecules, such as fibroblast growth factor (FGF), support neural induction (2), whereas WNT, SHH, Notch, transforming growth factor beta (TGFβ) and BMP hinder neural differentiation (3). On the other hand, intrinsic transcription factors, such as Sox1, have been found to control neural determination in mice (4). Sox1 was further demonstrated to influence mouse neural specification by regulating expression of neural genes, including Pax6 (5). These findings suggest that the differentiation of pluripotent stem cells into neuroepithelia is regulated by the interplay between signaling factors and neural transcription factors.
Complicated ethical issues, and lack of a cellular system suitable for investigating the molecular mechanisms underlying early lineage differentiation, have hampered studies into early human development; however, the establishment of human embryonic stem cells (hESCs) has helped to mitigate these challenges (6). Human ESCs are pluripotent, capable of unlimited proliferation in vitro. They thus provide an alluring and powerful pipeline through which to study the molecular basis of the transition from pluripotency to the neural lineage. To date, obligated primitive stage neural cells (7,8) and functional transplantable neurons have been generated from hESCs in vitro under chemically defined neural induction culture conditions (9,10). Using this neural induction procedure, the genetic factors that contribute to the regulation of early human neural differentiation have begun to be unearthed (4,5,7,11).
Transcriptional activator LIM homeobox 2 (Lhx2) has roles in neural induction and morphogenesis, as well as diverse functions among species (12). Lhx2 homologs are found in Drosophila (apterous), zebrafish (bel), Xenopus and mouse (12–15), where their sequences are largely conserved, but show some divergence in spatial expression in the central nervous system (12,16). During mouse embryogenesis, Lhx2 expression initiates before neurulation, and it is highly expressed in the neuroepithelium and ventricular zone (12,17,18). In Lhx2 mutant mice, central nervous system defects are detected in the dorsal telencephalon, hippocampus, thalamus, optic vesicle and olfactory bulb, suggesting that Lhx2 plays a critical role in brain morphogenesis (18–26). Although recent studies indicate that Lhx2 participates in the determination of regional fate decisions specifying cortical identity (18,25), the functional role of LHX2 in transcriptional control of early neural differentiation in human is not well understood.
The expression pattern and role of LHX2 in human neural differentiation have not yet been investigated. Here, we report that LHX2 is one of the early genes switched on in differentiating hESCs, before PAX6 and SOX1, and its expression is maintained with that of other neural genes in hESC-derived neural progenitors (hESC-NPs). Additionally, gain- and loss-of-function experiments revealed that overexpression of LHX2 enhances neural differentiation, whereas its knockdown impairs neural differentiation in hESCs. We have further demonstrated that LHX2 directly regulates expression of the neural transcription factor PAX6, and the WNT and BMP signaling antagonist Cerberus 1 (CER1). Taken together, our results demonstrate that LHX2 regulates the transcription of downstream intrinsic and extrinsic molecules that control early neural differentiation.
MATERIALS AND METHODS
Cultivation and neural differentiation of hESCs
Human ESCs H9 (WiCell, Madison, WI) (6) and NTU1 (from C.F. Chen, NTUH) (27), and human induced pluripotent stem cells (iPSCs) iCFB46 (28), were used at a passage number below 50. Undifferentiated hESCs were routinely maintained and subcultivated as previously described (29). For neural differentiation (Figure 1A), hESCs were dissociated with 1 mg/ml dispase (Sigma), and then left to form embryoid bodies (EBs) in ultra-low attachment dishes (Corning) for 4 days in hESC media [Dulbecco's modified Eagle's medium (DMEM)/F12, 20% knockout serum replacement, 1% non-essential amino acids, 2 mM l-glutamine, 100 µM β-mercaptoethanol and 5 ng/ml basic fibroblast growth factor (bFGF)]. EBs were transferred for further neural differentiation in N2 media [DMEM/F12, N2 supplement, 1% non-essential amino acids, 2 mM l-glutamine and 20 ng/ml bFGF (all purchased from Invitrogen)]. EBs were plated on tissue culture dishes (Nunc) in N2 media for 6 days of in vitro differentiation (IVD). For gain-of-function experiments, cells were separated from feeder cells using 1 mg/ml dispase (Sigma) and re-plated on Matrigel coated dishes. One day after plating, cells were treated with 2 µg/ml doxycyline (Sigma) in DMEM, 3% FBS, 1% non-essential amino acid (NEAA) and 2 mM l-glutamine (Invitrogen) for 3 days.
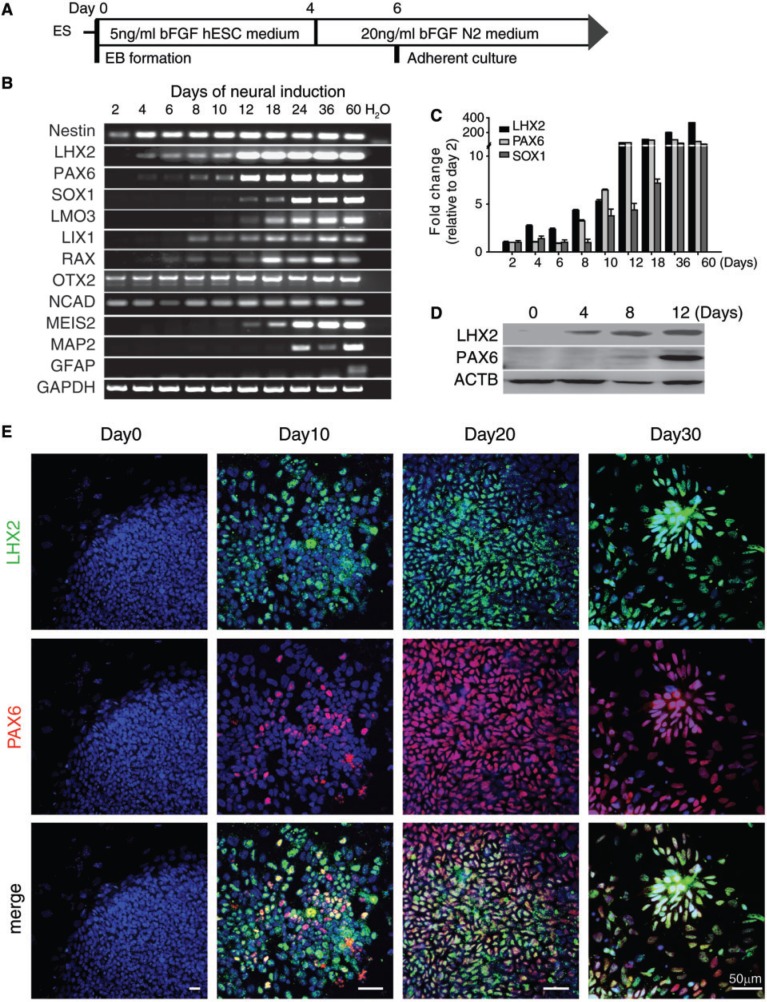
Figure 1.
LHX2 is expressed in neural lineage cells derived from hESCs. (A) Procedure of in vitro neural induction from hESCs. (B) RT-PCR and (C) qRT-PCR analysis of neural genes using mRNA isolated from differentiating H9 hESCs at the indicated days. LHX2 transcripts could be detected earlier (at day 4) than PAX6 and SOX1 transcripts. Error bars represent the mean ± SEM (n = 3). (D) Western blot of proteins isolated from differentiating hESCs at the indicated days with specific antibodies against LHX2 and PAX6. (E) Immunocytochemical analysis of differentiating hESCs at the indicated days with specific antibodies against PAX6 and LHX2. LHX2 was co-localized with PAX6 in differentiating hESCs at various differentiation stages.
Generation of inducible overexpression and constitutive knockdown hESCs
For generation of Tet-On inducible overexpression hESCs, EF1α and TetR (from pcDNA6-TR, Invitrogen) were cloned into pcDNA3.1 as pTetR; and EF1a, TetO, LHX2 coding sequences cloned from hESC-derived neural progenitors, and puroTK (from Y.T. Chen, NTU) were cloned into pcDNA3.1 as pTetOLHX2. Electroporation of DNA constructs into hESCs and establishment of stable clones was performed as described previously (30). Briefly, 107 undifferentiated hESCs were prepared for each electroporation. After pretreatment with Y-27632 (688000, Merk) for 2 h, cells were trypsinized into single cells, and used for electroporation or lentiviral infection. Electroporation involved 25 µg plasmid and the following conditions: 230 V, 500µF. For generation of conditional overexpression hESCs, TetR-H9 was established by transfecting pTetR into H9, followed by drug selection using G418 (50 µg/ml; Invitrogen). After establishment of TetR-H9, pTetOLHX2 or pTetOGFP was transfected into TetR-H9 by electroporation and selected by puromycin (400 µg/ml; Sigma) for establishment of iLHX2 or iGFP stable clones, respectively. Two iLHX2 clones with higher expression levels of LHX2 after 2 µg/ml doxycycline (Sigma) induction were selected for subsequent overexpression experiments. For generation of constitutive knockdown hESCs carrying shLHX2, shPAX6, shCER1 or shLuc, shRNA on pLKO_TRC001, constructs were obtained from the RNAi Core, National Science Council, Taiwan, and lentiviral particles were produced using 293T cells following standard procedures. The shRNA constructs were named according to the targeted gene and the position of the first shRNA targeting nucleotide in the gene coding sequence (number). The targeting sequences were as follows: shLHX2-831 (GCTTCGGACCATGAAGTCTTA), shLHX2-968 (GCAACCTCTTACGGCAGGAAA), shPAX6-284 (CGTCCATCTTTGCTTGGGAAA), shCER1-338 (CCAGCCGATAGATGGAATGAA) and shCER1-460 (GCCATGAAGTACATTGGGAGA). Lentiviral particles were generated carrying shLHX2, shPAX6, shCER1 or shLuc. Titers of resulting lentiviral particles were examined following standard MTT procedures, and 0.01 R.I.U. viral particles were introduced into undifferentiated hESCs; cells were then selected for with puromycin (400 µg/ml; Sigma) for stable clone establishment. After selection, two to three single colonies were selected in mixed culture as constitutive knockdown hESCs. Each stable clone was examined for pluripotency gene expression by reverse transcriptase-polymerase chain reaction (RT-PCR) and AP staining (Supplementary Figure S2).
Transient overexpression in hESCs
For transient overexpression of LHX2 and PAX6 in hESCs, GFP, LHX2 and PAX6 were cloned into FUW to generate FUW-GFP, FUW-LHX2 and FUW-PAX6, respectively. Transient transfection of hESCs with 2 µg FUW-GFP, FUW-LHX2 or FUW-PAX6 was performed with Lipofectamine 2000 (Invitrogen), following the standard Lipofectamine 2000 transfection protocol.
Reverse transcription-polymerase chain reaction
RNA was extracted with TRIzol reagent following the standard extraction protocol (Molecular Research Center). Extracted RNA was reverse-transcribed into cDNA with SuperScript III Reverse Transcriptase (Invitrogen). Each PCR used 25 ng of cDNA. GoTaq Green Master Mix (Promega) was used for RT-PCR. For quantitative PCR (qPCR), SYBR® FAST 2X qRT-PCR Master Mix (KAPA) and a 7900HT Fast Real-Time PCR System (Applied Biosystems) were used. Primers are listed in Supplementary Table S1.
