Abstract
Discovery of the TET/JBP family of dioxygenases that modify bases in DNA has sparked considerable interest in novel DNA base modifications and their biological roles. Using sensitive sequence and structure analyses combined with contextual information from comparative genomics, we computationally characterize over 12 novel biochemical systems for DNA modifications. We predict previously unidentified enzymes, such as the kinetoplastid J-base generating glycosyltransferase (and its homolog GREB1), the catalytic specificity of bacteriophage TET/JBP proteins and their role in complex DNA base modifications. We also predict the enzymes involved in synthesis of hypermodified bases such as alpha-glutamylthymine and alpha-putrescinylthymine that have remained enigmatic for several decades. Moreover, the current analysis suggests that bacteriophages and certain nucleo-cytoplasmic large DNA viruses contain an unexpectedly diverse range of DNA modification systems, in addition to those using previously characterized enzymes such as Dam, Dcm, TET/JBP, pyrimidine hydroxymethylases, Mom and glycosyltransferases. These include enzymes generating modified bases such as deazaguanines related to queuine and archaeosine, pyrimidines comparable with lysidine, those derived using modified S-adenosyl methionine derivatives and those using TET/JBP-generated hydroxymethyl pyrimidines as biosynthetic starting points. We present evidence that some of these modification systems are also widely dispersed across prokaryotes and certain eukaryotes such as basidiomycetes, chlorophyte and stramenopile alga, where they could serve as novel epigenetic marks for regulation or discrimination of self from non-self DNA. Our study extends the role of the PUA-like fold domains in recognition of modified nucleic acids and predicts versions of the ASCH and EVE domains to be novel ‘readers’ of modified bases in DNA. These results open opportunities for the investigation of the biology of these systems and their use in biotechnology.
INTRODUCTION
Diverse modifications of bases in DNA have been identified in viruses and cellular organisms across the three superkingdoms of life (1,2). The best known modifications are methylations of cytosine at the C5 or N4 position and adenine at the N6 position, catalyzed by 5-cytosine (5C), N4-cytosine (N4C) and N6-Adenine (N6A)-methyltransferases, respectively (3). One of their key roles is as epigenetic marks, which are important in DNA replication, repair, recombination, chromatin organization and hypermutation (3–8). A remarkable panoply of hypermodified bases have been observed in bacteriophages including 5-hydroxymethylpyrimidines and their glycosylated derivatives, α-putrescinylated and α-glutamylated thymines, sugar-substituted 5-hydroxypentyl uracil, N6-Mom-modified adenine and 7-methylguanine (2,4). In contrast to the base methylations that are catalyzed in situ in DNA, in several phages such as T4, the hydroxymethyl deoxypyrimidines are produced from free nucleotides by phage-encoded deoxypyrimidine hydroxymethylases before incorporation into DNA (5,6). On incorporation into DNA, these hydroxymethylpyrimidines are often further modified in situ by phage-encoded enzymes such as glycosyltransferases (Figure 1) (2,7) or those that generate α-putrescinylated and α-glutamylated thymines (8,9). Another in situ DNA modification known from phages and also predicted to occur in bacterial mobile elements is the acyl modification of the N6 position of adenine (Momylation) catalyzed by a member of the GCN5-like acetyltransferase superfamily (10–12). Most of these diverse modifications in phage DNA are implicated in evasion of host restriction enzymes (2) in addition to replication and packaging of phage DNA (2,13). In situ deamination of cytosine, catalyzed by the AID/APOBEC family of deaminases, is another DNA modification involved in generating variability in antigen receptors of vertebrates and as a defense mechanism against retroviruses (14). Related deaminases found in secreted prokaryotic toxins are predicted to modify DNA as part of their toxicity (15–17).
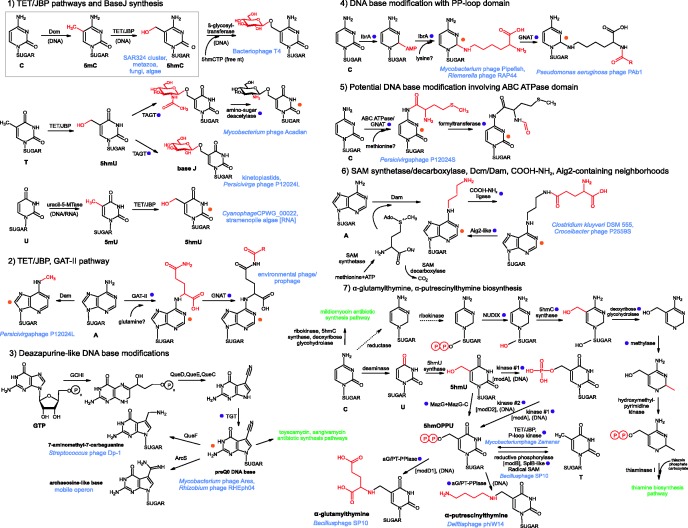
Figure 1.
Pathways are numbered in the order of discussion in the ‘Results and Discussion’ section. Names of enzymes are given above reaction arrows. Location of the reaction in DNA or free bases is indicated in parentheses below reaction arrows. Chemical groups added during a step are highlighted in red in the ensuing product. Green labels indicate entry points for pathway intermediates and products into non-base modification pathways. The orange dot indicates a compound and the purple dot a DNA-modification enzyme predicted here for the first time.
The explosion of genome sequence data, development of sensitive protein sequence and structure analysis methods and contextual analysis tools have greatly aided the computational discovery of novel DNA-modifying enzymes, leading to a renewed interest in their biochemistry and biology. One such discovery was that of the DNA-modifying hydroxylases of the TET/JBP family. These are 2-oxoglutarate-Fe+2-dependent dioxygenases (2OGFeDO) with a double-stranded β-helix fold catalytic domain that hydroxylate pyrimidines using Fe2+ and oxoglutarate as co-factors (12,18). Kinetoplastid representatives of this family, JBP1 and JBP2, hydroxylate thymine in DNA, which is further glucosylated by an unknown glycosyltransferase to yield the hypermodified base, β-D-glucopyranosyloxymethyluracil or base J (2,19,20) (Figure 1), an important regulatory epigenetic mark in these organisms (21,22). Metazoan TET proteins serially oxidize 5-methylcytosine to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine and 5-carboxycytosine (12,18,23,24), which either function as stable epigenetic marks or as a potential intermediate in the DNA demethylation pathway (25,26). In our earlier study on TET/JBP superfamily, we had reported at least four other TET/JBP subfamilies that were predicted to similarly modify DNA or RNA in other eukaryotes and also certain phages (12). We sought to exploit the increased genomic information available to elucidate poorly understood aspects of the natural history of the TET/JBP family (12), such as the role and catalytic specificity of the viral and bacterial TET/JBP enzymes. We were also interested in identifying the hitherto mysterious DNA glycosylases that act with the JBP enzymes in DNA modification (19). Moreover, enzymes catalyzing several of the complex modifications of DNA in phages still remain unknown. Hence, we developed a computational screen to identify these enzymes using genome sequences of these and related viruses and also discover previously unknown DNA-modification enzymes and biosynthetic pathways.
MATERIALS AND METHODS
Iterative sequence profile searches were performed using the PSI-BLAST (27) and web version of the JACKHMMER (http://hmmer.janelia.org/search/jackhmmer) (28) programs run against the non-redundant (NR) protein database of National Center for Biotechnology Information (NCBI). Multiple sequence alignments were built by the Kalign2 (29) and Muscle (30) programs, followed by manual adjustments on the basis of profile-profile and structural alignments. Similarity-based clustering for both classification and culling of nearly identical sequences was performed using the BLASTCLUST program (ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html). The HHpred program (31) was used for profile–profile comparisons. Structure similarity searches were performed using the DaliLite program (32). Secondary structures were predicted using the JPred program (33). For previously known domains, the Pfam database (34) was used as a guide and augmented by addition of newly detected divergent members. Phylogenetic analysis was conducted using an approximately maximum-likelihood method implemented in the FastTree 2.1 program under default parameters (35). Structural visualization and manipulations were performed using the PyMol (http://www.pymol.org) program. The in-house TASS package comprising Perl scripts was used to automate the analysis.
RESULTS AND DISCUSSION
Identification of novel TET/JBP proteins and reconstruction of their early evolutionary history
To retrieve novel members of the TET/JBP family, we initiated sequence profile and iterative HMM searches of the NR database and the database of microbial metagenomic sequences from environmental samples (env_nr) using previously identified TET/JBP protein sequences and a manually curated sequence alignment as queries (12). Novel proteins retrieved in these searches were further analyzed for their relationship with known subfamilies of TET/JBP proteins using single-linkage clustering with the BLASTCLUST program run using a range of sequence length and score thresholds. These sequences were then aligned with the curated alignment and used for further sequence and phylogenetic analyses (Supplementary Data).
We observed that several of the newly sequenced fungal genomes encode 3 to >50 copies of TET/JBP proteins belonging to the clade associated with transposable elements of basidiomycete fungi, chlorophytes and the three paralogous versions from Physcomitrella patens (12) (LMI, DZ, LA manuscript in preparation). We also recovered a member of this subfamily in the amoebozoan Acanthamoeba (gi: 440804588, Figure 2). One distinct eukaryotic TET/JBP protein that apparently did not fit into any previously identified subfamily is from the chlorophyte alga Coccomyxa subellipsoidea (gi: 384246050, Figure 2). Several novel prokaryotic TET/JBP proteins were recovered from the NR database, most of which were related to the actinophage/prophage gp2 subfamily. However, TET/JBP proteins from Legionella drancourtii, Perscivirga phage P12024L, the SAR324 cluster deltaproteobacterium JCVI-SC AAA005 and several uncultured environmental microbes were distinct from the gp2 family (Figure 2). Of the sequences from the env_nr database, at least 10 distinct TET/JBP proteins encoded by marine metagenomes are neighbors of genes coding for proteins closely related to signature proteins from nucleo-cytoplasmic large DNA viruses (NCLDV) that infect algae and amoebae (36), such as an ortholog of the NCLDV ERV1/2-like flavin-dependent thiol oxidoreductases, involved in maturation of surface viral proteins (37). This indicates the presence of TET family proteins in a subset of uncharacterized marine NCLDVs, where they could possibly catalyze synthesis of a 5hmC or hmU mark that could play an epigenetic role or help evade host defenses targeting viral DNA. Precedence for such modifications come from the earlier report of DNA methylases encoded by alga-parasitic NCLDVs, such as the Paramecium bursaria Chlorella Virus, that protect their genomes even as they restrict the host genome with associated restriction enzymes (38). Inclusion of the newly available sequences in the multiple sequence alignment also confirmed that the region unique to the metazoan TETs with nine conserved cysteines and one histidine in part constitutes a metal-chelating insert domain within the 2OGFeDO fold occurring N-terminal to the helix preceding the DSBH unit (Supplementary Data) (39).
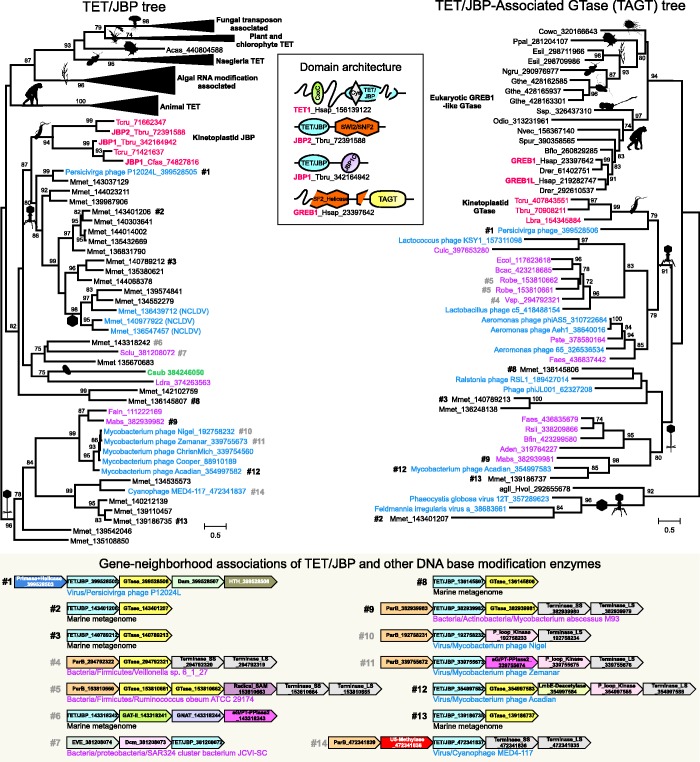
Figure 2.
Previously described and well-supported eukaryotic clades of TET/JBP proteins are collapsed. Nodes supported by bootstrap >75% are shown. Relevant domain architectures and operons of TET/JBP and TAGT families are shown between or below the trees. Proteins are either denoted by a species abbreviation (eukaryotes and bacteria) or by the complete name (phages/viruses) followed by GenBank GIs. Branch coloring: Phage sequences- blue, bacteria- pink, kinetoplastid- red and C. subellipsoidea- green. See Supplementary Data for species abbreviations.
