Abstract
The three-dimensional structure of the 81-residue mercury transporter MerF determined in liquid crystalline phospholipid bilayers under physiological conditions by Rotationally Aligned (RA) solid-state NMR has two long helices, which extend well beyond the bilayer, with a well-defined inter-helical loop. Truncation of the N-terminal 12 residues, which are mobile and unstructured when the protein is solubilized in micelles, results in a large structural rearrangement of the protein in bilayers. In the full-length protein, the N-terminal helix is aligned nearly parallel to the membrane normal and forms an extension of the first transmembrane helix. By contrast this helix adopts a perpendicular orientation in the truncated protein. The close spatial proximity of the two Cys-containing metal binding sites in the three-dimensional structure of full-length MerF provides insights into possible transport mechanisms. These results demonstrate that major changes in protein structure can result from differences in amino acid sequence (e.g. full-length vs. truncated proteins) as well as the use of a non-native membrane mimetic environment (e.g. micelles) vs. liquid crystalline phospholipid bilayers. They provide further evidence of the importance of studying unmodified membrane proteins in near-native bilayer environments in order to obtain accurate structures that can be related to their functions.
The three-dimensional structure of a protein is determined by two principal factors: its sequence of amino acids and its environment1. For globular proteins that are soluble in aqueous solution the focus of the ‘folding problem’ has been on the sequence of amino acids, while the environmental contribution has been largely ignored because various solvent conditions and even crystallization generally result in minimal perturbation of their structures, as demonstrated by many examples where essentially the same structures have been determined by X-ray crystallography and solution NMR. For membrane proteins, in contrast, drastic modifications have been made to both the amino acid sequences and the choice of membrane mimicking environments to facilitate structure determination. For example, to enhance crystallization, amino acid sequences have been subject to multiple mutations, truncations of termini and loops, and insertions of globular proteins, such as T4 lysozyme, in the middle of the protein2. A wide range of membrane mimetic environments has been used in studies of membrane proteins by both X-ray crystallography and solution NMR3, including mixed organic solvents, detergent micelles, isotropic bicelles, nanodiscs, and lipid cubic phases of monoolein and cholesterol4. Relatively little is known about the effects of these environments on membrane protein structures because so few structures have been determined in liquid crystalline phospholipid bilayers for comparisons.
Rotationally aligned (RA) solid-state NMR5,6 merges elements of Oriented Sample (OS) solid-state NMR5, as performed on stationary, aligned samples, with those of Magic Angle Spinning (MAS) solid-state NMR of unoriented ‘powder’ samples7. The robustness of RA solid-state NMR has been demonstrated by the structure determination of a 350-residue G-protein coupled receptor (GPCR) in phospholipid bilayers8. The most distinctive feature of the method is its reliance on a universal property of membrane proteins, which is that they undergo rapid rotational diffusion about the bilayer normal in liquid crystalline phospholipid bilayers9. In early high resolution solid-state NMR experiments it was shown that motion about a single axis yields an axially symmetric powder pattern with its frequency breadth scaled by the angle between the principal axis of the nuclear spin interaction, e.g., the bond axis in a two-spin dipolar couplings (DC)10. The same spectroscopic principles applied to chemical shift anisotropy (CSA) in 31P NMR spectra of phospholipids11 and 13C’ NMR spectra of bacteriorhodopsin12 demonstrated that both lipid and protein components undergo rapid rotational diffusion in liquid crystalline bilayers. The frequency of the parallel edge of the motionally scaled powder patterns is exactly the same as that measured from the single-line resonance signals observed from stationary, aligned samples in OS solid-state NMR spectra. Thus, it is possible to obtain the same angular restraints for the calculation of the three-dimensional structures of the membrane proteins from stationary mechanically or magnetically aligned, or unoriented proteoliposome samples5. The spectral resolution for the unoriented samples results from the application of MAS solid-state NMR to the uniformly 13C and 15N labeled proteins, which is followed by recoupling of the motionally averaged powder patterns13,14.