Teratoma generation and ethics statement
For teratoma generation, 5 × 106 undifferentiated hESCs were subcutaneously injected into NOD-SCID mice. After 7 weeks, mice were sacrificed and teratomas were isolated. All the animal experiments were approved by the Animal Care and Use Committee of Academia Sinica, and performed in accordance with the Institutional Animal Care and Use Committee guidelines of Academia Sinica
Immunocytochemistry, immunohistochemistry and microscopy
The immunostaining procedure was performed as previously described (29). The primary antibodies were as follows: Rabbit anti-LHX2 (1:200, AB5756, Chemicon); Mouse anti-PAX6 (1:50, Hybridoma bank); Mouse anti-NESTIN (1:100, AB5326, Chemicon); Mouse anti-Tuj1 (1:200, MAB1637, Chemicon); Mouse anti-NEUN (1:50, MAB377, Chemicon); Mouse anti-OCT4 (1:50, sc-9081, Santa Cruz); Mouse anti-SSEA4 (1:200, MAB4304, Millipore); Goat anti-OTX2 (1:200, AF1979, R&D), and; Goat anti-CER1 (1:50, sc-15134, Santa Cruz). The secondary antibodies were as follows: Alexa594-donkey anti-mouse (Molecular Probe); Alexa488-donkey anti-rabbit (Molecular Probe); Alexa594-donkey anti-goat (Molecular Probe). For histochemical analysis, H&E staining was performed using an Autostainer XL Leica ST5010 (Leica Microsystem). The number of rosette-like structures per section was counted within 60 out of 200 serial sections in 1000 mm3 teratomas, n = 3 (Figure 3F). All images were recorded with a Leica TCS SP5 confocal microscope.
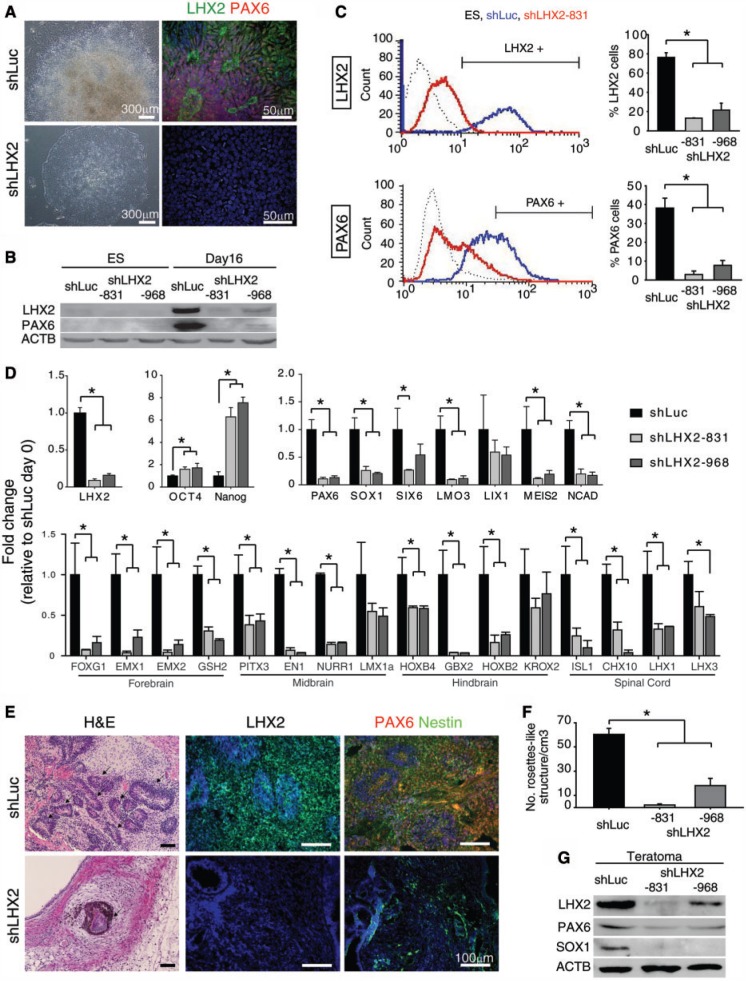
Figure 3.
Reduction of LHX2 expression impaired in vitro and in vivo neural developmental potential of hESCs. (A) Morphology and immunocytochemical analysis of differentiating shLuc- (upper panel) or shLHX2-expressing (lower panel) hESCs at IVD day 16 using antibodies against LHX2 and PAX6. Nuclei were counterstained with DAPI (blue). (B) Western blot analysis of LHX2 and PAX6 protein expression in shLHX2 and shLuc-expressing hESCs at the indicated days. Undifferentiated hESCs were used as a negative control. (C) Flow cytometric analysis of the number of LHX2+ or PAX6+ cells in differentiating shLHX2- or shLuc-expressing hESCs at IVD day 16. Undifferentiated H9 ES cells were used as a negative control. The average percentage of LHX2+ or PAX6+ cells is indicated (right panels, n = 3). (D) qRT-PCR for the indicated genes using cDNA derived from mRNA isolated from shLuc- or shLHX2-expressing hESCs at IVD day 12. ACTIN was used as the internal control for normalization (n = 3). (E) Histological examination of neural rosette-like structures with H&E staining and specific antibodies against LHX2, PAX6 and Nestin (arrows indicate neural rosette-like structures). Nuclei were counterstained with DAPI (blue). (F) Quantitative analysis of the number of neural rosette-like structures in teratomas derived from shLHX2 (831 and 968) or shLuc-expressing hESCs (n = 3). (G) Western blot analysis of the teratomas derived from shLHX2- or shLuc-expressing hESCs with specific antibodies against LHX2, PAX6 and SOX1. Error bars represent the mean ± SEM. Significance: *P < 0.05. ShRNA were named according to the targeted gene and the first shRNA targeting nucleotide in the gene coding sequence (number). For the targeting sequences of shLHX2-831 shLHX2-968, please see the ‘Materials and Methods’ section.
Flow cytometry
Cells were trypsinized, and then resuspended in 0.1% paraformaldehyde-phosphate buffered saline (PBS) for 10 min on ice. Cells were subsequently transferred to 90% methanol for 30 min on ice. After blocking with 2% normal donkey serum-PBS, cells were incubated with diluted primary antibodies for 1 h at room temperature. The primary antibodies were as follows: Rabbit anti-LHX2 (1:200, AB5756, Chemicon) and Mouse anti-PAX6 (1:50, Hybridoma bank). After washing with 0.05% Tween 20-PBS, cells were incubated with diluted secondary antibodies for 30 min at room temperature. The secondary antibodies were as follows: Alexa488-donkey anti-mouse (1:500, Molecular Probe) and Alexa488-donkey anti-rabbit (1:500, Molecular Probe). After washing three times with 0.05% Tween 20-PBS, cells were suspended in 2% normal goat serum, 0.1% sodium azide and 0.05% Tween 20-PBS .The results were recorded and analyzed with a BD FACSCalibur flow cytometer.
Western blot
Western blotting was performed as previously described (29), and separate blots were used for individual antibodies. The primary antibodies were as follows: Rabbit anti-LHX2 (1:2000, AB5756, Chemicon); Mouse anti-PAX6 (1:500, Hybridoma bank); Mouse anti-SOX1 (1:1000, LS-C39194, Life span); Rabbit anti-SMAD1 (1:1000, 6944, Cell Signaling); Rabbit anti-phosphor-SMAD1 (1:1000, 9511, Cell Signaling); Rabbit anti-SMAD2 (1:1000, 3102, Cell Signaling); Rabbit anti-phosphor-SMAD2 (1:1000, 3101, Cell Signaling); Rabbit anti-β-catenin (1:1000, 9581, Cell Signaling); Rabbit anti-phosphor-β-catenin (Ser33/37/Thr41) (1:1000, 9561, Cell Signaling); Mouse anti-β-Actin (1:100 000, A5441, Sigma). The secondary antibodies were as follows: HRP-donkey anti mouse (1:6000, Sigma); HRP-goat anti rabbit (1:6000, Sigma); HRP-donkey anti goat (1:3000, Jackson). Signals were detected and recorded with an ECL detection system (Thermo) and a luminescent image analyzer (LAS-4000 mini, Fujifilm).
Microarray analysis
Total RNA was extracted from iLHX2 cells at day 4 and day 16 after doxycycline treatment under conditions unfavorable to neural differentiation (3% FBS-DMEM), and from shLHX2 cells at day 12 of IVD in N2 media (11), using TRIzol reagent (Molecular Research Center). Two biological replicates per condition were used. All gene expression results were obtained by the Illumina Gene Expression Service Laboratory at National Taiwan University, Taiwan, using the Illumina microarray platform. A HumanHT-12 v4 Expression BeadChip was scanned with a BeadStation 450 Scanner, and data were analyzed using GeneSpring X software (Agilent) and Ingenuity Pathway Analysis (http://www.ingenuity.com/). Raw data were normalized independently for each experiment using Robust Multichip Average. Three pairwise comparisons were performed with GeneSpring software: day 4 doxycyline-treated and non-treated iLHX2 cells; day 16 doxycyline-treated and non-treated iLHX2 cells; day 12 differentiating shLuc- and shLHX2-expressing hESCs. Changes in gene expression found to have an adjusted P < 0.05 and a fold change >2 were considered to be significantly different. Gene Ontology (GO) enrichment was determined by annotating each gene with the relevant lineage (ectoderm, mesoderm or endoderm); neural-related genes were identified by GO descriptions on the NCBI Web site (http://www.ncbi.nlm.nih.gov/gene). For signal pathway analysis, the results of two groups (day 4 doxycyline-treated and non-treated iLHX2 cells, and day 12 differentiating shLuc- and shLHX2-expressing hESCs) were analyzed using Ingenuity Pathway Analysis Software. Genes were annotated with signal pathways related to neural development (Wnt/β-catenin, TGFβ, FGF, BMP, Notch and Sonic Hedgehog signal pathways) using Ingenuity Pathway Analysis Software. Pathways with P < 0.01 [−log(P value) >2] were considered significantly different. The NCBI accession number for the microarray data reported in this article is GES31810.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as previously described (31). Briefly, undifferentiated H9 hESCs and H9 derived neural precursors isolated from rosettes at day 16 of IVD were treated with 1% (v/v) formaldehyde for 8 min at room temperature, and the cross-linking reaction was then quenched with 125 mM glycine. Chromatin was sonicated to an average size of 100–150 bp. Each reaction contained 5 × 105 cells and 2 μg antibody (goat anti-LHX2 antibody, SC-19344, Santa Cruz Biotechnology; rabbit anti-H3K4me1 antibody, rabbit anti-H3K27ac antibody, Abcam). The isolated DNA was purified using a QIAquick PCR purification kit (Qiagen). The purified DNA was quantified by qPCR using SYBR® FAST 2X qPCR Master Mix (KAPA) and an ABI PRISM© 7900 Sequence Detection System (Applied Biosystems), with H9 hESC DNA as standard. Enrichment (% Input) was determined as ChIP DNA/ input DNA × 100%. Region positions are listed in Supplementary Table S2 and primers are listed in Supplementary Table S1. Primers for qPCR were confirmed to be specific by the presence of a single band on electrophoresis on a 2% agarose gel, and of a single peak in the dissociation curve. Each experiment was repeated at least three times, with each replicate producing a similar pattern. The results are expressed as means ± SE of three qPCR triplicates.