A maximum-likelihood phylogenetic tree showed that the originally defined kinetoplastid JBP family now expands into a larger clade, where it groups specifically with the Persicivirga phage P12024L TET/JBP protein (gi: 399528505, #1 in Figure 2) and its homologs from environmental metagenomes. The precise higher-order relationship of the animal TET clade with remaining clades could not be established owing to rapid divergence concomitant with the acquisition of the large cysteine-rich insert. Nevertheless, this phylogenetic tree supports the idea that the structurally simple (i.e. lacking any major inserts or elaborations) bacteriophage/bacterial versions represent the ancestral versions that were transferred on at least three independent occasions to eukaryotes (Figure 2). The deep nesting of kinetoplastid JBP proteins and the C. subellipsoidea TET/JBP protein within clades with bacteriophage and bacterial homologs along with their relatively limited phyletic patterns are indicative of them being late transfers to eukaryotes. In this regard, identification of a TET/JBP protein in the strictly intracellular Acanthamoeba parasite, L. drancourtii (40), suggests that such intracellular bacterial parasites/symbionts could have served as conduits for the repeated transfer of the TET/JBP genes to eukaryotes (Figure 2). Based on its architecture, we propose that it could potentially function as a host-directed effector and could represent a parallel to the earlier identified histone methylase H3K79 methylase Dot1 homolog, which is a potential effector deployed by several Legionellae (41). Remarkably, on each independent occasion, transfer from prokaryotes has resulted in the TET/JBP protein being apparently recruited as a generator of epigenetic marks in the recipient eukaryotes. This is suggested by their fusion to distinct domains characteristic of eukaryotic chromatin proteins, such as the SWI2/SNF2 ATPase module and JBP1C in kinetoplastids (12,19), the methylated H3K4-binding PHDX/Zf-CW domain in C. subellipsoidea and the hemi-modified or unmodified CpG-binding CXXC domain in animals (Figure 2) (25).
Other DNA-modifying enzymes associated with prokaryotic TET/JBP proteins
Genes present in conserved gene neighborhoods, or domains present in conserved domain architectures, are likely to function in a common pathway or participate in the same biochemical complex, thereby enabling functional predictions for uncharacterized proteins and domains (42,43). With the prokaryotic TET/JBP proteins as the starting nodes, we systematically investigated the biochemical implications of their entire network of gene neighborhood and domain associations.
Genomic linkage of a novel GT-A/fringe-like glycosyltransferase to TET/JBP genes in bacteriophages
The prokaryotic TET/JBP genes from the Perscivirga phage P12024L and certain mycobacteriophages (e.g. Mycobacterium tuberculosis phage Acadian), a prophage in Mycobacterium abscessus and phages from environmental samples were found to show an operonic association with a gene coding for a conserved protein prototyped by phage P12024L B618_gp27 (gi: 399528506, #1 in Figure 2). PSI-BLAST searches initiated with B618_gp27 retrieved prophage, phage and kinetoplastid proteins with significant expectation (e)-values (e < 10−5) in the first two iterations. Further iterations recovered significant hits to the C-terminal region of eukaryotic GREB1-like proteins (Homo sapiens GREB1, iteration 2, e = 10−7), the DNA β-glucosyl-hmC-α-glucosyltransferase (iteration 4, e ∼ 10−31) from bacteriophages T2 and T6 and the Haloferax AglI hexuronic acid transferase involved in protein asparagine (N)-glycosylation (44) (iteration 5, 7 × 10−20) and the so-called family-2 glycosyltransferases (PFAM: PF00535). Profile-profile searches using HHPRED with the phage P12024L B618_gp27 protein as query against a panel of HMMs derived using PDB structures as search seeds, significantly retrieved GT-A/fringe-like glycosyltransferases such as the Mycobacterial UDP-galactofuranosyl transferase GLFT2 (PDB: 4fixA, probability 98.98, P ∼ 10−10) and chondroitin synthase (PDB: 2z87A; probability 98.66, P ∼ 10−08), among several others. These results confirm that B618_gp27 and related proteins belong to the GT-A/fringe-like glycosyltransferase superfamily (45). Owing to their operonic association with the TET/JBP encoding genes (#1, 2, 3, 8, 9, 12, 13 in Figure 2), we termed these the TET/JBP-associated glycosyltrasferases (TAGTs; Figures 2 and 3A).
Figure 3.
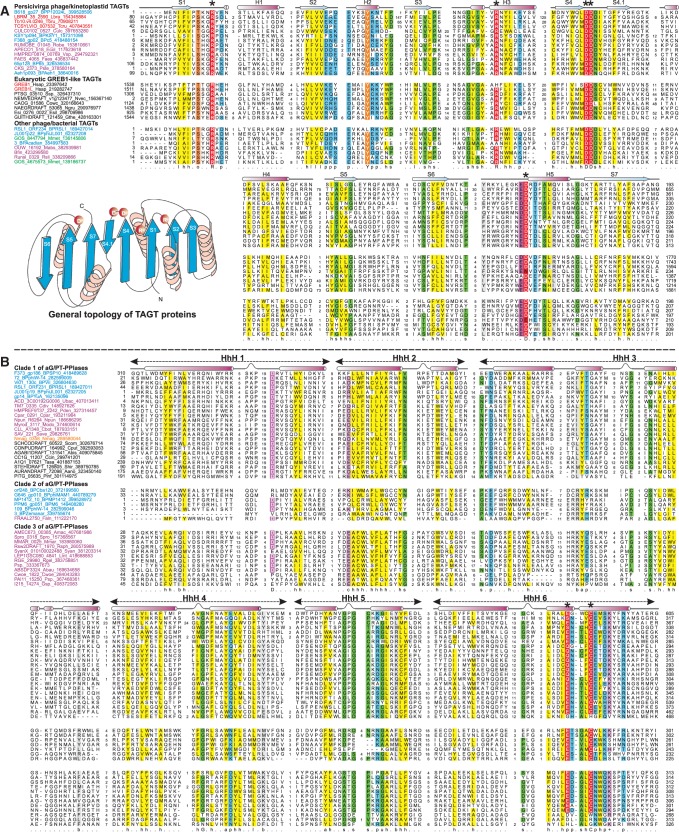
Multiple sequence alignment of the DNA base glycosyltransferases (A) and the aG/P-T-pyrophosphorylases (B). Protein sequences are labeled by gene names followed by species abbreviation and Genbank GIs. Phage protein names are colored blue, bacterial ones in pink, archaea in orange and eukaryotes in black. Predicted catalytic residues for both TAGT and aG/PT-PPlase are indicated by asterisks, with secondary structure assignments shown above the alignment. Alignment columns are colored based on the 80% conservation consensus. Topology of the glycosyltransferase domain is adjacent to the alignment. See Supplementary Data for species abbreviations.
A phylogenetic analysis based on a multiple sequence alignment of the TAGTs showed that the kinetoplastid homologs (e.g. Trypanosoma brucei protein Tb10.v4.0246) specifically group with Perscivirga phage B618_gp27 glycosyltransferase in a clade comprised of phage, prophage and prokaryotic glycosyltransferases (Figure 2). The remaining eukaryotic homologs cluster together into the GREB1-like clade with a gene-duplication at the base of the jawed vertebrate lineage. TAGTs possess all structural elements and catalytic residues characteristic of the Rossmannoid nucleotide-diphospho-sugar binding fold typical of the GT-A/fringe superfamily (Figure 3A): an arginine between strand-1 and helix-1 contacting the diphosphate backbone of the nucleotidyl-sugar diphosphate substrate, a conserved aspartate (usually present as a DD motif) at the end of strand-4 coordinating a divalent cation critical for catalysis and a conserved acidic residue (usually D) at the beginning of helix-5 functioning as the proton-accepting catalytic base (Figure 3A) (45,46).
The operonic association with TET/JBP enzymes suggests that the TAGTs could glycosylate in situ in DNA hydroxylated methylpyrimidines generated by the former enzymes. In some phages (e.g. mycobacteriophage Acadian), the TAGT gene is followed by an additional gene for an amino-sugar deacetylase, which could possibly act on an acetylated amino-sugar that is transferred by the TAGT (Figure 1 and #12 in Figure 2). Genome-wide analysis revealed that phages encoding the TET/JBP-TAGT pair lack other hydroxypyrimidine biosynthesis enzymes, such as those of the thymidylate synthase superfamily implying that the TET/JBP enzyme are the sole generators of hydroxypyrimidine in these phages. In contrast, nearly all other glycosyltransferase-containing phages lacking the TET/JBP enzyme have a gene for a hydroxypyrimidine synthase. Neither phages encoding both TET/JBP and TAGT nor those that contain only the TET/JBP gene possess DNA 5-C methyltransferases, a pre-requisite for 5hmC generation by the TET/JBP enzymes. These observations suggest that the nucleotide substrate hydroxylated by the TET/JBP enzymes of phages is likely to be a thymine. This proposal is also consistent with the close relationship between the thymine-modifying kinetoplastid JBP proteins to a subset of the phage enzymes predicted to modify this base. In certain phages such as the cyanophage MED4-117, the TET/JBP gene is linked to a uracil-5-methyltransferase (Figure 2, #14). Here, the TET/JBP gene might generate 5hmU from T generated by the methyltransferase from U, which is incorporated in place of thymine (Figure 1) (2). Similar uracil methyltransferases are fused to the earlier-reported stramenopile RNA-modifying TET/JBP proteins, which together might convert uracil in RNA to 5hmU (12). In contrast, in the SAR324 cluster bacterium (Figure 2, #7) and other uncultured marine bacteria, the TET/JBP gene is juxtaposed in a predicted operon with a DNA-5C methyltransferase; hence, these versions might oxidize 5-methylcytosine to generate its oxidized derivatives (Figures 1 and 2).
Prediction of the glycosyltransferase involved in Base J synthesis and other eukaryotic TAGT homologs
The close relationship between TAGTs from kinetoplastids and the Perscivirga phage P12024L in phylogenetic analysis and also their corresponding TET/JBP enzymes suggests that the ancestor of kinetoplastids acquired both these genes simultaneously via lateral transfer from the same phage source (Figure 2). Given these strong associations, we predict that the kinetoplastid TAGT prototyped by T.brucei Tb10.v4.0246 (gi: 70908211) is the elusive glycosyltransferase involved in base J synthesis. The second clade of eukaryotic TAGTs are typified by the human GREB1 and GREB1L proteins, which contain a circularly permuted superfamily II (SFII) helicase module fused to the C-terminal glycosyltransferase domain (Figure 2). Orthologs of this protein are present in chordates and sporadically in metazoans, choanoflagellates, Capsaspora, Naegleria, dictyostellids, cryptophyte red algae and stramenopile algae. In metazoans, based on the conservation pattern, the SFII module is predicted to be catalytic inactive while retaining nucleic-acid binding capability. Human GREB1 is a nuclear protein that is induced on estrogen treatment and has been shown to function as a transcriptional co-activator that binds the estrogen receptor at transcriptionally active cis-elements (47). GREB1 is also implicated in proliferation of estrogen and androgen receptor positive breast and prostate cancer cells, respectively, and as a risk factor for endometriosis (48–51). The C-terminal TAGT domain provides new leads regarding the function of the GREB1-like proteins. One obvious possibility is that like the phage and kinetoplastids TAGTs, they target 5hmC generated by TET/JBP enzymes for further glycosylation. However, at least in mammals, there is no clear evidence in mass-spectroscopic analysis for such a modification of 5hmC being present in large amounts. Furthermore, GREB1-like proteins are present in dictyostellids and cryptophyte red algae that apparently lack the apparatus to synthesize hydroxymethylpyrimidines. Hence, GREB1-like enzymes might alternatively have undergone a functional shift to operate as protein glycosyltransferases targeting chromatin proteins similar to the Ogt glycosyltransferase (52,53).
TET/JBP genes are associated with genes for enzymes catalyzing distinct modifications of other DNA bases
In the Perscivirga phage P12024L, the TET/JBP-TAGT operon contains a third gene coding for a DNA N6A-methylase (Dam; #1 in Figure 2). Another distinct operonic association combines the TET/JBP genes with genes coding for a class-II glutamine amidotransferase protein (GAT-II) of the NTN-hydrolase fold and a GCN5-like acetyltransferase (GNAT) superfamily (Figure 2; #6). The GAT-II-like proteins are related to versions found in phage operons previously implicated in the non-ribosomal biosynthesis of peptides and are predicted to function as peptidases or glutamine transamidases (54). GNAT superfamily proteins typically transfer an acyl chain to an amino group to form amide linkages (55). Based on reactions catalyzed by such enzymes, we speculate that the GAT-II enzyme could link a glutamate to a base with an amino group (A, G or C) using glutamine as a substrate for a transamidation reaction followed by acylation of the amino group of the linked glutamate by the GNAT (Figure 1). Indeed, such hypermodified bases that are formed via the amino group of adenine are known in phages such as Mu (Momylation, see later in the text) (10). Thus, the aforementioned genes linked to the TET/JBP gene specify modification of a second base distinct from the hydroxylation catalyzed by the TET/JBP.
