Abstract
The homeodomain-containing transcription factor, NKX3.1, plays an important role in the suppression of prostate tumorigenesis. Herein, we identify the receptor activity–modifying protein 1 (RAMP1) as a direct NKX3.1 target gene through analysis of chromatin immunoprecipitation coupled to massively parallel sequencing and gene expression data. RAMP1 is a coreceptor for certain G-protein–coupled receptors, such as the calcitonin gene-related peptide receptor, to the plasma membrane. We found that RAMP1 expression is specifically elevated in human prostate cancer relative to other tumor types. Furthermore, RAMP1 mRNA and protein levels are significantly higher in human prostate cancer compared with benign glands. We identified multiple NKX3.1 binding sites in the RAMP1 locus in human prostate cancer cells and in the normal mouse prostate. Analyses of Nkx3.1 knockout mice and human prostate cancer cell lines indicate that NKX3.1 represses RAMP1 expression. Knockdown of RAMP1 by shRNA decreased prostate cancer cell proliferation and tumorigenicity in vitro and in vivo. By using gene expression profiling and pathway analyses, we identified several cancer-related pathways that are significantly altered in RAMP1 knockdown cells, including the mitogen-activated protein kinase signaling pathway. Further experiments confirmed a reduction in MAP2KI (MEK1) expression and phosphorylated-extracellular signal–regulated kinase 1/2 levels in RAMP1 knockdown cells. These data provide novel insights into the role of RAMP1 in promoting prostate tumorigenesis and support the potential of RAMP1 as a novel biomarker and possible therapeutic target in prostate cancer.
Receptor activity–modifying proteins (RAMPs) 1, 2, and 3 are a small family of single transmembrane spanning proteins that were first identified during an attempt to clone the human calcitonin gene-related peptide (CGRP) receptor.1 Since their initial discovery, RAMPs have been shown to interact with G-protein–coupled receptors, such as calcitonin receptor-like receptor, vasoactive intestinal peptide receptor (VIPR), calcitonin receptor, calcium-sensing receptor, parathyroid hormone receptor, glucagon receptor, and corticotrophin releasing factor receptor 1, to regulate their trafficking, pharmacological characteristics, and signaling capabilities.1–5 Although the cellular biological and biochemical characteristics of these G protein-coupled receptors and their ligands are well studied, the functional significance and roles of RAMPs in human diseases remain undefined.
RAMP1 is expressed in many tissues, including the heart, uterus, bladder, brain, pancreas, skeletal muscle, and gastrointestinal tract.1 The expression of RAMP1 in these various tissues indicates that it may have diverse physiological functions. RAMP1 mRNA is also expressed in human adrenal tumors and meningiomas, and is up-regulated in prostate tumors.6–8 However, neither the underlying mechanism for up-regulation of RAMP1 expression in these tumors nor the functional significance of RAMP1 in tumorigenesis is fully understood.
NKX3.1 is an androgen-regulated, prostate-specific homeodomain transcription factor that plays critical roles in prostate development and in suppressing tumorigenesis.9–13 Throughout development and adulthood, Nkx3.1 functions to maintain prostate cellular homeostasis, and in mice, targeted deletion of Nkx3.1 in the prostate leads to developmental defects in ductal branching morphogenesis, secretory protein production, and growth.11,14 In addition, conditional deletion of one or both alleles of Nkx3.1 in the adult mouse prostate has been shown to promote the formation of premalignant lesions termed prostatic intraepithelial neoplasia.11–13 In humans, loss of heterozygosity at the NKX3.1 locus has been observed in a significant fraction of early-stage prostate cancer specimens.15,16 Furthermore, loss of NKX3.1 protein has been observed in approximately 20% of human prostatic intraepithelial neoplasia lesions and 40% of prostate tumors, which correlate with prostate tumor progression.17 NKX3.1 function can also be impaired by mutations in the NKX3.1 gene that can decrease its expression or affect the stability of the homeodomain structure.18–21 Although loss of Nkx3.1 does not result in invasive carcinoma, the introduction of additional mutations, such as loss of the tumor suppressor Pten in the Nkx3.1 mutant mouse prostate, can lead to invasive adenocarcinoma and, in some cases, metastatic disease.22,23 We and others have also shown that loss of Nkx3.1 and Myc overexpression can cooperate to promote prostate carcinogenesis.24–26 Furthermore, our laboratory showed that the cooperativity of these oncogenic mutations was the result of coregulation of shared target genes between Nkx3.1 and Myc that were observed in patients with prostate cancer and in a mouse model. Thus, a full understanding of the role of NKX3.1 in tumorigenesis requires the identification of genes directly regulated by NKX3.1 that may play a role in transformation.
We have recently used chromatin immunoprecipitation coupled to massively parallel sequencing (ChIP-Seq) to identify genomic loci bound by NKX3.1 in the human and mouse genomes.26 These data were integrated with gene expression profiling data from Nkx3.1 mutant mouse prostates27,28 to yield a core set of direct NKX3.1 target genes.26 In the present study, we identified RAMP1 as a novel NKX3.1 target gene and, for the first time to our knowledge, have characterized its functional role in prostate tumorigenesis.
Materials and Methods
Cell Lines and Constructs
Human prostate cancer cell lines, PC-3 and LNCaP, were obtained from ATCC (Manassas, VA). Cells were cultured in RPMI 1640 medium or Dulbecco’s modified Eagle’s medium/F-12 medium supplemented with 10% fetal bovine serum. Lentiviral-mediated gene transfer was used to generate stable knockdown of RAMP1 in PC-3 and LNCaP cells. The 293FT packaging cells were transfected with shRAMP1 (V2LHS_196808 or VLHS_180867) or pGIPZ vector control (RHS4346), provided by the Vanderbilt Genome Sciences Resource (Vanderbilt University Medical Center, Nashville, TN), along with Δ8.9 and vesicular stomatitis virus-G (provided by Dr. David Baltimore, Caltech, Pasadena, CA) to produce lentivirus as described.29 At 2 days after transfection, medium containing viral particles was collected and added to PC-3 cells for infection with15 μg/mL polybrene. At 48 hours after infection, medium was changed and 6 μg/mL puromycin or 800 μg/mL G418 was added for selection of stable clones. Transient transfection of PC-3 and DU145 was performed using polyethylenimine, with FUGW-GFP or FUGW-Nkx3.1 plasmid (a kind gift from Dr. Hong Wu, University of California, Los Angeles).
Human Tissue Arrays
Histological slides of radical prostatectomy specimens from patients with prostatic carcinoma were reviewed to identify areas of prostatic carcinoma of various grades and areas with benign prostatic glands remote from site(s) of carcinoma. The corresponding paraffin blocks were used to generate a TMA containing 38 foci of benign prostatic tissue and 23 foci of prostatic adenocarcinoma with different Gleason scores. The TMA was produced using a manual tissue microarrayer model MTA-1 from Beecher Instruments, Inc. (Sun Prairie, WI). An additional TMA (PR483a) was obtained from US Biomax (Rockville, MD) and contained 16 foci of benign prostatic tissue and 37 foci of prostatic adenocarcinoma. RAMP1 staining in each focus on the TMAs was scored semiquantitatively using the H-score, as previously described.30 The H-score was determined as the intensity of expression (0, none; 1, weak; 2, moderate; 3, strong) multiplied by the percentage of cells (0% to 100%) stained at each intensity level to provide a score ranging from 0 to 300. Anti-RAMP1 staining was evaluated between the individual cores, and within the same core, because some cores contained both benign and malignant tissues. Two pathologists (S.T.S. and O.H.) evaluated the TMAs for RAMP1 staining, and any potential discrepancies were resolved by joint review on a double-headed microscope.
Oncomine Database Analysis
We used the Oncomine (Compendia Bioscience, Ann Arbor, MI) Cancer Microarray database31 to query the profile of RAMP1 expression in multicancer data sets and in prostate cancer data sets.
Cell Cycle Analysis
Cell cycle analysis was performed as previously described.32 Cells were harvested, washed thoroughly with PBS, fixed with cold 70% ethanol, and stained with propidium iodide (0.05 mg/mL) in a solution containing 0.1% Triton X-100 and 0.005 mg/mL RNase A. DNA content was analyzed by flow cytometry.
ChIP-qPCR
ChIP was performed using the ChIP Assay kit (Millipore, Billerica, MA), as described by the manufacturer, with the following modifications. LNCaP cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin and streptomycin, and 1 nmol/L dihydrotestosterone (Sigma, St. Louis, MO) for 48 hours. Cells were fixed in 1% formaldehyde at room temperature for 10 minutes to cross-link protein-DNA complexes. Next, cells were thoroughly washed with ice-cold PBS, pelleted, and resuspended in SDS lysis buffer (1% SDS, 10 mmol/L EDTA, and 50 mmol/L Tris at pH 8.1). Chromatin was sheared to a size of approximately 300 to 500 bp and diluted 1:10 with ChIP dilution buffer. An aliquot of the diluted sample (1%) was saved as input. Samples were precleared and precipitated overnight at 4°C with anti-NKX3.1 (Santa Cruz Biotechnology, Santa Cruz, CA) or normal goat IgG (Santa Cruz Biotechnology) antibodies. Antibody complexes were collected with Protein A Agarose/Salmon Sperm DNA (Millipore, Billerica, MA) for 2 hours and washed extensively, per manufacturer’s instructions. Samples were reverse cross-linked at 65°C overnight with 0.3 mol/L NaCl and 30 μg of RNase A (Qiagen, Germantown, MD). Input and bound DNAs were purified with a PCR Purification kit (Qiagen) and analyzed by quantitative PCR (qPCR; model 7300; Applied Biosystems, Grand Island, NY) using SYBR Green (Applied Biosystems). Primers were designed to flank the three NKX3.1 binding sites in the RAMP1 gene in LNCaP cells. The following primers were used for qPCR: RAMP1 (site 1), 5′-ACGATCACATATAAAGACCTTCCTTGT-3′ (forward) and 5′-AGGCCACTCAAAATAACGTTAAAATT-3′ (reverse); RAMP1 (site 2), 5′-ATTTCAGGGCCTCCTTTTCTAAG-3′ (forward) and 5′-AATGACTCAGCTGTGGCAGAAG-3′ (reverse); and RAMP1 (site 3), 5′-CACTCACCCGTAGGAGTTCCA-3′ (forward) and 5′-ATGAAAAGCACTTCAGCACACTGT-3′ (reverse). Immunoprecipitated DNA was normalized to 1% input.