Enhancer reporter assay
PAX6 P0 (1.5 kb) (32) and P1 (1.4 kb) (33) promoters, CER1 promoter (0.85 kb) (34) and putative enhancers (region of each enhancer: primer −1 to +1) (listed in Supplementary Table S2) were cloned into pGL3 (Promega) to generate reporter plasmids, as shown in Figures 4E and 7G. pGal containing the β-galactosidase gene was used as an internal standard to control for transfection efficiency.
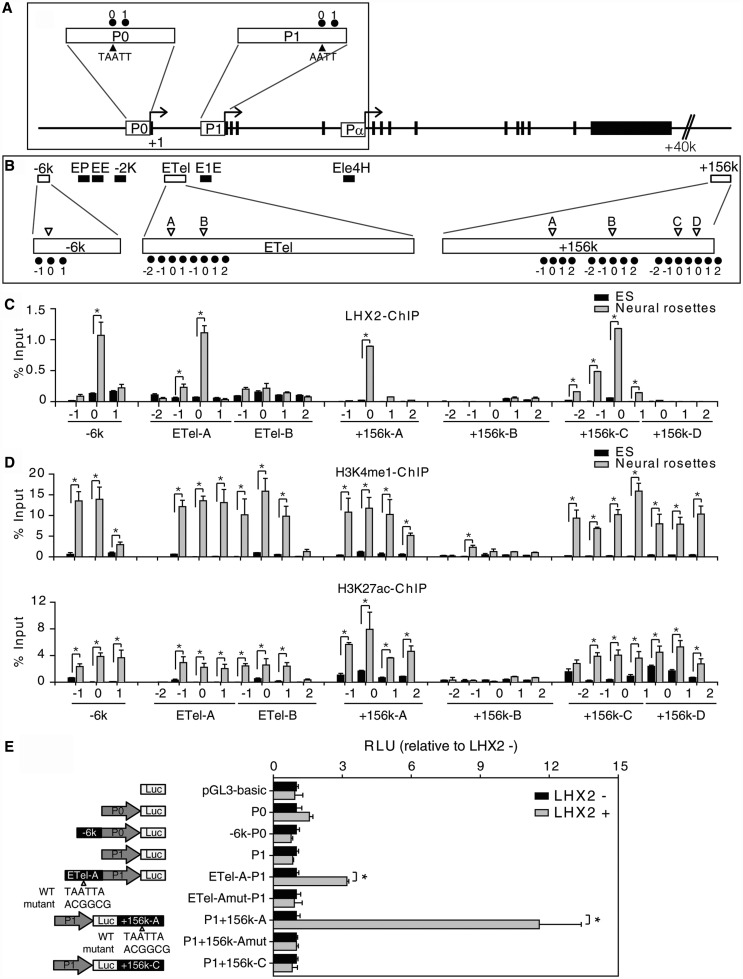
Figure 4.
LHX2 regulates PAX6 expression through its enhancers in hESC-NPs. (A) Schematic representing the genomic organization of the PAX6 promoter regions, P0 and P1, from which full-length PAX6 mRNA is transcribed. P0 spans positions −1450 to −1 and P1 spans positions +6630 to +8083 with respect to the transcription starting site (TSS). The filled triangles indicate incomplete LHX2 binding sites. ChIP primers used in the study are indicated by black dots. ‘0’ indicate regions containing incomplete LHX2 binding sites and ‘1’ indicates regions located at the 5′ end of ‘0’. (B) A schematic representation of the putative enhancers of PAX6. Unfilled boxes indicate PAX6 enhancers containing putative LHX2 binding sites (−6K, ETel and +156K). Filled boxes indicate PAX6 enhancers without putative binding sites (EP, EE, −2K, E1E and Ele4H). Unfilled triangles indicate complete LHX2 binding sites (TAATTA) on PAX6 enhancers. The positions of ChIP primers used in this study are indicated by black dots. ‘0’ indicates regions containing a complete LHX2 binding site. ‘1’ and ‘2’ indicate regions located at the 5′ end of ‘0’, and ‘−2’ and ‘−1’ indicate regions located at the 3′ end of ‘0’. (C) Combined ChIP and qPCR analysis of DNA isolated from hESC-NPs using a specific antibody against LHX2 revealed that LHX2 exhibits enhanced binding to certain PAX6 enhancers in hESC-NPs, as compared with undifferentiated hESCs (DNA from the latter was used as a control). (D) Combined ChIP and qPCR analyses of hESC-NP DNA using specific antibodies against H3K4me1 (upper panel) or H3K27ac (lower panel) revealed enhanced occupancy of these chromatin marks on LHX2 binding sites of certain PAX6 enhancers. (E) Mutational analysis of LHX2 binding sites in PAX6 enhancers in iLHX2 hESCs with or without doxycycline induction. Reporter constructs containing wild-type or mutated LHX2 binding sites in enhancer −6k (E-6k), ETel-A (ETel-A) or +156K (E+156K-A and -C) and PAX6 promoters P0 and P1 were generated, and used to evaluate the role of the enhancers in regulating activation of PAX6 promoters and PAX6 expression under LHX2 overexpression (LHX2+). Error bars represent the mean ± SEM (n = 3). Significance: *P < 0.05.
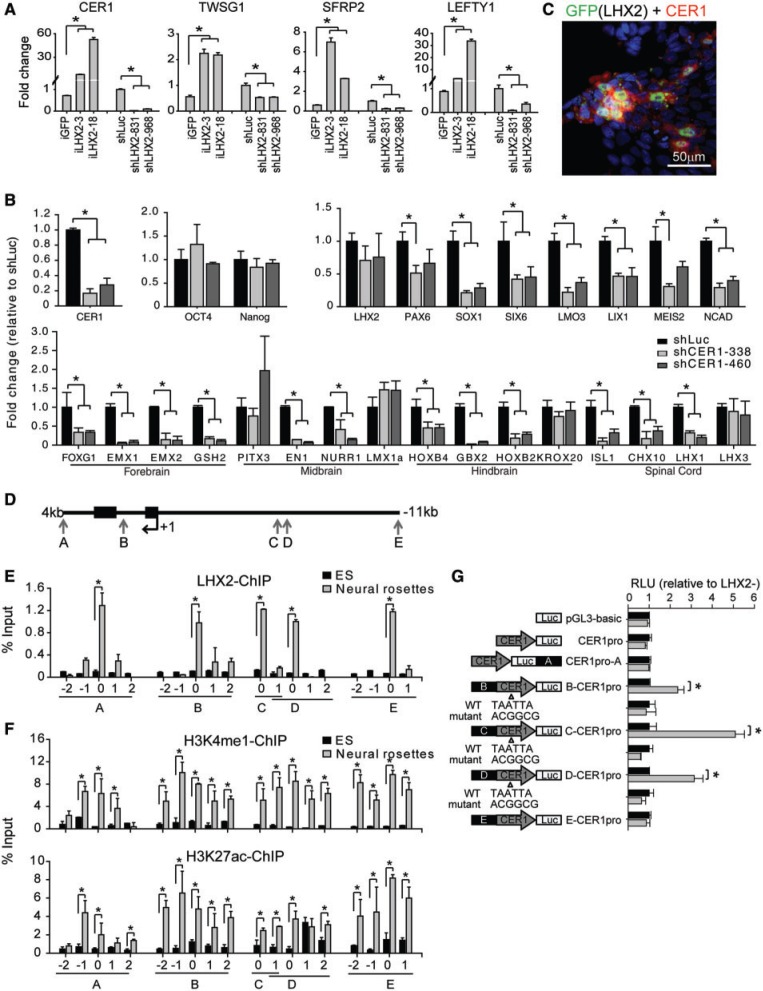
Figure 7.
LHX2 attenuates BMP and WNT signaling by augmenting CER1 expression. (A) qRT-PCR analysis of the expression of CER1, TWSG1, SFRP2 and LEFTY1 genes using mRNA isolated from iLHX2 hESCs (iLHX2-3 and -18, induced by doxycycline for 3 days) or shLuc- or shLHX2-expressing hESCs (IVD day 12). ACTIN was used as the internal control for normalization. Fold change was normalized independently using iGFP expression or shLuc expression in parallel as a control (n = 3). (B) qRT-PCR analysis of indicated genes using mRNA isolated from differentiating shCER1-expressing hESCs at IVD day 12 (n = 3). (C) Immunocytochemical analysis using specific antibodies against CER1 and GFP in hESCs transiently expressing LHX2-eGFP for 3 days by lentivirus infection. CER1 and LHX2 (eGFP) were co-expressed in hESCs. (D) Schematic representation of the identified LHX2 binding sites (arrows, Supplemental Figure S6B) neighboring the CER1 gene. (E) Combined ChIP and qPCR analyses of DNA isolated from hESC-NPs and hESCs with a specific antibody against LHX2 revealed that LHX2 binds to the indicated sites neighboring the CER1 gene in hESC-NPs (n = 3). (F) Combined ChIP and qPCR analyses revealed enhanced occupancy of H3K4me1 (upper panel) and H3K27ac (lower panel) at LHX2 binding sites in the indicated CER1 enhancers in hESCs-NPs, as compared with hESCs (n = 3). (G) Reporter constructs containing the indicated wild-type or mutant LHX2 binding sites and CER1 promoters were generated, and used to evaluate the role of the enhancers in regulating CER1 promoter activation and CER1 gene expression in iLHX2 hESCs (n = 3). ‘0’ indicates regions containing complete LHX2 binding sites. ‘1’ and ‘2’ indicate regions located at the 5′ end of ‘0’ and ‘−2’ and ‘−1’ indicate regions located at the 3′ end of ‘0’. Error bars represent the mean ± SEM. Significance: *P < 0.05.
Reporter plasmid (0.5 μg) and pGal (0.1 μg) were co-transfected into iLHX2 undifferentiated cells with Lipofectamine 2000 (Invitrogen), following the standard Lipofectamine 2000 transfection protocol. Two days after transfection, the iLHX2 cells were separated from the feeder cells, and treated with 2 μg/ml doxycycline. After doxycycline induction for 3 days, cells were lysed with Reporter Lysis Buffer (Promega). Luciferase activity was examined using the Luciferase Assay System (E4030, Promega) according to the manufacturer’s standard protocol, and luminescence was measured using a Luminescence Counter (PerkinElmer). The internal control (β-galactosidase) was examined using the β-galactosidase Enzyme Assay System (E2000, Promega) according to the manufacturer’s standard protocol, and signals were measured by TopCount NXT Microplate Scintillation. Relative luciferase units (RLU) were calculated as the activity of luciferase/β-galactosidase normalized to cells untreated with doxycycline.
Statistical analyses
All experiments were performed in triplicate and the results are shown as the mean ± SE. Student’s t test was used for statistical analysis; P < 0.05 was considered significant.
