Abstract
HIV-infected individuals currently cannot be completely cured because existing antiviral therapy regimens do not address HIV provirus DNA, flanked by long terminal repeats (LTRs), already integrated into host genome. Here, we present a possible alternative therapeutic approach to specifically and directly mediate deletion of the integrated full-length HIV provirus from infected and latently infected human T cell genomes by using specially designed zinc-finger nucleases (ZFNs) to target a sequence within the LTR that is well conserved across all clades. We designed and screened one pair of ZFN to target the highly conserved HIV-1 5′-LTR and 3′-LTR DNA sequences, named ZFN-LTR. We found that ZFN-LTR can specifically target and cleave the full-length HIV-1 proviral DNA in several infected and latently infected cell types and also HIV-1 infected human primary cells in vitro. We observed that the frequency of excision was 45.9% in infected human cell lines after treatment with ZFN-LTR, without significant host-cell genotoxicity. Taken together, our data demonstrate that a single ZFN-LTR pair can specifically and effectively cleave integrated full-length HIV-1 proviral DNA and mediate antiretroviral activity in infected and latently infected cells, suggesting that this strategy could offer a novel approach to eradicate the HIV-1 virus from the infected host in the future.
INTRODUCTION
Although highly active antiretroviral treatment can substantially improve the survival rate of HIV-infected individuals, it cannot completely cure the patient (1). This is largely because current antiviral drugs are limited in their scope in that they can only inhibit the replication of HIV-1 virus. Although these therapies can target the viral proteins or receptors for viral entry, they cannot specifically delete the integrated HIV proviral DNA. Without eradicating this proviral DNA, the infected individual cannot be cured. The integrated HIV provirus, located between the so-called long terminal repeats (LTRs), is not only used as template for viral gene transcription in the infected cells but also as the basis for the long incubation period of HIV in latency cells (2–5). Therefore, one promising strategy to eradicate HIV provirus from the host is to treat the disease at the root cause and specifically target and delete the integrated HIV provirus within these cells. Recently, Buchholz and colleagues developed a Cre-recombinase mutant, called Tre-recombinase, which recognizes a sequence (LoxLTR) located on the LTR of HIV strain TZB0003 (6). Remarkably, this enzyme effectively deleted integrated HIV proviral DNA from infected cells. Unfortunately, the LTR sequence of strain TZB0003 is atypical for HIV and was chosen for these studies because of its similarity to the DNA sequence recognized by the wild-type Cre recombinase. The corresponding LTR sequences from other HIV strains are much more divergent from the wild-type Cre recognition sequence, and thus it is unlikely that this approach can ever be generalized to clinically relevant HIV strains (7). Although substrate-linked protein evolution could be used to construct additional sequence-specific enzymes, execution would be impractical in many circumstances (8), for example, multiple enzymes per patient could be required, as HIV-1 diversifies to quasispecies as patients progress toward AIDS. This approach would therefore not be practical in clinical applications treating a broad spectrum of patients infected with different HIV subtypes (7,8).
Zinc-finger nucleases (ZFNs) are artificially modified endonucleases comprising two components: the DNA-binding zinc-finger domain specific to the DNA of interest and the non-specific FokI restriction endonuclease DNA cleavage domain (9,10). Because they can direct DNA cleavage to specific sequences, ZFNs are now being used to target specific genes for gene modification purposes in various species (11–22). An area of great promise for ZFNs is in human gene therapy. For example, Perez et al. (13), using engineered ZFNs targeting human CCR5 gene, encoding a receptor necessary for HIV entry into the T cell, previously demonstrated the establishment of HIV-1 resistance in CD4+ T cells through generation of a double-strand break (DSB) at pre-determined sites in the CCR5 coding region upstream of the natural CCR5D32 mutation. More recently, Holt et al. (14) demonstrated control of HIV-1 infection within non-obese diabetic/severe combined immunodeficient/interleukin 2rγnull mice transplanted with human hematopoietic stem/progenitor cells modified by ZFN targeting CCR5. Owing to its relative simplicity, ZFN-mediated genome surgery in primary human cells has become a reality: two phase I clinical trials (NCT00842634; NCT01044654) are underway to determine whether destruction of the CCR5 gene can confer protection from HIV infection when ZFNs are used to target and destroy CCR5 in autologous T cells from HIV patients. There are also studies using zinc-finger proteins to act as repressors of HIV (23,24) and to interfere with integration (25,26). However, none of the current strategies can directly target integrated HIV provirus genomes for cleavage using ZFNs. Here, unlike previous ZFN-mediated gene disruption strategies aiming to cure HIV by targeting the host coreceptor CCR5 gene pathways, we present a possible alternative therapeutic approach to specifically and directly mediate deletion of the integrated full-length HIV provirus from the host genome by one pair of ZFNs to target a sequence within the LTR that is well conserved across all clades.
MATERIALS AND METHODS
Generation of ZFNs
The ZFN expression plasmids were obtained from Sigma’s CompoZr product line. The ZFNs were designed, assembled, and validated by the company similar to what is described elsewhere (16). Sixteen ZFN pairs (target sites in Supplementary Table S1) were assembled by overlapping PCR and cloned into a vector upstream of a FokI cleavage domain. Two pairs of ZFNs with confirmed activity in a yeast Mel-1 assay (16) were obtained commercially from Sigma-Aldrich (Supplementary Figure S1). Based on in vitro cleavage efficiency, one ZFN-LTR pair (ZFN-LTR-L and ZFN-LTR-R) was chosen for further testing in proof-of-principle experiments. ZFN-LTR pair used in this experiment had 11 zinc-finger domains recognizing 33 bases, respectively (Figure 1A). To make catalytically inactive ZFN monomers, we introduced the D450A mutation (numbered relative to the native FokI enzyme) (27) at the active site of the FokI domains by site-directed mutagenesis using the oligonucleotides: D450A-forward: 5′-GAAGCAGAAAGCCTGCCGGCGCCATCTATAC-3′; D450A-reverse: 5′-GTATAGATGGCGCCGGCAGGCTTTCTGCTTC-3′.
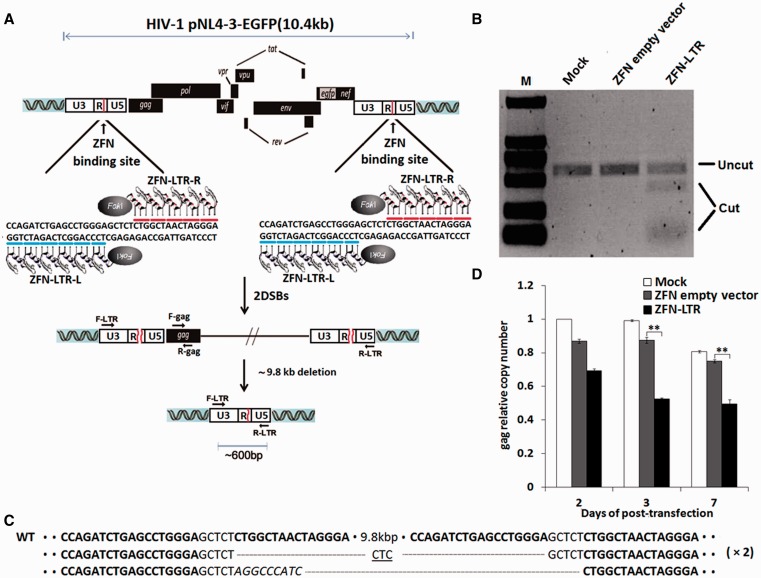
Figure 1.