Real-Time RT-qPCR Analyses
RNA isolation, reverse transcription, and subsequent RT-qPCR (TaqMan; Applied Biosystems) using SYBR Green dye were performed as described previously.33 All RT-qPCRs were performed in triplicate. The following primers were used: 18S forward, 5′-CGCCGCTAGAGGTGAAATTCT-3′; 18S reverse, 5′-CGAACCTCCGACTTTCGTTCT-3′; Ramp1 forward, 5′-CGCACACGATTGGCTGTTT-3′; Ramp1 reverse, 5′-TGGACAGCGATGAAGAATCTGT-3′; GAPDH forward, 5′-ATGGAAATCCCATCACCATCTT-3′; GAPDH reverse, 5′-CGCCCCACTTGATTTTGG-3′; RAMP1 forward, 5′-CCTCACCCAGTTCCAGGTAG-3′; RAMP1 reverse, 5′-CATGTGCCAGGTGCAGTC-3′; RAMP2 forward, 5′-CCTTATAGCACCCTGCGAGATT-3′; RAMP2 reverse, 5′-GGGAAGCCCAGGTCAAACA-3′; RAMP3 forward, 5′-GGAAGGCTTTCGCAGACATG-3′; RAMP3 reverse, 5′-CGGACAGGTTGCACCACTT-3′; MAP2K1 forward, 5′-TTCTTGCTGGGCATACTTTCTCT-3′; and MAP2K1 reverse, 5′-CATGCACTGCCTGTGAAGGA-3′.
Western Blot Analyses
Western blot analysis was performed, as described previously,34 using the following antibodies: anti-Ramp1 (rabbit, 1:1000; Santa Cruz Biotechnology), anti–β-actin (goat, 1:1000; Santa Cruz Biotechnology), anti-Nkx3.1 (rabbit, 1:2000; provided by Dr. Charles Bieberich, University of Maryland, Baltimore County, Baltimore), and anti–phosphorylated-extracellular signal regulated kinase (ERK) 1/2 (rabbit, 1:2000; Cell Signaling, Boston, MA).
IHC and Immunocytochemistry
Immunohistochemistry (IHC) was performed as previously described.35 For antigen retrieval, slides were steamed for 15 minutes in Tris-EDTA buffer (pH 9.0). Slides were incubated overnight at 4°C with the following antibodies: anti-RAMP1 (rabbit, 1:150; Santa Cruz Biotechnology) for mouse tissue or 1:40 for human prostate TMAs; anti–Ki-67 (rabbit, 1:1000; Abcam, Cambridge, MA), phosphorylated-histone H3 (pHH3; rabbit, 1:500; Millipore), and activated caspase-3 (rabbit, 1:100; Cell Signaling). Slides were stained with 3,3′-diaminobenzidine tablets (Sigma) and counterstained with hematoxylin. For immunocytochemistry, cells were plated on coverslips and incubated overnight at 4°C with anti-RAMP1 (1:50), pHH3 (rabbit, 1:500; Millipore), and activated caspase-3 (rabbit, 1:100; Cell Signaling). Slides were counterstained with DAPI (Vector Labs, Burlingame, CA). The number of cells staining positive for pHH3 and caspase-3, of a total of 500 cells in three independent fields, was recorded. For cell block preparation, cells were grown to 80% confluency, harvested, and washed thoroughly with PBS. Cells were fixed for 5 minutes in 10% neutral-buffered formalin. Cell block was processed by the Vanderbilt Translational Pathology Shared Resource, and slides were stained for anti-RAMP1 (1:50).
Cell Growth Assay
Cell growth was determined by cell count. A total of 1 × 104 cells were plated in a 6-well plate in triplicate in complete medium and counted at the indicated time points.
Soft Agar Assay
A soft agar colony formation assay was performed as previously described.36 Briefly, 1.5 × 104 PC-3 or LNCaP cells, stably expressing shControl or shRAMP1, were mixed with 0.35% soft agar and plated on top of a 0.5% bottom agar in a 6-well plate. Cells were incubated at 37°C for 2 to 3 weeks to allow colony formation. Each cell line was plated in triplicate. Three random low-power view fields were chosen, and the total number of colonies (cutoff sizes or more) was counted.
Xenograft Studies
Xenograft studies were performed as previously described.37 Male nude (nu/nu) mice were obtained from Charles River Laboratories (Wilmington, MA). A total of 8 × 106 PC-3 cells stably expressing shControl or shRAMP1 were injected s.c. in both flanks of nude mice. Tumor volumes were calculated as previously described.37 Animal care and experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee at Vanderbilt University.
Gene Expression Profiling and Analysis
Total RNA was extracted from PC-3 cells stably expressing shControl or shRAMP1, according to the TRIzol manufacturer’s protocol (Invitrogen, Grand Island, NY). Three independent sample preparations for shControl and shRAMP1 cells were performed. RNA was treated with DNase I (Qiagen, Gaithersburg, MD), according to the manufacturer’s protocol, followed by purification using the RNA Clean Up protocol from the RNeasy Mini Kit (Qiagen). RNA processing and microarray analysis were performed by the Vanderbilt Genome Sciences Resource Core, as previously described.38 cDNA was hybridized to the GeneChip PrimeView Human Gene Expression Array (Affymetrix, Santa Clara, CA). CEL files were imported to R, version 2.15.0 (http://www.r-project.org), for quality control and preprocessing. Quality control was implemented in Bioconductor with affyPLM, version 1.32, and simpleaffy, version 2.32, packages. Arrays for three independent shControl and shRAMP1 samples passed quality control tests. By using the Affy package, version 1.34.0,39 raw intensity scores were normalized by quartiles, background corrected with robust multi-array average,40 and summarized by mean polish using perfect match-only probes. The C2 (curated) and C6 (oncogenic signatures) gene sets of Molecular Signatures Database, version 3.1 (Broad Institute, Cambridge, MA), were queried using Gene Set Enrichment Analysis (GSEA) (Broad Institute), version 2.07,41 to test for differences between shControl and shRAMP1 cells. All microarray and GSEA analyses were performed on a node running Debian Linux Squeeze, version 6.0.6 (http://www.debian.org/releases).
Statistical Analysis
To compare groups, we used the unpaired Student’s t-test. Data are expressed as means ± SD or SEM for each group. Data were considered statistically significant at P < 0.05.
Results
RAMP1 Is Overexpressed in Human Prostate Cancer
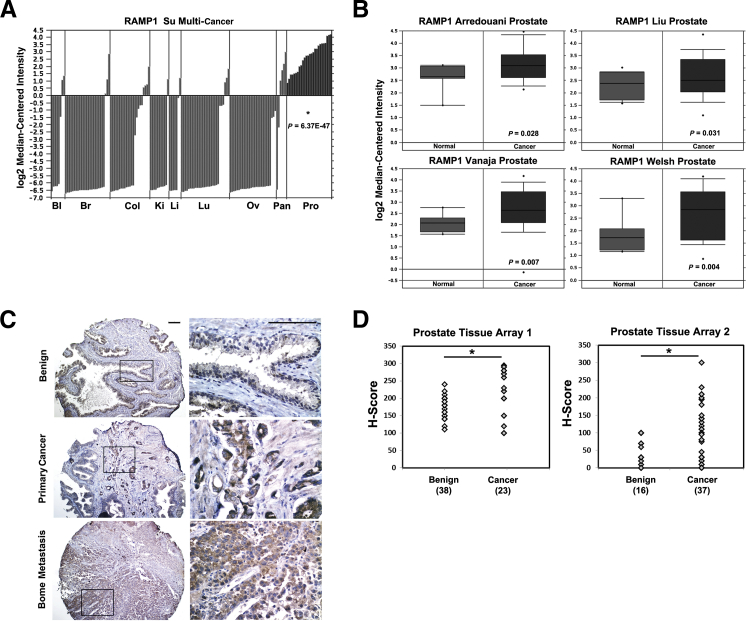
A previous study identified RAMP1 mRNA as one of several mRNAs with potential to be a biomarker in prostate cancer.8 To determine whether RAMP1 is up-regulated in prostate or other cancers, we queried the Oncomine database for a comprehensive evaluation of RAMP1 expression in human cancers. Oncomine is a cancer-specific database that identifies genes, pathways, and networks deregulated across gene expression microarrays.31 Notably, RAMP1 was specifically up-regulated in prostate cancer (P = 6.36 × 10−47) compared with other human cancers that include bladder, breast, colorectal, kidney, liver, lung, ovarian, and pancreatic in the Su Multicancer data set42 (Figure 1A). Next, we examined the expression of RAMP1 in normal prostate versus prostate carcinoma. RAMP1 was significantly up-regulated in prostate carcinoma compared with normal prostate in multiple data sets43–46 (Figure 1B). To determine whether RAMP1 protein expression is also altered during prostate tumorigenesis, we performed RAMP1 IHC on two independent human prostate cancer TMAs. RAMP1 expression was detected in the cytoplasm and membrane of prostatic epithelial cells with limited expression in the stroma (Figure 1C). RAMP1 protein expression was also significantly up-regulated in human prostate cancer compared with benign glands (Figure 1D). Overall, these findings show that RAMP1 is specifically up-regulated in prostate cancer and may be relevant for prostate tumorigenesis.