RESULTS
LHX2 is one of the earliest markers for hESC-derived neural progenitors
To investigate the expression pattern of LHX2 in human neural differentiation, H9 hESCs were plated and cultured in neural differentiation media (Figure 1A), and then subjected to qRT-PCR and western blotting analysis of LHX2 and various neural markers. LHX2 mRNA was readily induced in hESCs at day 4 of differentiation, and its expression preceded the induction of other neural progenitor marker genes, such as PAX6, SOX1, LMO3, LIX1, MEIS2 and RAX, and the neuronal gene MAP2 (Figure 1B). Expression of LHX2 plateaued at IVD day 12, as demonstrated by qRT-PCR (Figure 1C). Western blotting analysis revealed that the expression level of LHX2 protein was also significantly increased at IVD day 4, before the upregulation of PAX6 (Figure 1D). To examine the spatial expression pattern of LHX2, we stained the differentiating hESCs with antibodies against LHX2 and other neural markers. Immunocytochemical staining revealed that LHX2 was co-expressed with PAX6, Nestin or OTX2 in the cells within or around the emerging early neural rosette-like structures (Figure 1E, Supplementary Figure S1A and B). The number of LHX2+ and LHX2+/PAX6+ cells increased as differentiation progressed (Supplementary Figure S1E), and most of the PAX6+ cells also expressed LHX2 at IVD day 10 (Figure 1E). After 4 weeks of IVD, certain cells at the periphery of the neural rosette structures started to lose LHX2 and/or PAX6 expression (Figure 1E and Supplementary Figure S1A), and began to exhibit the morphological features of neurons (Supplementary Figure S1C and D). After further differentiation, these cells expressed the mature neuronal markers Tuj1 and NeuN; LHX2 was found to be expressed in some of the Tuj1+ (Supplementary Figure S1C) and NeuN+ neurons (Supplementary Figure S1D). This is in agreement with previous reports that Lhx2 is expressed in a subpopulation of postmitotic neurons (18,35). Together, these results demonstrate that LHX2 expression starts at an early stage of neural induction in differentiating hESCs, and precedes the expression of PAX6 and SOX1 in hESC-NPs.
LHX2 promotes neural differentiation
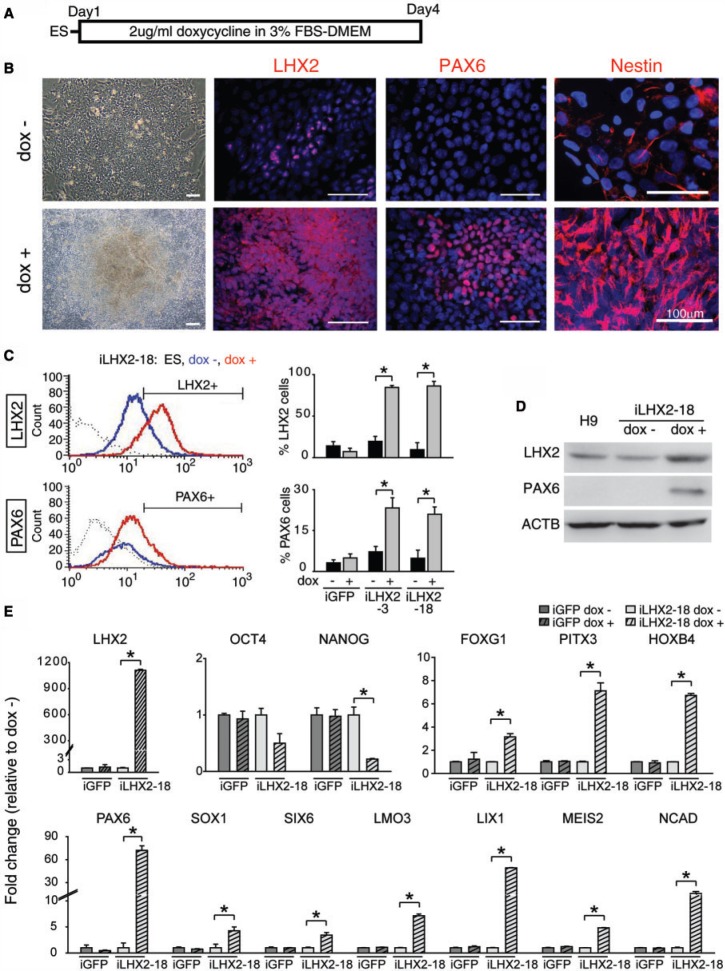
Next, we performed conditional gain-of-function studies to investigate if expression of LHX2 is sufficient for neural fate induction in hESCs. To this end, we established hESCs that ectopically overexpressed LHX2 (iLHX2 hESCs) by using electroporation and antibiotic selection to introduce the inducible Tet-On constructs TetR and TetO-LHX2 into undifferentiated H9 hESCs (Supplementary Figure S2). After doxycycline induction (dox+) for 4 days (Figure 2A), the differentiating iLHX2 hESCs contained a large number of emerging Nestin+ cells and rosette-like structures containing LHX2+ and PAX6+, whereas iLHX2 hESCs without doxycycline induction (dox−) maintained the typical morphological traits of hESCs without rosette-like structures (Figure 2B). Furthermore, immunocytochemical and flow cytometric analyses demonstrated that the number of LHX2- and PAX6-expressing cells was significantly increased in the dox+ iLHX2 hESCs, as compared with the dox− iLHX2 hESCs (Figure 2B and C). The increased level of PAX6 protein in the LHX2-overexpressing cells was also confirmed by western blot analysis (Figure 2D). Furthermore, qRT-PCR analysis showed that expression of the pluripotency gene, NANOG, was decreased, whereas expression of the early neural precursor marker genes PAX6, SOX1, SIX6 (36,37), LMO3 (38,39), LIX1 (39,40), MEIS2 (41,42) and NCAD (8,43) [which are highly expressed in hESC-derived neural progenitors (7,8) and upregulated by PAX6 (11)], and the regional neural genes, including forebrain gene FOXG1, midbrain gene PITX3 and hindbrain gene HOXB4, were significantly increased in dox+ as compared with dox− iLHX2 hESCs (Figure 2E). Similar effects of LHX2 overexpression on neural gene expression were also observed in several other hESC lines and iPSC lines (Supplementary Figure S3). Together, these data indicate that LHX2 overexpression is sufficient to promote and facilitates neural differentiation in pluripotent stem cells.
Figure 2.
Conditional overexpression of LHX2 promotes the transition of hESCs into the neuroectoderm lineage. (A) Procedure of doxycycline induction in iLHX2 hESCs. (B) Immunocytochemical analysis of iLHX2 hESCs treated with (dox+, lower panel) or without (dox−, upper panel) doxycycline for 3 days with specific antibodies against LHX2, PAX6 and Nestin. Nuclei were counterstained with DAPI (blue). (C) Flow cytometric analysis of a number of LHX2+ or PAX6+ cells during IVD of iLHX2 hESCs treated with doxycycline for 3 days. Gating was determined using H9 ES cells and iLHX2 hESCs without doxycycline treatment in parallel as controls. The average percentages of LHX2+ or PAX6+cells are indicated (right panels, n = 4). (D) Western blot of proteins isolated from iLHX2 hESCs after doxycycline induction for 3 days using specific antibodies against LHX2 and PAX6. Differentiating H9 ESCs and dox− iLHX2 hESCs were used as controls. (E) qRT-PCR for the indicated genes using mRNA isolated from iLHX2 or iGFP hESCs after doxycycline induction for 3 days. dox− iLHX2 hESCs were used as a control. ACTIN was used as the internal control for normalization (n = 3). Error bars represent the mean ± SEM. Significance: *P < 0.05.
Disruption of LHX2 expression compromised neural differentiation in hESCs
To examine whether LHX2 is required for neural differentiation in hESCs, LHX2 was knocked down by constitutive overexpression of LHX2 shRNA. Two lentiviral shRNA vectors targeting the LHX2 homeodomain sequence, or shLuc as a control targeting the luciferase gene, were introduced into hESCs, and puromycin selection was performed to establish stable hESC clones (Supplementary Figure S2). After neural induction for 16 days, distinct neural rosette-like structures were found in shLuc-expressing hESCs, whereas the shLHX2-expressing hESCs retained undifferentiated phenotypes (Figure 3A). In addition, the expression level of LHX2 and PAX6 proteins, and the number of LHX2 and PAX6-expressing cells, were significantly decreased in differentiating shLHX2-expressing hESCs, as revealed by western blot and flow cytometry (Figure 3B and C), respectively. To examine whether knockdown of LHX2 affected the expression of pluripotency and neural genes, qRT-PCR analysis was performed on RNA extracted from shLuc and shLHX2-expressing hESCs at day 0 and day 12 of neural differentiation (Figure 3D). shLHX2-expressing hESCs presented with significantly upregulated expression of the pluripotency genes OCT4 and NANOG, and significantly reduced expression of neural progenitor genes, such as PAX6, and regional neural genes, especially, forebrain associated genes, as compared with shLuc-expressing hESCs (Figure 3D).
In addition to the above in vitro experiments, we also examined whether knockdown of LHX2 affected neural differentiation in vivo by teratoma formation. To generate teratomas, hESCs constitutively expressing shLHX2 or shLuc were subcutaneously injected into NOD/SCID mice. There was no difference in the efficiency of generation or the weight of the LHX2 knockdown (shLHX2) or control (shLuc) hESC-derived teratomas (data not shown). By subjecting a section of teratomas to H&E staining, we found that neural rosette-like structures were significantly more abundant in teratomas derived from shLuc-expressing hESCs than in teratomas derived from shLHX2-expressing hESCs (Figure 3E and F). Importantly, the levels of LHX2, PAX6 and SOX1, and the numbers of LHX2+, PAX6+ and Nestin+ cells in teratomas derived from shLHX2-expressing hESCs were also reduced as compared with shLuc-expressing hESCs (Figure 3E and G). Taken together, these results suggest that LHX2 is critical for the neural differentiation of hESCs.
LHX2 regulates PAX6 expression by binding to its enhancers during neural differentiation
Our gain- and loss-of-function studies revealed that LHX2 is both necessary and sufficient for neural differentiation in hESCs. We therefore went on to investigate the molecular mechanisms by which LHX2, in its role as a transcription factor, controls neural differentiation. Based on the earlier finding that PAX6 is a human neuroectoderm determinant (11), and our finding that LHX2 expression precedes that of PAX6 (Figure 1), we postulated that LHX2 may regulate PAX6 to direct neural differentiation in hESCs. To test this hypothesis, we first examined whether LHX2 binds to the PAX6 promoters directly. To this end, we screened for LHX2-binding sites (TAATTA) (44) in the PAX6 promoters P0 and P1 (responsible for the expression of PAX6a and PAX6a and b, respectively). However, we did not find complete LHX2 binding motifs in the PAX6 promoters (Figure 4A and Supplementary Figure S4). Furthermore, ChIP and luciferase reporter assays showed that LHX2 does not bind to either of the PAX6 promoters (Supplementary Figures S4C) and has no effect on the activities of the PAX6 promoters (Figure 4E).
Because recent studies have shown that enhancers are highly cell-type specific (45–47), we further tested whether LHX2 binds to PAX6 enhancers (48–52). We screened for potential LHX2-binding sites by searching for LIM HD-sensitive regulatory regions, which contain a LHX2 consensus binding sequence (TAATTA) close to a second LIM HD-binding sequence (TTAATAAA) (44); this resulted in the identification of three PAX6 enhancers containing putative LHX2-binding sites (Figure 4B). Furthermore, ChIP using an anti-LHX2 antibody and chromatin prepared from undifferentiated hESCs and hESC-NPs demonstrated that LHX2 bound to sites residing in these three PAX6 enhancer regions (E-6k, ETel and E+156k) in hESC-NPs (Figure 4C and Supplementary Table S2). As it has been recently reported that mono-methylation of H3 lysine 4 (H3K4me1) and acetylation of H3 lysine 27 (H3K27ac) serve as chromatin signatures of active enhancers and can predict the developmental state of a given enhancer (46,47), we next used ChIP to investigate the appearance of these chromatin marks at LHX2-binding sites in PAX6 enhancers in hESCs and hESC-NPs (Figure 4D). We found that the LHX2-binding sites in E-6 kb, ETel (site A) and E+156 kb (site A and C) enhancers were all enriched with both H3K4me1 and H3K27ac marks in hESC-NPs, but not in hESCs. This suggests that the LHX2 binding sites on PAX6 enhancers may be inactive in ESCs and activate in neural progenitors, and as such these signatures may be indicative of a direct role for LHX2 in activating PAX6 transcription during neural development.