Genomic diversity at the ParB-Terminase large subunit (Tls) locus of bacteriophages helps identify several novel DNA base-modifying enzymes
Syntactical logic of the ParB-Tls locus and its predictive value
The so-called cluster B of mycobacteriophages (56), while closely related in overall genomic organization and gene content, showed considerable diversity in the region of genome between the ParB gene, which encodes for a DNA-processing nuclease (57) and the downstream Terminase large subunit (Tls) gene coding for the ATPase required for packaging of the phage head (58). For instance, in several cluster B mycobacteriophages, such as phage Acadian (Figure 2, #12; Supplementary Data), the region between the ParB and Tls genes contains genes for the TET/JBP, TAGT, amino-sugar deacetylase and a distinctive P-loop kinase (see later in the text) (59). In other cluster B mycobacteriophages, no TET/JBP genes were found in their inter-ParB-Tls regions but the locus contained genes for other potential DNA-modification enzymes (Figure 4; see later in the text for details). This observation prompted us to systematically analyze the inter-ParB-Tls region (hereinafter the ParB-Tls locus) of phages and prophages. We found that not only cluster B mycobacteriophages and related actinobacterial phages but also a wide-range of caudoviruses (tailed phages) infecting firmicutes, actinobacteria, spirochetes, bacteroidetes, proteobacteria and haloarchaea showed a comparable organization of the ParB-Tls locus.
Figure 4.
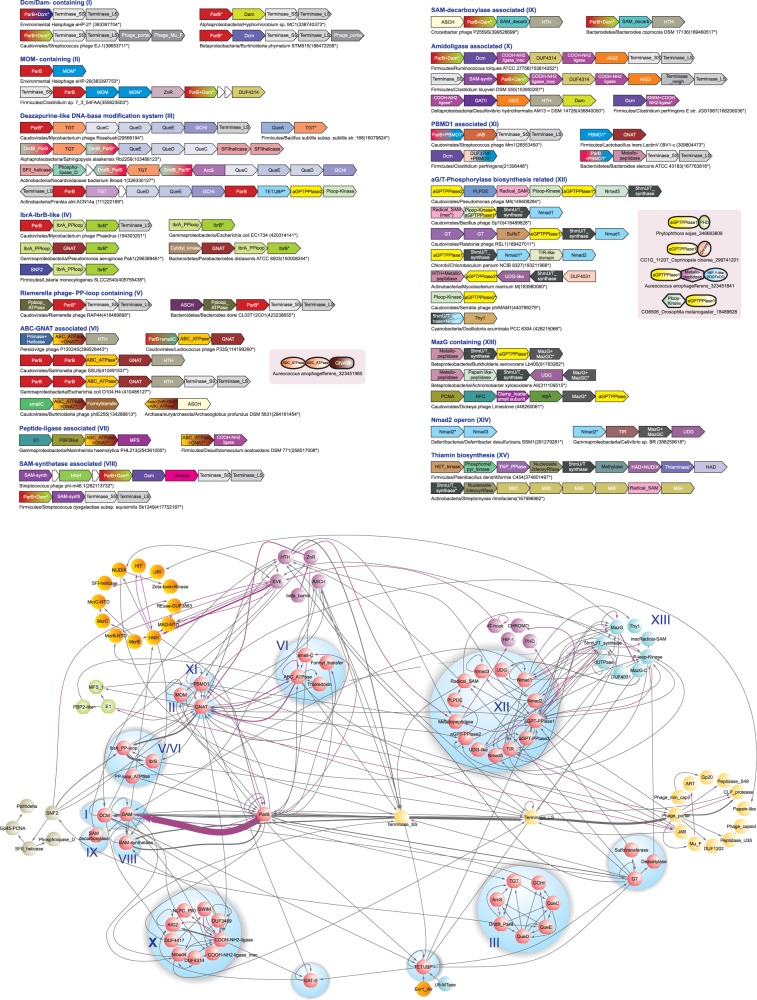
Gene neighborhoods and contextual information network of domains involved in DNA modification. Genes are shown as arrows pointing from the 5′ to the 3′ end. Representative gene-neighborhoods are labeled with taxonomic lineage and species name of origin and an anchor GI number of the gene marked with an asterisk. Operons are grouped by the type of predicted DNA modification they catalyze and also by Roman numerals to match corresponding nodes in the network. Domain architectures are shown as insets. In the network, gray edges connect neighboring genes (5′–>3′), and magenta-colored edges connect neighboring domains (N–>C terminus). Nodes are clustered either by their common function or by the predicted DNA modifications they catalyze. Thickness of edges reflects the frequency of associations between two nodes. The graph was calculated from 1751 gene neighborhoods, including 101 nodes, 377 pairwise connections and 5922 genes.
The most frequently observed genes sandwiched in the ParB-Tls locus of diverse phages are DNA methyltransferases of either the DNA N6A-methylase (Dam) or DNA 5C-methylase (Dcm) families (Dcm/Dam-containing operons in Figure 4). Dams are usually fused C-terminal to the ParB domain, whereas Dcms are present as neighbors of the ParB gene. Presence of two distinct enzymes in the same ParB-Tls locus, such as the Dam and Dcm, suggests that the locus might specify modifications targeting different bases in DNA. Another gene coding for a previously characterized base-modifying enzyme that we recovered in ParB-Tls loci from prophages of firmicutes and γ-proteobacteria and in the halophages eHP-29 and eHP-D7 was Mom (Mom-containing operons in Figure 4). In phages coding for a thymidylate synthase family enzyme for hydroxypyrimidine biosynthesis (2) (e.g. Lactobacillus phage c5), we found that the ParB-Tls locus contained a glycosyltransferase gene that is likely to glycosylate the hydroxypyrimidine generated by the former enzyme (Figures 1 and 3). Presence of genes coding for at least five distinct DNA-modifying enzymes, Dam, Dcm, Mom, TET/JBP and glycosyltransferases, sandwiched in the ParB-Tls locus of a wide range phages and prophages indicated that this locus is a privileged site for embedding genes for DNA-modifying enzymes. Hence, we reasoned that systematic analysis of genes in the ParB-Tls locus could be used to identify genes for DNA-modification enzymes and predict new modifications by combining the contextual network with case-by-case sequence profile analysis.
A deazapurine-like DNA base modification
In certain cluster B mycobacteriophages, such as Rosebush, AnnaL29, Hedgerow and Ares, as well as phages/prophages from actinobacteria and bacteriodetes, the ParB-Tls locus sandwiches a conserved cluster of five genes (Figure 4). Sequence profile searches with the encoded proteins established them to be related to those involved in biosynthesis of hypermodified deazapurines such as queuine found in the wobble position of asparaginyl, tyrosyl, histidyl and aspartyl tRNA, archaeosine found in the D-loop of several archaeal tRNAs, and antibiotics such as sangivamycin and toyocamycin (60). These genes maintain a strict order between the ParB gene at the 5′ end and the Tls gene at the 3′ end (Figure 4): (i) a previously unrecognized divergent member of the tRNA:guanine transglycosylase family (TGT; Supplementary data), (ii) QueC-like PP-loop ATPase, (iii) QueD-like 6-pyruvoyl-tetrahydropterin synthase, (iv) QueE-like radical S-adenosyl methionine (SAM) superfamily protein and (v) GTP cyclohydrolase-I (GCHI). A previous study of these genes in mycobacteriophages proposed that they are involved in the biosynthesis of the hypermodified base queuosine in tRNA, given that the mycobacteria lack this modification (56). However, systematic analysis revealed several contradictions to this hypothesis: (i) the biochemical pathway reconstructed based on these mycobacteriophages ParB-Tls loci (60) can only synthesize 7-cyano-7-deazaguanine (PreQ0) from a GTP precursor (Figure 1). Queuosine synthesis requires the further action of the reductase QueF and the SAM:tRNA ribosyltransferase-isomerase QueA (60), both of which are absent in these loci and also in mycobacterial genomes (56). Genes for QueA and TGT are typically tightly linked in genomes as the action of QueA is completed only after the TGT incorporates PreQ0 into tRNA by exchanging it for guanine. Absence of QueA and QueF in the ParB-Tls locus (Figure 4, Supplementary Material) suggests that the products of this locus are unlikely to synthesize queuosine seen in bacterial tRNAs but only PreQ0 similar to the one seen in toyocamycin (60). (ii) Although mycobacteria lack genes involved in queuosine modification, all bacteroidetes genomes infected by phages with a related deazapurine biosynthesis ParB-Tls locus, also possess a canonical apparatus for tRNA-queuosine biosynthesis genes, rendering no particular advantage to the phage genes in modifying the tRNA. (iii) The novel transglycosylase that we identified in this study is highly divergent from its cellular counterpart, which acts on tRNA, whereas the PreQ0 biosynthesis genes are not. This suggests that the target for the TGT is likely to be distinct from tRNA. (iv) Finally, in a Frankia alni prophage, the ParB-Tls locus containing deazapurine biosynthesis genes is disrupted, and the gene encoding the Tls is displaced by a second ParB-containing operon involved in modified base synthesis containing genes for a TET/JBP dioxygenase, a P-loop kinase and a pyrophosphorylase (Figure 4; see later in the text). These observations taken together lead to the prediction that PreQ0 synthesized by these gene products is incorporated into phage DNA in place of specific guanines by the action of the linked TGT (Figure 1). Genes for this biosynthetic system are also found in genomes of classes of phages that do not have a ParB-Tls locus, such as Streptococcus phage Dp-1, the Rhizobium phage RHEph04 and related prophages (Figure 4), suggesting that it might be a more widely used DNA modification across phages. Phage Dp-1 additionally codes for a QueF-like reductase, suggesting that here the base is further modified to 7-aminomethyl-7-carbaguanine (Figure 1) (61).
Outside phage genomes, a related mobile operon is found across several proteobacteria, planctomycetes, actinobacteria, cyanobacteria and fusobacteria, most of which possess their regular tRNA queuine biosynthesis genes (Figure 4). These operons code for a divergent TGT (Supplementary Material), a subset or all proteins involved in deazapurine biosynthesis (i.e. GCHI, QueD, QueC and QueE) and, in most instances, a divergent version of the archaeosine synthase (ArcS)-like amidinotransferase (60), which lacks the C-terminal PUA domain characteristic of euryarchaeal homologs (Figure 4) (62). Presence of the ArcS homolog suggests that unlike phage-encoded systems, which terminate at PreQ0, these systems add an amidino group to the deazapurine resulting in a base identical or similar to archaeosine (60) (Figure 1). These operons also code for two ParB proteins of the DndB family (17), which are also found in the mobile Dnd system that replaces the non-bridging oxygen atom in the backbone of DNA with sulfur (DNA phosphorothioation) (63,64). The DndB family of ParB proteins is best characterized in the DNA phosphorothioate modification system (63). DndB regulates efficiency and specificity of DNA modification by the Dnd system by potentially recognizing sequences flanking the site of phosphorothioate modification and controlling access to DNA by the Dnd modification apparatus (64). Embedding of DndB-like ParB genes between genes for deazapurine synthesis in the aforementioned systems parallels the lodging of DndB in Dnd operons amidst genes involved in generation of sulfur for DNA phosphorothioation. In most bacteria, these gene neighborhoods might also code for a RapA-related SWI2/SNF2 ATPase, a RecQ-like SF-II helicase, and a nuclease of the phospholipase D superfamily and components of the DNA-modifying phage-growth-limitation system (Figure 4). This, along with the absence of the RNA-binding PUA domain in the ArcS homolog, supports a role for this system in modifying DNA rather than RNA. We predict that the mobile deazapurine biosynthesis operon functions comparable with Dnd operons, but probably replaces guanines in DNA with an archaeosine-like moiety, with the DndB-like ParB along with the associated helicases and nucleases perhaps directing this complex to specific sequences (63,64).
Predicted DNA-base-modification systems using PP-loop ATPases
In cluster B mycobacteriophages, such as Pipefish, Phaedrus and Daisy and a prophage in M. abscessus, the ParB-Tls locus encloses a conserved pair of genes encoding the IbrA-like pyrophosphate-binding (PP)-loop ATPase and the ParB family IbrB protein. A previous study had shown the IbrAB locus to be required for expression of immunoglobulin-binding proteins encoded by a phage integrated into the Escherichia coli strain ECOR-9 (65), but their functions remain unknown. Outside phage genomes the IbrAB locus is a mobile operon sporadically distributed across most major bacterial clades with the gene order of IbrA followed by IbrB being strongly maintained (Supplementary Data). These non-phage versions often show additional linked genes such as those coding for a SWI2/SNF2 ATPase, a GNAT superfamily acyltransferase or a P-loop kinase (Figure 4). Comparable with IbrA is another distinct PP-loop ATPase gene encoded in the ParB-Tls locus of the Riemerella phage RAP44 and several related prophages from diverse bacterial lineages.
All PP-loop ATPases share a two-step reaction mechanism where the functional group being modified is activated by adenylation with an AMP in the first step. In the second step, the adenylated intermediate is attacked by a nucleophile such as an amine/ammonia or sulfur (66–68). Indeed, PP-loop enzymes catalyze several base modifications of nucleic acids via ligation of lysine or ammonia (tRNAIle lysidine synthetase and PreQ0 synthetase QueC) or insertion of sulfur (thiouridine synthetases, e.g. ThiI) and backbone phosphorothioation of DNA (DndC) (60,63,67,69,70). The sulfur-inserting PP-loop ATPases (e.g. ThiI and DndC) are characterized by associated sulfur-transfer domains (e.g. rhodanese domain) or contain conserved cysteines that participate in sulfur transfer (67,71). Absence of sulfur-transferring accessory domains or conserved cysteines in IbrA and the phage RAP44-like PP-loop ATPases suggests that the nucleophile in the second step of the reaction catalyzed by them is likely to be an amine. Based on these observations, we propose that IbrA and the phage RAP44-like PP-loop ATPase probably function similar to lysidine- or 2-agmatinyl cytosine synthetases, which first adenylate the 2-oxo group of a pyrimidine followed by conjugation of a NH2 group from an amino acid such as lysine or agmatine to the base (69,72) (Figure 1). The presence of a GNAT superfamily acyltransferase in a subset of these systems suggests that the modifying moiety contains an additional free NH2 (e.g. lysine), which might be protected by acylation (Figures 1 and 4). Their predicted role in DNA modification is further supported, at least in the case of the IbrAB systems, by presence of IbrB of the ParB DNase superfamily—which could function analogous to DndB that directs the DNA-modifying PP-loop enzyme DndC from the Dnd system to facilitate access of IbrA to appropriate target DNA sequences (64). This is also reinforced by the encoding of SWI2/SNF2 ATPases, which have a strong DNA preference (73), in several non-phage IbrAB systems.