Mercury transport membrane proteins play a crucial role in the bacterial mercury detoxification system15–17. They transport ionic mercury from the periplasmic protein MerP18–20 across the bilayer to MerA21,22, the enzyme that reduces Hg(II) to Hg(0). The 81-residue MerF23 has two trans-membrane helices and two mercury-binding sites, both of which contain a pair of cysteine residues. We have expressed MerF without the N-terminal methionine and with the cysteines mutated to serines (Figure 1). In principle, this is the same as studying an unmodified MerF protein because the spectra of polypeptides with and without Ser substituted for Cys are identical. However, the absence of cysteines increases the stability of the samples; future studies will compare these and other mutations of MerF to understand structure-function relationships. Since the initial solution NMR spectra of MerF in micelles24 were dominated by intense, narrow resonances from mobile residues near the N-and C-termini, a total of 12-residues from the N-terminus and 9-residues from the C-terminus were removed, eliminating all of the mobile residues to form a 60-residue truncated protein, which retained the helix-loop-helix core of the native protein. This was the first example of a membrane protein with more than a single trans-membrane helix used in the development of many NMR methods. We have determined the three-dimensional structure of MerFt (truncated) in micelles by solution NMR24, in magnetically aligned bilayers by OS solid-state NMR25, and in proteoliposomes by RA solid-state NMR6.
FIGURE 1.

Amino acid sequences of full-length 81-residue MerF and truncated 60-residue MerFt. The last residue of both polypeptides is homoserine (denoted as S*) that results from cleavage from the fusion protein.
Here we describe the structure determination of the full-length MerF protein by RA solid-state NMR in its native phospholipid bilayer environment. This structure provides some of the first insights into its mechanism of transporting metal ions across into the cell. It also enables direct comparisons of the structures of the full-length and truncated forms of the protein under identical experimental conditions. Remarkably, we find drastic structural perturbations caused by removal of the N-terminal residues. These residues are mobile and unstructured in micelles, but are differently structured in the full-length and truncated forms of the protein in bilayers.
MerF was isolated and purified from bacteria, and reconstituted into 1,2-di-O-tetradecyl-sn-glycero-3-phosphocholine (14-o-PC) proteoliposomes. The significant reduction of the 13C’ powder pattern due to motional averaging verifies that the protein is undergoing rotational diffusion fast enough (>106 Hz) to fully average the powder patterns of the 13C and 15N nuclear spin interactions at 25°C, a temperature above the gel to liquid crystalline phase transition of protein-containing 14-o-PC bilayers.