To further examine whether these newly identified neural-specific enhancers play a direct role in controlling PAX6 expression, we set up an enhancer reporter assay in inducible LHX2 overexpression hESCs (iLHX2 hESCs). The LHX2-binding sites in E-6 kb, ETel (site A) and E+156 kb (site A and C) enhancers (Figure 4C) were used for the luciferase reporter assay. The reporter constructs were generated by fusing a luciferase reporter with the selected enhancers and their corresponding promoters (48,52) (Figure 4E), and were then introduced into iLHX2 hESCs. Reporters containing the ETel-A and E+156 kb-A PAX6 enhancer regions resulted in significantly greater luciferase activity in dox+ iLHX2 cells than reporters containing the promoter alone (Figure 4E). However, mutation of the LHX2-binding motif in these enhancers abolished the effect (Figure 4E). Together, these results suggest that LHX2 is recruited to active PAX6 enhancers, and promotes PAX6 expression during neural lineage differentiation.
LHX2 regulates neural differentiation through PAX6
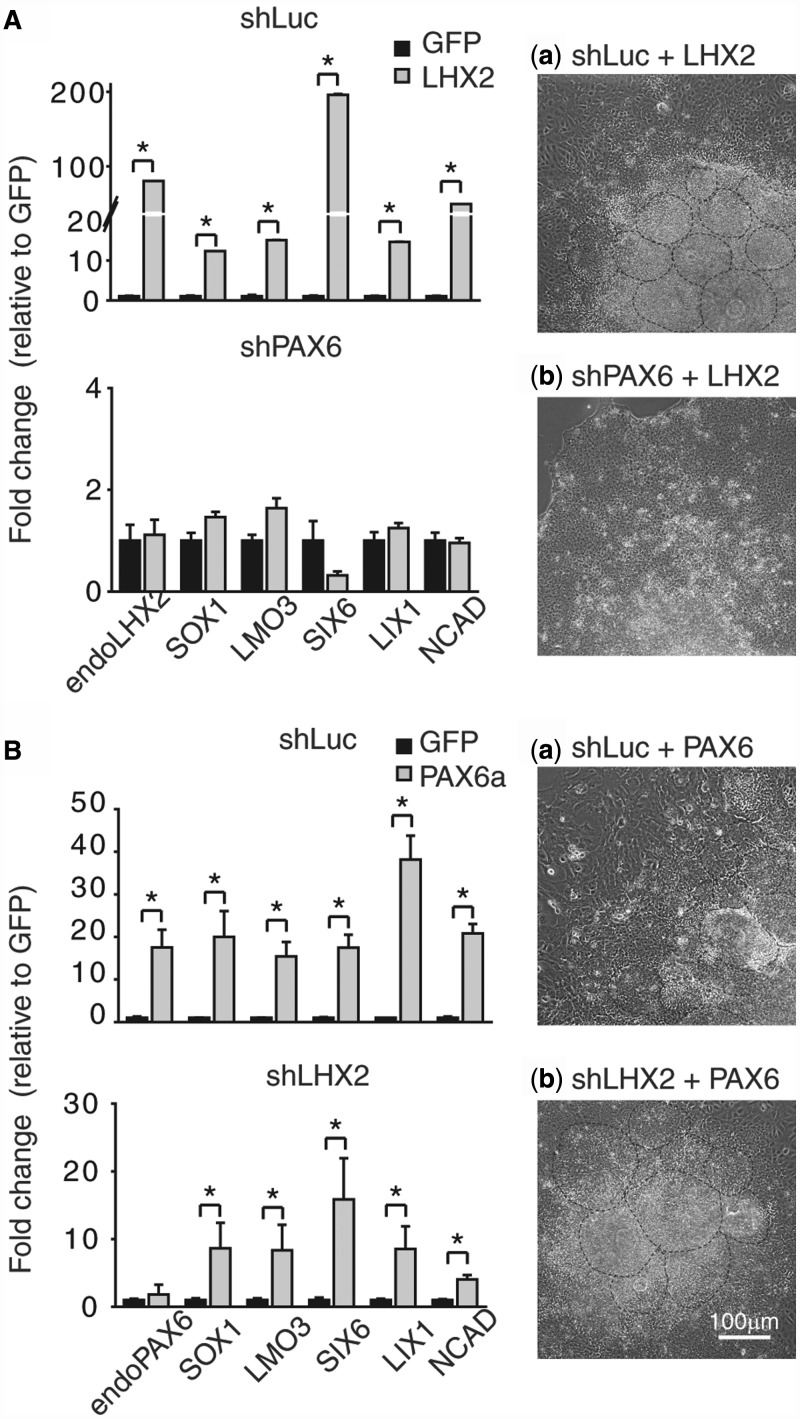
The ChIP and enhancer luciferase assays demonstrated that LHX2 modulates the neural differentiation of hESCs through regulating PAX6 expression; however, whether LHX2 primarily targets PAX6 to induce neural differentiation in hESCs is unclear. We hypothesized that if LHX2 promotes neural differentiation primarily through PAX6, a reduction of PAX6 expression in hESCs would compromise LHX2-mediated neural differentiation. To test this hypothesis, we manipulated expression of LHX2 and PAX6 in hESCs simultaneously, and investigated the effects on morphology and the expression profile of a panel of neural genes, including SOX1, LMO3, SIX6, LIX1 and NCAD (as determined by qRT-PCR). PAX6 knockdown overrode the effect of LHX2 overexpression on both neural rosette formation and neural gene induction (Figure 5A). On the contrary, the same set of neural genes was upregulated, and abundant neural rosettes could still be observed when PAX6 was overexpressed in LHX2 knockdown cells (Figure 5B). Therefore, these results demonstrate that LHX2 is an essential neural transcription factor upstream of PAX6, and LHX2 primarily targets PAX6 to regulate the neural differentiation of hESCs.
Figure 5.
PAX6 is a major downstream target of LHX2 for regulation of neural differentiation. (A) qRT-PCR analysis of the indicated genes using mRNA isolated from hESCs constitutively expressing shLuc or shPAX6 and transiently expressing GFP or LHX2 by lentiviral infection for 3 days. Bright field images show the morphological change in hESCs 6 days after viral infection (compare a and b). The number of neural rosette-like structures and the expression level of the indicated neural genes were dramatically reduced by PAX6 knockdown. (B) qRT-PCR analysis of the indicated genes using mRNA isolated from hESCs constitutively expressing shLuc or shLHX2 and transiently expressing GFP or PAX6 by lentiviral infection for 3 days (compare a and b). Bright field images show the morphological changes in the hESCs 6 days after viral infection. The number of neural rosette-like structures and the expression level of the indicated neural genes were not affected by knockdown of LHX2 in hESCs overexpressing PAX6. Error bars represent the mean ± SEM (n = 3). Significance: *P < 0.05. Dotted lines indicate rosette-like structures.
LHX2 affects transcription factor expression and signaling pathways involved in neural differentiation
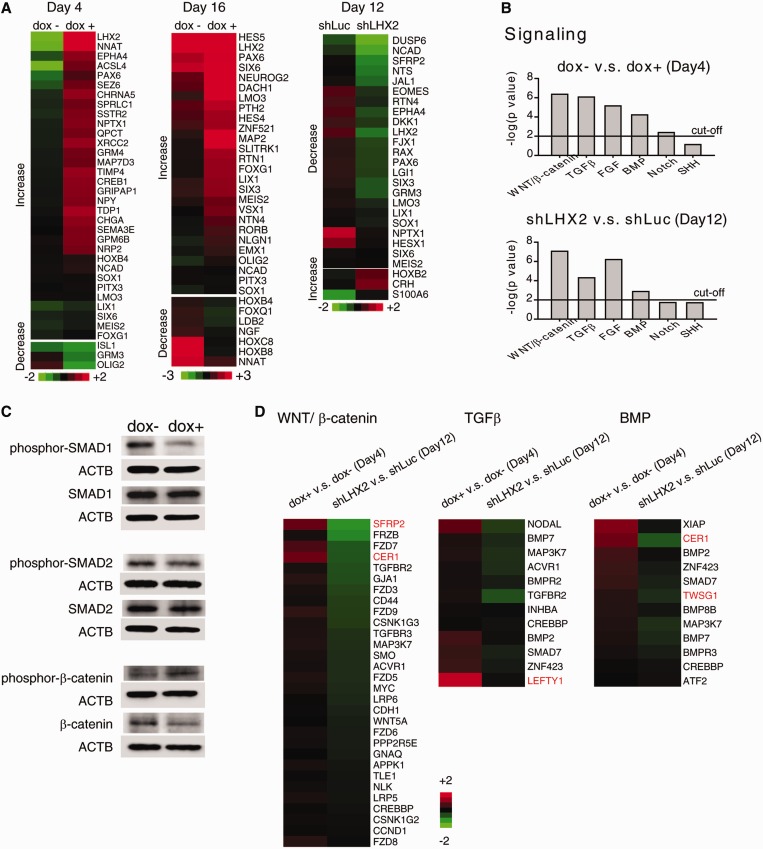
To determine the effect of manipulating LHX2 expression on the global gene expression profile of hESCs, we performed transcriptome analysis using a cDNA microarray, and mRNA extracted from iLHX2 cells treated with or without doxycycline to induce LHX2 overexpression. Using the fold change analysis tool of GeneSpring software, 209 genes at day 4 and 661 genes at day 16 with at least a 2-fold average expression change between dox− and dox+ iLHX2 hESCs were selected and subjected to GO analysis. Intriguingly, the majority of the neural genes in dox+ iLHX2 hESCs showed increased expression at both day 4 and day 16 (Figure 6A). Some of the neural genes with increased expression at day 16 in dox+ iLHX2 hESCs are postmitotic neuronal genes (MAP2, NEUROG2), and others are involved in regional patterning of the central nervous system (CNS) (FOXG1, PTH2, EMX1, OLIG2). These results confirmed our earlier observation in dox+ iLHX2 hESCs that LHX2 induces expression of neural genes (Figure 2), and thereafter activates the neuronal program. We subsequently performed transcriptome analysis to investigate the effects of LHX2 knockdown in hESCs. We focused on 41 out of 86 embryonic germ layer–related genes, which have at least a 2-fold average expression change between shLuc- and shLHX2-expressing hESCs at day 12 of IVD. Interestingly, the expression of most of the neural genes was reduced in differentiating shLHX2-expressing hESCs (Figure 6A), confirming our observation in Figure 3. Overall, the findings of the genome-wide gene expression analysis of iLHX2 and shLHX2 hESCs suggest that LHX2 expression has a profound impact on transcriptional regulation during neural differentiation.
Figure 6.