ZFN-LTR-induced excision of integrated HIV-1 proviral DNA in HIV-1-infected cells. (A) Schematic representation of ZFN-LTR-mediated genome deletions. Red lines in the 5′- and 3′-LTR of HIV-1 genome indicate ZFN-LTR target sites. ZFN-LTR-L and ZFN-LTR-R are pairs of zinc-finger proteins that have been customized to specifically recognize the left and right half-sites (indicated by blue and red lines) that are separated by a 5 bp spacer. F-LTR and R-LTR are PCR primers (arrows) located in the 5′- and 3′-LTRs, respectively, which were used for the detection of genome deletion events. (B) T7 endonuclease 1 assay of ZFN cleavage in HIV-1 infected cells. Jurkat T cells were infected with the HIV-1 pNL4-3-EGFP virus, and EGFP-positive cells were sorted by flow cytometry as HIV-1-infected cells. Then, the HIV-1-infected cells were untransfected (mock) or nucleofected with 2.5 μg of ZFN empty vector (negative control) or ZFN-LTR plasmid. At 3 days post-transfection, cells were collected for extraction of genomic DNA, and PCR was performed with the F-LTR and R-LTR primers to detect ZFN-LTR-mediated HIV-1 genomic DNA deletions. Amplicons encompassing the target sites were digested with the T7E1. The expected positions of the resulting DNA bands are indicated by an horizontal line (uncut ∼583 bp) and a bracket (cut ∼450 and ∼130 bp) at the left of the gel panels. (C) DNA sequences of PCR products. The PCR products were cloned and sequenced. ZFN-LTR target sites are shown in boldface letters. Dashes indicate deleted bases relative to the wild-type sequence. Inserted bases are shown in italics. In cases where a deletion sequence was detected more than once, the number of instances is given on the right in parentheses. (D) Analysis of HIV-1 proviral DNA deletions by quantitative real-time PCR. HIV-1-infected cells were untransfected (mock) or nucleofected with 2.5 μg of ZFN empty vector or ZFN-LTR plasmid. Genomic DNA was isolated at the indicated times and subjected to quantitative real-time PCR using gene-specific primers for HIV-1 gag and human β-globin. The relative copy numbers of gag were calculated based on the standard curve obtained by serial dilution (10–160 ng) of an infected cell DNA on the same plate. Normalization was carried out by division of gag gene amplicons in mock group of 2 days post-transfection. Data are representative of three independent experiments, and error bars represent standard errors (SD). *P < 0.05, **P < 0.01, ***P < 0.001; paired t-test.
Establishment of the HIV-1-infected cells and latently cells
Jurkat T cells were cultured in RPMI 1640 containing 100 units/ml of penicillin and streptomycin and supplemented with 10% fetal bovine serum at 37°C under 5% CO2. To establish the HIV-1-infected and latently infected cell models, we used an HIV-1 pseudovirus that expresses enhanced green-fluorescent protein (EGFP) (pNL4-3-EGFP) to infect Jurkat T cells, as described by Jordan et al. (28). Briefly, 106 TCID50 pseudoviruses were used to infect Jurkat T cells. The EGFP-positive and EGFP-negative cell populations were then sorted by flow cytometry, and the EGFP-positive cells were used as the HIV-1-provirus-infected cell lines. Potential latently infected clones were identified by screening for EGFP reactivation and p24 antigen expression before and after stimulation. Under the stimulation of 200 nM trichostatin A (TSA) or 10 ng/ml phorbol 2-myristate 13-acetate, EGFP was expressed in some cells that were originally EGFP-negative. These EGFP reactivated cell populations were isolated by flow cytometry. Several clones were acquired by a limiting dilution assay. Further testing was performed using an HIV p24 enzyme-linked immunosorbent assay (ELISA) assay. Five HIV-1 latently infected cell lines were ultimately identified (29). Alu-LTR nest-PCR assay confirmed that the HIV-1 genome had been integrated in the genome of these clone lines. The latently infected cell line clone that showed the best reactivation effects was named C11 and chosen for use in our experiments.
Isolation of primary peripheral blood lymphocyte and primary CD4+T cells
Peripheral blood mononuclear cells (PBMCs, from one blood unit, 200 ml) isolated from healthy donors were purchased from the Shanghai Blood Center (Shanghai, China). CD4+ T cells were further purified from PBMCs by using anti-CD4-antibodies-coated magnetic beads (Miltenyi Biotech), according to the manufacturer’s instructions, and peripheral blood lymphocyte (PBLs) were isolated from PBMCs by depleting CD14+ monocytes, via using anti-CD14-antibodies-coated magnetic beads (Miltenyi Biotech). PBLs or CD4+ T cells were stimulated for 3 days with phytohemagglutinin-P(PHA-P) (5 μg/ml) and cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum and recombinant human interleukin-2 (20 IU/ml; Roche Applied Science).
HIV-1 infection of human primary T cells and ELISA detection of antigen p24 levels
Replication competent HIV-1/NL4-3 viruses (CXCR4 tropism) were generated by using calcium phosphate-mediated transfection of HEK293T cells with the plasmid of HIV-1/NL4-3. Harvested supernatants of transfected cells containing HIV-1 particles were filtered and titrated with p24 capture ELISA. Viral stock was stored at −80°C. For infection, PHA-P-activated-PBLs or -CD4+ T Cells were inoculated with 10 ng p24 amount of HIV-1/NL4-3 for overnight and further cultured for 3 days after washing off cell-free viruses. In all, 1 × 107 of infected PBLs or 5 × 106 CD4+ T Cells were nucleofected with 5 μg of ZFN-LTR or ZFN empty vector plamids using the Amaxa Human T Cell Nucleofector kit (Lonza). At the indicated times after transfection, viral replication were monitored via quantifying the amounts of p24 produced in culture supernatant by using HIV-1 p24 Antigen ELISA kit (ZeptoMetriX), according to the manufacturer’s instructions.