Novel DNA modification systems using an ABC-ATPase and a GNAT superfamily enzyme
The Persicivirga phage P12024L is closely related to the phage P12024S—they share 47 open reading frames with over 90% amino acid identity and several regions of synteny (74). The TET/JBP-TAGT-Dam locus in phage P12024L described earlier in the text (Figure 2, #1) is flanked by a DNA primase-helicase gene at its 5′ end and a gene encoding a small protein with a helix-turn helix (HTH) domain (P12024L_29, match with TetR HTH domain, PDB: 3on4, HHPRED probability 68.9%, P-value 6.1 × 10−4) at the 3′ end. In phage P12024S, the region between the primase-helicase and HTH genes, which are nearly identical to their phage P12024L counterparts, curiously lacks the TET/JBP-TAGT-Dam operon. Instead, it is replaced by an unrelated gene coding for a protein with an N-terminal ABC superfamily ATPase domain and a C-terminal GNAT superfamily domain (Figure 4). This suggested that this gene product could possibly encode an alternative DNA-modification enzyme that has taken the place of the P12024L TET/JBP-TAGT-Dam modification system in phage P12024S. We found further support for this conjecture through detection of homologous proteins encoded in the ParB-Tls loci of several phages (e.g. Salmonella phage SSU5) and prophages infecting diverse bacteria (e.g. a prophage in E. coli O104:H4; Figure 4, Supplementary Data). Thus, on two independent occasions, these genes appear to have been incorporated in loci that usually encode DNA-modifying enzymes. Systematic analysis of gene neighborhoods containing the ABC ATPase-GNAT gene revealed that in phages/prophages from diverse bacteria (e.g. Burkholderia phage phiE255), it might be linked to two additional genes. The first of these encodes a formyltransferase and the second a small protein with a conserved C-terminal cysteine, which on occasions might also be fused to adjacent ParB genes (Figure 4). In certain proteobacterial phages, such as Salmonella phage SSU5 and prophages of planctomycetes, the ABC ATPase and GNAT domains are encoded by separate closely linked genes (Figure 4). These ParB-Tls loci additionally contain a gene encoding a paralogous ParB domain next to the 5′ most ParB gene and also a gene encoding a HTH domain before the genes coding for the terminase subunits (Figure 4). Interestingly, these ABC ATPase-GNAT proteins appear to have been laterally transferred to two photosynthetic eukaryotic lineages: chlorophyte and stramenopile algae (Supplementary Data). Outside phages, we detected proteins related to the phage/prophage ABC ATPase-GNAT protein in several thermophilic archaea, proteobacteria, bacteroidetes, chloroflexi and deinococci. However, in these proteins, the ABC ATPase is to the C-terminus, and in addition to the N-terminal GNAT domain, they also contain a β-strand rich domain followed by a zinc ribbon (Figure 4).
Presence of a GNAT domain suggests that a base containing an amino group is being modified by an acyl group to result in an amide linkage, perhaps comparable with the Momylation of adenine seen in phage Mu (11,12,75). The ABC domain might function as an ATP-dependent conformational switch that activates the GNAT enzyme, or might reorganize DNA structure to facilitate modification. The formyltransferase encoded by these gene neighborhoods is related but distinct from the methionyl-tRNA transformylase that transfers a formyl group from a folate carrier to the NH2 group of methionine (76). Hence, it is likely that the moiety conjugated by the GNAT contains a free NH2 group that is further formylated (Figures 1 and 4). A subset of the predicted operons with ABC-GNAT genes from firmicutes and gammaproteobacteria has domain architectures similar to the phage versions but are associated with ligases such as those belonging to the glutamine synthetase/COOH-NH2 ligase superfamily or the E1-like superfamily (54,77) (Figure 4). As these E1-like genes lack the thiolating cysteine residue of the classical E1 and ThiF enzymes and show no associated Ub-like proteins, they likely adenylate a carboxyl group to enable its conjugation, potentially to an amino group (77). Additionally, the neighborhood contains genes encoding an MFS family transporter and a protein of the periplasmic-binding protein-2 (PBP2) superfamily (Figure 4). The latter associations suggest that this system is likely to synthesize a modified base or nucleoside-like secondary metabolite that is then exported from the cell (Figure 1). Thus, it could represent a parallel to the toyocamycin/sangivamycin biosynthesis pathways in which paralogs of base-modifying queuosine biosynthesis genes are used to generate an antibiotic (60).
Possible use of S-adenosylmethionine derivatives in synthesis of modified bases
ParB-Tls loci of certain phages (e.g. Streptococcus phages phi-SsUD.1 and phi-m46.1) and prophages of firmicutes, actinobacteria and spirochaetes, in addition to either both Dcm and Dam or only Dam genes, are associated with a gene for the SAM synthetase; canonical versions conjugate a methionine to adenosine (Figure 1 and SAM-synthetase-associated operons in Figure 4). Although SAM is the co-factor for methylases, there is no evidence that SAM is a limiting metabolite for phages during infection (78), and majority of phages with DNA methylation enzymes do not encode associated SAM synthetase genes. Bacterial hosts of phages containing the SAM synthetase code for the canonical SAM synthetase. SAM synthetases from phage/prophage ParB-Tls loci form a distinct monophyletic clade in phylogenetic analyses. These observations raise the possibility that rather than synthesizing SAM, the phage versions generate SAM derivatives or analogs that enable synthesis of distinct modified bases such as S-adenosyl ethionine (79). Alternatively, these enzymes could generate decarboxylated SAM derivatives, such as S-adenosyl methioninamine, which is used in synthesis of spermidine from putrescine and spermine from SAM. In agreement with such a possibility, we found that certain phages (e.g. Croceibacter phage P2559S and prophages from bacteroidetes) contain a gene for SAM decarboxylase, which generates S-adenosyl methioninamine from SAM, adjacent to a ParB-Dam gene (SAM decarboxylase-associated operons in Figure 4). Phage-induced SAM decarboxylase activity has been proposed to generate polyamines for neutralizing the charge of DNA during packaging (80); however, we cannot rule out a role in DNA modification (Figure 1).
Other gene-neighborhood associations from the ParB-Tls loci of phages/prophages from firmicutes, actinobacteria, synergistetes, spirochaetes and chloroflexi also provides circumstantial support for such polyamine modifications dependent on the phage-encoded SAM synthetases. These are distinguished by 1–2 copies of a gene encoding an amidoligase of the COOH-NH2 ligase superfamily (54) and genes for two small uncharacterized proteins, namely, Nucleotide-modification associated domain 4 (Nmad4) and another labeled DUF4314 in the Pfam database (Amidoligase associated operons in Figure 4). They are typically linked to Dam, Dcm and SAM synthetase genes. The DUF4314 family also show fusions to DNA-associated domains, such as the phage antirestriction ArdA domain and restriction-endonuclease domains (Supplementary Data), suggesting that they function as the DNA-binding component of this base-modifying system. Analysis of these neighborhoods showed that in instances where two copies of the genes are present, one of them codes for an active enzyme and the other is inactive. Operons with a single copy of the amidoligase typically possess an inactive copy (Figure 4), whereas those with an active copy also contain a neighboring gene encoding a cyclotransferase Aig2 superfamily protein (54). A parallel for the action of these enzymes is suggested by the biosynthetic pathway for the antibiotic butirosin (81). Here, a free NH2 group is protected by conjugation of a glutamyl moiety. In the terminal stages of this pathway BtrG, an Aig2-like cyclotransferase removes the protecting glutamate via a cyclotransfer reaction that releases it as oxoproline (81). Based on this precedent, we predict that the amidoligase is likely to protect one of the NH2 groups of the polyamine being used to modify a base by conjugating glutamate to it, whereas the Aig2-like enzyme eventually removes it (Figure 1). When only an inactive amidoligase is present, consistent with the absent accompanying Aig2, it probably protects the NH2 by merely binding it. In bacterial genomes (e.g. firmicutes, bacteroidetes and deltaproteobacteria), related amidoligase and Aig2 family genes are linked to Dcm, Dam, SAM-synthetase and GAT-II family genes (Figure 4). The firmicute amidoligases are fused to an N-terminal DNA-binding SWIM domain, supporting action on a DNA substrate (Figure 4) (54).
We suggest that the aforementioned gene neighborhoods specify novel base-modifying systems: Nmad4 might act as an enzyme similar to SAM decarboxylase to generate a methionine derivative, which the associated SAM-synthetase could possibly use to generate a SAM derivative with a free NH2 group in turn protected by the amidoligase-Aig2 pair or the inactive amidoligase. This NH2-bearing moiety is probably transferred to a base by the methyltransferase encoded by the adjacent gene in a reaction comparable with the transfer of aminocarboxypropyl from SAM by the Tyw2 methyltranferase in biosynthesis of the tRNA base wybutosine (82).
Other obscure DNA-modification genes associated with the ParB-Tls loci
The roles of few of the ParB-Tls loci were refractory to our analytical approaches, and we could only assign tentative role for them in DNA modifications. One such conserved but uncharacterized gene, found in the ParB-Tls loci of Streptococcus phage MM1, and prophages from firmicutes and bacteroidetes encodes a protein henceforth labeled Predicted base modifying domain 1 (PBMD1) (overlaps with the model for DUF4417 in the Pfam database; PBMD1 associated operons in Figure 4). PBMD1 is often found fused C-terminal to the ParB domain. Secondary structure prediction based on a multiple sequence alignment of PBMD1 predicts an α + β fold with a conserved core of seven strands and four helices and several conserved polar residues suggestive of an enzymatic function. PBMD1 is also found in non-phage contexts associated with Dcm genes, supporting a DNA-modifying role for these proteins. Versions in Lactobacillus species are operonic with a GNAT-encoding gene, suggesting a further modification of an amino group by an acyl group. Hence, it is possible that PBMD1 catalyzes modification of bases by transfer of an amino group-containing moiety. These ParB-Tls loci also often contain a gene encoding a distinctive JAB domain peptidase, or in its place those coding papain-like or metallopeptidase superfamilies (Figure 4), which could be involved in processing phage structural proteins during virion maturation (58,83).
Identification of further DNA-base-modifying enzymes using previously studied bacteriophage models
Enzymes encoded by Bacillus phage SP10 and Delftia phage phiW-14 catalyze synthesis of hypermodified thymines
Pioneering studies on phage DNA modifications identified two hypermodified thymine species, α-glutamylthymine (agT) and α-putrescinyl thymine (apT), respectively, in the DNA of the Bacillus subtilis phage SP10 and the Delftia acidovorans (formerly Pseudomonas acidovorans) phage phiW-14 (8,84). The identity of the enzymatic apparatus for these modifications has remained unknown, although the genomes of these viruses were sequenced recently (85). Early genetic and biochemical studies in both these phages indicated that they used a common biochemical pathway for their respective modifications (9,86). 15% of the T in SP10 and 50% of the T in phiW-14 were shown to be replaced by agT and apT respectively (2,9,86), with both T and the hypermodified T being synthesized from a 5hmU intermediate in situ in DNA. The 5hmU itself is synthesized as a free nucleotide from uracil and incorporated during DNA replication. The hydroxyl group of 5hmU was observed to be pyrophosphorylated to 5-(hydroxymethyl-O-pyrophosphoryl)deoxyuridylate (5hmOPPU) (2,9,86). A further nucleophilic attack reduces 5hmOPPU through a reactive methylene-containing intermediate to thymine. Alternatively, 5hmOPPU is either ligated to the α-amino group of glutamic acid to give agT or the amino group of putrescine to yield apT (9,86). Genetic studies in SP10 and phiW-14 recovered mutations at multiple distinct loci (mod mutants) that partly or entirely failed to complete the modification of 5hmU and accumulated different ratios of 5hmU, T or the 5hmOPPU intermediate (9,86).
Analysis of modified base ratios and epistastatic relationships of SP10 mod mutants allowed reconstruction of the following biochemical pathway: ModA is absolutely required for all T and agT production from 5hmU and is predicted to function as a kinase catalyzing two successive 5hmU phosphorylations to generate 5hmOPPU for T biosynthesis and the initial phosphorylation of 5hmU to 5hmOPU for eventual agT synthesis (Figure 1). ModB is inferred to function as the pyrophosphorylase that reductively processes 5hmOPPU to generate T as modB mutants are entirely deficient in T (Figure 1). Two mutations were recovered at the modD locus, of which modD2 was consistent with the kinase that phosphorylates 5hmOPU to generate 5hmOPPU for agT synthesis and modD1 with a predicted pyrophorylase that conjugates glutamate to 5hmOPPU to form agT (Figure 1). ModC is predicted to perform a regulatory function modulating the action of the previous mod products to determine the ratio of 5hmUs, which are converted to T to those which are converted to agT (9). Similarly, genetic analysis of phage phiW-14 suggested that the locus encoding a predicted kinase generating 5hmOPPU was needed before the loci encoding the predicted pyrophosphorylases that generates apT by putrescine conjugation or T by reduction (Figure 1) (86).