Transcriptome profiling of hESCs with altered LHX2 expression revealed that LHX2 affects the expression of neural genes and signaling pathways. (A) The heat map shows filtered neural genes with expression significantly affected by manipulation of LHX2 expression in hESCs. (B) Identification of signaling pathways affected by manipulation of LHX2 expression in hESCs by Ingenuity Pathway Analysis software. P < 0.01 was used as the cutoff value for selecting neural signaling pathways significantly affected by manipulation of LHX2 expression in hESCs. (C) Western blot analysis of proteins isolated from dox+ iLHX2 hESCs with antibodies against ACTB and proteins downstream of the BMP4 (phosphor-SMAD1 and SMAD1), TGFβ (phosphor-SMAD2 and SMAD2) and WNT (phosphor-β-catenin and β-catenin) signaling pathways. Proteins isolated from dox− iLHX2 hESCs were used as a control. (D) The heat map shows genes annotated to the indicated signaling pathways by Ingenuity Pathway Analysis software with increased expression in dox+ iLHX2 cells (fold change >1.3), and decreased expression in differentiating shLHX2-expressing hESCs (fold change >1.3). Green and red values represent fold change for down- and upregulation, respectively (A and D). The fold change of gene expression was calculated using GeneSpring software as iLHX2 hESC with doxycycline induction to without (iLHX2 hESC dox+/dox−) or shLHX2- to shLuc-expression hESCs (shLHX2/shLuc) on the days indicated.
We also investigated whether LHX2 directs neural differentiation by affecting relevant extrinsic signaling pathways, in addition to its role in regulating transcriptional factors. To this end, we performed Ingenuity Pathways Analysis on the gene expression profiles of hESCs with LHX2 overexpression or knockdown. Through this, we identified four pathways affected in hESCs by altered LHX2 expression: the WNT/β-catenin, TGFβ, FGF and BMP signaling pathways (Figure 6B). We confirmed downregulation of the WNT/β-catenin, TGFβ and BMP signaling pathways by western blot: phosphor-SMAD1 (a downstream effector of BMP) and phosphor-SMAD2 (a downstream effector of TGFβ) were downregulated, and phosphor-β-catenin (a downstream effector of WNT) was upregulated, when LHX2 was overexpressed in differentiating hESCs (Figure 6C). We then examined the gene expression profiles of hESCs with LHX2 overexpression or knockdown to identify the genes through which LHX2 affects these signaling pathways (Figure 6D). Based on GO terms, extracellular components and transmembrane activity, we identified four candidate genes with expression strongly correlated to that of LHX2: CER1 (encoding a BMP, nodal and WNT antagonist), TWSG1 (encoding a BMP antagonist), SFRP2 (encoding a negative regulator of the WNT receptor) and LEFTY1 (encoding a TGFβ antagonist) (Figures 6D and 7A).
LHX2 attenuates BMP and WNT signaling and inhibits non-neural differentiation by augmenting expression of CER1
To test our hypothesis that LHX2 promotes neural differentiation by suppressing the non-neural program through targeting signaling antagonists, we examined whether LHX2 directly targets CER1, the expression of which was most significantly affected by manipulation of LHX2 expression (Figure 7A). We first examined the temporal expression pattern of CER1 during neural differentiation using RT-PCR and qRT-PCR. We found that CER1 expression was initiated in differentiating hESCs at day 6 of IVD, and peaked at days 8–10, when neural progenitors were evidently emerging in the culture (Supplementary Figure S5A); this suggests that expression of LHX2 is followed by that of CER1. In addition, CER1 was observed to be co-expressed with LHX2 at day 10 and day 20 of IVD, suggesting that initiation of CER1 expression is highly correlated with that of LHX2 during neural differentiation of hESCs (Supplementary Figure S5B).
To establish whether CER1 plays a role in human neural differentiation, loss-of-function studies were conducted with hESCs constitutively expressing shCER1. The use of qRT-PCR revealed that CER1 knockdown significantly decreased expression of both neural progenitor genes (SOX1, SIX6, LMO3, LIX1, MEIS2 and NCAD) and other CNS regional-specific genes [with the exception of midbrain-specific genes (PITX3 and LMX1a), a hindbrain-specific gene (KROX20) and a spinal cord–specific gene (LHX3)] (Figure 7B); these results suggest that knockdown of CER1 in hESCs affects the differentiation of multiple neural lineages, although its role may be more pronounced in forebrain differentiation. In addition, mesoderm- and endoderm-related genes (MIXL1, T, GSC and SOX17) were increased at day 12 of IVD in shCER1-expressing hESCs, suggesting that CER1 may function as a repressor of mesoderm- and endoderm-related genes during IVD of hESCs (Supplementary Figure S6A). To confirm that CER1 is sufficient to mediate LHX2-dependent regulation of both WNT and BMP (Figure 6C), we overexpressed CER1 in shLHX2-expressing hESCs, and investigated the effect on the downstream proteins of BMP and WNT signaling by western blot (Supplementary Figure S6B). This revealed that overexpression of CER1 is sufficient to attenuate BMP and WNT signaling.
Next, we examined whether overexpression of LHX2 (through transient infection with lentivirus encoding LHX2-eGFP) results in upregulation of CER1 expression in hESCs. Immunostaining revealed that CER1 was expressed in LHX2-overexpressing cells 4 days after viral infection, suggesting that LHX2 may regulate CER1 expression (Figure 7C). These results, combined with our finding that PAX6 is the primary downstream target of LHX2, raise the possibility that LHX2 may regulate CER1 expression through PAX6. To address this, we transiently overexpressed shPAX6 in doxycycline-treated iLHX2 hESCs by lentiviral infection (Supplementary Figure S5C). CER1 expression continued to be significantly increased in iLHX2 hESCs, even on PAX6 knockdown. This result suggests that LHX2 may regulate CER1 through a PAX6-independent pathway in hESCs.
To elucidate the direct transcriptional mechanisms that govern CER1 expression by LHX2, we sought to identify cis-acting regulatory elements by aligning the CER1 region (Chr9:14673331-14769701) of the human genome to various species, including rhesus monkey, dog, horse, mouse, rat and chicken. Through using the VISTA browser to identify the degree of evolutionary constraint in the CER1 gene, we identified nine conserved regions, seven of which contained putative LHX2 binding sites (Supplementary Figure S7B). LHX2-ChIP analysis further revealed that LHX2 specifically targeted 5 out of 20 predicted sites in the 7 conserved regions in hESC-NPs (Figure 7D and E), but not the CER1 promoter (34) (Supplementary Figure S7A). Furthermore, these regions were also enriched with the active enhancer marks H3K4me1 and H3K27ac in hESC-NPs (Figure 7F). To further test whether these five newly identified regions serve as putative enhancers to regulate CER1 promoter activities, we performed enhancer reporter assays in iLHX2 hESCs. We report that three out of the five putative enhancers of CER1 were activated by LHX2, and acted as enhancers to regulate the CER1 promoter, while mutation of the LHX2-binding site eliminated this effect (Figure 7G). Collectively, these results suggest that LHX2 may also contribute to neural differentiation via upregulation of CER1, which itself encodes an antagonist of the BMP and WNT/β-catenin signaling pathways.
DISCUSSION
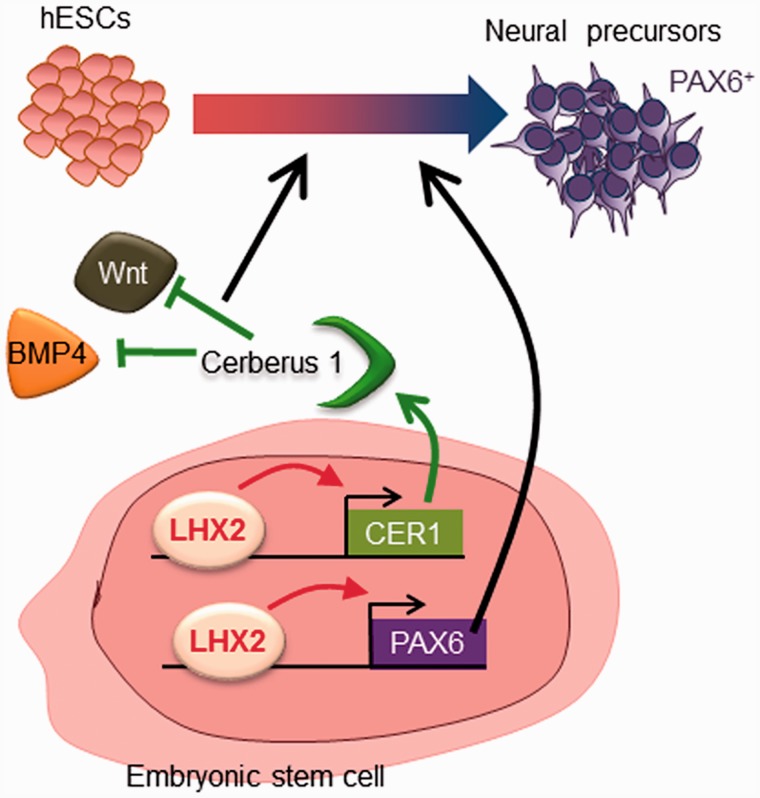
Previous studies have reported that the extrinsic signals WNT, SHH, BMP and FGF, and the intrinsic factor PAX6, regulate neural differentiation (2,3,11). However, the transcriptional regulation underlying early neural lineage differentiation remains poorly understood. The aim of the current study was to increase our understanding of the transcriptional regulatory network governing human early neural lineage differentiation. We provide evidence that LHX2 is expressed in the early neural lineage derived from human pluripotent stem cells. By combining global gene expression profiling with biochemical analysis of hESCs with altered expression levels of LHX2, we found that LHX2 also affects extracellular signaling pathways, including the WNT, TGFβ and BMP pathways. This reveals that LHX2 is likely to be involved in regulating the neural differentiation of hESCs through multiple mechanisms. We demonstrate here that expression of LHX2 leads to neural fate acquisition at two levels (Figure 8). First, we report that LHX2 regulates PAX6 (a transcription factor critical for human early neural differentiation) through binding to its enhancers (enriched with active chromatin markers), and through this mechanism LHX2 drives neural lineage differentiation. Second, we have demonstrated that LHX2 suppresses the WNT and BMP signaling pathways in differentiating hESCs through regulation of CER1 expression; CER1 acts as an antagonist to attenuate the BMP and WNT signaling pathways. Therefore, we have identified LHX2 as a key factor for regulating the interplay between intrinsic and extrinsic mechanisms required for neural differentiation of hESCs.
Figure 8.
Model for LHX2-mediated neural differentiation in hESCs. LHX2 directly regulates PAX6 expression by binding to its enhancers, and promotes neural differentiation of hESCs. On the other hand, LHX2 can also regulate CER1 expression by directly binding to the putative enhancer region of CER1 and attenuating Wnt and BMP signaling, further suppressing the mesodermal and endodermal potential of hESCs.
We demonstrate here that LHX2 binds to enhancers, rather than promoters, of both PAX6 and CER1. All of the identified LHX2 binding sites in the PAX6 (E-6k, ETel-A, E+156k-A and C) and CER1 enhancers (A, B, C, D and E) are marked by H3K4me1 and H3K27ac, chromatin marks of active enhancers. These findings are in agreement with a previous study that indicated that human enhancers are enriched with active chromatin signatures, and that enhancers, not promoters, reflect cell-type–specific gene expression (45). We also noted that two of the identified LHX2 binding sites in PAX6 enhancers, E-6k and E+156k-C, are not active in hESC-NPs. It has been previously reported that multiple PAX6 promoters are regulated by diverse enhancers to transcribe different PAX6 isoforms in a cell-type–specific manner. For example, the enhancer located 6 kb upstream from the Pax6 P0 promoter (enhancer −6k) is conserved in mice and humans, and regulates P0 promoter activity for transcription of PAX6a mRNA in neuroretina (49); an enhancer located in intron 1 (ETel), which is conserved in fugu and humans, regulates P1 promoter activity for transcription of PAX6a and PAX6b mRNA in telencephalon, lens and pancreas (48); and enhancers located 156 kb distal from PAX6 P0 promoter (enhancer +156k), which are conserved in mice and humans, regulate P1 promoter activity for transcription of PAX6a and PAX6b mRNA in the central nervous system (52). Therefore, the likely function of the ETel-A and E+156k-A enhancers is to regulate neural differentiation in hESCs, and the role of the other two identified LHX2-binding enhancers (E-6k and E+156k-C) may involve later regional specification.