Extraction of genomic DNA and quantitative real-time PCR
To detect changes in HIV-1-encoded gag gene copy number after ZFN-LTR-mediated recombination, quantitative real-time PCR analysis was performed as described previously (6). Briefly, total genomic DNA was isolated from cells using a Blood & Cell Culture DNA Midi Kit (Qiagen) according to the manufacturer’s protocol. Quantitative PCR specific to HIV-1 gag gene (gag-forward: 5′-ATCAATGAGGAAGCTGCAGAA-3′, gag-reverse: 5′-GATAGGTGGATTATGTGTCAT-3′, gag-probe: 5′-FAM- ATTGCACCAGGCCAGATGAGAGAA-TAMRA-3′) was performed as follows: 40 cycles containing 100 ng genomic DNA, 500 nM forward primer, 500 nM reverse primer, and 250 nM hybridization probe were performed using Platinum qPCR Super Mix UDG (Invitrogen) in a Roche Real-Time PCR System. Each sample was tested in triplicate, and results were normalized using amplification of the same genomic DNA with human β-globin primers (HBG-forward: 5′-CTTAATGCCTTAACATTGTGTATAA-3′, HBG-reverse: 5′-GAATATGCAAATAAGCACACATATAT-3′, HBG-probe: 5′-FAM-ACTTTACACAGTCTGCCTAGTACATTAC-TAMRA-3′). Total HIV-1 DNA levels were measured as the relative copy numbers of gag gene. The relative copy numbers of gag were calculated based on the standard curve obtained by serial dilution (10–160 ng) of an infected cell DNA on the same plate. Normalization was carried out by division of gag gene amplicons in mock group. The standard curve of the human β-globin gene as a control locus was used for exact quantification of the input sample DNAs, in which HIV gag copies were measured.
PCR and sequencing analysis
To detect genomic deletions, PCR and sequencing analysis were performed as described previously (11,14). Briefly, genomic DNA was subjected to PCR analysis using Taq DNA polymerase (Promega) and appropriate primers. The primers F-LTR and R-LTR were used for DNA template from pNL4-3-EGFP-infected cells or HIV-based lentiviral infected 293T cells transfected with ZFN-LTR (Primers F-LTR-forward: 5′- GATCCTTGATCTGTGGATCTACCA -3′, R-LTR-reverse: 5′- ACACTGACTAAAAGGGTCTGAGGG -3′); F-HG and R-HG for DNA template from latent cell clone 11 (C11) transfected with ZFN-LTR (F-HG-forward: 5′- TGCCACCCGAAACTATTCACAAG -3′, R-HG -reverse: 5′- CCGGCATGGATTCCAGTTCTTAG -3′). The PCR program (94°C, 3 min; 30 cycles of 94°C, 30 s; 55°C, 30 s; and a final 72° C, 1 min) took place in a 25 μl of PCR reaction chamber containing 1 × PCR buffer and 250 μM dNTP, 3 mM MgCl2 and 0.5 units of BioTaq DNA polymerase. PCR products were analyzed by agarose gel electrophoresis. For sequencing analysis, PCR products corresponding to genomic deletions were purified using a Gel Extraction Kit (Tiangen) and cloned into the T-Blunt vector using the T-Blunt PCR Cloning Kit (TaKaRa). Cloned plasmids were sequenced using M13 primers.
T7 endonuclease 1 assay
To detect genomic modification mediated by ZFN-LTR, PCR analysis were performed as described previously (13,14). Briefly, the HIV-1-infected cells were not transfected (mock) or nucleofected with 2.5 μg of ZFN empty vector (negative control) or ZFN-LTR plasmid. At 3 days post-transfection, genomic DNA from ZFN-treated and control cells was extracted. The genomic region encompassing the ZFN target site was amplified, melted and annealed to form heteroduplex DNA. The annealed DNA was treated with 5 units of the mismatch-sensitive T7 endonuclease 1 (T7E1) (New England BioLabs) for 15 min at 37°C and then precipitated by addition of 2.5 volumes of ethanol. The precipitated DNA was analyzed by agarose gel electrophoresis.
Visualization of EGFP and flow cytometry assay
To analyze EGFP fluorescence after ZFN-LTR treatment in the cell model infected with the pNL4-3-EGFP virus containing the HIV-1 provirus, cells were mock-treated or nucleofected with ZFN-LTR or ZFN empty vector using the Amaxa Cell line Nucleofector kit V (Lonza). Expression of EGFP as a marker for activation of HIV-1 in infected cell lines was observed by fluorescence microscopy. At the indicated times after transfection, all three groups of cells were collected and washed with phosphate buffered saline (PBS). They were kept in PBS before analysis on a BD LSRII flow cytometer for EGFP expression.
Immunocytochemical analysis of double-strand-break-induced 53BP1 foci
53BP1 immunocytochemical analysis was performed as described previously (14). Briefly, Jurkat T cells were collected at the indicated times after nucleofection with 2.5 μg of ZFN-LTR expression plasmid or ZFN empty vector. As a positive control, the cells were exposed to 1 mM etoposide for 60 min 2 h before harvest. Cells were attached on slides treated with poly-l-lysine and then fixed with 4% paraformaldehyde. The cells were then permeabilized by treatment with 0.5% Triton X-100 buffer (0.5% Triton X-100, 1% bovine serum albumin (BSA), 0.02% NaN3 and 98.5% PBS) for 15 min and then blocked with 10% goat serum for 60 min at room temperature. Cells were incubated after blocking with anti-53BP1 rabbit polyclonal antibodies [Bethyl Laboratories, diluted 1:500 in PBS containing 1% BSA] overnight at 4°C followed by incubation with Alexa Fluor 594-conjugated secondary antibodies (Invitrogen-Molecular Probes, diluted 1:2000 in PBS containing 1% BSA) at 37°C in the dark for 2 h. Slides were mounted in the presence of 1 mg/ml 4′, 6-diamidino-2-phenylindole (DAPI) (Sigma) to counter-stain cell nuclei and examined under confocal microscope (Zeiss LSM-710). Individual regions of 53BP1 fluorescence were then enumerated and measured. Only red fluorescent regions that colocalized with nucleus-specific DAPI fluorescence were included in the final analyses.
Statistical analysis
Data are representative of three independent experiments, and error bars represent standard errors (SD). Paired samples t-tests were performed with use of SPSS version 13.0 (SPSS Inc., Chicago), and statistical significance was indicated at *P < 0.05, **P < 0.01 or ***P < 0.001.
Bioinformatics tools
The ZFN-LTR binding sequence and the LoxLTR sequence were aligned with all HIV-1 genome sequences in the Los Alamos HIV Sequence Database (http://www.hiv.lanl.gov/) using the web alignment tool (http://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html). Then the alignments were used to highlight mismatches by similarity using Highlighter for Nucleotide Sequences v2.1.1 online (http://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/HIGHLIGHT_XYPLOT/highlighter.html). The off-target cleavage events of ZFN-LTR in human genome were predicted using an online tool called ZFN-Site (http://ccg.vital-it.ch/tagger/targetsearch.html).