Figure 1.
RAMP1 is up-regulated in human prostate cancer. A: RAMP1 mRNA expression in various human cancers in a multicancer data set from the Oncomine cancer microarray database shows significantly higher RAMP1 levels in prostate relative to other tumors. Bl, bladder; Br, breast; Cl, colon; Ki, kidney; Li, liver; Lu, lung; Ov, ovarian; Pan, pancreatic; Pro, prostate. B: RAMP1 mRNA expression in normal prostate versus prostate cancer from four different prostate cancer microarray data sets from the Oncomine database. C: Examples of immunostaining for anti-RAMP1 in benign prostate, primary prostate cancer, and bone metastasis from a human prostate TMA. Original magnifications: ×10 (left panel); ×40 (right panel). Scale bar = 0.1 mm. D: H-score analysis of anti-RAMP1 staining of samples from two independent human prostate TMAs. The expression levels of RAMP1 in benign and malignant prostatic tissue were compared using the unpaired Student’s t-test. ∗P < 0.05.
RAMP1 Is a Direct NKX3.1 Target Gene
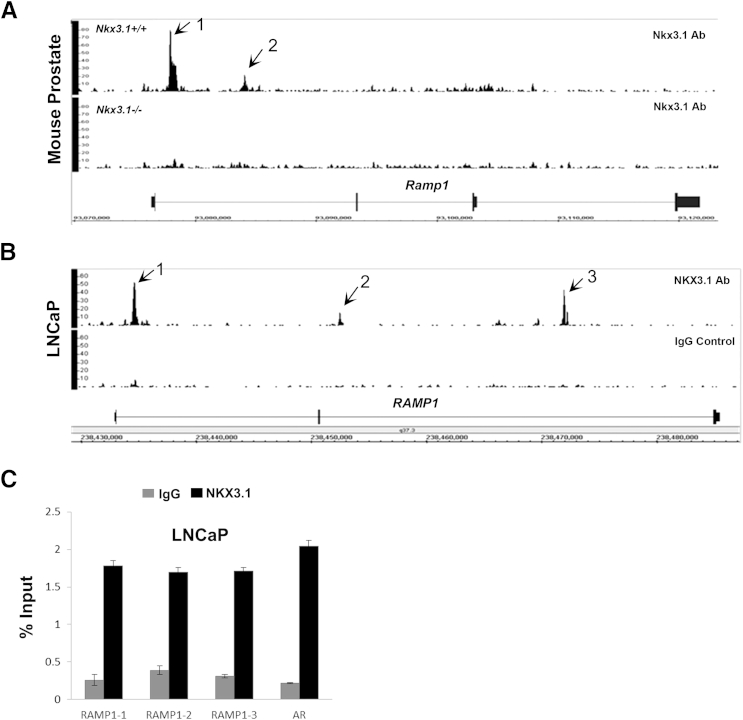
We have previously used ChIP-Seq to identify genomic loci bound by NKX3.1 in the prostate cancer cell line, LNCaP (M.L., P.D.A., and S.A.A., unpublished data) and in the normal mouse prostate.26 Analysis of data from these ChIP-Seq studies showed that Nkx3.1 binds Ramp1 downstream of the transcription start site and within the gene in the mouse prostate (Figure 2A). In LNCaP cells, NKX3.1 binds the RAMP1 locus at three different sites (Figure 2B). NKX3.1 binding to the RAMP1 gene was confirmed by ChIP, followed by qPCR, using primers flanking the three NKX3.1 binding sites within the RAMP1 gene in LNCaP cells (Figure 2C). An androgen receptor promoter region containing a consensus NKX3.1 binding site was used as a positive control. All three binding sites exhibited significant enrichment for NKX3.1 compared with IgG control. These results suggest that the prostate tumor suppressor NKX3.1 might be involved in the regulation of RAMP1 expression in the prostate.
Figure 2.
NKX3.1 binds to the RAMP1 gene in human and mouse prostate cells. Integrated Genome Browser screenshots of NKX3.1 binding sites in the mouse (A) and human (LNCaP) (B) genomes. Binding of NKX3.1 is indicated by arrows. IgG and Nkx3.1 knockout tissue served as a negative control. C: ChIP-qPCR confirmation of the three NKX3.1 binding sites in the RAMP1 gene in LNCaP cells. An androgen receptor (AR) promoter region known to bind NKX3.1 was used as a positive control, whereas IgG served as a negative control.
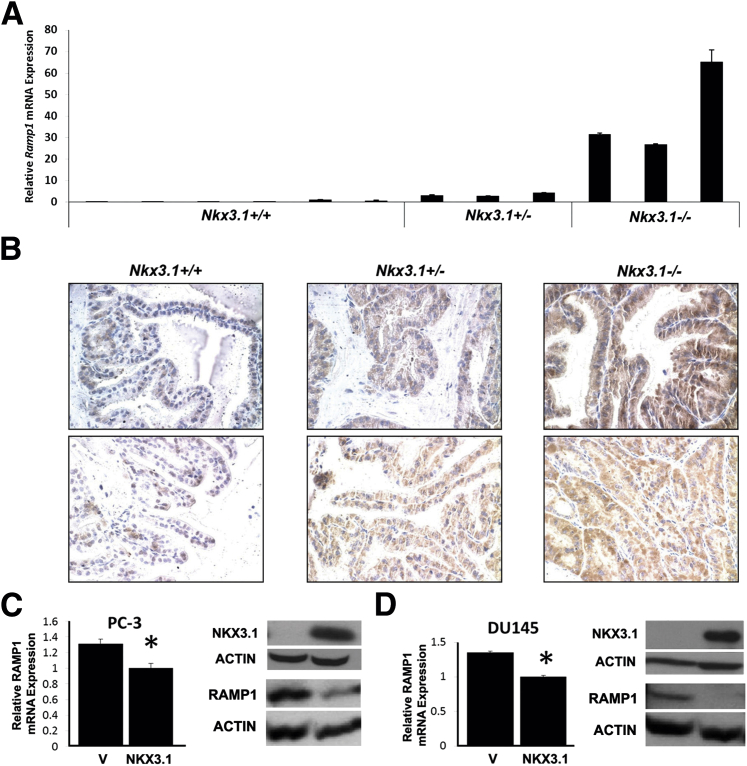
To explore the functional significance of Nkx3.1 binding to the Ramp1 gene, we examined the expression of Ramp1 in Nkx3.1 knockout mice. RT-qPCR analysis of the prostates of Nkx3.1 mutant mice showed a dose-dependent increase in Ramp1 mRNA in Nkx3.1+/− and Nkx3.1−/− mouse prostates (Figure 3A). We observed a similar effect of loss of Nkx3.1 on Ramp1 protein expression in the prostate by IHC (Figure 3B). Nkx3.1 knockout mice expressed high levels of Ramp1 protein in the prostatic epithelium with limited expression in the stroma, consistent with the known cellular localization of Nkx3.1 protein in the prostate.12 These results indicate that Nkx3.1 represses Ramp1 expression in a dose-dependent manner in the mouse prostate. We extended these mouse results to human prostate cells by assessing the effect of exogenous NKX3.1 expression in PC-3 and DU145 cells on RAMP1 expression. We used lentiviral-mediated gene transfer to express mouse Nkx3.1 in PC-3 and DU145 cells, which normally lack NKX3.1 expression (Figure 3, C and D). The levels of exogenous Nkx3.1 expression achieved are comparable to the endogenous NKX3.1 levels in LNCaP cells (data not shown). In both PC-3 and DU145 cells, Nkx3.1 expression repressed RAMP1 mRNA and protein expression, as determined by RT-qPCR and Western blot analyses, respectively (Figure 3, C and D). The results presented, when taken together, indicate that NKX3.1 binds to and represses the RAMP1 gene in the prostate.
Figure 3.
NKX3.1 represses RAMP1 expression in mouse and human prostate cells. A: Real-time RT-qPCR analysis of Ramp1 expression in the Nkx3.1-deficient mouse prostate. Total RNA was isolated from anterior prostates of 16–week-old Nkx3.1+/+ (n = 6), Nkx3.1+/− (n = 3), and Nkx3.1−/− (n = 3) mice. Expression levels were normalized to 18S. The results shown are representative of two independent experiments performed in triplicate. Data are reported as means ± SD. B: Immunostaining for anti-Ramp1 in the anterior prostate of 16–week-old Nkx3.1+/+, Nkx3.1+/−, and Nkx3.1−/− mice shows up-regulation of Ramp1 (brown) in Nkx3.1+/− and Nkx3.1−/− mouse prostates. Images from two different mice are shown for each genotype. Original magnification, ×40. C and D: Lentiviral-mediated expression of Nkx3.1 represses RAMP1 mRNA and protein levels in PC-3 and DU145 cells. RT-qPCR for RAMP1 and Western blot analyses for NKX3.1, RAMP1, and ACTIN control are shown. Results are representative of two independent experiments. RT-qPCR data are reported as means ± SD, with RAMP1 expression levels normalized to GAPDH. ∗P < 0.05.