Several studies have demonstrated the important role of Lhx2 in regulating the extracellular signaling pathways involved in neural development. For example, Lhx2 has been suggested to couple with BMP signaling to promote optic cup ventral identity in mice (21). Recently, it has been suggested that Lhx2 may play a role in the regionalization of thalamus formation, through attenuating WNT signaling emanating from the epithalamus in zebrafish (53). Additionally, expression of Lhx2 in the presumptive hippocampus during mouse neocortex formation attenuates WNT and BMP signaling emanating from the cortical hem, thereby restricting cortical hem extension and thereafter forming a sharp border between the hippocampus and cortical hem (18). These observations establish Lhx2 as an instructive factor that attenuates various signaling pathways during forebrain formation. However, the mechanism through which Lhx2 regulates its associated signaling pathways to control forebrain formation is not clear. We identified and confirmed that expression of CER1, a gene encoding a secretory ligand that binds to WNT and BMP to inhibit the downstream signaling cascade, is directly regulated by LHX2 in differentiating hESCs. These findings therefore provide a possible mechanism by which Lhx2 regulates downstream signaling and controls forebrain patterning.
The dual roles of Cer1 in neural induction and suppression of mesodermal or endodermal lineages are well documented (54–57). In mice, Cer1 is expressed in the anterior mesoendoderm, and antagonizes the BMP signal emitted from the ventrolateral mesoderm to promote the onset of neurulation at early gastrulation (57). Additionally, Cer1 was also found to induce expression of anterior neural genes, such as Ncam and Otx2 in frogs (57). Moreover, ectopic expression of CER1-S (a truncated form of CER1, which can only target nodal proteins) in hESCs resulted in early onset of neural rosette formation, and an increase in neural gene expression at the expense of non-neural lineages, such as the mesoderm and endoderm (56). In line with these observations, we found that several mesoderm- and endoderm-related genes are increased in differentiating CER1 knockdown hESCs (Supplementary Figure S6A), and WNT and BMP signaling is attenuated (Supplementary Figure S6B) and neural genes are activated in CER1-overexpressing hESCs (data not shown). These results indicate that upregulation of CER1 by LHX2 may be responsible for the activation of the neural program and suppression of the non-neural program during early hESC differentiation.
In addition to CER1, our results indicate that expression of TWSG1 (encoding a BMP antagonist), SFRP2 (encoding a WNT antagonist) and LEFTY1 (encoding a TGFβ antagonist) are affected by LHX2 expression levels in hESCs. WNT, BMP and TGFβ signaling have been previously implicated in the repression of neural differentiation, and inactivation of these pathways is essential for neural induction (3). Thus, an analogous mechanism, whereby LHX2 activates antagonists of WNT and BMP proteins in certain areas to restrict these signaling pathways, may explain the earlier observation that the presence of LHX2 in the developing forebrain prevents excessive expansion of misplaced tissue (to enable appropriate regionalization) (18).
Through gene-expression profiling and manipulation of LHX2 expression in hESCs, we found that expression of several neural transcription factor genes, including MEIS2 and RAX, overlaps with expression of LHX2 and/or PAX6 at differentiation stages, and is strongly correlated with LHX2 expression. This suggests that MEIS2 and RAX are likely to be involved in the LHX2 and/or PAX6 transcriptional networks that determine neural fate. MEIS2, a member of the TALE family, is expressed in the proliferation zone of the human fetal brain, and is co-expressed with PAX6 during the development of the ganglionic eminence (42). Regulation of PAX6 also contributes to mouse lens morphogenesis (58). In our gain-of-function studies, ectopic LHX2 or PAX6 expression in hESCs activated MEIS2 expression (Figure 2), and ectopic MEIS2 expression in hESCs increased the expression of LHX2 and PAX6 (Hou et al., unpublished observation). Thus, it is tempting to suggest that MEIS2 may initially be activated by LHX2 and PAX6, and its expression may subsequently serve as a positive feedback cue to enhance LHX2 and PAX6 expression. In mice, RAX is expressed in the anterior neural plate of E8.5 embryos, and is later expressed in the optic cup and ventral forebrain of E10.5 mouse embryos; RAX activates PAX6 expression during retina formation (59), suggesting it may also play a role in regulating PAX6 expression in neural development in mice. Therefore, we propose that RAX may also be involved in the LHX2-PAX6-mediated regulatory network controlling neural differentiation. Understanding of the functional roles of these factors in neural induction/differentiation, and their relationship to LHX2 and/or PAX6, will be critical for further dissection of the genetic regulatory network controlling neural differentiation in hESCs.
Although a reduction in LHX2 expression in hESCs significantly compromises neural differentiation, we still observed expression of neural markers and the formation of neural rosette-like structures during differentiation of LHX2-knockout hESCs, both in vitro and in vivo (teratoma formation). This may be because LHX2 is not absolutely required for neural differentiation of hESCs, or because shLHX2 does not completely eliminate LHX2 protein expression (Figure 3B and G). As regards the first possibility, (an)other related gene(s) may result in some degree of functional redundancy for LHX2 in the control of neural differentiation. Such functional redundancy has been described between Lhx2 and Lhx9 in spinal cord development in mice (60). Another possible explanation is that LHX2 may only be critical for differentiation of certain neural cell types, such as cells with anterior neural cell identity that constitute the majority of IVD neural populations under the current neural induction protocol (Supplementary Figure S8A) (7). Our observations on forebrain-related genes on the effect of LHX2 loss of function (Supplementary Figure S8B) further support this possibility. Dissection of the distinct role of LHX2 in regional neural specification in humans will require further examination of its effect on the generation of specific neural subtypes from hESCs.
The kinetics of PAX6 and SOX1 expression during neural differentiation are inconsistent across the literature (11,61). Such discrepancies may arise from two possibilities. The first is the neural induction method, as there are two procedures for in vitro neural differentiation from hESCs. The key difference between these procedures is the presence or absence of SMAD inhibitors, such as SB431542 for the TGFβ pathway, and Noggin for the BMP pathway. Studies that did not use such inhibitors (such as the present study) have reported that PAX6 precedes that of SOX1 (11,39,62). On the contrary, the sequential expression of SOX1 and PAX6 was reported to be reversed in the presence of inhibitors (61). Further, it has been reported that initiation of SOX1 expression requires inhibition of the TGFβ pathway, and initiation of PAX6 expression requires the absence of FGF signaling (63). These findings suggest that the kinetic expression of PAX6 and SOX1 is influenced by the culture condition.
Taken together, our findings indicate that LHX2 mediates neural differentiation not only through intrinsic transcriptional regulation of PAX6, but also through suppression of extrinsic factors of key signaling pathways that are essential for early lineage differentiation in human.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–8.
FUNDING
At an intramural grant from Academia Sinica, Stem Cell Priority Grants from the National Science Council [100-2321-B-001-035 to H.C.K., and 100-2321-B-002-075 to C.L.C.]. Funding for open access charge: Academia Sinica.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank You-Tzung Chen for the kind gift of puroTK, and Yi-Shien Su, Jr-Kai Yu, and Sheng-Ping Huang for their comments on this work.
REFERENCES
- 1.Stern CD. Neural induction: 10 years on since the ‘default model’. Curr. Opin. Cell Biol. 2006;18:692–697. doi: 10.1016/j.ceb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Guillemot F, Zimmer C. From cradle to grave: the multiple roles of fibroblast growth factors in neural development. Neuron. 2011;71:574–588. doi: 10.1016/j.neuron.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Munoz-Sanjuan I, Brivanlou AH. Neural induction, the default model and embryonic stem cells. Nat. Rev. Neurosci. 2002;3:271–280. doi: 10.1038/nrn786. [DOI] [PubMed] [Google Scholar]
- 4.Pevny LH, Sockanathan S, Placzek M, Lovell-Badge R. A role for SOX1 in neural determination. Development. 1998;125:1967–1978. doi: 10.1242/dev.125.10.1967. [DOI] [PubMed] [Google Scholar]
- 5.Aubert J, Stavridis MP, Tweedie S, O'Reilly M, Vierlinger K, Li M, Ghazal P, Pratt T, Mason JO, Roy D, et al. Screening for mammalian neural genes via fluorescence-activated cell sorter purification of neural precursors from Sox1-gfp knock-in mice. Proc. Natl Acad. Sci. USA. 2003;100(Suppl. 1):11836–11841. doi: 10.1073/pnas.1734197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 7.Pankratz MT, Li XJ, Lavaute TM, Lyons EA, Chen X, Zhang SC. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells. 2007;25:1511–1520. doi: 10.1634/stemcells.2006-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabar V, Panagiotakos G, Greenberg ED, Chan BK, Sadelain M, Gutin PH, Studer L. Migration and differentiation of neural precursors derived from human embryonic stem cells in the rat brain. Nat. Biotechnol. 2005;23:601–606. doi: 10.1038/nbt1088. [DOI] [PubMed] [Google Scholar]
- 10.Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Huang CT, Chen J, Pankratz MT, Xi J, Li J, Yang Y, Lavaute TM, Li XJ, Ayala M, et al. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7:90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachy I, Vernier P, Retaux S. The LIM-homeodomain gene family in the developing Xenopus brain: conservation and divergences with the mouse related to the evolution of the forebrain. J. Neurosci. 2001;21:7620–7629. doi: 10.1523/JNEUROSCI.21-19-07620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez-Esteban C, Schwabe JW, Pena JD, Rincon-Limas DE, Magallon J, Botas J, Izpisua Belmonte JC. Lhx2, a vertebrate homologue of apterous, regulates vertebrate limb outgrowth. Development. 1998;125:3925–3934. doi: 10.1242/dev.125.20.3925. [DOI] [PubMed] [Google Scholar]
- 14.Seth A, Culverwell J, Walkowicz M, Toro S, Rick JM, Neuhauss SC, Varga ZM, Karlstrom RO. belladonna/(Ihx2) is required for neural patterning and midline axon guidance in the zebrafish forebrain. Development. 2006;133:725–735. doi: 10.1242/dev.02244. [DOI] [PubMed] [Google Scholar]