RESULTS
ZFN-LTR is designed to specifically target the highly conserved HIV-1 LTR region in HIV-1 provirus DNA
HIV-1 LTR is a highly conserved region in the HIV genome, therefore making it an attractive and ideal target for the development of new anti-HIV therapies. To design zinc-finger motifs that specifically target the HIV-1 of 5′-LTR and 3′-LTR DNA sequences, the DNA sequence of HIV-1 strain NL4-3 was submitted to Sigma-Aldrich for the design and construction of ZFNs. Sixteen candidate DNA sequences in HIV-1 LTR were obtained (Supplementary Table S1) and sixteen ZFN pairs targeted these sites were assembled. Two pairs of ZFNs with confirmed activity in a yeast Mel-1 assay were obtained commercially from Sigma-Aldrich (Supplementary Figure S1). Based on in vitro cleavage efficiency, the ZFN pair, PZFN1/PZFN2 (Set 1), with the highest relative Mel-1 activity (which was named ZFN-LTR), was chosen for further testing in proof-of-principle experiments. The ZFN-LTR (ZFN-LTR-L and ZFN-LTR-R, containing five or six zinc-fingers, full amino acid sequences in Supplementary Figure S2) binding sequence (Figure 1A), which corresponds to 471–508 bp of HXB2 reference isolate of HIV-1 (GenBank accession number K03455), was aligned with all HIV-1 genome sequences in the Los Alamos HIV Sequence Database, and mismatches were highlighted using an online tool (http://www.hiv.lanl.gov/). Conservation analysis showed that the total number of sequences analyzed was 344, and the average similarity to the ZFN-LTR binding sequence was 0.937 (Supplementary Figure S3A). This 0.937 conservation of the sequence recognized by ZFN-LTR among various HIV subtypes was much higher than for the LoxLTR sequence recognized by Tre-recombinase, whose average similarity was only 0.775 over a total of 269 subtype sequences (Supplementary Figure S3B), indicating that ZFN-LTR is likely to be more effective than Tre-recombinase in removing HIV-1 proviral DNA.
ZFN-LTR mediated targeted HIV-1 proviral DNA deletions in infected cell lines
Next, we determined whether ZFN-LTR could delete HIV-1 provirus from HIV-infected cell lines. We infected Jurkat T cells with pNL4-3 virus, an HIV-1 pseudovirus, carrying EGFP reporter genes (pNL4-3-EGFP). EGFP-positive cells were sorted and used to express ZFN-LTR plasmids by nucleofection. As controls, HIV-infected cells also were untransfected (mock) or nucleofected with parental ZFN empty vectors. After 3 days, genomic DNA was extracted from HIV-infected cell pools and analyzed by PCR. To probe for deletion of the provirus, good primer design should be located at the two ends of the integrated HIV genome, but it is not easy to get information of all proviral integration sites throughout chromosomes in the respective infected cell pools. We thus used forward primer (F-LTR) located the U3 region of upstream of ZFN-LTR target site in HIV-1 5′-LTR and reverse primmer (R-LTR) located the U5 region of downstream of ZFN-LTR target site in HIV-1 3′-LTR (Figure 1A). We found that the large 9.8 kb intervening DNA segment between the primers, containing the HIV provirus, was too long to amplify under conventional PCR conditions. However, a 583-bp PCR product was observed not only in the ZFN-LTR plasmid transfection group but also in the control ZFN empty vectors and mock groups. This is because PCR primers are also present in the HIV-1 5′-LTR U5 region and HIV-1 3′-LTR U3 region; therefore, they should give a 583-bp band even in the unmodified proviral DNA. To distinguish which PCR product result from HIV-LTR modified by ZFN-LTR, amplicons encompassing the target sites were digested with the T7E1. If the DNA amplicons contain both wild-type and mutated DNA sequences, heteroduplexes would be formed. T7E1 recognizes and cleaves heteroduplexes, but not homoduplexes (13,14). The DNA fragments were assessed by agarose gel electrophoresis. We found that the 583-bp PCR amplicon from the ZFN-LTR plasmid transfection group is cleaved into ∼450- and∼130-bp fragments, butnot in the control ZFN empty vectors and mock groups (Figure 1B). Next, we cloned the PCR product from the ZFN-LTR plasmid transfection group and performed sequence analysis. The breakpoint junction sequences showed that small deletions or insertions regions were present (Figure 1C), which is consistent with imprecise repair of ZFN-LTR nuclease-mediated DSB via non-homologous end joining (NHEJ). To further address the deletion of the provirus, we measured the integrated HIV-1-encoded gag gene copy numbers in the genomes of infected cells treated with ZFN-LTR or controls by quantitative real-time PCR. The results showed significant decreases in gag gene copies in infected cells transfected with ZFN-LTR relative to controls at 3 and 7 days after treatment (Figure 1D). Collectively, these data demonstrate that a single ZFN-LTR pair can target two sites with identical sequences and cleave integrated HIV-1 proviral DNA in infected cells.
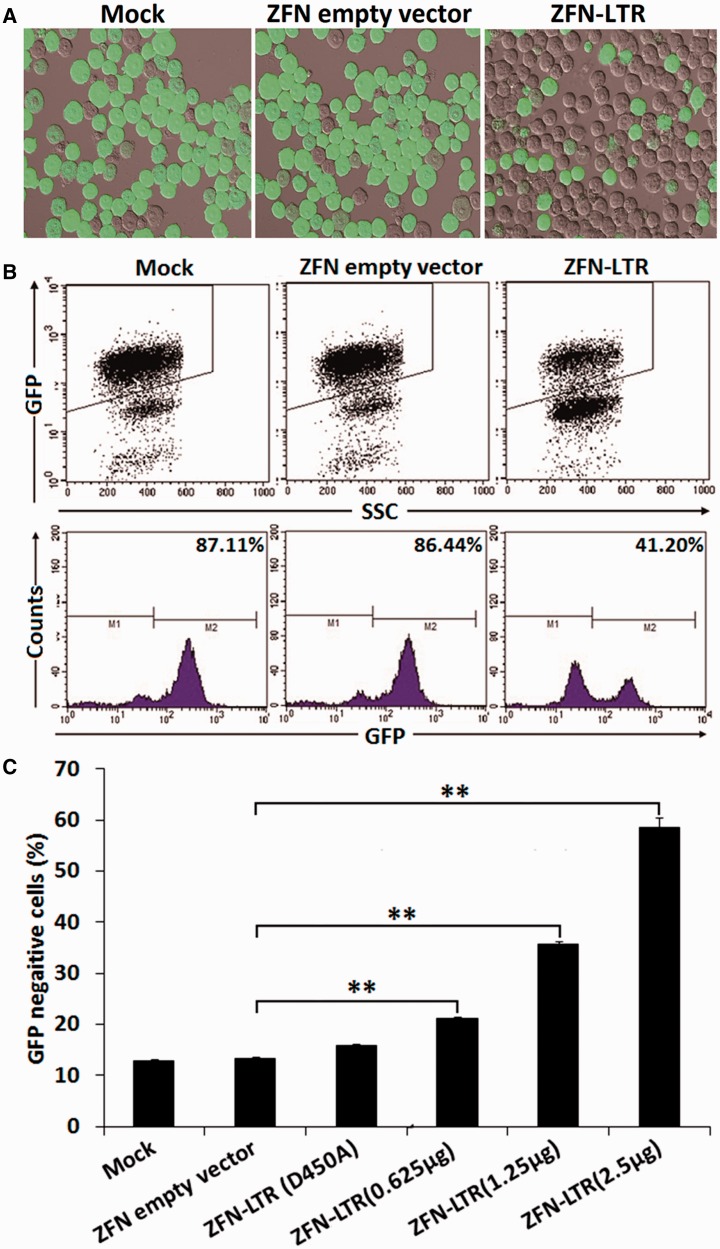
To visually determine whether ZFN-LTR treatment of infected cells removed the viral infection from the Jurkat T cells, we took advantage of the viral EGFP reporter system. Recombination by NHEJ repair after ZFN-LTR-mediated cleavage and DNA deletion also cause disruptions in EGFP-coding sequences, which can be used to monitor the loss of EGFP expression in infected cells. We observed by confocal microscopy that a proportion of the ZFN-LTR-treated infected cells were negative for EGFP reporter expression (Figure 2A), indicating that these cells were no longer virally infected. To quantify the frequency of ZFN-LTR-mediated HIV-1 proviral DNA deletions in HIV-1-infected cell lines, we assessed the percentage of EGFP-positive cells by flow cytometry. We observed a loss of EGFP expression in 45.9% of the cell lines 3 days after treatment with ZFN-LTR at the 2.5 μg of dose compared with controls (Figure 2B and C); significant loss was also observed even at lower doses of ZFN-LTR (Figure 2C). When using the transfection efficiency (∼80% for nucleofection in Jurkat cells), we obtained the correct excision frequency, which could be up to 57.4%. To test whether ZFN-LTR could interfere with expression from the LTR promoter, which could contribute to the decrease in EGFP expression without actually excising the viral genome, we introduced a previously described mutation (D450A) at the active site of the FokI domains, which inactivates the catalytic activity of the FokI (27). We found that there was almost no difference in the percentage of EGFP-positive cells between the inactive FokI group and the empty vector group (Figure 2C), indicating that ZFN-LTR binding to the LTR did not contribute to the decrease in EGFP expression. Similar results were observed in the HIV-based lentiviral infected 293T cells treated with ZFN-LTR (Supplementary Figure S4). These results suggest that ZFN-LTR treatment effectively excised HIV-1 proviral DNA and therefore mediated antiretroviral activity in viral infected cells.