Identification of key Bacillus phage SP10 and Delftia phage phiW-14 enzymes catalyzing synthesis of hypermodified thymines
As these phages do not belong to the subgroup of caudoviruses containing a ParB-Tls, we used an alternative strategy to identify the enzymes producing these modified bases. As the two phages are distantly related (with practically no large scale genomic synteny), we reasoned that comparative genomic analysis using sensitive sequence similarity searches could help identify shared proteins accounting for the common biochemistry of these modifications. Both SP10 and phiW-14 code for enzymes involved in salvage of dUTP (dUTPase), conversion of cytidine to uridine (deoxycytidylate deaminase) and an enzyme of the thymidylate synthase superfamily that catalyzes the hydroxymethylation of UMP (5hmU synthase) (85). Based on previous phage studies, these observations confirmed that SP10 and phiW-14 possess the apparatus to produce 5hmU for incorporation into DNA (2) (Figure 1). The aforementioned reconstruction of the modification pathway suggested to us that identification of potential kinases catalyzing phosphorylation of 5hmU would be critical to elucidate it. Kinases of the P-loop and the ribokinase-like superfamilies are known to catalyze successive phosphorylations to generate pyrophosphates (59,87). As only members of the former are known from phages, we initiated sequence profile searches for small molecule kinases of the P-loop superfamily and recovered two distinct kinases from the SP10 genome, consistent with the genetic inference that there should be two distinct kinase activities (corresponding to ModD2 and ModA). The first of these (F373_gp186) displayed an N-terminal domain (residues: 1–228) characterized by a conserved EG in the Walker B motif belonging to the uridine/cytidine kinase, phosphoribulokinase, pantothenate kinase (UCPP) clade of P-loop kinases (Supplementary Data) (59) and a large conserved C-terminal region. Homologs of both this version of the kinase domain and the C-terminal region fused to it occur as separate proteins in phiW-14. Hence, we surmised that they are probably components of the hypermodified T biosynthetic pathway.
This was further supported by several independent lines of evidence: (1) Secondary structure prediction indicated that the C-terminal region this protein (F373_gp186), which occurs as a standalone protein in phiW-14 (DP-phiW-14_gp072), contains a conserved globular domain. Profile–profile comparisons revealed that it is comprising multiple DNA-binding HhH motifs related to the α-helical DNA glycosylases (e.g. Mag1 DNA glycosylase PDB: 3s6i; probability = 93.3%, P = 8.4 × 10−5; Figure 3B) (3). The multiple sequence alignment showed a distinct set of polar residues mapping to the C-terminal most HhH domain that are likely to constitute its catalytic active site (usually glutamate/glutamine and a cysteine; Figure 3). This suggested that it bound DNA and probably acted on a flipped out base similar to the HhH-containing DNA glycosylases (88). (2) This gene (F373_gp186) in SP10 is linked to the 5hmU synthase of the thymidylate synthase superfamily (F373_gp188) (aG/PT-Pyro phosphorylase biosynthesis related operons in Figure 4). This neighborhood association is also preserved across several other phages such as MP1412, YuA, M6, which infect Pseudomonas aeruginosa, and RSL1, which infects Ralstonia, indicating that it operates in conjunction with 5hmU synthesis (Figure 4 and Supplementary Data). (3) The structure of the SP10 protein with an N-terminal kinase domain and the C-terminal domain described earlier in the text matched the genetic evidence for two associated mutations modD2 and modD1, which respectively catalyze phosphorylation of 5hmU and glutamate addition through a pyrophorylase reaction. Hence, we concluded that the N-terminal kinase domain corresponds to modD2 activity and the conserved C-terminal domain of F373_gp186 corresponds to the modD1 activity or α-glutamyl/putrescinyl thymine phosphorylase activity (aG/PT-pyrophoshorylase). In phages where the kinase and C-terminal predicted aG/PT-pyrophoshorylase are coded by standalone genes, they are often linked in the same predicted operon (Figure 4). By comparable reasoning, the second standalone kinase protein (F373_gp218) that we detected in SP10 possibly corresponds to ModA activity.
Based on sequence features, we defined three distinct clades of aG/PT-pyrophoshorylases (clades 1–3, Figure 3B). Clade-1 is prototyped by the aG/PT-pyrophoshorylase described earlier in the text from SP10 and Delftia phiW-14. Clade-2 pyrophoshorylases are mainly found mainly in caudoviruses and prophages, where they are usually linked to a P-loop kinase. Delftia phiW-14 and several phages (e.g. MP1412, YuA, M6, RSL1 and phiJL001) possess both a clade-1 aG/PT-pyrophoshorylase domain and a clade-2 protein (e.g. phiW-14 gp109; in Figure 3B). In these viruses, it could potentially function as the second pyrophosphorylase that reductively generates T from 5hmOPPU (Figure 1) (84,86). Together, these observations lead to the testable hypothesis that all phages containing the aG/PT-pyrophoshorylase genes such as MP1412, YuA, M6, RSL1 and phiJL001 (89) should contain hypermodified thymine in their DNA and generate T via 5hmOPPU. In Mycobacterium phage Zemanar and in a Frankia prophage, clade-2 enzymes are found between the TET/JBP gene and the P-loop kinase gene in the ParB-Tls loci (Figure 2). Given the relationship of the UCPP clade P-loop kinase from these phages to those from SP10 and phiW-14 predicted earlier in the text to phosphorylate the OH group of 5hmU (Supplementary Data), it is possible that they do so downstream of the hydroxylase activity of the TET/JBP encoded by the neighboring gene (Figure 1, #11). The associated clade-2 pyrophosphorylase could then use the 5hmOPPU for further modification just as in SP10 and phiW-14.
The locus in SP10 which combines F373_gp186 (kinase + aG/PT-pyrophoshorylase) and the 5hmU synthase contains two other linked genes. One of these (F373_gp189) contains an all-alpha helical domain with a strictly conserved pair of lysines, a pair of tyrosines and an arginine residue, suggestive of an enzymatic function (Figure 4, Nmad1; overlaps with DUF1599 in the Pfam database). Nmad1 shows a strong connection to the 5hmU synthase domain of the thymidylate synthase superfamily, to which it is either fused at the C-terminus or linked as a 3′ gene neighbor in caudoviruses and prophages of cyanobacteria and chlorobi. On occasions, when the 5hmU synthase in this neighborhood is inactive, we observed the presence of an additional gene encoding the non-homologous Thy1 superfamily protein with a similar catalytic activity (Figure 4) (90). This supports its role in modified base biosynthesis in conjunction with 5hmU. Given that SP10 lacks a second pyrophosphorylase gene (unlike PhiW-14), Nmad1 could possibly takes its place as the SP10 modB activity that generates thymine from 5hmOPPU. The other gene (F373_gp185) linked to the predicted aG/PT-pyrophoshorylase in SP10 encodes an N-terminal divergent, catalytically active domain of the thymidylate synthase superfamily (HHPRED probability 41.45%, P = 7 × 10−4) fused to a C-terminal inactive radical-SAM superfamily domain related to the DNA photolyase SplB (Figure 4). In phages such as MP1412, YuA, M6 the equivalent radical-SAM domain occurs independently of the thymidylate synthase-like domain and is catalytically active. It could function downstream of the clade-2 phosphorylase to generate thymine from the methylene intermediate using a reaction similar that used by SplB to restore thymine from photo-dimers (91). It is also accompanied by a further gene coding for a pyridoxal 5′-Phosphate Dependent (PLPDE) superfamily enzyme that could potentially function as an aminotransferase needed to synthesize the amino-group-containing moiety that is ligated to the methyl group of thymine (Figure 4) (92). Additionally, RSL1 codes for two linked glycosyltransferases and a sulfotransferase (Figure 4); hence, a subset of the hydroxylated pyrimidines in this phage might also be hypermodified by glycosylation and possibly even sulfatation.
Predicted thymine modification systems in bacterial genomes and links to thiamin and mildiomycin biosynthesis
Homologs of the clade-1 predicted aG/PT-pyrophoshorylase are also found sporadically in proteobacteria, cyanobacteria, actinobacteria, bacteroidetes and euryarchaea in a distinct mobile operon with several other linked genes. Its most common association is with a distinctive member of the thymidylate synthase superfamily that we predict to function as a pyrimidine hydroxymethytransferase like the phage members of this superfamily (MazG-containing operons in Figure 4, Supplementary Data). Unlike their phage counterparts, these neighborhoods do not code for a P-loop kinase needed to generate the pyrophosphorylated intermediate required by the aG/PT-pyrophoshorylase. However, most of these operons code for a distinctive version of the MazG superfamily pyrophosphatase, distinguished by a unique C-terminal α + β domain beyond the core MazG domain (MazG-C domain) (Figure 4, Supplementary Data). The latter domain has an absolutely conserved DxYRxHDxxH motif and other charged residues suggesting that it might possess a distinct catalytic activity (Supplementary Data). As the MazG domain can release a pyrophosphate from ATP (93), it is possible that the fused MazG-C domain acts in conjunction with the former to transfer the pyrophosphate moiety to the 5-hydroxmethylpyrimidine, thereby taking up the role of the P-loop kinase (Figure 1). This might also explain the role of comparable MazG domain proteins found in certain phages with the thymine hypermodification apparatus such as phiW-14 and Dickeya phage Limestone. These operons might on occasions also code for a further predicted β-strand-rich protein with conserved charged residues and a cysteine (Nmad2), and a metallopeptidase (Figure 4). These distinctive thymidylate synthase and MazG superfamily proteins also show additional associations in mobile gene neighborhoods, primarily from proteobacteria, independently of the aG/PT-pyrophoshorylase (Figure 4). Here, they are linked with a gene coding for an α/β fold uracil DNA glycosylase superfamily protein (UDG) (94). These neighborhoods also often encode Nmad2 and TIR domains; the latter could possibly function in a DNA-associated capacity reminiscent of those from previously described restriction-modification-like loci, where they are functionally associated with MORC-like ATPases (95). Nmad2 shows several of its own distinctive associations with other genes (e.g. Nmad3, which contains a highly conserved HxD motif and an aspartate suggestive of enzymatic activity) that could define other, more obscure modified nucleotide biosynthesis systems (Figure 4).
An analogous set of associations is seen in clade-3 aG/PT-pyrophoshorylases encoded by mobile operons from proteobacteria, actinobacteria, spirochaetes and bacteroidetes. Here, the gene encoding the aG/PT-pyrophoshorylase is linked to a predicted 5hmU synthase of the thymidylate synthase superfamily (Figure 4). The latter is often further fused or operonically linked to an UDG in some spirochaetes, gamma-proteobacteria and actinobacteria (Figure 4, Supplementary Data). Additionally, these neighborhoods contain a gene coding for a metallopeptidase fused to HTH domains. In a subset of these bacteria, the operons further contain an uncharacterized domain with an α + β fold (DUF4031 in the Pfam database), with a conserved glutamate and motifs with HxxxD and HxD signatures. It also occurs fused to HD hydrolases and an inactive NUDIX domain in some bacteria (e.g. gi: 84498502 from Janibacter) leading the prediction that it might be an enzyme that acts on nucleotides (Figure 4, Supplementary Data). In systems with UDGs, the DNA glycosylase activity might help restore the regular pyrimidine after the function of the modified base is fulfilled.
Strikingly, clade-1 aG/PT-pyrophoshorylases are also found in basidiomycete fungi, and several stramenopile lineages such as diatoms, phaeophyte, pelagophyte and oomycetes. These eukaryotic aG/PT-pyrophoshorylase homologs are fused to either a chromodomain in basidiomycetes or a PHD domain in stramenopiles (Figure 4), both which bind methylated histones (41). Further, in Aureococcus, it is also fused to a metallopeptidase and Hif1-like 2OGFeDO domains. These fusions indicate that they are likely to function in the context of chromatin and could catalyze formation of hypermodified pyrimidines. In fungi, their phyletic patterns closely mirror those of the TET/JBP hydroxylases (12); hence, they could potentially use the 5-hydroxymethylpyrimidines generated by these enzymes for further modifications.