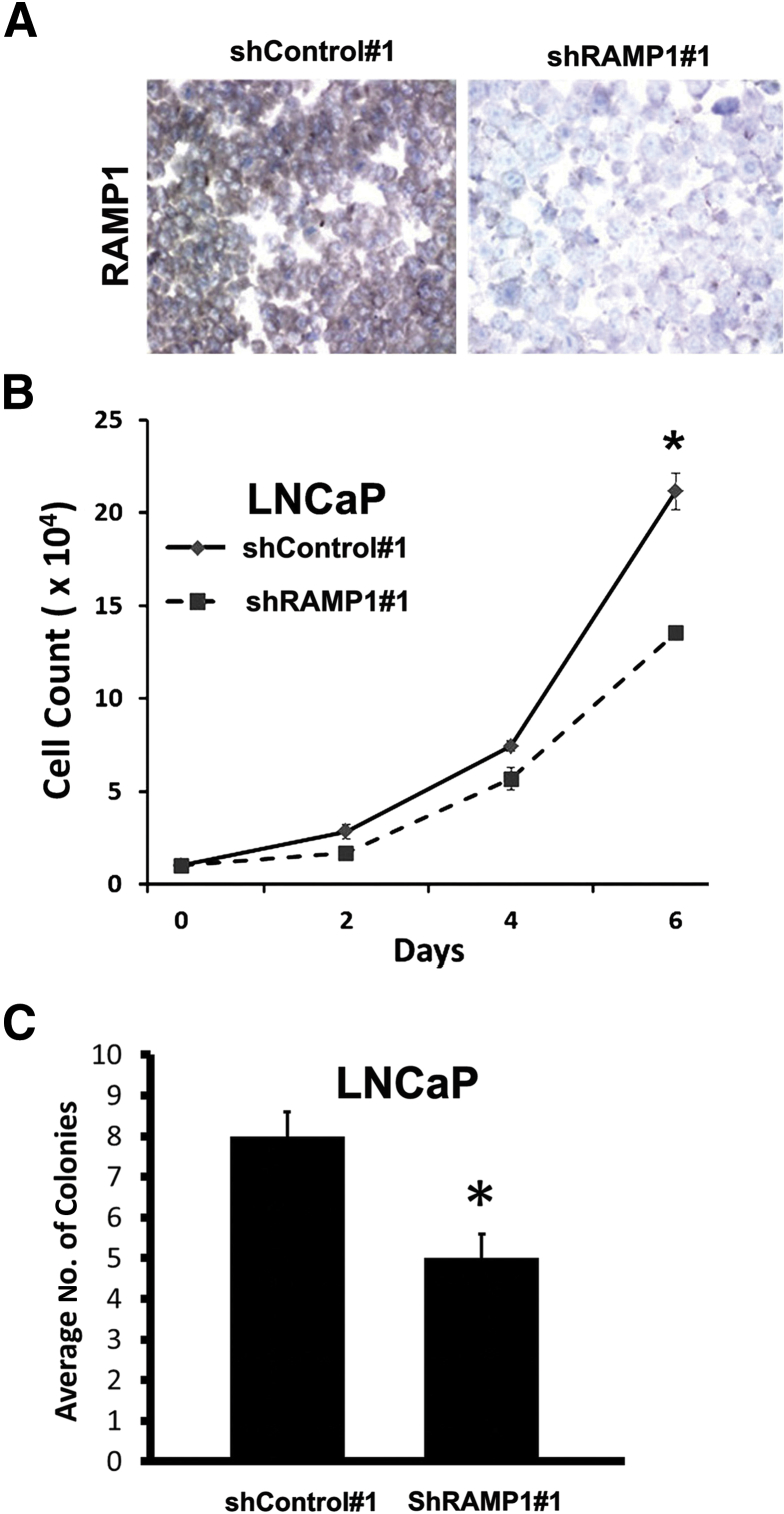
Down-Regulation of RAMP1 Suppresses Cell Proliferation and in Vitro Tumorigenicity in PC-3 and LNCaP Cells
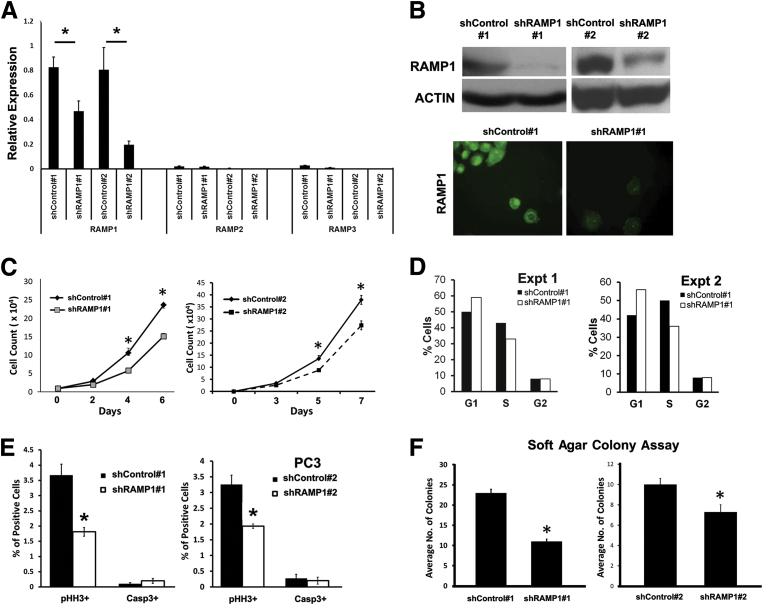
To examine the functional significance of the RAMP1 up-regulation in human prostate cancer, we stably knocked down RAMP1 in PC-3 cells using two independent shRNAs, shRAMP1 1 and shRAMP1 2 (Figure 4, A and B). Knockdown efficiency was evaluated by RT-qPCR, Western blot, and immunocytochemistry analyses (Figure 4, A and B). To further confirm specificity of the shRAMP1 constructs, we evaluated the expression of RAMP2 and RAMP3 and found these to be expressed at low levels in these cells without being affected by RAMP1 knockdown (Figure 4A). Functionally, knockdown of RAMP1 significantly decreased cell proliferation in both shRAMP1 1 and shRAMP1 2 PC-3 cells, as determined by cell counting and the pHH3 index (Figure 4, C and E). Similarly, we observed a decrease in the percentage of cells in the S phase of the cell cycle in RAMP1-deficient cells compared with shControl cells (Figure 4D). There was no significant change in apoptosis, as determined by staining for activated caspase-3 (Figure 4E). We next examined the effect of RAMP1 depletion in prostate cancer cells on in vitro tumorigenicity using the soft agar colony formation assay. Consistent with the proliferation assays, a significant decrease in colony formation was observed in PC-3 cells with stable knockdown of RAMP1 (Figure 4F). These findings were replicated in LNCaP prostate carcinoma cells with stable depletion of RAMP1 by shRNA, which also showed significant reductions in proliferation and tumorigenicity (Figure 5, A–C). These results establish a functional role for RAMP1 in promoting the proliferation and in vitro tumorigenicity of prostate cancer cells.
Figure 4.
RAMP1 promotes prostate cancer cell proliferation and tumorigenicity in PC-3 cells. A: RT-qPCR analysis of RAMP1, RAMP2, and RAMP3 expression in PC-3 cells stably expressing shControl and shRAMP1 confirms specificity of RAMP1 knockdown. B: Western blot (top panel) and immunocytochemical (bottom panel) analyses confirm knockdown of RAMP1 in PC-3 cells. C: Cell growth was determined by cell count. D: Cell cycle analysis of RAMP1 knockdown PC-3 cells by flow cytometry. E: Quantitation of proliferation and apoptosis in shControl and shRAMP1 cells after staining for pHH3 and caspase 3, respectively. Data are reported as percentage of cells positive for the marker. F: Analysis of in vitro tumorigenicity by soft agar assay. Results are representative of at least two independent experiments performed in triplicate. Data are reported as means ± SEM. ∗P < 0.05.
Figure 5.
Knockdown of RAMP1 in LNCaP cells decreases cell proliferation and tumorigenicity in vitro. A: IHC analysis of cell blocks shows reduced RAMP1 expression (reddish brown staining) in LNCaP cells expressing shRAMP1 compared with shControl cells. B: Cell growth was determined by cell count. C: Analysis of in vitro tumorigenicity by soft agar assay. Results are representative of at least two independent experiments performed in triplicate. Data are reported as means ± SEM. ∗P < 0.05.
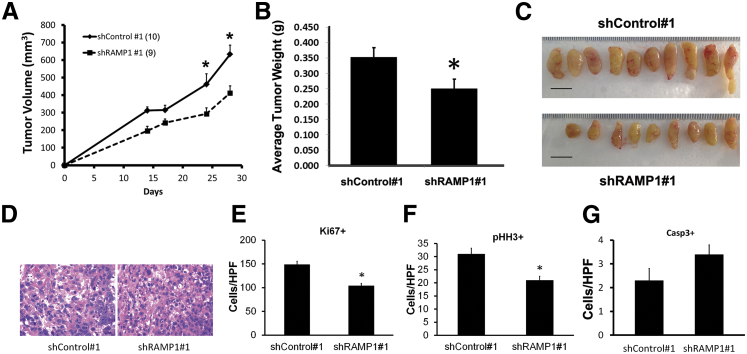
Knockdown of RAMP1 Decreases Prostate Cancer Cell Tumorigenicity in Vivo
Next, we performed xenograft studies in nude mice using PC-3/shControl 1 and PC-3/shRAMP1 1 cells to access the effect of RAMP1 knockdown in vivo. Tumors in mice injected with shRAMP1-expressing cells grew at a slower rate and were smaller in size and weight at the end of the experiment compared with tumors in mice injected with shControl cells (Figure 6, A–C). We analyzed proliferation in these tumors by IHC staining for Ki-67 and pHH3. Proliferation was significantly reduced in RAMP1-depleted tumors relative to the control tumors (Figure 6, D–F). Staining for activated caspase-3 showed a trend for increased apoptosis that did not achieve statistical significance (Figure 6G). These results are consistent with our in vitro data showing that RAMP1 knockdown mainly affects prostate cancer cell proliferation.
Figure 6.
Depletion of RAMP1 reduces tumorigenicity and proliferation in vivo. A–C: Xenografts of shRAMP1 1 PC3 cells grown s.c. in nude mice show reduced tumor growth (A) and final tumor weight (B) and size (C). D: Representative images of H&E-stained sections of tumor grafts. E: Ki-67 proliferation index. F: pHH3, phosphorylated-histone H3 serine 10 index. G: Apoptotic index by activated caspase-3 staining. The total number of cells staining positive for either Ki-67, pHH3, or activated caspase-3 in five random high-power field (HPFs) were counted for each graft section. Data are reported as means ± SEM. ∗P < 0.05.