Figure 2.
ZFN-LTR-mediated excision efficiency of HIV-1 proviral DNA in infected cells. (A) Detection of EGFP expression in infected cells treated with 2.5 μg of ZFN empty vector (negative control) or ZFN-LTR plasmid or mock-treated by fluorescence microscopy. (B and C) The efficiency of HIV-1 proviral DNA excision in infected cells untransfected or transfected with 2.5 μg of ZFN empty vector or 2.5 μg of ZFN-LTR with inactive FokI (D450A) or ZFN-LTR plasmid dose titration (0.625–2.5 μg). At 3 days after transfection, the percentage of EGFP-negative cells was measured by flow cytometry. Results are presented as fluorescence histograms. Data are representative of three independent experiments, and error bars represent SD. *P < 0.05, **P < 0.01, ***P < 0.001; paired t-test.
ZFN-LTR-induced excision of integrated HIV-1 proviral DNA in human primary T cells
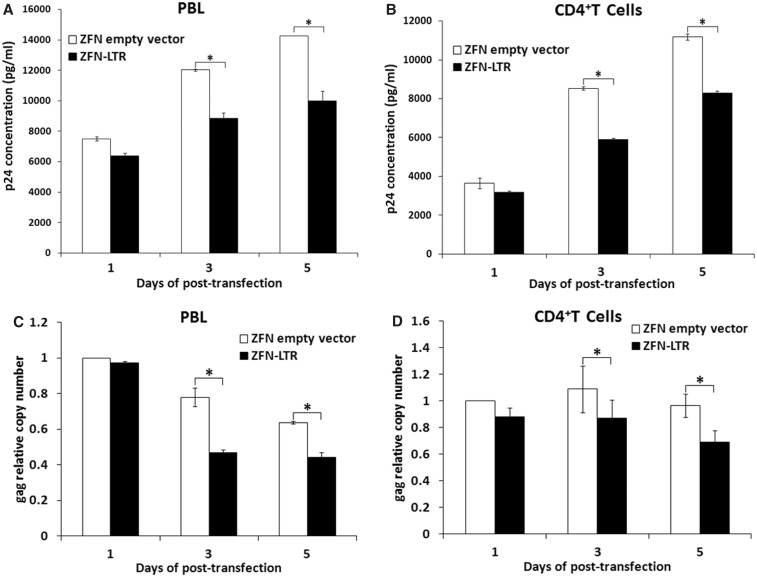
To determine whether ZFN-LTR could induce excision of integrated HIV-1 proviral DNA in viral infected human primary cells, human PBL or CD4+ T cells were infected with HIV-1/NL4-3 viruses and then nucleofected with ZFN-LTR or ZFN empty vector, followed by ELISA of culture supernatants for p24 antigen content at the indicated times. We found that the transfection of ZFN-LTR, compared with empty vector-transfected control, decreased HIV-1 p24 production maximally by ∼29% in PBLs (Figure 3A) and by ∼31% in CD4+ T cells (Figure 3B). To detect whether the decrease of viral production was due to ZFN-LTR-mediated excision of integrated HIV-1 proviral DNA, the copy number of integrated HIV-1/NL4-3-encoded gag gene was quantified by real-time PCR. As expected, integrated-gag genes showed significant decrease in ZFN-LTR-treated-, compared with empty vector treated-, primary T cells, and ∼38% decrease in infected PBL cells (Figure 3C) and ∼20% decrease in infected CD4+ T cells (Figure 3D) were observed at 3 or 5 days post-transfection. These results confirmed the excision of integrated HIV-1 proviral DNA mediated by ZFN-LTR in viral infected human primary T cells.
Figure 3.
ZFN-LTR-induced excision of integrated HIV-1 proviral DNA in HIV-1-infected PBL or CD4+ T cells. HIV-1/NL4-3 infected-PBLs and -CD4+ T Cells were nucleofected with 5 μg of ZFN-LTR plasmid or ZFN empty vector, and at the indicated post-transfection time points, (A and B) culture supernatants were harvested for detecting HIV-1 p24 production; (C and D) the relative copy numbers of gag were calculated based on the standard curve obtained by serial dilution (10–160 ng) of an infected cell DNA on the same plate. Normalization was carried out by division of gag gene amplicons in ZFN empty vector group of 1 day post-transfection. Data are representative of three independent experiments, and error bars represent SD. *P < 0.05, **P < 0.01, ***P < 0.001; paired t-test.