The analysis of three-dimensional HnNCo, HnNCa and HcCxCx MAS solid-state NMR spectra provided simultaneous resonance assignments and measurements of structural restraints in the form of DCs (Supplementary Figure 1). The first benefit of the approach is the improvement of the spectral resolution through the inclusion of a DC frequency dimension in each experiment. The site-to-site variation of the motionally averaged DC frequencies is able to resolve signals from the same types of amino acids even in situations where their 13Cα, 13C’ and amide 15N shifts have similar frequencies. The second benefit is that only two magnetization transfer steps are required, compared to the three or more transfer steps in resonance assignment methods based on the observation of isotropic chemical shifts26,27. Not only is the sensitivity significantly improved, but also the total number of three-dimensional experiments is reduced. Multiple-contact cross polarization28 was incorporated into MAS pulse sequences capable of efficient dipolar recoupling using R187 129. Notably, the approach is applicable only to membrane proteins undergoing rotational diffusion, and not polycrystalline or other immobile proteins.
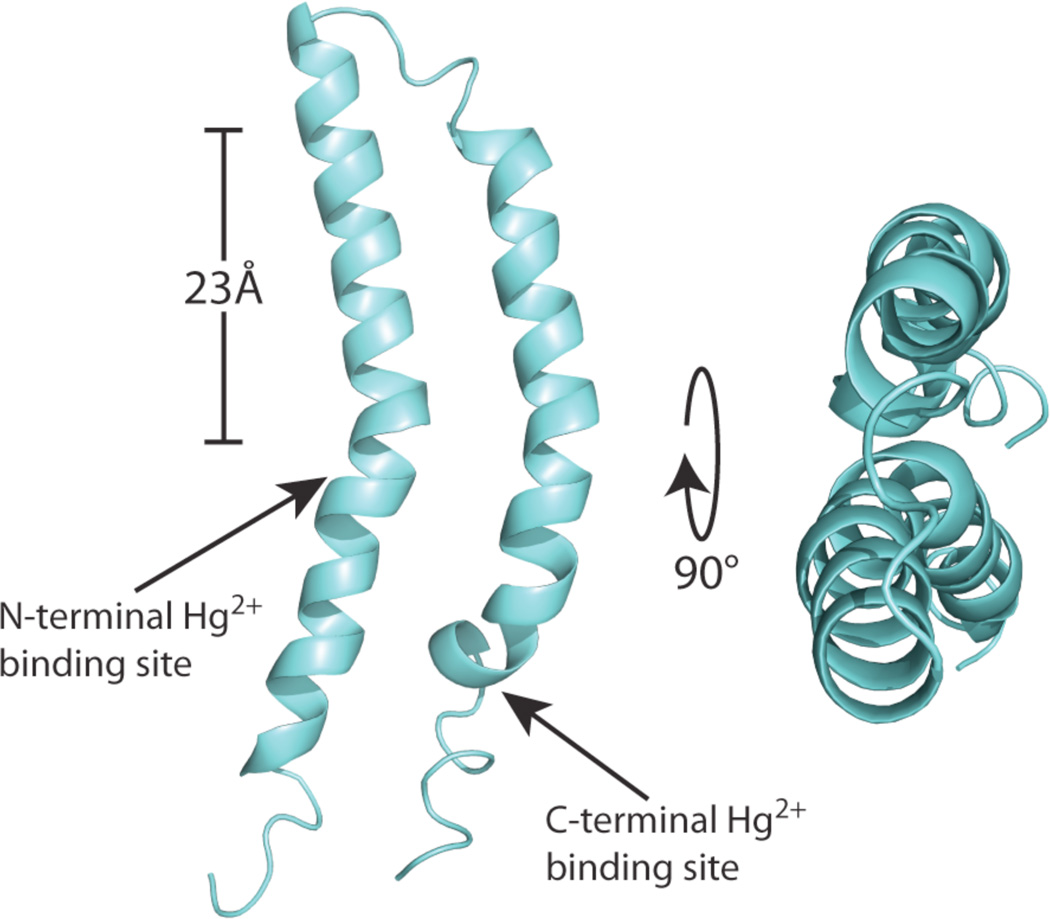
A long-range inter-helical distance restraint between F23 and Q63 or Q67 unambiguously assigned from 13C-13C correlation spectra due to the distinctive chemical shifts of carbons in aromatic rings and Cγ of Gln (Supplementary material) was incorporated along with the DC frequencies in the calculation of the protein structure following a protocol similar to that described previously6. The structure of MerF in phospholipid bilayers is shown in Figure 2.
FIGURE 2.
The three-dimensional structure of MerF is shown as a ribbon diagram in aqua with the two mercury-binding sites labeled with arrows. The average backbone pairwise rmsd is 1.5 Å. The scale bar corresponds to the 23Å thickness of the hydrocarbons in 14-o-PC lipid bilayer. Both termini of the protein are in the cytosol. The coordinates are deposited in the PDB as 2m67 and the associated data in the BMRB as 19115.
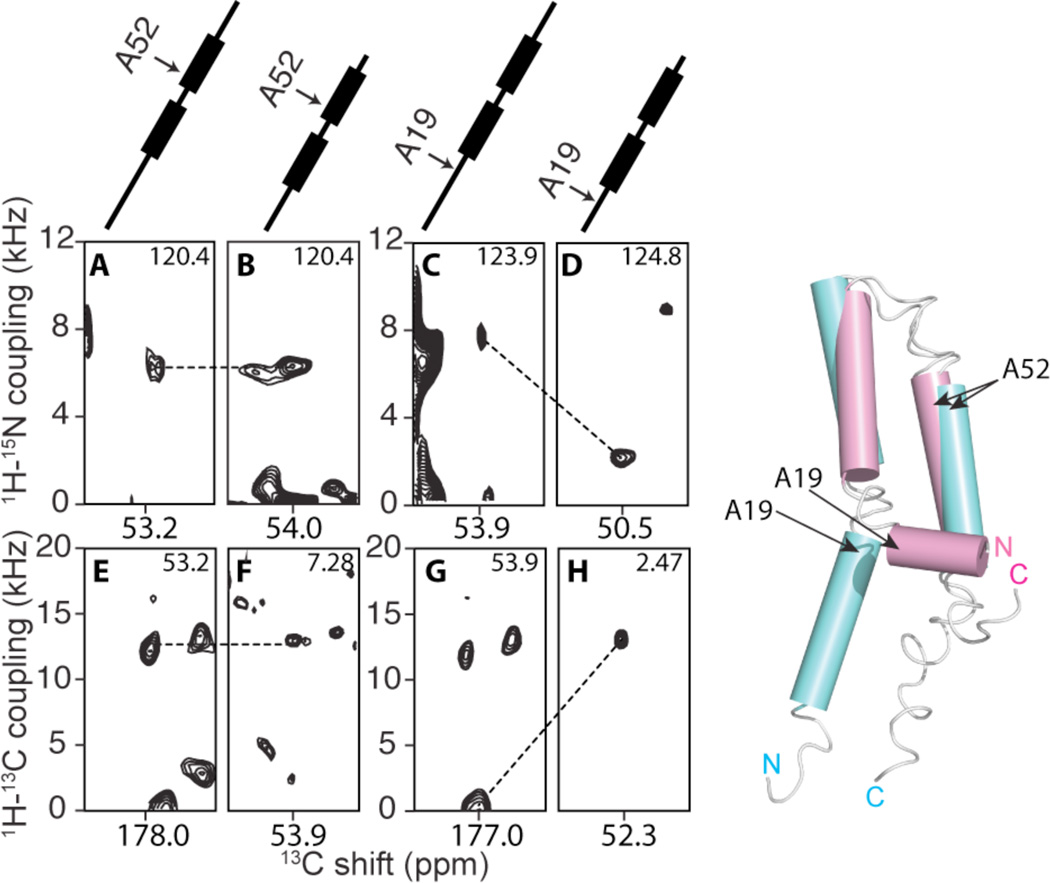
The motionally averaged DC values provide a direct measurement of the orientations of individual bonds in the polypeptide relative to a common axis, the bilayer normal. Comparing the DCs of the full-length protein to those of the truncated protein6, it is clear that the N-terminal portion of the protein, residue I13 through Pro25, has a significantly different structure as a consequence of the truncation performed 12 residues away from Pro25. In contrast, the core of the protein, including the two transmembrane helices and the inter-helical loop, is nearly identical in the full-length and truncated proteins. As examples, the DCs for residues Ala52 (unchanged) and Ala19 (dramatically changed) are compared in Figure 3.
FIGURE 3.