Down-Regulation of RAMP1 Modulates MAPK Signaling in Prostate Carcinoma Cells
To explore possible molecular pathways by which RAMP1 down-regulation modulates the tumorigenicity of prostate cancer cells, we performed gene expression profiling using a GeneChip PrimeView Human Gene Expression Array (Affymetrix). We found 936 genes to be significantly altered (α = 0.05) before a multiple testing penalty in RAMP1 knockdown cells. By using these genes to interrogate WebGestalt, we identified several significantly altered pathways, including the epidermal growth factor receptor 1 (EGFR1), mitogen-activated protein kinase (MAPK), and IL-6 signaling pathways (Table 1). Notable among the genes altered in the EGFR1 and MAPK signaling pathways is the MAPK signaling pathway regulator, MAP2K1 (MEK1) (Supplemental Table S1). Some of the IL-6 signaling pathway genes identified as altered in RAMP1-depleted cells include CCND, JAK1, IL6, IL6R, and SOS2.
Table 1.
Pathway Analysis of Genes Dysregulated in RAMP1 Knockdown PC3 Cells
| Variable | No. of genes | Expected | Enriched | Adjusted P value |
|---|---|---|---|---|
| KEGG | ||||
| Pathways in cancer | 23 | 5.62 | 4.09 | 1.37 × 10−6 |
| MAPK signaling pathway | 14 | 4.58 | 3.06 | 0.0016 |
| Endocytosis | 16 | 3.18 | 5.02 | 6.52 × 10−6 |
| Spliceosome | 11 | 2.18 | 5.05 | 0.0004 |
| Neurotrophin signaling pathway | 10 | 2.15 | 4.66 | 0.001 |
| Prostate cancer | 8 | 1.52 | 5.28 | 0.001 |
| Small-cell lung cancer | 8 | 1.43 | 5.59 | 0.001 |
| Chronic myeloid leukemia | 8 | 1.28 | 6.26 | 0.0009 |
| Pancreatic cancer | 7 | 1.23 | 5.71 | 0.0016 |
| Glioma | 7 | 1.11 | 6.32 | 0.001 |
| WIKI Pathways | ||||
| EGFR1 signaling pathway | 20 | 3.03 | 6.6 | 1.95 × 10−9 |
| Insulin signaling | 10 | 2.72 | 3.67 | 0.0024 |
| B-cell receptor signaling pathway | 12 | 2.69 | 4.46 | 0.0002 |
| Regulation of actin cytoskeleton | 10 | 2.43 | 4.11 | 0.0013 |
| mRNA processing | 11 | 2.32 | 4.75 | 0.0002 |
| Adipogenesis | 9 | 2.26 | 3.97 | 0.0027 |
| Androgen receptor signaling pathway | 11 | 1.96 | 5.62 | 8.22 × 10−5 |
| IL-6 signaling pathway | 10 | 1.67 | 5.99 | 9.31 × 10−5 |
| Senescence and autophagy | 10 | 1.02 | 9.79 | 1.79 × 10−6 |
| Id signaling pathway | 6 | 0.87 | 6.91 | 0.0013 |
| Pathway Commons | ||||
| Glypican pathway | 23 | 7.42 | 3.1 | 0.0001 |
| Glypican 1 network | 23 | 6.88 | 3.34 | 4.95 × 10−5 |
| IFN-γ pathway | 19 | 5.82 | 3.26 | 0.0004 |
| TRAIL signaling pathway | 17 | 5.06 | 3.36 | 0.0006 |
| Signaling by GPCR | 13 | 3.32 | 3.92 | 0.0008 |
| EGFR1 | 14 | 2.45 | 5.71 | 2.33 × 10−5 |
| Sphingosine 1-phosphate pathway | 10 | 1.92 | 5.2 | 0.0007 |
| Signaling by EGFR | 10 | 1.12 | 8.9 | 2.33 × 10−5 |
| Synaptic transmission | 7 | 0.92 | 7.61 | 0.0008 |
| IL-6–mediated signaling events | 7 | 0.77 | 9.14 | 0.0004 |
GPCR, G protein-coupled receptor; Id, inhibitor of DNA binding; IFN, interferon; KEGG, Kyoto Encyclopedia of Genes and Genomes; TRAIL, TNF-related apoptosis-inducing ligand.
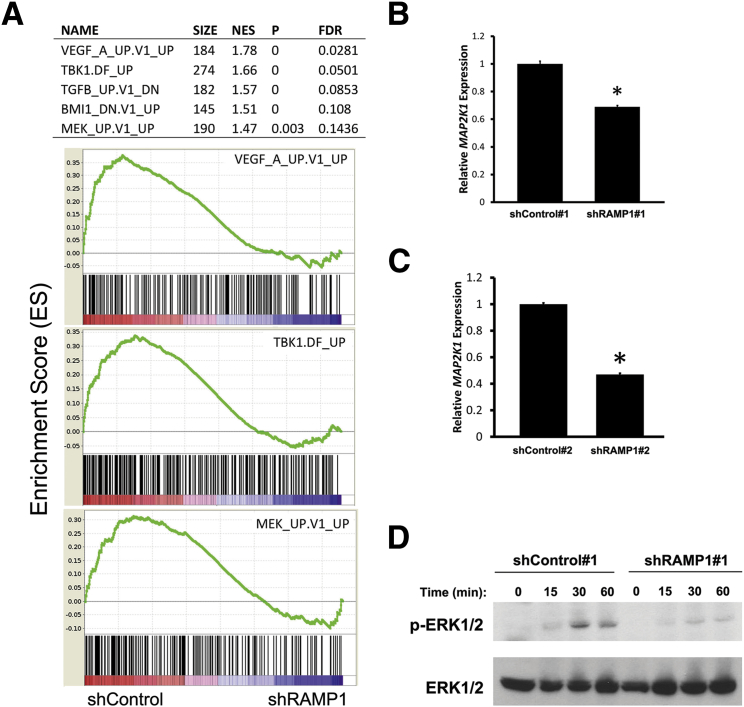
We next performed GSEA to define gene sets significantly enriched or depleted in RAMP1 knockdown cells. GSEA is a comprehensive database of genes, pathways, and regulatory and functional networks activated and repressed in human cancer.31,47 We interrogated the curated cancer-relevant database C6 of the Molecular Signatures Database, a collection of gene sets curated from gene expression profiles in cancer-relevant experiments, to identify possible cancer-relevant pathways altered in shRAMP1 cells. Interestingly, among the top gene sets enriched in control relative to shRAMP1 cells is MEK_UP.V1_UP, a gene set found to be activated in cells with enhanced MAPK signaling (Figure 7A and Supplemental Table S2). This gene set consists of genes up-regulated in MCF-7 breast cancer cells stably overexpressing constitutively active MAP2K1 (MEK1).48 Other gene sets identified in this analysis were proangiogenic genes, VEGF-A and TBK1 (Figure 7A). The VEGF-A gene set consists of genes that were found to be up-regulated on vascular endothelial growth factor (VEGF)-A treatment, whereas the TBK1 gene set is composed of genes that were up-regulated after overexpression of the RAS oncogene.
Figure 7.
Down-regulation of RAMP1-altered gene expression and MAPK signaling in PC-3 cells. A: GSEA shows a list of selected gene sets enriched in shControl relative to shRAMP1 cells (top panel). GSEA enrichment score plots for selected gene sets (bottom panel). Normalized enrichment score (NES), nominal P value (statistical significance of enrichment), and false-discovery rate (FDR) are presented. B and C: RT-qPCR analysis of MAP2K1 expression in RAMP1 knockdown cells. Results are representative of at least two independent experiments performed in triplicate. Data are reported as means ± SEM. ∗P < 0.05. D: Western blot analysis of ERK1/2 activation in shControl and shRAMP1 cells. Cells were serum starved for 24 hours, induced with 10% fetal bovine serum, and harvested at 0-, 15-, 30-, and 60-minute time points. Results are representative of at least two independent experiments.
Because MEK encodes a protein that phosphorylates and activates ERK1/2, we hypothesized that depletion of RAMP1 may inhibit prostate cell tumorigenicity by suppressing the MAPK signaling pathway. We confirmed reduced expression of MAP2K1 (MEK1) in prostate cells depleted of RAMP1 using both shRAMP1 1 and shRAMP1 2 (Figure 7, B and C). Next, we performed Western blot analysis to determine the effect of RAMP1 knockdown on ERK1/2 activation. As expected, RAMP1 knockdown suppressed activation of ERK1/2 in PC3 cells, as revealed by reduced ERK1/2 phosphorylation (Figure 7D). These results suggest suppression of MAPK signaling as a mechanism for the reduced prostate tumorigenicity observed in RAMP1-deficient cells.
Discussion
Loss of NKX3.1 expression is often associated with the initiation of prostate tumorigenesis. In this study, we identified RAMP1 as a direct NKX3.1 target gene involved in promoting prostate tumorigenesis and have, for the first time to our knowledge, reported a functional role for RAMP1 in this disease. NKX3.1 binds multiple sites in the RAMP1 locus in both the human and mouse genomes. Furthermore, deletion of the Nkx3.1 gene in mice led to the up-regulation of Ramp1 in a dose-dependent manner, whereas exogenous Nkx3.1 suppressed RAMP1 expression in human prostate cancer cells. Our studies suggest that loss of NKX3.1 during human prostate tumorigenesis may be a key factor in the up-regulation of RAMP1 in tumors.