ZFN-LTR treatment also mediated effective HIV-1 proviral DNA excision in latently infected T cells
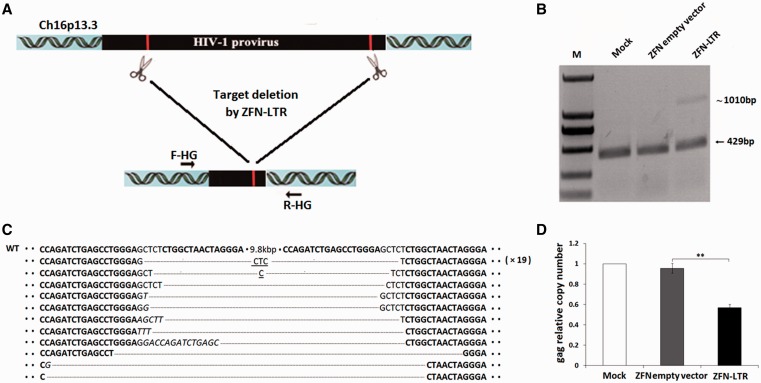
The persistence of latent HIV-infected cellular reservoirs represents the major barrier to virus eradication in patients treated with highly active antiretroviral therapy. In latently infected cells, the replication-competent integrated HIV-1 provirus remains transcriptionally silent, resulting in the absence of viral protein expression and the evasion of host immune surveillance and antiretroviral drugs. The molecular mechanisms by which integrated HIV-1 is repressed during latency include transcriptional interference, insufficient levels of transcriptional activators, the presence of transcriptional repressors, epigenetics, nucleosome positioning, insufficient Tat activity, blocks to mRNA splicing or nuclear export, cellular microRNA (miRNA), and homeostatic proliferation of latently infected cells (1,5). The gene therapy approaches to eradicate integrated HIV-1 might be an alternative strategy. Therefore, we next asked whether similar results could be obtained in latently infected T cells. To address this, we established an HIV-1 latently infected cell line by transducing the Jurkat lymphocytic cell line with a full-length HIV genome containing EGFP open reading frames under the control of the viral 5′-LTR. Cells that exhibited the latent phenotype, which at first represented EGFP-negative but could be reactivated by TSA treatment, were cloned and further characterized. To perform the following experiments, we selected the latent cell C11, which was found to carry a single integrated HIV-1 vector at position Ch16p13.3 and showed low spontaneous HIV-1 promoter activity (29). We transfected the C11 with ZFN-LTR or control vectors; to probe for deletion of the provirus, we used primers located at the two ends of the integrated HIV genome normally separated by 10.9 kb when the provirus is integrated into the host genome (Figure 4A). The PCR product size we observed after ZFN-LTR treatment was ∼1010 bp (Figure 4B), a band size expected if the DNA segments between the two ZFN target sites were deleted from the chromosome. No 1010-bp band in the control groups were observed (Figure 4B), indicating that deletion occurred only in the presence of ZFN-LTR.
Figure 4.
ZFN-LTR mediated deletions of HIV-1 proviral DNA in latently infected cell clone C11. (A) Schematic representation of ZFN-LTR-mediated genome deletions. The red lines in the 5′- and 3′-LTR of HIV-1 genome indicate ZFN-LTR target sites. F-HG and R-HG are PCR primers (arrows) located at the two ends of the integrated HIV genome, which were used for the detection of genome deletion events. (B) PCR products validating ZFN-LTR-induced HIV-1 proviral deletions. Jurkat T cells were infected with the HIV-1 pNL4-3-EGFP virus; originally, EGFP-negative cells, which can be reactivated by TSA stimulation, were then sorted by FACS and identified as HIV-1 latently infected cells. The latently infected clone C11 cells were untransfected (mock) or nucleofected with 2.5 μg of ZFN empty vector or ZFN-LTR plasmid. Genomic DNA was extracted 3 days post-transfection. PCR was performed with the F-HG and R-HG primers. (C) DNA sequences of PCR products. The PCR products were cloned and sequenced. ZFN-LTR target sites are shown in boldface letters. Dashes indicate deleted bases relative to the wild-type sequence (WT). Microhomologies are underlined, and inserted bases are shown in italics. In cases where a deletion sequence was detected more than once, the number of instances is given on the right in parentheses. (D) Quantification of integrated provirus. At 3 days post-transfection, genomic DNA from infected cells untransfected (mock) or nucleofected with 2.5 μg of ZFN empty vector or ZFN-LTR plasmid was isolated and subjected to quantitative real-time PCR using gene-specific primers for HIV-1 gag and human β-globin. The relative copy numbers of gag were calculated based on the standard curve obtained by serial dilution (10–160 ng) of an infected cell DNA on the same plate. Normalization was carried out by division of gag gene amplicons in mock group. Data are representative of three independent experiments, and error bars represent SD. *P < 0.05, **P < 0.01, ***P < 0.001; paired t-test.
To ensure that the 1010-bp band was produced by rejoining DNA at the endonuclease cut sites, we cloned the PCR product and performed sequence analysis. Indeed, the ZFN half-site at the 5′-LTR locus is directly linked to the ZFN half-site at 3′-LTR locus, indicating that the integrated HIV 5′- and 3′-LTR sites had been joined, and the intervening 9.8 kb DNA segment had been deleted (Supplementary Figure S5). The breakpoint junction sequences showed small deletions and insertions (Figure 4C). These mutagenic patterns are characteristic of DSB DNA repair via NHEJ. In addition, microhomologies (shown as underlined letters in Figure 4C) of three bases often were observed at the junctions, which also strongly supports that the DSB repair was mediated by the NHEJ system.
To analyze the frequency of ZFN-LTR-mediated HIV-1 proviral DNA deletions in latently HIV-infected cells, we also assessed the gag gene copy numbers in the genome from cells treated with ZFN-LTR or control group by quantitative real-time PCR. The results showed significant decrease in latently infected cells transfected with ZFN-LTR relative to the control groups at 3 days post-transfection (Figure 4D). Taken together, these results indicate that ZFN-LTR could also excise HIV-1 proviral DNA and mediate antiretroviral activity in latently infected cells.
Specificity of ZFN-LTR in Jurkat T cells
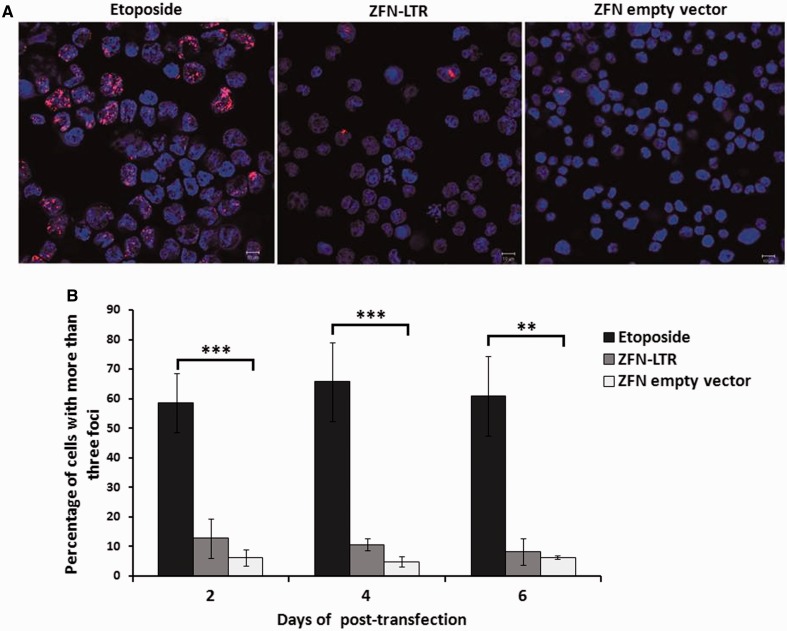
To determine the safety of ZFN-LTR and ensure that there are minimal off-target effects that could lead to catastrophic DSBs in the host genome, we quantified DNA damage levels by analyzing the number of DSBs in ZFN-LTR treated cells using an antibody against p53-binding protein 1 (53BP1), a protein that is recruited to DSBs (13,30). Genomic integrity was enumerated by counting the 53BP1 foci per nucleus at several time points after transduction with ZFN-LTR-expressing plasmids by confocal microscopy (Figure 5A). Etopside, a topoisomerase inhibitor that causes widespread DNA breaks leading to cell cytotoxicity, was used as a positive control. Etoposide-treated cells exhibited an ∼10-fold increase in 53BP1 staining compared with ZFN-empty-vector-transduced cells that persisted for at least 6 days (P < 0.01), with an average of ∼60% of the etopside-treated cells containing more than three foci (Figure 5B). In contrast, we observed that only ∼10% of ZFN-LTR-transduced cells exhibited more than three 53BP1 foci on days 2 and 4. This gradually decreased over time, as the average percentage dropped to near control background levels (∼5–6%) on day 6 (Figure 5B). These data show that ZFN-LTR causes minimal genotoxicity in treated cells and suggest that ZFN-LTR is specific to HIV-1 provirus.