Left: Observed changes in orientationally dependent frequencies demonstrate drastic changes in the structure of MerF caused by truncation of residues at the N-terminus. At the top are schematic drawings of the MerF secondary structure (thicker lines are trans-membrane helices) marked with the positions of the residues contributing to the signals. A, C, E and G are slices from spectra of MerF. B, D, F and H are slices from spectra of MerFt. The left panels (A, B, E and F) are from Ala52 located in the second trans-membrane helix. Both the 1H-15N and 1H-13C DCs of Ala52 are very similar for the full-length and truncated proteins. The right panels (C, D, G and H) are from Ala19, located in the N-terminal region, where the conformational change occurs and consequently large changes in DC values are observed. All slices are extracted from three-dimensional spectra at the noted values of the third dimension. Right: The structure of the truncated 60-residue protein (magenta) is superimposed on the structure of the full-length 81-residue protein (aqua).
The superposition of the structures of the full-length (aqua) and truncated (magenta) proteins in Figure 3 is highly informative. The truncation of the N-terminal residues, which are mobile and unstructured in micelles, results in a dramatic rearrangement of the amphipathic helix (I13-Pro25). These residues change from an orientation parallel to the bilayer surface to one that is nearly perpendicular to bilayer surface and appears to be a continuation of the first trans-membrane helix that extends out of the bilayer. Both structures were determined under identical sample conditions and methods; therefore, the observed structural rearrangement must result solely from the alteration of the amino acid sequence of the protein. In contrast to the N-terminal truncation, the C-terminal truncation does not result in significant structural changes. Clearly, this strengthens the requirement of caution in the use of truncations and other protein modifications of membrane proteins.
A key question in the study of the mercury transport mechanism is the location of the N-terminal Hg(II) binding site, which is shown in Figure 2 to be in close proximity to the second Hg(II) binding site, both of which are located at the bilayer’s hydrophobic to hydrophilic interface. Previous biochemical studies and predictions on various mercury transporter proteins are inconsistent in identifying the location of this binding site, ranging from the interface on the cytosolic side30 to the middle of the first transmembrane helix23. The structure in Figure 2 supports the existence of the former topology. The second important observation is the proximity of the two metal binding sites on MerF, which supports the possibility of direct intramolecular Hg(II) transfers. Notably, the existence of direct contact has been hypothesized from various studies on mercury ligand exchange22 and mercury coordination chemistry16,31. In any transport mechanism tight control of Hg(II) is essential because of its high reactivity in solution, and the only way to transfer Hg(II) while maintaining tight control is through direct contact and ligand exchange between Hg(II) binding sites.