RAMP1 is well known for regulating the trafficking and pharmacological features of G protein-coupled receptors. In addition, overexpression of RAMP1 has been shown to play a protective role in cardiovascular disease by attenuating angiotensin II–induced hypertension and vascular dysfunction.49,50 Herein, we described another functionally significant role for RAMP1 in human prostate cancer. Previously, RAMP1 mRNA was shown to be up-regulated in human prostate tumors, and it was considered to be a novel potential biomarker for prostate cancer.8 We observed up-regulation of RAMP1 protein in prostate tumors, and that depletion of RAMP1 decreased cell proliferation and tumorigenicity in prostate cancer cells. These findings indicate that RAMP1 may play a pivotal role in prostate tumorigenesis.
RAMP1 has been shown to interact with VIPR1 and CALCRL, two receptors that bind ligands that have been shown to promote tumorigenesis.1,4 RAMP1 interaction with the CALCRL receptor yields the CGRP receptor, which has high affinity for the neuropeptide, CGRP.1 CGRP has been shown to increase the invasiveness and migration of human prostate cancer cells, and angiogenesis and tumor growth in a xenograft model.51–53 Equally important, CGRP has been reported to be elevated in the serum of untreated patients with prostate cancer.54 It has previously been reported that CGRP can regulate VEGF expression in human HaCaT keratinocytes by activation of ERK1/2.55 VEGF is a proangiogenic gene that is overexpressed in prostate cancer and in NKX3.1-deficient cells.56–58 Interestingly, our GSEA analysis shows VEGF-A as the top gene altered in RAMP1-deficient cells. TANK-binding kinase-1 (TBK1) is another proangiogenic gene that is altered in RAMP1-knockdown cells. TBK1 has been shown to induce cell proliferation and secretion of angiogenesis-associated factors,59 possibly by mediating insulin and AKT signaling pathways.60 Taken together, our data suggest that RAMP1 may regulate tumor growth and tumor-associated angiogenesis by modulating cell survival pathways.
Likewise, VIPR1 binds the neuropeptide, VIP. In vitro, VIP induced the malignant transformation of nontumorigenic prostate epithelial cells and increased migration of LNCaP cells, an androgen-sensitive human prostate cancer cell.52,61 VIP has also been shown to enhance angiogenesis and tumor growth in in vivo models of human prostate cancer.61–63 These studies are important considering that the prostate gland contains autonomic and sensory neurons and neuroendocrine cells that secrete these neuropeptides. In fact, it has been reported that, in prostatic adenocarcinomas, neuroendocrine cells tend to occur in close proximity to proliferating cells.64 Thus, this may explain why both CGRP and VIP may cause prostate tumor growth. However, it is possible that there are other unknown RAMP1-interacting partners that may also promote prostate tumorigenesis.
Our gene expression profiling data indicated that altered MAPK signaling is one of the functional consequences of RAMP1 depletion. We identified MAP2K1, an upstream kinase in the MAPK signaling pathway as a gene whose expression is impaired in RAMP1-depleted cells. Previous studies have shown that both activation of CGRP and VIP can induce ERK1/2 activation,55,65 suggesting that lack of RAMP1 leads to reduced MAPK signaling by attenuating signal transduction via RAMP1 coreceptors. Another pathway altered in RAMP1-deficient cells is the EGFR1 signaling pathway. EGFR1 is frequently overexpressed in hormone-refractory and metastatic prostate cancer.66,67 EGFR1 signaling can also lead to activation of downstream cell survival pathways, MAPK, and phosphatidylinositol 3-kinase.68 In addition, activation of IL-6 signaling was also suppressed in RAMP1-depleted cells. IL-6 is a pleiotropic cytokine with diverse roles in immune and inflammatory responses and in mediating growth of normal and tumor cells.69 It mediates its effects through activation of the MAPK, phosphatidylinositol 3-kinase, and Janus activated-kinase/signal transducers and activators of transcription signaling pathways that promote survival of cancer cells.70–72 Several studies have observed elevated IL-6 in the sera of patients with castration-resistant prostate cancer compared with men with a normal-functioning prostate, benign prostatic hyperplasia, prostatitis, and localized or recurrent disease.73–76 Also, IL-6 is expressed in metastatic prostate cancer cells, PC-3, which do not express NKX3.1.71 Interestingly, IL-6 is undetectable in LNCaP cells that express NKX3.1.71,77 Our studies suggest that loss of NKX3.1 results in dysregulation of RAMP1, which leads to activation of cell survival pathways.
RAMP1 has been proposed as a potential biomarker in prostate cancer.8 Because RAMP1 presents G protein-coupled receptors that bind ligands known to promote prostate tumorigenesis to the cell membrane, it could potentially be a molecular target for the diagnosis and treatment of prostate cancer. The use of a monoclonal antibody to therapeutically target RAMP1-expressing cells may prove beneficial in treating patients with prostate cancer, or may be ideal for imaging prostate lesions.
Acknowledgments
We thank Drs. Meejeon Roh and Josiah Ochieng for technical assistance and members of the Abdulkadir laboratory for helpful discussions.
Footnotes
Supported by National Cancer Institute grant R01 CA094858 (S.A.A.), Vanderbilt Ingram Cancer Center Support grant P30 CA068485, National Heart, Lung, and Blood Institute grant 2T32H007735-17 (M.L.), and Department of Defense Postdoctoral Fellowship Award W81XWH-11-1-0230 (P.D.A.).
Supplemental Data
Supplemental Data
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2013.05.021.
References
- 1.McLatchie L.M., Fraser N.J., Main M.J., Wise A., Brown J., Thompson N., Solari R., Lee M.G., Foord S.M. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature. 1998;393:333–339. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- 2.Bouschet T., Martin S., Henley J.M. Regulation of calcium-sensing-receptor trafficking and cell-surface expression by GPCRs and RAMPs. Trends Pharmacol Sci. 2008;29:633–639. doi: 10.1016/j.tips.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouschet T., Martin S., Henley J.M. Receptor-activity-modifying proteins are required for forward trafficking of the calcium-sensing receptor to the plasma membrane. J Cell Sci. 2005;118:4709–4720. doi: 10.1242/jcs.02598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christopoulos A., Christopoulos G., Morfis M., Udawela M., Laburthe M., Couvineau A., Kuwasako K., Tilakaratne N., Sexton P.M. Novel receptor partners and function of receptor activity-modifying proteins. J Biol Chem. 2003;278:3293–3297. doi: 10.1074/jbc.C200629200. [DOI] [PubMed] [Google Scholar]
- 5.Wootten D., Lindmark H., Kadmiel M., Willcockson H., Caron K.M., Barwell J., Drmota T., Poyner D.R. Receptor activity modifying proteins (RAMPs) interact with the VPAC2 receptor and CRF1 receptors and modulate their function. Br J Pharmacol. 2013;168:822–834. doi: 10.1111/j.1476-5381.2012.02202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morimoto R., Satoh F., Murakami O., Hirose T., Totsune K., Imai Y., Arai Y., Suzuki T., Sasano H., Ito S., Takahashi K. Expression of adrenomedullin 2/intermedin in human adrenal tumors and attached non-neoplastic adrenal tissues. J Endocrinol. 2008;198:175–183. doi: 10.1677/JOE-08-0103. [DOI] [PubMed] [Google Scholar]
- 7.Aarhus M., Bruland O., Bredholt G., Lybaek H., Husebye E.S., Krossnes B.K., Vedeler C., Wester K., Lund-Johansen M., Knappskog P.M. Microarray analysis reveals down-regulation of the tumour suppressor gene WWOX and up-regulation of the oncogene TYMS in intracranial sporadic meningiomas. J Neurooncol. 2008;88:251–259. doi: 10.1007/s11060-008-9569-6. [DOI] [PubMed] [Google Scholar]
- 8.Romanuik T.L., Ueda T., Le N., Haile S., Yong T.M., Thomson T., Vessella R.L., Sadar M.D. Novel biomarkers for prostate cancer including noncoding transcripts. Am J Pathol. 2009;175:2264–2276. doi: 10.2353/ajpath.2009.080868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He W.W., Sciavolino P.J., Wing J., Augustus M., Hudson P., Meissner P.S., Curtis R.T., Shell B.K., Bostwick D.G., Tindall D.J., Gelmann E.P., Abate-Shen C., Carter K.C. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics. 1997;43:69–77. doi: 10.1006/geno.1997.4715. [DOI] [PubMed] [Google Scholar]
- 10.Bieberich C.J., Fujita K., He W.W., Jay G. Prostate-specific and androgen-dependent expression of a novel homeobox gene. J Biol Chem. 1996;271:31779–31782. doi: 10.1074/jbc.271.50.31779. [DOI] [PubMed] [Google Scholar]