Figure 5.
Genotoxicity assay of ZFN-LTR based on the formation of nuclear repair foci. (A) Intranuclear 53BP1 immunostaining and confocal microscopy 2 days after Jurkat T cells were treated with 1 μM DNA-damaging agent etoposide (positive control) or nucleofected with 2.5 μg of ZFN-LTR plasmid or ZFN empty vector (negative control). Representative images of cells stained with DAPI (blue) and antibodies to 53BP1 (red) are shown. (B) The relative number of cells containing more than three foci was assessed at the indicated times after Jurkat T cells were treated with 1 μM etoposide or nucleofected with 2.5 μg of ZFN-LTR plasmid or ZFN empty vector. Error bars represent SD, *P < 0.05, **P < 0.01, ***P < 0.001; paired t-test.
To further validate the specificity of ZFN-LTR, we searched for off-target cleavage events within the human genome using the ZFN-Site online tool (31). After entering the left and right half-site sequences, we defined the parameters as follows: two mismatches per half-site with a 5 or 6 bp spacer and allowed for left and right protein homodimerization. No off-target site for ZFN-LTR was found in the human genome.
DISCUSSION
The integrated form of HIV-1, also known as the provirus, is ∼9.8 kb in length. Both ends of the provirus are flanked by a repeated sequence known as the LTRs. The genes of HIV are located in the central region of the proviral DNA and encode at least nine proteins. Once the provirus has been integrated into the host chromosome, termination of the infection process is almost impossible. There are no current therapies that directly target integrated HIV provirus genomes for cleavage. In this study, we demonstrate that ZFN-LTR can specifically and effectively excise the integrated HIV-1 provirus 9.8 kb DNA segments and join together the HIV-1 5′-LTR and 3′-LTR site in several infected and latently infected cell types and HIV-1-infected human primary T cells using ZFN-mediated targeted deletion strategies, indicating ZFN-LTR mediated antiretroviral activity.
The ZFN-LTR-mediated deletion of integrated HIV-1 provirus strategies may have important advantages. First, ZFN can be adapted to recognize many different DNA sequences via mutation of the key surface residues within the recognition α-helix. Moreover, multiple fingers may be linked in tandem to recognize a correspondingly extended DNA target. These features permit the design and engineering of ZFN with high affinity and specificity for a given DNA sequence. Second, ZFN can be targeted specifically against any gene in HIV genome, especially the target site conserved across different HIV-1 strains. Although HIV-1 does have a high mutation rate, certain sequences in the viral genome must be conserved for proper replication of the virus, and targeting these sequences for the development of new therapies is the goal of many researchers. There are some regions of the 5′-LTR that show remarkably high sequence conservation across all clades, probably owing to their role in the viral life cycle. In this study, selected ZFN-LTR-binding sequence is extremely well conserved, perhaps owing to its location to the highly conserved trans-activation-responsive region. Conservation analysis results show that our elaborately chosen ZFN-LTR target site is more conserved than the LoxLTR sequence recognized by Tre recombinase among different isolates and subtypes, indicating a higher usefulness of ZFN-LTR than Tre recombinase.
Third, the excision efficiency of HIV-1 proviral DNA mediated by ZFN-LTR is relatively high. In our study, our results showed that ZFN-LTR was able to excise the integrated HIV-1 provirus 9.8 kb DNA segments and join together the HIV-1 5′-LTR and 3′-LTR site in HIV-infected and latently infected cell lines and viral infected human primary T cells. This result is consistent with a number of studies reporting that large genomic deletions in human cells mediated by ZFNs targeting two different sites (11,32). We observed that the frequency of excision was 45.9% in infected human cell lines after treatment with ZFN-LTR, which is closed to the decrease in gag gene copy number in infected human cell lines (Figure 1D). These results are similar to those of other studies in which the CCR5 disruption by ZFN is found in ∼50% of the transduced model cell lines and primary human CD4+ T cells (13). Lee et al. (11) have demonstrated that the interjacent sequence of 15 kb between CCR5 and CCR2 was deleted in up to 10% of cells. Though these results are not comparable across different cell lines subjected to various delivery strategies, the excision efficiency of ZFN-LTR for HIV-1 proviral DNA is relatively high.
Finally, nuclease specificity is an essential part of the successful biotechnological application of ZFN. An off-target cleavage site may confound the interpretation of the intended genome-editing event or, worse, lead to an adverse event. Based on the consideration of safety issue, we took into account of ZFN specificity initially in the target site selection and ZFN design. The selected target site of ZFN-LTR was so carefully chosen, that even using the most stringent definitions available for the ZFN-Site online bioinformatics tool (31), we did not find any homologous or similar sequence of the target site in the human genome. On the other hand, we obtained a commercially designed pair of ZFNs: one containing six zinc fingers and the other containing five zinc fingers that we determined to specifically recognize the highly conserved ZFN-LTR target site. A combination of six fingers recognizes a DNA sequence of 18 nt that have a probability of occurring after 418 nt, much more than the human genome size (3000 MB). In addation, we used the heterodimer variants of the FokI cleavage domain in ZFN-LTR, which significantly reduce the homodimer-associated off-target events (33,34). To address this concern, we detected the formation of nuclear repair foci induced by genome-wide DSBs via intranuclear staining of 53BP1 (13,30). We observed only a transient, approximately 1–1.5-fold increase in cells containing more than three intranuclear 53BP1 foci in ZFN-LTR-transduced Jurkat T cells compared with ZFN empty-vector-transduced cells on days 2 and 4 of culture. In contrast, etoposide-treated positive control cells had ∼10-fold increase in 53BP1 staining over control cells that persisted for at least a week. Perez et al. also found similar results, where a transient 1.4–1.6-fold increase was observed in the mean number of intranuclear 53BP1 foci in ZFN-224 transduced cells as compared with untransduced or GFP-transduced CD4+ T cells on days 2 and 3 of culture (14). Collectively, these data show that ZFN-LTR can target and cleave integrated HIV-1 provirus DNA with high selectivity and minimal genotoxicity in treated cells. We propose that this minimal toxicity can be further reduced in the following ways: by the lowest ZFN-LTR concentrations necessary to cleave the target sequence to the desired extent, adjusting the DNA-binding specificity of the zinc-finger, or modifying the linker either between the zinc-finger and FokI or between individual fingers (35–40).