Outside of these thymine modifications, pyrophosphorylation-dependent activation of hydroxymethylpyrimidine bases for conjugation of an amine moiety has only been reported in thiamin biosynthesis (87). Here, ThiC synthesizes a 5-hydroxymethylpyrimidine, which is then converted to 5-methylpyrimidine-OPP by the kinase ThiD, just as in phage DNA modifications. The ThiE pyrophosphorylase then conjugates the thiazole phosphate group with the release of the pyrophosphate group analogous to the reaction proposed for the aG/PT-pyrophoshorylases (84,86,87). This parallel base pyrophosphorylation-dependent modification mechanism seen in the DNA base hypermodification and thiamin biosynthesis systems is reflective of the energetic requirement of a pyrophosphate intermediate for further modifications of that base position. Strikingly, we found that the thymidylate synthase superfamily genes found in the clade 3 DNA base pyrophophorylases gene-neighborhoods described earlier in the text are alternatively linked to a gene for a methylase, a NUDIX phosphoesterase, base deoxyribose glycohydrolase and thiamin biosynthesis genes such as ThiD, ThiE and ThiL in certain firmicutes, betaproteobacteria and deltaproteobacteria (Figure 4). We predict that here the thymidylate synthase superfamily gene encodes a 5hmU synthase, whose product is then further modified by the methylase, dephosphorylated by the NUDIX enzyme and separated from the deoxyribose by the base deoxyribose glycohydrolase to generate the 5-hydroxymethylpyrimidine used for thiamin biosynthesis (Figure 1). Thus, these neighborhoods appear to code an alternative mechanism for synthesis of the methylpyrimidine moiety of thiamin by salvaging cytosine nucleotides. Consistent with this proposal, a closely related thymidylate synthase superfamily is also observed in the mildiomycin biosynthesis operon where it synthesizes a hydroxymethylpyrimidine. The base is then further modified by other linked genes to synthesize the peptidyl-nucleoside antibiotic mildiomycin (96).
Prediction of DNA-binding domains involved in discrimination of modified DNA: role of the PUA-like fold
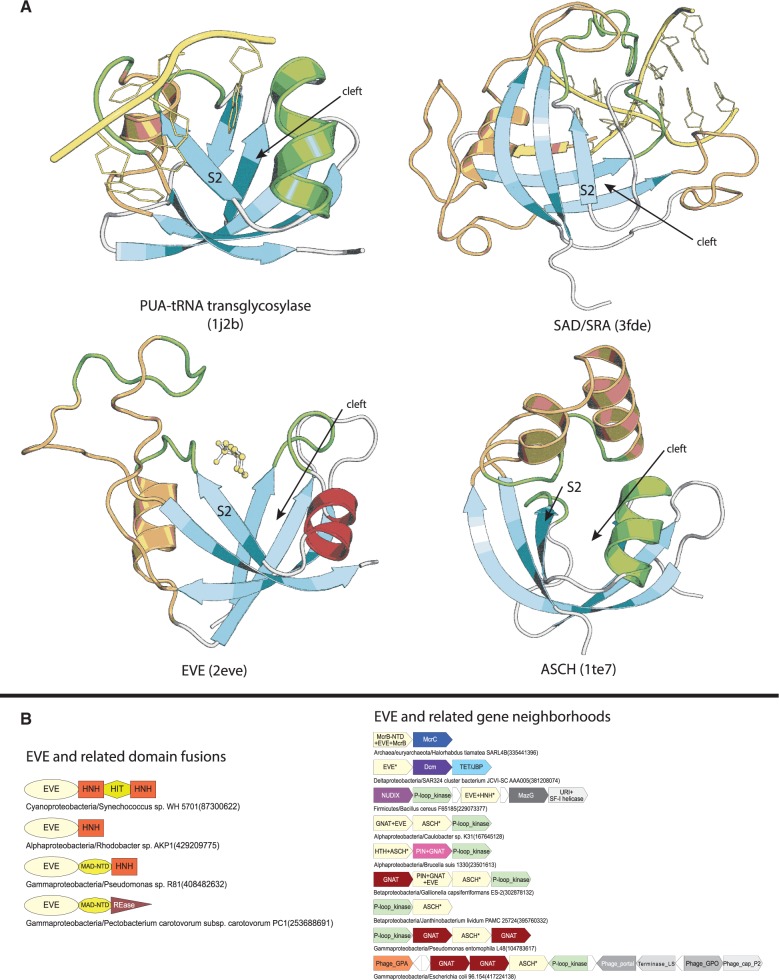
We uncovered at least four independent sets of contextual connections in the form of conserved gene neighborhoods and domain architectures between known and predicted DNA-modification enzymes and proteins containing a domain with the PUA-like fold (62,97,98) (Figure 5). Multiple members of the PUA-like fold have been implicated in recognition of the nucleic acids including those with modified bases (3,62,99–101): the founding member of this fold, the PUA domain, is fused to or associates as a subunit with several RNA-modifying enzymatic modules such as pseudouridine synthases, tRNA guanine transglycosylases and thiouridine synthases, where it might bind or discriminate modified RNA bases (62,67). We have also shown that the SAD/SRA domain, which binds DNA with hemimethylated cytosine and hmC, contains the PUA-like fold (3,99,102). PUA-like fold domains from aforementioned gene neighborhoods belong to two functionally poorly understood clades of this fold, namely, ASCH (98) and EVE (97). Accordingly, we performed a systematic survey of the ASCH and EVE clades of the PUA-like fold to identify novel connections that might throw light on their functions vis-à-vis DNA modification.
Figure 5.
Structures, domain architectures and operons of representative members of the PUA-like fold. (A) Structures are centered on the conserved binding cleft with regions contributing to it colored in green. Family-specific inserts are colored in orange. The EVE structure was crystallized with a 3[N-morpholino]propane sulfonic acid molecule in the predicted binding site. (B). Genes in operons are represented and labeled as described in Figure 4. Fusions of the EVE-like domain are shown to the left.
EVE domains as discriminators of methylated pyrimididines and their oxidized derivatives
The operonic linkage of an EVE domain-encoding gene with the TET/JBP and DNA 5C-methyltransferase genes indicated that the EVE domain might recognize DNA with 5hmC or other oxidized mC species (Figures 1 and 5). Across phylogenetically diverse bacteria, we found EVE domains fused to nucleases domains of the HNH/Endo-VII superfamily (Figure 5), which includes the McrA-like restriction nucleases that cleave methylated or hydroxymethylated DNA bases from phage DNA (103). We had also earlier reported similar fusions between HNH restriction nucleases and another PUA-like fold domain, the SAD/SRA domain (3). In certain archaea and diverse bacterial lineages, the EVE domain is fused to a McrB-like GTPase of the AAA+ superfamily and, in some cases, the MAD-NTD domain (104), which is normally fused to McrB AAA+ domains (Figure 5B). These are usually linked to a McrC-like gene that contains the McrB-interacting McrC-NTD domain and the C-terminal McrC-like restriction endonuclease domain (104). McrBC restriction systems are involved in GTP-dependent cleavage of phage DNA containing methylated, hemi-methylated cytosine or hmC (103,105). The aforementioned restriction systems with these EVE proteins typically do not encode any methylases, though they might encode additional nucleases such as an URI nuclease domain (106,107) fused to a superfamily-I helicase (Figure 5B). EVE domains are far more frequently encoded by cellular than in phage genomes. Together, these observations support the possibility that the EVE domain might help the associated restriction endonucleases to discriminate cellular DNA specifically from modified phage DNA, such as those with oxidized methylpyrimidines.
Restriction systems with EVE proteins often additionally code for proteins with one or more phosphoesterase domains such as the HIT, NUDIX and MazG and also zeta toxin-like kinase domains (Figure 5B). Given that several phages produce 5hmC and 5hmU as nucleotides that are then incorporated into DNA during replication (2,8), these phosphoesterases could form a second line of defense by hydrolyzing 5hmCTP and 5hmUTP and limiting replication. Thus, these associations circumstantially support a role of these prokaryotic EVE domains functioning in recognition of phage DNA with oxidized methylpyrimidines. Finally, zeta toxin-like kinases are known to inhibit cell-wall synthesis by phosphorylating precursor sugars resulting in cell lysis (108). This could represent the third and final line of defense, namely cell-suicide if the mechanisms to target modified phage DNA fail (8,104,109).
Eukaryotic EVE domains are mostly found in species possessing modified bases such as mC, hmC or Base J, such as animals, plants, fungi and kinetoplastids and are typified by the human Thy28 protein (Supplementary Data) (97). Fusion to DNA minor-groove targeting AT-hook motifs in certain fungal orthologs of Thy28 further support the role of the EVE domain in DNA binding (97). As we were preparing this manuscript for submission, a mass-spectroscopic study to identify human proteins binding 5hmC containing DNA uncovered Thy28 as a potential 5hmC binder (102). This provides further support to our hypothesis that these domains play an important role in discrimination of DNA with oxidized pyrimidines. The PUA-like fold contains a structurally equivalent cleft, which is however highly divergent at the sequence level between different representatives of the fold. Previously available structures suggest that residues lining this cleft are likely to be critical for recognition of modified bases (3,98) (Figure 5A).
Possible role for EVE and ASCH domains in recognition of other hypermodified bases
We also uncovered evidence for EVE and ASCH domains playing a role in recognizing other modified bases. The phage DNA-modifying RAP44-like PP-loop ATPase gene described earlier in the text is sometimes preceded by a gene encoding a ASCH superfamily protein (98) (Figure 4, panel V). Certain non-phage mobile systems with the GNAT + ABC protein predicted to synthesize a hypermodified base contain a linked or fused ASCH domain (Figures 4 and 5B). Our systematic survey of EVE proteins recovered another striking conserved gene neighborhood in certain proteobacteria, bacteroidetes, firmicutes and prophages. This predicted operon codes for a protein with a GNAT acyltransferase domain fused to an EVE domain, sometimes a second GNAT protein, a P-loop kinase of the archaeal adenylate kinase clade, and an ASCH domain protein, which might be further fused to a HTH domain (Figure 5B). In certain E. coli prophages, this operon is positioned close to the Terminase large subunit and other packaging proteins, analogous to the DNA modification genes of the ParB-Tls locus (Figure 5B). This observation, together with fusion of the ASCH domain to HTH domains related to prokaryotic transcription factors, suggests that this system might operate on DNA. However, in several instances proteins with the EVE and GNAT domains additionally contain a PIN RNase domain (110) at its N-terminus raising the possibility of RNA acting either as a substrate or as a guide for DNA modification (Figure 5B). In either case, based on the GNAT domain, we propose that this system might acylate the amino group in a base.
Biological and evolutionary implications of DNA modification systems
Role of specialized DNA modification loci in phages and prokaryotes
The biological roles for DNA modifications can be broadly interpreted along two lines, namely, as purveyors of epigenetic information and as strategies in genome conflicts (2,3,19,103,111–113). Modifications, such as 5hmC and its glycosylated derivatives in T4, agT in SP10 and α-putrescinylthymine in phiW-14, have been interpreted as primarily playing a role in genomic conflicts, i.e. to evade restriction by the host (2,4,103). However, agT in SP10 is also required for effective replication and packaging of DNA into the head (9). Similarly, studies on the temperate phage P1 have shown that adenine methylation at Pac sites catalyzed by its own Dam is required for headful packaging of concatenated phage DNA by facilitating cleavage of Pac sites by the Terminase-associated nuclease (114). Adenine methylation of phage P1 replication origin (oriR) was also shown to be required for regulation and accurate initiation of DNA replication and control of copy number in its plasmid form (115,116). Mutations in the phage Dam severely compromise both functions. Genome-wide analysis of Dam sites in Phage P1 further suggested that expression of several genes might be regulated by DNA methylation (13). These observations suggest that phage DNA modifications are not just restricted to evasion of host defenses but may also have an epigenetic role.
Studies on phage P1 provide the rationale for the embedding of DNA modification genes in the ParB-Tls locus. Like the P1 Dam, genes in this locus are likely to be expressed late in the lifecycle along with the neighboring terminase genes. They might similarly provide an epigenetic mark at Pac-like sites to specify one genome’s worth of phage DNA to be packaged into the head (13). Members of the ParB superfamily are nucleases and ATPases (17) that might show sequence preferences (64). However, phages containing the ParB-Tls locus do not encode a ParA that couples with ParB to mediate symmetric chromosome segregation in bacterial cells or plasmids. Hence, we predict that the ParB proteins in ParB-Tls loci, rather than mediating chromosome segregation, by analogy with DndB (64), direct the DNA-modification apparatus to specific chromosomal sites during packaging. In a considerable group of phages lacking the ParB-Tls locus, the DNA modification genes are usually in the proximity of DNA replication genes (Supplementary Data). Here, modifications might serve as epigenetic marks at the origin (again as in P1) for efficient replication initiation (115,116). These predicted roles would imply that several DNA modifications might be restricted to a few nucleotides at specific sites, thereby minimizing the primary constraint against complex DNA modifications, i.e. the need to maintain Watson–Crick base pairing. Moreover, our study allows us to distinguish, to an extent, epigenetic DNA modifications from those used in an anti-restriction strategy. Phages using TET/JBP enyzmes to generate hydroxymethyl pyrimidines are likely to generate them in situ at specific locations in the DNA, thus favoring a primarily epigenetic function. In contrast, phages using members of the thymidylate synthetase superfamily to generate precursor nucleotides with hydroxymethyl pyrimidines are likely to incorporate modified nucleotides throughout the genome (2,4,9,103), thus favoring a primarily anti-restriction function. However, selective in situ hypermodification of some of these hydroxymethylpyrimidines might restore a degree of epigenetic function as seen in the phages SP10 and PhiW-14 (agT or apT). In either case, the great diversity of DNA modification systems in phage genomes appears to have been driven by the constant pressure to escape host defense mechanisms that evolve to discriminate non-self DNA based on these modifications.