- 11.Bhatia-Gaur R., Donjacour A.A., Sciavolino P.J., Kim M., Desai N., Young P., Norton C.R., Gridley T., Cardiff R.D., Cunha G.R., Abate-Shen C., Shen M.M. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 1999;13:966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdulkadir S.A., Magee J.A., Peters T.J., Kaleem Z., Naughton C.K., Humphrey P.A., Milbrandt J. Conditional loss of Nkx3.1 in adult mice induces prostatic intraepithelial neoplasia. Mol Cell Biol. 2002;22:1495–1503. doi: 10.1128/mcb.22.5.1495-1503.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim M.J., Bhatia-Gaur R., Banach-Petrosky W.A., Desai N., Wang Y., Hayward S.W., Cunha G.R., Cardiff R.D., Shen M.M., Abate-Shen C. Nkx3.1 mutant mice recapitulate early stages of prostate carcinogenesis. Cancer Res. 2002;62:2999–3004. [PubMed] [Google Scholar]
- 14.Tanaka M., Komuro I., Inagaki H., Jenkins N.A., Copeland N.G., Izumo S. Nkx3.1, a murine homolog of Drosophila bagpipe, regulates epithelial ductal branching and proliferation of the prostate and palatine glands. Dev Dyn. 2000;219:248–260. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1054>3.3.co;2-5. [DOI] [PubMed] [Google Scholar]
- 15.Asatiani E., Huang W.X., Wang A., Rodriguez Ortner E., Cavalli L.R., Haddad B.R., Gelmann E.P. Deletion, methylation, and expression of the NKX3.1 suppressor gene in primary human prostate cancer. Cancer Res. 2005;65:1164–1173. doi: 10.1158/0008-5472.CAN-04-2688. [DOI] [PubMed] [Google Scholar]
- 16.Vocke C.D., Pozzatti R.O., Bostwick D.G., Florence C.D., Jennings S.B., Strup S.E., Duray P.H., Liotta L.A., Emmert-Buck M.R., Linehan W.M. Analysis of 99 microdissected prostate carcinomas reveals a high frequency of allelic loss on chromosome 8p12-21. Cancer Res. 1996;56:2411–2416. [PubMed] [Google Scholar]
- 17.Bowen C., Bubendorf L., Voeller H.J., Slack R., Willi N., Sauter G., Gasser T.C., Koivisto P., Lack E.E., Kononen J., Kallioniemi O.P., Gelmann E.P. Loss of NKX3.1 expression in human prostate cancers correlates with tumor progression. Cancer Res. 2000;60:6111–6115. [PubMed] [Google Scholar]
- 18.Akamatsu S., Takata R., Ashikawa K., Hosono N., Kamatani N., Fujioka T., Ogawa O., Kubo M., Nakamura Y., Nakagawa H. A functional variant in NKX3.1 associated with prostate cancer susceptibility down-regulates NKX3.1 expression. Hum Mol Genet. 2010;19:4265–4272. doi: 10.1093/hmg/ddq350. [DOI] [PubMed] [Google Scholar]
- 19.Gelmann E.P., Steadman D.J., Ma J., Ahronovitz N., Voeller H.J., Swope S., Abbaszadegan M., Brown K.M., Strand K., Hayes R.B., Stampfer M.J. Occurrence of NKX3.1 C154T polymorphism in men with and without prostate cancer and studies of its effect on protein function. Cancer Res. 2002;62:2654–2659. [PubMed] [Google Scholar]
- 20.Kwon E.M., Holt S.K., Fu R., Kolb S., Williams G., Stanford J.L., Ostrander E.A. Androgen metabolism and JAK/STAT pathway genes and prostate cancer risk. Cancer Epidemiol. 2012;36:347–353. doi: 10.1016/j.canep.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng S.L., Ju J.H., Chang B.L., Ortner E., Sun J., Isaacs S.D., Sun J., Wiley K.E., Liu W., Zemedkun M., Walsh P.C., Ferretti J., Gruschus J., Isaacs W.B., Gelmann E.P., Xu J. Germ-line mutation of NKX3.1 cosegregates with hereditary prostate cancer and alters the homeodomain structure and function. Cancer Res. 2006;66:69–77. doi: 10.1158/0008-5472.CAN-05-1550. [DOI] [PubMed] [Google Scholar]
- 22.Abate-Shen C., Banach-Petrosky W.A., Sun X., Economides K.D., Desai N., Gregg J.P., Borowsky A.D., Cardiff R.D., Shen M.M. Nkx3.1; Pten mutant mice develop invasive prostate adenocarcinoma and lymph node metastases. Cancer Res. 2003;63:3886–3890. [PubMed] [Google Scholar]
- 23.Kim M.J., Cardiff R.D., Desai N., Banach-Petrosky W.A., Parsons R., Shen M.M., Abate-Shen C. Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci U S A. 2002;99:2884–2889. doi: 10.1073/pnas.042688999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellwood-Yen K., Graeber T.G., Wongvipat J., Iruela-Arispe M.L., Zhang J., Matusik R., Thomas G.V., Sawyers C.L. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 25.Iwata T., Schultz D., Hicks J., Hubbard G.K., Mutton L.N., Lotan T.L., Bethel C., Lotz M.T., Yegnasubramanian S., Nelson W.G., Dang C.V., Xu M., Anele U., Koh C.M., Bieberich C.J., De Marzo A.M. MYC overexpression induces prostatic intraepithelial neoplasia and loss of Nkx3.1 in mouse luminal epithelial cells. PLoS One. 2010;5:e9427. doi: 10.1371/journal.pone.0009427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson P.D., McKissic S.A., Logan M., Roh M., Franco O.E., Wang J., Doubinskaia I., van der Meer R., Hayward S.W., Eischen C.M., Eltoum I.E., Abdulkadir S.A. Nkx3.1 and Myc crossregulate shared target genes in mouse and human prostate tumorigenesis. J Clin Invest. 2012;122:1907–1919. doi: 10.1172/JCI58540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouyang X., DeWeese T.L., Nelson W.G., Abate-Shen C. Loss-of-function of Nkx3.1 promotes increased oxidative damage in prostate carcinogenesis. Cancer Res. 2005;65:6773–6779. doi: 10.1158/0008-5472.CAN-05-1948. [DOI] [PubMed] [Google Scholar]
- 28.Magee J.A., Abdulkadir S.A., Milbrandt J. Haploinsufficiency at the Nkx3.1 locus: a paradigm for stochastic, dosage-sensitive gene regulation during tumor initiation. Cancer Cell. 2003;3:273–283. doi: 10.1016/s1535-6108(03)00047-3. [DOI] [PubMed] [Google Scholar]
- 29.Wang J. Pim1 kinase is required to maintain tumorigenicity in MYC-expressing prostate cancer cells. Oncogene. 2012;31:1794–1803. doi: 10.1038/onc.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X., Zayzafoon M., Zhang X., Hameed O. Is there a role for fatty acid synthase in the diagnosis of prostatic adenocarcinoma? a comparison with AMACR. Am J Clin Pathol. 2011;136:239–246. doi: 10.1309/AJCP0Y5QWWYDKCJE. [DOI] [PubMed] [Google Scholar]
- 31.Rhodes D.R., Kalyana-Sundaram S., Mahavisno V., Varambally R., Yu J., Briggs B.B., Barrette T.R., Anstet M.J., Kincead-Beal C., Kulkarni P., Varambally S., Ghosh D., Chinnaiyan A.M. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roh M., van der Meer R., Abdulkadir S.A. Tumorigenic polyploid cells contain elevated ROS and ARE selectively targeted by antioxidant treatment. J Cell Physiol. 2012;227:801–812. doi: 10.1002/jcp.22793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roh M., Gary B., Song C., Said-Al-Naief N., Tousson A., Kraft A., Eltoum I.E., Abdulkadir S.A. Overexpression of the oncogenic kinase Pim-1 leads to genomic instability. Cancer Res. 2003;63:8079–8084. [PubMed] [Google Scholar]
- 34.Roh M., Song C., Kim J., Abdulkadir S.A. Chromosomal instability induced by Pim-1 is passage-dependent and associated with dysregulation of cyclin B1. J Biol Chem. 2005;280:40568–40577. doi: 10.1074/jbc.M509369200. [DOI] [PubMed] [Google Scholar]
- 35.Abdulkadir S.A., Qu Z., Garabedian E., Song S.K., Peters T.J., Svaren J., Carbone J.M., Naughton C.K., Catalona W.J., Ackerman J.J., Gordon J.I., Humphrey P.A., Milbrandt J. Impaired prostate tumorigenesis in Egr1-deficient mice. Nat Med. 2001;7:101–107. doi: 10.1038/83231. [DOI] [PubMed] [Google Scholar]
- 36.Mogal A.P., van der Meer R., Crooke P.S., Abdulkadir S.A. Haploinsufficient prostate tumor suppression by Nkx3.1: a role for chromatin accessibility in dosage-sensitive gene regulation. J Biol Chem. 2007;282:25790–25800. doi: 10.1074/jbc.M702438200. [DOI] [PubMed] [Google Scholar]
- 37.Kim J., Roh M., Abdulkadir S.A. Pim1 promotes human prostate cancer cell tumorigenicity and c-MYC transcriptional activity. BMC Cancer. 2010;10:248. doi: 10.1186/1471-2407-10-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez E.E., Anderson P.D., Logan M., Abdulkadir S.A. Antioxidant treatment promotes prostate epithelial proliferation in nkx3.1 mutant mice. PLoS One. 2012;7:e46792. doi: 10.1371/journal.pone.0046792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gautier L., Cope L., Bolstad B.M., Irizarry R.A. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 40.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 41.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su A.I., Welsh J.B., Sapinoso L.M., Kern S.G., Dimitrov P., Lapp H., Schultz P.G., Powell S.M., Moskaluk C.A., Frierson H.F., Jr., Hampton G.M. Molecular classification of human carcinomas by use of gene expression signatures. Cancer Res. 2001;61:7388–7393. [PubMed] [Google Scholar]
- 43.Welsh J.B., Sapinoso L.M., Su A.I., Kern S.G., Wang-Rodriguez J., Moskaluk C.A., Frierson H.F., Jr., Hampton G.M. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 2001;61:5974–5978. [PubMed] [Google Scholar]
- 44.Vanaja D.K., Cheville J.C., Iturria S.J., Young C.Y. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res. 2003;63:3877–3882. [PubMed] [Google Scholar]