ZFN-mediated deletion of integrated HIV-1 provirus technology could be further researched in an HIV-1-infected organism. Although HIV transgenic mice or humanized mice provide a possibility for the monitoring of ZFN-LTR antiretroviral activity in a small animal system, targeting ZFNs to the relevant cells comprising the HIV is a major issue and will require significant improvements on our existing delivery systems (41).
In summary, our findings demonstrate that a single ZFN-LTR pair can specifically and effectively cleave integrated full-length HIV-1 proviral DNA and mediate antiretroviral activity in infected and latently infected cells in vitro; the strategy that target and excise integrated HIV-1 like the one described here will be a useful basis for the development of HIV gene therapy in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–5.
FUNDING
National Natural Science Funding of China (NSFC) [31271418 and 31171247 to H.Z.]. Funding for open access charge: NSFC [31271418].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Prof. Jianqing Xu (Shanghai Medical College of Fudan University) for pNL4-3-EGFP plasmid.
REFERENCES
- 1.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 2.Nisole S, Saïb A. Early steps of retrovirus replicative cycle. Retrovirology. 2004;1:9. doi: 10.1186/1742-4690-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl Acad. Sci. USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 5.Trono D, Van Lint C, Rouzioux C, Verdin E, Barré-Sinoussi F, Chun TW, Chomont N. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science. 2010;329:174–180. doi: 10.1126/science.1191047. [DOI] [PubMed] [Google Scholar]
- 6.Sarkar I, Hauber I, Hauber J, Buchholz F. HIV-1 proviral DNA excision using an evolved recombinase. Science. 2007;316:1912–1915. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 7.Aubert M, Ryu BY, Banks L, Rawlings DJ, Scharenberg AM, Jerome KR. Successful targeting and disruption of an integrated reporter lentivirus using the engineered homing endonuclease Y2 I-AniI. PLoS One. 2011;6:e16825. doi: 10.1371/journal.pone.0016825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelman A. AIDS/HIV: a reversal of fortune in HIV-1 integration. Science. 2007;316:1855–1857. doi: 10.1126/science.1145015. [DOI] [PubMed] [Google Scholar]
- 9.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 10.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 11.Lee HJ, Kim E, Kim JS. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20:81–89. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 13.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, Malani N, Anguela XM, Sharma R, Ivanciu L. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang D, Yang H, Li W, Zhao B, Ouyang Z, Liu Z, Zhao Y, Fan N, Song J, Tian J. Generation of PPARγ mono-allelic knockout pigs via zinc-finger nucleases and nuclear transfer cloning. Cell Res. 2011;21:979–982. doi: 10.1038/cr.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui X, Ji D, Fisher DA, Wu Y, Briner DM, Weinstein EJ. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat. Biotechnol. 2010;29:64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- 20.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ochiai H, Sakamoto N, Fujita K, Nishikawa M, Suzuki K, Matsuura S, Miyamoto T, Sakuma T, Shibata T, Yamamoto T. Zinc-finger nuclease-mediated targeted insertion of reporter genes for quantitative imaging of gene expression in sea urchin embryos. Proc. Natl Acad. Sci. USA. 2012;109:10915–10920. doi: 10.1073/pnas.1202768109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merlin C, Beaver LE, Taylor OR, Wolfe SA, Reppert SM. Efficient targeted mutagenesis in the monarch butterfly using zinc-finger nucleases. Genome Res. 2013;23:159–168. doi: 10.1101/gr.145599.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segal DJ, Gonçalves J, Eberhardy S, Swan CH, Torbett BE, Li X, Barbas CF., III Attenuation of HIV-1 replication in primary human cells with a designed zinc finger transcription factor. J. Biol. Chem. 2004;279:14509–14519. doi: 10.1074/jbc.M400349200. [DOI] [PubMed] [Google Scholar]
- 24.Eberhardy SR, Goncalves J, Coelho S, Segal DJ, Berkhout B, Barbas CF., III Inhibition of human immunodeficiency virus type 1 replication with artificial transcription factors targeting the highly conserved primer-binding site. J. Virol. 2006;80:2873–2883. doi: 10.1128/JVI.80.6.2873-2883.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakkhachornphop S, Jiranusornkul S, Kodchakorn K, Nangola S, Sirisanthana T, Tayapiwatana C. Designed zinc finger protein interacting with the HIV-1 integrase recognition sequence at 2-LTR-circle junctions. Protein Sci. 2009;18:2219–2230. doi: 10.1002/pro.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakkhachornphop S, Barbas CF, III, Keawvichit R, Wongworapat K, Tayapiwatana C. Zinc finger protein designed to target 2-long terminal repeat junctions interferes with human immunodeficiency virus integration. Hum. Gene Ther. 2012;23:932–942. doi: 10.1089/hum.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding D, Qu X, Li L, Zhou X, Liu S, Lin S, Wang P, Liu S, Kong C, Wang X, et al. Involvement of histone methyltransferase GLP in HIV-1 latency through catalysis of H3K9 dimethylation. Virology. 2013;440:182–189. doi: 10.1016/j.virol.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 30.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cradick TJ, Ambrosini G, Iseli C, Bucher P, McCaffrey AP. ZFN-Site searches genomes for zinc finger nuclease target sites and off-target sites. BMC Bioinformatics. 2011;12:152. doi: 10.1186/1471-2105-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen F, Pruett-Miller SM, Huang Y, Gjoka M, Duda K, Taunton J, Collingwood TN, Frodin M, Davis GD. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat. Methods. 2010;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 34.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 35.Szczepek M, Brondani V, Büchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 36.Gupta A, Christensen RG, Rayla AL, Lakshmanan A, Stormo GD, Wolfe SA. An optimized two-finger archive for ZFN-mediated gene targeting. Nat. Methods. 2012;9:588–590. doi: 10.1038/nmeth.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cornu TI, Thibodeau-Beganny S, Guhl E, Alwin S, Eichtinger M, Joung JK, Cathomen T. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol. Ther. 2007;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 38.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schierling B, Dannemann N, Gabsalilow L, Wende W, Cathomen T, Pingoud A. A novel zinc-finger nuclease platform with a sequence-specific cleavage module. Nucleic Acids Res. 2012;40:2623–2638. doi: 10.1093/nar/gkr1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhakta MS, Henry IM, Ousterout DG, Das KT, Lockwood SH, Meckler JF, Wallen MC, Zykovich A, Yu Y, Leo H. Highly active zinc-finger nucleases by extended modular assembly. Genome Res. 2013;23:530–538. doi: 10.1101/gr.143693.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF., 3rd Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat. Method. 2012;9:805–807. doi: 10.1038/nmeth.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.