On the cellular side, at least five types of predicted DNA modification loci corresponding to their phage counterparts, namely, TET/JBP-dependent methyl pyrimidine hydroxylation, thymine hypermodification, the GNAT + ABC system, IbrAB and the deazapurine incorporation system, were observed. This greatly extends the types of cellular DNA modifications beyond the well-known DNA methylation and DNA-phosphorothioating Dnd systems. Like them, the phyletic patterns of these new loci indicate considerable lateral mobility. This implies that they are likely to provide specific selective advantages to unrelated organisms, which probably stems from a role in facilitating discrimination of self from non-self DNA in the course of inter-genomic conflicts. The current study also suggests that such gene clusters from cellular genomes are also the ‘nurseries’ for the innovation of diverse PUA-fold domains, such as SAD/SRA, ASCH and EVE, which are proposed to play a critical role in discrimination of modified DNA; i.e. ‘readers’ of epigenetic marks.
The evolutionary origins of viral, prokaryotic and eukaryotic DNA modification systems
At least 12–15 different types of enzymes, including TET/JBP, aG/PT-pyrophoshorylase, glycosyltransferases and Mom, among others, appear to have been first recruited for DNA modification in loci from phages and prokaryotic genomes. Some novel DNA modification systems identified in this study appear to have been derived from enzymes originally used for RNA modifications. For example, enzymes involved in synthesis and incorporation of deazapurines appear to have been derived from the ancient pathway for hypermodified RNA base biosynthesis that was already present in the last universal common ancestor (60,67). Likewise, the PUA-like fold might have been recruited from RNA base recognition systems, some of which were already present in the last universal common ancestor (67).
A major revelation of comparative genomics has been the role of prokaryotic genomic conflict systems in supplying components for key regulatory innovations of eukaryotes (3,41,111). This is most clearly illustrated by epigenetic regulators in eukaryotic chromatin: eukaryotic DNA methyltransferases, MORC-like ATPases, SWI2/SNF2 ATPases, SAD/SRA, HIRAN and HARE-HTH found in the Asxl proteins of the Polycomb-repressive complex can all be traced back to genomic conflict systems such as restriction-modification (3,38,95,112,117,118). We now add multiple independent transfers of TET/JBP proteins, predicted glycosyltransferases such as GREB1 and the predicted base J synthesis enzyme from phage DNA modification systems. Likewise, we also find evidence for transfer of Mom, aG/PT pyrophorylases and the ABC + GNAT to different eukaryotic lineages such as stramenopiles, chlorophytes and mushrooms from phages or mobile bacterial operons (12). Although Mom might have been recruited for modification of silaffin and related proteins in eukaryotes (Supplementary Data) (12), the aG/PT pyrophorylases display domain architectures clearly suggestive of a role in DNA modification in fungi and stramenopiles (Figure 4). The previously described HARE-HTH and SAD/SRA domains are now joined by EVE domains as potential discriminators of modified pyrimidines in eukaryotes.
CONCLUSIONS
We predict at least 12 novel, viral and cellular DNA modification systems as well as ‘readers’ of modified bases in DNA. Even among previously characterized systems, we identify candidate enzymes catalyzing key steps, such as the J-base-generating glycosyltransferase and determine the catalytic specificity of phage TET/JBP proteins. Likewise, we identified enzymes involved in synthesis of hypermodified bases agT and apT, which have remained elusive for over 40 years (2,4,8,84). Description of these systems opens new vistas for use their use in biotechnology, parallel to conventional modification methylases and phage glycosyltransferases. We hope that this work inspires further laboratory studies in this regard.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online. The supplementary data may also be accessed from: ftp://ftp.ncbi.nih.gov/pub/aravind/tet_glycosylase/supplementary.html.
ACKNOWLEDGEMENTS
As this article was being produced there was new publication showing the presence of novel SAM derivatives in modification of hydroxylated uracil in tRNA (Kim J et. al., (2013) Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function Nature, 498, 123–126). This work provides additional support for our prediction regarding the use of novel SAM derivatives in DNA modifications.
FUNDING
Intramural funds of the US Department of Health and Human Services (National Library of Medicine, National Institutes of Health (NIH)). Funding for open access charge: The intra-mural funds of the NIH.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Bloomfield VA, Crothers DM, Tinomo IJ. Nucleic Acids: Structures, Properties and Functions. Sausalito, CA: University Science Books; 2000. [Google Scholar]
- 2.Gommers-Ampt JH, Borst P. Hypermodified bases in DNA. FASEB J. 1995;9:1034–1042. doi: 10.1096/fasebj.9.11.7649402. [DOI] [PubMed] [Google Scholar]
- 3.Iyer LM, Abhiman S, Aravind L. Natural history of eukaryotic DNA methylation systems. Prog. Mol. Biol. Transl. Sci. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]
- 4.Warren RA. Modified bases in bacteriophage DNAs. Annu. Rev. Microbiol. 1980;34:137–158. doi: 10.1146/annurev.mi.34.100180.001033. [DOI] [PubMed] [Google Scholar]
- 5.Greenberg GR, He P, Hilfinger J, Tseng MJ. In: Molecular Biology of Bacteriophage T4. Karam JD, editor. Washington, DC: American Society of Microbiology; 1994. pp. 14–27. [Google Scholar]
- 6.Song HK, Sohn SH, Suh SW. Crystal structure of deoxycytidylate hydroxymethylase from bacteriophage T4, a component of the deoxyribonucleoside triphosphate-synthesizing complex. EMBO J. 1999;18:1104–1113. doi: 10.1093/emboj/18.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vrielink A, Ruger W, Driessen HP, Freemont PS. Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. EMBO J. 1994;13:3413–3422. doi: 10.1002/j.1460-2075.1994.tb06646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelln RA, Warren RA. Studies on the biosynthesis of alpha-putrescinylthymine in bacteriophage phi W-14-infected Pseudomonas acidovorans. J. Virol. 1973;12:1427–1433. doi: 10.1128/jvi.12.6.1427-1433.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Witmer H, Wiatr C. Polymer-level synthesis of oxopyrimidine deoxynucleotides by Bacillus subtilis phage SP10: characterization of modification-defective mutants. J. Virol. 1985;53:522–527. doi: 10.1128/jvi.53.2.522-527.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swinton D, Hattman S, Crain PF, Cheng CS, Smith DL, McCloskey JA. Purification and characterization of the unusual deoxynucleoside, alpha-N-(9-beta-D-2′-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc. Natl Acad. Sci. USA. 1983;80:7400–7404. doi: 10.1073/pnas.80.24.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaminska KH, Bujnicki JM. Bacteriophage Mu Mom protein responsible for DNA modification is a new member of the acyltransferase superfamily. Cell Cycle. 2008;7:120–121. doi: 10.4161/cc.7.1.5158. [DOI] [PubMed] [Google Scholar]
- 12.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lobocka MB, Rose DJ, Plunkett G, 3rd, Rusin M, Samojedny A, Lehnherr H, Yarmolinsky MB, Blattner FR. Genome of bacteriophage P1. J. Bacteriol. 2004;186:7032–7068. doi: 10.1128/JB.186.21.7032-7068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith HC, editor. RNA and DNA Editing: Molecular Mechanisms and Their Integration into Biological Systems. Hoboken, New Jersey: John Wiley & Sons, Inc.; 2008. [Google Scholar]
- 15.Zhang D, Iyer LM, Aravind L. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 2011;39:4532–4552. doi: 10.1093/nar/gkr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer LM, Zhang D, Rogozin IB, Aravind L. Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic Acids Res. 2011;39:9473–9497. doi: 10.1093/nar/gkr691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct. 2012;7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borst P, Sabatini R. Base J: discovery, biosynthesis, and possible functions. Annu. Rev. Microbiol. 2008;62:235–251. doi: 10.1146/annurev.micro.62.081307.162750. [DOI] [PubMed] [Google Scholar]
- 20.Cliffe LJ, Hirsch G, Wang J, Ekanayake D, Bullard W, Hu M, Wang Y, Sabatini R. JBP1 and JBP2 proteins are Fe2+/2-oxoglutarate-dependent dioxygenases regulating hydroxylation of thymidine residues in trypanosome DNA. J. Biol. Chem. 2012;287:19886–19895. doi: 10.1074/jbc.M112.341974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Luenen HG, Farris C, Jan S, Genest PA, Tripathi P, Velds A, Kerkhoven RM, Nieuwland M, Haydock A, Ramasamy G, et al. Glucosylated hydroxymethyluracil, DNA base J, prevents transcriptional readthrough in Leishmania. Cell. 2012;150:909–921. doi: 10.1016/j.cell.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekanayake D, Sabatini R. Epigenetic regulation of polymerase II transcription initiation in Trypanosoma cruzi: modulation of nucleosome abundance, histone modification, and polymerase occupancy by O-linked thymine DNA glucosylation. Eukaryot. Cell. 2011;10:1465–1472. doi: 10.1128/EC.05185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pastor WA, Aravind L, Rao A. TETonic shift: Biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 29.Lassmann T, Frings O, Sonnhammer EL. Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 2009;37:858–865. doi: 10.1093/nar/gkn1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Senkevich TG, White CL, Koonin EV, Moss B. A viral member of the ERV1/ALR protein family participates in a cytoplasmic pathway of disulfide bond formation. Proc. Natl Acad. Sci. USA. 2000;97:12068–12073. doi: 10.1073/pnas.210397997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Etten JL, Dunigan DD. Chloroviruses: not your everyday plant virus. Trends Plant Sci. 2012;17:1–8. doi: 10.1016/j.tplants.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iyer LM, Abhiman S, de Souza RF, Aravind L. Origin and evolution of peptide-modifying dioxygenases and identification of the wybutosine hydroxylase/hydroperoxidase. Nucleic Acids Res. 2010;38:5261–5279. doi: 10.1093/nar/gkq265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.La Scola B, Birtles RJ, Greub G, Harrison TJ, Ratcliff RM, Raoult D. Legionella drancourtii sp. nov., a strictly intracellular amoebal pathogen. Int. J. Syst. Evol. Microbiol. 2004;54:699–703. doi: 10.1099/ijs.0.02455-0. [DOI] [PubMed] [Google Scholar]
- 41.Aravind L, Abhiman S, Iyer LM. Natural history of the eukaryotic chromatin protein methylation system. Prog. Mol. Biol. Transl. Sci. 2011;101:105–176. doi: 10.1016/B978-0-12-387685-0.00004-4. [DOI] [PubMed] [Google Scholar]
- 42.Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 43.Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl Acad. Sci. USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yurist-Doutsch S, Abu-Qarn M, Battaglia F, Morris HR, Hitchen PG, Dell A, Eichler J. AglF, aglG and aglI, novel members of a gene island involved in the N-glycosylation of the Haloferax volcanii S-layer glycoprotein. Mol. Microbiol. 2008;69:1234–1245. doi: 10.1111/j.1365-2958.2008.06352.x. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Mushegian A. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 2003;12:1418–1431. doi: 10.1110/ps.0302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 47.Mohammed H, D'Santos C, Serandour AA, Ali HR, Brown GD, Atkins A, Rueda OM, Holmes KA, Theodorou V, Robinson JL, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3:342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghosh MG, Thompson DA, Weigel RJ. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 2000;60:6367–6375. [PubMed] [Google Scholar]
- 49.Liu M, Wang G, Gomez-Fernandez CR, Guo S. GREB1 functions as a growth promoter and is modulated by IL6/STAT3 in breast cancer. PLoS One. 2012;7:e46410. doi: 10.1371/journal.pone.0046410. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Nyholt DR, Low SK, Anderson CA, Painter JN, Uno S, Morris AP, MacGregor S, Gordon SD, Henders AK, Martin NG, et al. Genome-wide association meta–analysis identifies new endometriosis risk loci. Nat. Genet. 2012;44:1355–1359. doi: 10.1038/ng.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rae JM, Johnson MD, Cordero KE, Scheys JO, Larios JM, Gottardis MM, Pienta KJ, Lippman ME. GREB1 is a novel androgen-regulated gene required for prostate cancer growth. Prostate. 2006;66:886–894. doi: 10.1002/pros.20403. [DOI] [PubMed] [Google Scholar]
- 52.Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, Roberto A, Christensen J, Bonaldi T, Helin K, et al. Tet proteins connect the O-Linked N-acetylglucosamine transferase ogt to chromatin in embryonic stem cells. Mol. Cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 54.Iyer LM, Abhiman S, Maxwell Burroughs A, Aravind L. Amidoligases with ATP-grasp, glutamine synthetase-like and acetyltransferase-like domains: synthesis of novel metabolites and peptide modifications of proteins. Mol. Biosyst. 2009;5:1636–1660. doi: 10.1039/b917682a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neuwald AF, Landsman D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 56.Hatfull GF. The secret lives of mycobacteriophages. Adv. Virus Res. 2012;82:179–288. doi: 10.1016/B978-0-12-394621-8.00015-7. [DOI] [PubMed] [Google Scholar]