- 15.Xu Y, Baldassare M, Fisher P, Rathbun G, Oltz EM, Yancopoulos GD, Jessell TM, Alt FW. LH-2: a LIM/homeodomain gene expressed in developing lymphocytes and neural cells. Proc. Natl Acad. Sci. USA. 1993;90:227–231. doi: 10.1073/pnas.90.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rincon-Limas DE, Lu CH, Canal I, Calleja M, Rodriguez-Esteban C, Izpisua-Belmonte JC, Botas J. Conservation of the expression and function of apterous orthologs in Drosophila and mammals. Proc. Natl Acad. Sci. USA. 1999;96:2165–2170. doi: 10.1073/pnas.96.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulchand S, Subramanian L, Tole S. Dynamic spatiotemporal expression of LIM genes and cofactors in the embryonic and postnatal cerebral cortex. Dev. Dyn. 2003;226:460–469. doi: 10.1002/dvdy.10235. [DOI] [PubMed] [Google Scholar]
- 18.Mangale VS, Hirokawa KE, Satyaki PR, Gokulchandran N, Chikbire S, Subramanian L, Shetty AS, Martynoga B, Paul J, Mai MV, et al. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science. 2008;319:304–309. doi: 10.1126/science.1151695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D, et al. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development. 1997;124:2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- 20.Bulchand S, Grove EA, Porter FD, Tole S. LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech. Dev. 2001;100:165–175. doi: 10.1016/s0925-4773(00)00515-3. [DOI] [PubMed] [Google Scholar]
- 21.Yun S, Saijoh Y, Hirokawa KE, Kopinke D, Murtaugh LC, Monuki ES, Levine EM. Lhx2 links the intrinsic and extrinsic factors that control optic cup formation. Development. 2009;136:3895–3906. doi: 10.1242/dev.041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagglund AC, Dahl L, Carlsson L. Lhx2 is required for patterning and expansion of a distinct progenitor cell population committed to eye development. PLoS One. 2011;6:e23387. doi: 10.1371/journal.pone.0023387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saha B, Hari P, Huilgol D, Tole S. Dual role for LIM-homeodomain gene Lhx2 in the formation of the lateral olfactory tract. J. Neurosci. 2007;27:2290–2297. doi: 10.1523/JNEUROSCI.5571-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian L, Sarkar A, Shetty AS, Muralidharan B, Padmanabhan H, Piper M, Monuki ES, Bach I, Gronostajski RM, Richards LJ, et al. Transcription factor Lhx2 is necessary and sufficient to suppress astrogliogenesis and promote neurogenesis in the developing hippocampus. Proc. Natl Acad. Sci. USA. 2011;108:E265–E274. doi: 10.1073/pnas.1101109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou SJ, Perez-Garcia CG, Kroll TT, O'Leary DD. Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat. Neurosci. 2009;12:1381–1389. doi: 10.1038/nn.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakagawa Y, O'Leary DD. Combinatorial expression patterns of LIM-homeodomain and other regulatory genes parcellate developing thalamus. J. Neurosci. 2001;21:2711–2725. doi: 10.1523/JNEUROSCI.21-08-02711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen HF, Kuo HC, Chien CL, Shun CT, Yao YL, Ip PL, Chuang CY, Wang CC, Yang YS, Ho HN. Derivation, characterization and differentiation of human embryonic stem cells: comparing serum-containing versus serum-free media and evidence of germ cell differentiation. Hum. Reprod. 2007;22:567–577. doi: 10.1093/humrep/del412. [DOI] [PubMed] [Google Scholar]
- 28.Huang HP, Chen PH, Hwu WL, Chuang CY, Chien YH, Stone L, Chien CL, Li LT, Chiang SC, Chen HF, et al. Human Pompe disease-induced pluripotent stem cells for pathogenesis modeling, drug testing and disease marker identification. Hum. Mol. Genet. 2011;20:4851–4864. doi: 10.1093/hmg/ddr424. [DOI] [PubMed] [Google Scholar]
- 29.Chuang CY, Lin KI, Hsiao M, Stone L, Chen HF, Huang YH, Lin SP, Ho HN, Kuo HC. Meiotic competent human germ cell-like cells derived from human embryonic stem cells induced by BMP4/WNT3A signaling and OCT4/EpCAM (epithelial cell adhesion molecule) selection. J. Biol. Chem. 2012;287:14389–14401. doi: 10.1074/jbc.M111.338434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen YT, Furushima K, Hou PS, Ku AT, Deng JM, Jang CW, Fang H, Adams HP, Kuo ML, Ho HN, et al. PiggyBac transposon-mediated, reversible gene transfer in human embryonic stem cells. Stem Cells Dev. 2010;19:763–771. doi: 10.1089/scd.2009.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dahl JA, Collas P. Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay, unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells. Stem Cells. 2007;25:1037–1046. doi: 10.1634/stemcells.2006-0430. [DOI] [PubMed] [Google Scholar]
- 32.Okladnova O, Syagailo YV, Mossner R, Riederer P, Lesch KP. Regulation of PAX-6 gene transcription: alternate promoter usage in human brain. Brain Res. Mol. Brain Res. 1998;60:177–192. doi: 10.1016/s0169-328x(98)00167-3. [DOI] [PubMed] [Google Scholar]
- 33.Xu ZP, Saunders GF. Transcriptional regulation of the human PAX6 gene promoter. J. Biol. Chem. 1997;272:3430–3436. doi: 10.1074/jbc.272.6.3430. [DOI] [PubMed] [Google Scholar]
- 34.Katoh M. CER1 is a common target of WNT and NODAL signaling pathways in human embryonic stem cells. Int. J. Mol. Med. 2006;17:795–799. [PubMed] [Google Scholar]
- 35.Abellan A, Menuet A, Dehay C, Medina L, Retaux S. Differential expression of LIM-homeodomain factors in Cajal-Retzius cells of primates, rodents, and birds. Cereb. Cortex. 2010;20:1788–1798. doi: 10.1093/cercor/bhp242. [DOI] [PubMed] [Google Scholar]
- 36.Fasano CA, Chambers SM, Lee G, Tomishima MJ, Studer L. Efficient derivation of functional floor plate tissue from human embryonic stem cells. Cell Stem Cell. 2010;6:336–347. doi: 10.1016/j.stem.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Tetreault N, Champagne MP, Bernier G. The LIM homeobox transcription factor Lhx2 is required to specify the retina field and synergistically cooperates with Pax6 for Six6 trans-activation. Dev. Biol. 2009;327:541–550. doi: 10.1016/j.ydbio.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 38.Aoyama M, Ozaki T, Inuzuka H, Tomotsune D, Hirato J, Okamoto Y, Tokita H, Ohira M, Nakagawara A. LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Cancer Res. 2005;65:4587–4597. doi: 10.1158/0008-5472.CAN-04-4630. [DOI] [PubMed] [Google Scholar]
- 39.Koch P, Opitz T, Steinbeck JA, Ladewig J, Brustle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc. Natl Acad. Sci. USA. 2009;106:3225–3230. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moeller C, Yaylaoglu MB, Alvarez-Bolado G, Thaller C, Eichele G. Murine Lix1, a novel marker for substantia nigra, cortical layer 5, and hindbrain structures. Brain Res. Gene Expr. Patterns. 2002;1:199–203. doi: 10.1016/s1567-133x(02)00018-2. [DOI] [PubMed] [Google Scholar]
- 41.Gunhaga L, Marklund M, Sjodal M, Hsieh JC, Jessell TM, Edlund T. Specification of dorsal telencephalic character by sequential Wnt and FGF signaling. Nat. Neurosci. 2003;6:701–707. doi: 10.1038/nn1068. [DOI] [PubMed] [Google Scholar]
- 42.Larsen KB, Lutterodt MC, Laursen H, Graem N, Pakkenberg B, Mollgard K, Moller M. Spatiotemporal distribution of PAX6 and MEIS2 expression and total cell numbers in the ganglionic eminence in the early developing human forebrain. Dev. Neurosci. 2010;32:149–162. doi: 10.1159/000297602. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Woodhead GJ, Swaminathan SK, Noles SR, McQuinn ER, Pisarek AJ, Stocker AM, Mutch CA, Funatsu N, Chenn A. Cortical neural precursors inhibit their own differentiation via N-cadherin maintenance of beta-catenin signaling. Dev. Cell. 2010;18:472–479. doi: 10.1016/j.devcel.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson SI, Shafer B, Lee KJ, Dodd J. A molecular program for contralateral trajectory: Rig-1 control by LIM homeodomain transcription factors. Neuron. 2008;59:413–424. doi: 10.1016/j.neuron.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 45.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA. 2010;14:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P. Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Dev. Biol. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- 49.Plaza S, Saule S, Dozier C. High conservation of cis-regulatory elements between quail and human for the Pax-6 gene. Dev. Genes Evol. 1999;209:165–173. doi: 10.1007/s004270050240. [DOI] [PubMed] [Google Scholar]
- 50.Xu ZP, Saunders GF. PAX6 intronic sequence targets expression to the spinal cord. Dev. Genet. 1998;23:259–263. doi: 10.1002/(SICI)1520-6408(1998)23:4<259::AID-DVG1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 51.Zheng JB, Zhou YH, Maity T, Liao WS, Saunders GF. Activation of the human PAX6 gene through the exon 1 enhancer by transcription factors SEF and Sp1. Nucleic Acids Res. 2001;29:4070–4078. doi: 10.1093/nar/29.19.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kleinjan DA, Seawright A, Schedl A, Quinlan RA, Danes S, van Heyningen V. Aniridia-associated translocations, DNase hypersensitivity, sequence comparison and transgenic analysis redefine the functional domain of PAX6. Hum. Mol. Genet. 2001;10:2049–2059. doi: 10.1093/hmg/10.19.2049. [DOI] [PubMed] [Google Scholar]
- 53.Peukert D, Weber S, Lumsden A, Scholpp S. Lhx2 and lhx9 determine neuronal differentiation and compartition in the caudal forebrain by regulating wnt signaling. PLoS Biol. 2011;9:e1001218. doi: 10.1371/journal.pbio.1001218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bouwmeester T, Kim S, Sasai Y, Lu B, De Robertis EM. Cerberus is a head-inducing secreted factor expressed in the anterior endoderm of Spemann's organizer. Nature. 1996;382:595–601. doi: 10.1038/382595a0. [DOI] [PubMed] [Google Scholar]
- 55.Kuroda H, Wessely O, De Robertis EM. Neural induction in Xenopus: requirement for ectodermal and endomesodermal signals via Chordin, Noggin, beta-Catenin, and Cerberus. PLoS Biol. 2004;2:E92. doi: 10.1371/journal.pbio.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith JR, Vallier L, Lupo G, Alexander M, Harris WA, Pedersen RA. Inhibition of Activin/Nodal signaling promotes specification of human embryonic stem cells into neuroectoderm. Dev. Biol. 2008;313:107–117. doi: 10.1016/j.ydbio.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 57.Biben C, Stanley E, Fabri L, Kotecha S, Rhinn M, Drinkwater C, Lah M, Wang CC, Nash A, Hilton D, et al. Murine cerberus homologue mCer-1: a candidate anterior patterning molecule. Dev. Biol. 1998;194:135–151. doi: 10.1006/dbio.1997.8812. [DOI] [PubMed] [Google Scholar]
- 58.Zhang X, Friedman A, Heaney S, Purcell P, Maas RL. Meis homeoproteins directly regulate Pax6 during vertebrate lens morphogenesis. Genes Dev. 2002;16:2097–2107. doi: 10.1101/gad.1007602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang L, Mathers PH, Jamrich M. Function of Rx, but not Pax6, is essential for the formation of retinal progenitor cells in mice. Genesis. 2000;28:135–142. [PubMed] [Google Scholar]
- 60.Retaux S, Rogard M, Bach I, Failli V, Besson MJ. Lhx9: a novel LIM-homeodomain gene expressed in the developing forebrain. J. Neurosci. 1999;19:783–793. doi: 10.1523/JNEUROSCI.19-02-00783.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosa A, Brivanlou AH. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Greber B, Wu G, Bernemann C, Joo JY, Han DW, Ko K, Tapia N, Sabour D, Sterneckert J, Tesar P, et al. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell. 2010;6:215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.