- 45.Liu P., Ramachandran S., Ali Seyed M., Scharer C.D., Laycock N., Dalton W.B., Williams H., Karanam S., Datta M.W., Jaye D.L., Moreno C.S. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006;66:4011–4019. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 46.Arredouani M.S., Lu B., Bhasin M., Eljanne M., Yue W., Mosquera J.M., Bubley G.J., Li V., Rubin M.A., Libermann T.A., Sanda M.G. Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin Cancer Res. 2009;15:5794–5802. doi: 10.1158/1078-0432.CCR-09-0911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Creighton C.J., Hilger A.M., Murthy S., Rae J.M., Chinnaiyan A.M., El-Ashry D. Activation of mitogen-activated protein kinase in estrogen receptor alpha-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor alpha-negative human breast tumors. Cancer Res. 2006;66:3903–3911. doi: 10.1158/0008-5472.CAN-05-4363. [DOI] [PubMed] [Google Scholar]
- 49.Chrissobolis S., Zhang Z., Kinzenbaw D.A., Lynch C.M., Russo A.F., Faraci F.M. Receptor activity-modifying protein-1 augments cerebrovascular responses to calcitonin gene-related peptide and inhibits angiotensin II-induced vascular dysfunction. Stroke. 2010;41:2329–2334. doi: 10.1161/STROKEAHA.110.589648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sabharwal R., Zhang Z., Lu Y., Abboud F.M., Russo A.F., Chapleau M.W. Receptor activity-modifying protein 1 increases baroreflex sensitivity and attenuates angiotensin-induced hypertension. Hypertension. 2010;55:627–635. doi: 10.1161/HYPERTENSIONAHA.109.148171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagakawa O., Ogasawara M., Fujii H., Murakami K., Murata J., Fuse H., Saiki I. Effect of prostatic neuropeptides on invasion and migration of PC-3 prostate cancer cells. Cancer Lett. 1998;133:27–33. doi: 10.1016/s0304-3835(98)00186-4. [DOI] [PubMed] [Google Scholar]
- 52.Nagakawa O., Ogasawara M., Murata J., Fuse H., Saiki I. Effect of prostatic neuropeptides on migration of prostate cancer cell lines. Int J Urol. 2001;8:65–70. doi: 10.1046/j.1442-2042.2001.00250.x. [DOI] [PubMed] [Google Scholar]
- 53.Toda M., Suzuki T., Hosono K., Hayashi I., Hashiba S., Onuma Y., Amano H., Kurihara Y., Kurihara H., Okamoto H., Hoka S., Majima M. Neuronal system-dependent facilitation of tumor angiogenesis and tumor growth by calcitonin gene-related peptide. Proc Natl Acad Sci U S A. 2008;105:13550–13555. doi: 10.1073/pnas.0800767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki K., Kobayashi Y., Morita T. Serum calcitonin gene-related peptide levels in untreated prostate cancer patients. Int J Urol. 2006;13:781–784. doi: 10.1111/j.1442-2042.2006.01402.x. [DOI] [PubMed] [Google Scholar]
- 55.Yu X.J., Li C.Y., Wang K.Y., Dai H.Y. Calcitonin gene-related peptide regulates the expression of vascular endothelial growth factor in human HaCaT keratinocytes by activation of ERK1/2 MAPK. Regul Pept. 2006;137:134–139. doi: 10.1016/j.regpep.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 56.Tomic T.T., Gustavsson H., Wang W., Jennbacken K., Welen K., Damber J.E. Castration resistant prostate cancer is associated with increased blood vessel stabilization and elevated levels of VEGF and Ang-2. Prostate. 2012;72:705–712. doi: 10.1002/pros.21472. [DOI] [PubMed] [Google Scholar]
- 57.Zhang P., Liu W., Zhang J., Guan H., Chen W., Cui X., Liu Q., Jiang A. Gene expression profiles in the PC-3 human prostate cancer cells induced by NKX3.1. Mol Biol Rep. 2010;37:1505–1512. doi: 10.1007/s11033-009-9549-8. [DOI] [PubMed] [Google Scholar]
- 58.Zhang H., Muders M.H., Li J., Rinaldo F., Tindall D.J., Datta K. Loss of NKX3.1 favors vascular endothelial growth factor-C expression in prostate cancer. Cancer Res. 2008;68:8770–8778. doi: 10.1158/0008-5472.CAN-08-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korherr C., Gille H., Schafer R., Koenig-Hoffmann K., Dixelius J., Egland K.A., Pastan I., Brinkmann U. Identification of proangiogenic genes and pathways by high-throughput functional genomics: TBK1 and the IRF3 pathway. Proc Natl Acad Sci U S A. 2006;103:4240–4245. doi: 10.1073/pnas.0511319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Helgason E., Phung Q.T., Dueber E.C. Recent insights into the complexity of Tank-binding kinase 1 signaling networks: the emerging role of cellular localization in the activation and substrate specificity of TBK1. FEBS Lett. 2013;587:1230–1237. doi: 10.1016/j.febslet.2013.01.059. [DOI] [PubMed] [Google Scholar]
- 61.Fernandez-Martinez A.B., Bajo A.M., Isabel Arenas M., Sanchez-Chapado M., Prieto J.C., Carmena M.J. Vasoactive intestinal peptide (VIP) induces malignant transformation of the human prostate epithelial cell line RWPE-1. Cancer Lett. 2010;299:11–21. doi: 10.1016/j.canlet.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 62.Fernandez-Martinez A.B., Bajo A.M., Valdehita A., Isabel Arenas M., Sanchez-Chapado M., Carmena M.J., Prieto J.C. Multifunctional role of VIP in prostate cancer progression in a xenograft model: suppression by curcumin and COX-2 inhibitor NS-398. Peptides. 2009;30:2357–2364. doi: 10.1016/j.peptides.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 63.Collado B., Carmena M.J., Clemente C., Prieto J.C., Bajo A.M. Vasoactive intestinal peptide enhances growth and angiogenesis of human experimental prostate cancer in a xenograft model. Peptides. 2007;28:1896–1901. doi: 10.1016/j.peptides.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 64.Bonkhoff H., Wernert N., Dhom G., Remberger K. Relation of endocrine-paracrine cells to cell proliferation in normal, hyperplastic, and neoplastic human prostate. Prostate. 1991;19:91–98. doi: 10.1002/pros.2990190202. [DOI] [PubMed] [Google Scholar]
- 65.Xie Y., Wolff D.W., Lin M.F., Tu Y. Vasoactive intestinal peptide transactivates the androgen receptor through a protein kinase A-dependent extracellular signal-regulated kinase pathway in prostate cancer LNCaP cells. Mol Pharmacol. 2007;72:73–85. doi: 10.1124/mol.107.033894. [DOI] [PubMed] [Google Scholar]
- 66.Hernes E., Fossa S.D., Berner A., Otnes B., Nesland J.M. Expression of the epidermal growth factor receptor family in prostate carcinoma before and during androgen-independence. Br J Cancer. 2004;90:449–454. doi: 10.1038/sj.bjc.6601536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zellweger T., Ninck C., Bloch M., Mirlacher M., Koivisto P.A., Helin H.J., Mihatsch M.J., Gasser T.C., Bubendorf L. Expression patterns of potential therapeutic targets in prostate cancer. Int J Cancer. 2005;113:619–628. doi: 10.1002/ijc.20615. [DOI] [PubMed] [Google Scholar]
- 68.Wells A. EGF receptor. Int J Biochem Cell Biol. 1999;31:637–643. doi: 10.1016/s1357-2725(99)00015-1. [DOI] [PubMed] [Google Scholar]
- 69.Culig Z., Bartsch G., Hobisch A. Interleukin-6 regulates androgen receptor activity and prostate cancer cell growth. Mol Cell Endocrinol. 2002;197:231–238. doi: 10.1016/s0303-7207(02)00263-0. [DOI] [PubMed] [Google Scholar]
- 70.Smith P.C., Hobisch A., Lin D.L., Culig Z., Keller E.T. Interleukin-6 and prostate cancer progression. Cytokine Growth Factor Rev. 2001;12:33–40. doi: 10.1016/s1359-6101(00)00021-6. [DOI] [PubMed] [Google Scholar]
- 71.Chung T.D., Yu J.J., Kong T.A., Spiotto M.T., Lin J.M. Interleukin-6 activates phosphatidylinositol-3 kinase, which inhibits apoptosis in human prostate cancer cell lines. Prostate. 2000;42:1–7. doi: 10.1002/(sici)1097-0045(20000101)42:1<1::aid-pros1>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 72.Wegiel B., Bjartell A., Culig Z., Persson J.T. Interleukin-6 activates PI3K/Akt pathway and regulates cyclin A1 to promote prostate cancer cell survival. Int J Cancer. 2007;122:1521–1529. doi: 10.1002/ijc.23261. [DOI] [PubMed] [Google Scholar]
- 73.Adler H.L., McCurdy M.A., Kattan M.W., Timme T.L., Scardino P.T., Thompson T.C. Elevated levels of circulating interleukin-6 and transforming growth factor-beta1 in patients with metastatic prostatic carcinoma. J Urol. 1999;161:182–187. [PubMed] [Google Scholar]
- 74.Hoosein N., Abdul M., McCabe R., Gero E., Deftos L., Banks M., Hodges S., Finn L., Logothetis C. Clinical significance of elevation in neuroendocrine factors and interleukin-6 in metastatic prostate cancer. Urol Oncol. 1995;1:246–251. doi: 10.1016/1078-1439(96)00012-9. [DOI] [PubMed] [Google Scholar]
- 75.Twillie D.A., Eisenberger M.A., Carducci M.A., Hseih W.S., Kim W.Y., Simons J.W. Interleukin-6: a candidate mediator of human prostate cancer morbidity. Urology. 1995;45:542–549. doi: 10.1016/S0090-4295(99)80034-X. [DOI] [PubMed] [Google Scholar]
- 76.Drachenberg D.E., Elgamal A.A., Rowbotham R., Peterson M., Murphy G.P. Circulating levels of interleukin-6 in patients with hormone refractory prostate cancer. Prostate. 1999;41:127–133. doi: 10.1002/(sici)1097-0045(19991001)41:2<127::aid-pros7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 77.Hobisch A., Ramoner R., Fuchs D., Godoy-Tundidor S., Bartsch G., Klocker H., Culig Z. Prostate cancer cells (LNCaP) generated after long-term interleukin 6 (IL-6) treatment express IL-6 and acquire an IL-6 partially resistant phenotype. Clin Cancer Res. 2001;7:2941–2948. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.