- 57.Johnson EP, Mincer T, Schwab H, Burgin AB, Helinski DR. Plasmid RK2 ParB protein: purification and nuclease properties. J. Bacteriol. 1999;181:6010–6018. doi: 10.1128/jb.181.19.6010-6018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burroughs AM, Iyer LM, Aravind L. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Genome Dyn. 2007;3:48–65. doi: 10.1159/000107603. [DOI] [PubMed] [Google Scholar]
- 59.Leipe DD, Koonin EV, Aravind L. Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol. 2003;333:781–815. doi: 10.1016/j.jmb.2003.08.040. [DOI] [PubMed] [Google Scholar]
- 60.McCarty RM, Bandarian V. Biosynthesis of pyrrolopyrimidines. Bioorg. Chem. 2012;43:15–25. doi: 10.1016/j.bioorg.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sabri M, Hauser R, Ouellette M, Liu J, Dehbi M, Moeck G, Garcia E, Titz B, Uetz P, Moineau S. Genome annotation and intraviral interactome for the Streptococcus pneumoniae virulent phage Dp-1. J. Bacteriol. 2011;193:551–562. doi: 10.1128/JB.01117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aravind L, Koonin EV. Novel predicted RNA-binding domains associated with the translation machinery. J. Mol. Evol. 1999;48:291–302. doi: 10.1007/pl00006472. [DOI] [PubMed] [Google Scholar]
- 63.Chen S, Wang L, Deng Z. Twenty years hunting for sulfur in DNA. Protein Cell. 2010;1:14–21. doi: 10.1007/s13238-010-0009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liang J, Wang Z, He X, Li J, Zhou X, Deng Z. DNA modification by sulfur: analysis of the sequence recognition specificity surrounding the modification sites. Nucleic Acids Res. 2007;35:2944–2954. doi: 10.1093/nar/gkm176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sandt CH, Hopper JE, Hill CW. Activation of prophage eib genes for immunoglobulin-binding proteins by genes from the IbrAB genetic island of Escherichia coli ECOR-9. J. Bacteriol. 2002;184:3640–3648. doi: 10.1128/JB.184.13.3640-3648.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bork P, Koonin EV. A P-loop-like motif in a widespread ATP pyrophosphatase domain: implications for the evolution of sequence motifs and enzyme activity. Proteins. 1994;20:347–355. doi: 10.1002/prot.340200407. [DOI] [PubMed] [Google Scholar]
- 67.Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aravind L, Anantharaman V, Koonin EV. Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: implications for protein evolution in the RNA. Proteins. 2002;48:1–14. doi: 10.1002/prot.10064. [DOI] [PubMed] [Google Scholar]
- 69.Suzuki T, Miyauchi K. Discovery and characterization of tRNAIle lysidine synthetase (TilS) FEBS Lett. 2010;584:272–277. doi: 10.1016/j.febslet.2009.11.085. [DOI] [PubMed] [Google Scholar]
- 70.Soma A, Ikeuchi Y, Kanemasa S, Kobayashi K, Ogasawara N, Ote T, Kato J, Watanabe K, Sekine Y, Suzuki T. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell. 2003;12:689–698. doi: 10.1016/s1097-2765(03)00346-0. [DOI] [PubMed] [Google Scholar]
- 71.Mueller EG, Palenchar PM, Buck CJ. The role of the cysteine residues of ThiI in the generation of 4-thiouridine in tRNA. J. Biol. Chem. 2001;276:33588–33595. doi: 10.1074/jbc.M104067200. [DOI] [PubMed] [Google Scholar]
- 72.Ikeuchi Y, Kimura S, Numata T, Nakamura D, Yokogawa T, Ogata T, Wada T, Suzuki T. Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea. Nat. Chem. Biol. 2010;6:277–282. doi: 10.1038/nchembio.323. [DOI] [PubMed] [Google Scholar]
- 73.Hopfner KP, Gerhold CB, Lakomek K, Wollmann P. Swi2/Snf2 remodelers: hybrid views on hybrid molecular machines. Curr. Opin. Struct. Biol. 2012;22:225–233. doi: 10.1016/j.sbi.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kang I, Jang H, Cho JC. Complete genome sequences of two Persicivirga bacteriophages, P12024S and P12024L. J. Virol. 2012;86:8907–8908. doi: 10.1128/JVI.01327-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hattman S. Specificity of the bacteriophage Mu mom+ -controlled DNA modification. J. Virol. 1980;34:277–279. doi: 10.1128/jvi.34.1.277-279.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blanquet S, Dessen P, Kahn D. Properties and specificity of methionyl-tRNAfMet formyltransferase from Escherichia coli. Methods Enzymol. 1984;106:141–152. doi: 10.1016/0076-6879(84)06013-4. [DOI] [PubMed] [Google Scholar]
- 77.Burroughs AM, Iyer LM, Aravind L. Natural history of the E1-like superfamily: implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins. 2009;75:895–910. doi: 10.1002/prot.22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sternberg N, Sauer B, Hoess R, Abremski K. Bacteriophage P1 cre gene and its regulatory region. Evidence for multiple promoters and for regulation by DNA methylation. J. Mol. Biol. 1986;187:197–212. doi: 10.1016/0022-2836(86)90228-7. [DOI] [PubMed] [Google Scholar]
- 79.Fisher JF, Mallette MF. The natural occurrence of ethionine in bactdria. J. Gen. Physiol. 1961;45:1–13. doi: 10.1085/jgp.45.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tabor H. Polyamine biosynthesis in Escherichia coli: construction of polyamine-deficient mutants. Med. Biol. 1981;59:389–393. [PubMed] [Google Scholar]
- 81.Llewellyn NM, Li Y, Spencer JB. Biosynthesis of butirosin: transfer and deprotection of the unique amino acid side chain. Chem. Biol. 2007;14:379–386. doi: 10.1016/j.chembiol.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 82.Umitsu M, Nishimasu H, Noma A, Suzuki T, Ishitani R, Nureki O. Structural basis of AdoMet-dependent aminocarboxypropyl transfer reaction catalyzed by tRNA-wybutosine synthesizing enzyme, TYW2. Proc. Natl Acad. Sci. USA. 2009;106:15616–15621. doi: 10.1073/pnas.0905270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu J, Mushegian A. Displacements of prohead protease genes in the late operons of double-stranded-DNA bacteriophages. J. Bacteriol. 2004;186:4369–4375. doi: 10.1128/JB.186.13.4369-4375.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Witmer H. Synthesis of deoxythymidylate and the unusual deoxynucleotide in mature DNA of Bacillus subtilis bacteriophage SP10 occurs by postreplicational modification of 5-hydroxymethyldeoxyuridylate. J. Virol. 1981;39:536–547. doi: 10.1128/jvi.39.2.536-547.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yee LM, Matsumoto T, Yano K, Matsuoka S, Sadaie Y, Yoshikawa H, Asai K. The genome of Bacillus subtilis phage SP10: a comparative analysis with phage SPO1. Biosci. Biotechnol. Biochem. 2011;75:944–952. doi: 10.1271/bbb.100921. [DOI] [PubMed] [Google Scholar]
- 86.Maltman KL, Neuhard J, Warren RA. 5-[(Hydroxymethyl)-O-pyrophosphoryl]uracil, an intermediate in the biosynthesis of alpha-putrescinylthymine in deoxyribonucleic acid of bacteriophage phi W-14. Biochemistry. 1981;20:3586–3591. doi: 10.1021/bi00515a043. [DOI] [PubMed] [Google Scholar]
- 87.Jurgenson CT, Begley TP, Ealick SE. The structural and biochemical foundations of thiamin biosynthesis. Annu. Rev. Biochem. 2009;78:569–603. doi: 10.1146/annurev.biochem.78.072407.102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hollis T, Ichikawa Y, Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lohr JE, Chen F, Hill RT. Genomic analysis of bacteriophage PhiJL001: insights into its interaction with a sponge-associated alpha-proteobacterium. Appl. Environ. Microbiol. 2005;71:1598–1609. doi: 10.1128/AEM.71.3.1598-1609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Myllykallio H, Lipowski G, Leduc D, Filee J, Forterre P, Liebl U. An alternative flavin-dependent mechanism for thymidylate synthesis. Science. 2002;297:105–107. doi: 10.1126/science.1072113. [DOI] [PubMed] [Google Scholar]
- 91.Li L. Mechanistic studies of the radical SAM enzyme spore photoproduct lyase (SPL) Biochim. Biophys. Acta. 2012;1824:1264–1277. doi: 10.1016/j.bbapap.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mehta PK, Hale TI, Christen P. Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 1993;214:549–561. doi: 10.1111/j.1432-1033.1993.tb17953.x. [DOI] [PubMed] [Google Scholar]
- 93.Zhang J, Zhang Y, Inouye M. Thermotoga maritima MazG protein has both nucleoside triphosphate pyrophosphohydrolase and pyrophosphatase activities. J. Biol. Chem. 2003;278:21408–21414. doi: 10.1074/jbc.M213294200. [DOI] [PubMed] [Google Scholar]
- 94.Aravind L, Koonin EV. The alpha/beta fold uracil DNA glycosylases: a common origin with diverse fates. Genome Biol. 2000;1:RESEARCH0007. doi: 10.1186/gb-2000-1-4-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Iyer LM, Abhiman S, Aravind L. MutL homologs in restriction-modification systems and the origin of eukaryotic MORC ATPases. Biol. Direct. 2008;3:8. doi: 10.1186/1745-6150-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li L, Xu Z, Xu X, Wu J, Zhang Y, He X, Zabriskie TM, Deng Z. The mildiomycin biosynthesis: initial steps for sequential generation of 5-hydroxymethylcytidine 5′-monophosphate and 5-hydroxymethylcytosine in Streptoverticillium rimofaciens ZJU5119. Chembiochem. 2008;9:1286–1294. doi: 10.1002/cbic.200800008. [DOI] [PubMed] [Google Scholar]
- 97.Bertonati C, Punta M, Fischer M, Yachdav G, Forouhar F, Zhou W, Kuzin AP, Seetharaman J, Abashidze M, Ramelot TA, et al. Structural genomics reveals EVE as a new ASCH/PUA-related domain. Proteins. 2009;75:760–773. doi: 10.1002/prot.22287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Iyer LM, Burroughs AM, Aravind L. The ASCH superfamily: novel domains with a fold related to the PUA domain and a potential role in RNA metabolism. Bioinformatics. 2006;22:257–263. doi: 10.1093/bioinformatics/bti767. [DOI] [PubMed] [Google Scholar]
- 99.Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ishitani R, Nureki O, Nameki N, Okada N, Nishimura S, Yokoyama S. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell. 2003;113:383–394. doi: 10.1016/s0092-8674(03)00280-0. [DOI] [PubMed] [Google Scholar]
- 101.Liang B, Zhou J, Kahen E, Terns RM, Terns MP, Li H. Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nat. Struct. Mol. Biol. 2009;16:740–746. doi: 10.1038/nsmb.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 103.Bickle TA, Kruger DH. Biology of DNA restriction. Microbiol. Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Anantharaman V, Iyer LM, Aravind L. Ter-dependent stress response systems: novel pathways related to metal sensing, production of a nucleoside-like metabolite, and DNA-processing. Mol. Biosyst. 2012;8:3142–3165. doi: 10.1039/c2mb25239b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pieper U, Groll DH, Wunsch S, Gast FU, Speck C, Mucke N, Pingoud A. The GTP-dependent restriction enzyme McrBC from Escherichia coli forms high-molecular mass complexes with DNA and produces a cleavage pattern with a characteristic 10-base pair repeat. Biochemistry. 2002;41:5245–5254. doi: 10.1021/bi015687u. [DOI] [PubMed] [Google Scholar]
- 106.Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 108.Mutschler H, Gebhardt M, Shoeman RL, Meinhart A. A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol. 2011;9:e1001033. doi: 10.1371/journal.pbio.1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Makarova KS, Anantharaman V, Aravind L, Koonin EV. Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol. Direct. 2012;7:40. doi: 10.1186/1745-6150-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Anantharaman V, Aravind L. The NYN domains: novel predicted RNAses with a PIN domain-like fold. RNA Biol. 2006;3:18–27. doi: 10.4161/rna.3.1.2548. [DOI] [PubMed] [Google Scholar]
- 111.Aravind L, Anantharaman V, Zhang D, de Souza RF, Iyer LM. Gene flow and biological conflict systems in the origin and evolution of eukaryotes. Front. Cell Infect. Microbiol. 2012;2:89. doi: 10.3389/fcimb.2012.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bestor TH. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 113.Cheng X, Blumenthal RM. Introduction—Epiphanies in epigenetics. Prog. Mol. Biol. Transl. Sci. 2011;101:1–21. doi: 10.1016/B978-0-12-387685-0.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sternberg NL, Maurer R. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 1991;204:18–43. doi: 10.1016/0076-6879(91)04004-8. [DOI] [PubMed] [Google Scholar]
- 115.Abeles A, Brendler T, Austin S. Evidence of two levels of control of P1 oriR and host oriC replication origins by DNA adenine methylation. J. Bacteriol. 1993;175:7801–7807. doi: 10.1128/jb.175.24.7801-7807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Abeles AL, Austin SJ. P1 plasmid replication requires methylated DNA. EMBO J. 1987;6:3185–3189. doi: 10.1002/j.1460-2075.1987.tb02630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Iyer LM, Babu MM, Aravind L. The HIRAN domain and recruitment of chromatin remodeling and repair activities to damaged DNA. Cell Cycle. 2006;5:775–782. doi: 10.4161/cc.5.7.2629. [DOI] [PubMed] [Google Scholar]
- 118.Aravind L, Iyer LM. The HARE-HTH and associated domains: novel modules in the coordination of epigenetic DNA and protein modifications. Cell Cycle. 2012;11:119–131. doi: 10.4161/cc.11.1.18475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.