The determination of the structure of the full-length MerF protein enables comparisons with the previously determined structure of the truncated MerFt protein. Drastic structural rearrangements are induced by truncation of terminal residues, and residues 5 – 12, which are mobile in micelles, are structured in phospholipid bilayers. Moreover, the structure of the full-length protein under near-native conditions provides insights into the mechanism of transport of mercury across from a mercury-binding protein in the periplasm to an enzyme in the cytoplasm.
Supplementary Material
ACKNOWLEDGMENT
We thank Chris Grant and Albert Wu for assistance with instrumentation. The research was supported by Grants RO1GM099986, R21GM075917, and P41EB002031 from the National Institutes of Health. It utilized the Biomedical Technology Resource for NMR Molecular Imaging of Proteins at the University of California, San Diego.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Experimental procedures, spectroscopic data, and data tables. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Anfinsen CB. Science (New York, N.Y.) 1973;181:223. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Rosenbaum D, Cherezov V, Hanson M, Rasmussen S, Thian F, Kobilka T, Choi H-J, Yao X-J, Weis W, Stevens R, Kobilka B. Science (New York, N.Y.) 2007;318:1266. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 3.Warschawski DE, Arnold AA, Beaugrand M, Gravel A, Chartrand E, Marcotte I. Biochim. Biophys. Acta -Biomembr. 2011;1808:1957. doi: 10.1016/j.bbamem.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Landau EM, Rosenbusch JP. Proc. Natl. Acad. Sci. 1996;93:14532. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park SH, Das BB, De Angelis AA, Scrima M, Opella SJ. J. Phys. Chem. B. 2010;114:13995. doi: 10.1021/jp106043w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das BB, Nothnagel HJ, Lu GJ, Son WS, Tian Y, Marassi FM, Opella SJ. J. Am. Chem. Soc. 2012;134:2047. doi: 10.1021/ja209464f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaefer J, Stejskal EO. J. Am. Chem. Soc. 1976;98:1031. [Google Scholar]
- 8.Park SH, Das BB, Casagrande F, Tian Y, Nothnagel HJ, Chu M, Kiefer H, Maier K, De Angelis AA, Marassi FM, Opella SJ. Nature. 2012;491:779. doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cone RA. Nature: New biology. 1972;236:39. doi: 10.1038/newbio236039a0. [DOI] [PubMed] [Google Scholar]
- 10.Gutowsky HS, Pake GE. J. Chem. Phys. 1950;18:162. [Google Scholar]
- 11.McLaughlin AC, Cullis PR, Hemminga MA, Hoult DI, Radda GK, Ritchie GA, Seeley PJ, Richards RE. FEBS Lett. 1975;57:213. doi: 10.1016/0014-5793(75)80719-8. [DOI] [PubMed] [Google Scholar]
- 12.Lewis BA, Harbison GS, Herzfeld J, Griffin RG. Biochemistry. 1985;24:4671. doi: 10.1021/bi00338a029. [DOI] [PubMed] [Google Scholar]
- 13.Griffin RG. Nat. Struct. Biol. 1998;5:508. doi: 10.1038/749. [DOI] [PubMed] [Google Scholar]
- 14.Chan JCC, Tycko R. J. Chem. Phys. 2003;118:8378. [Google Scholar]
- 15.Barkay T, Miller SM, Summers AO. FEMS Microbiol. Rev. 2003;27:355. doi: 10.1016/S0168-6445(03)00046-9. [DOI] [PubMed] [Google Scholar]
- 16.Melnick JG, Parkin G. Science (New York, N.Y.) 2007;317:225. doi: 10.1126/science.1144314. [DOI] [PubMed] [Google Scholar]
- 17.Parks JM, Johs A, Podar M, Bridou R, Hurt RA, Smith SD, Tomanicek SJ, Qian Y, Brown SD, Brandt CC, Palumbo AV, Smith JC, Wall JD, Elias DA, Liang L. Science (New York, N.Y.) 2013;339:1332. doi: 10.1126/science.1230667. [DOI] [PubMed] [Google Scholar]
- 18.Steele RA, Opella SJ. Biochemistry. 1997;36:6885. doi: 10.1021/bi9631632. [DOI] [PubMed] [Google Scholar]
- 19.Qian H, Sahlman L, Eriksson P-O, Hambraeus C, Edlund U, Sethson I. Biochemistry. 1998;37:9316. doi: 10.1021/bi9803628. [DOI] [PubMed] [Google Scholar]
- 20.Serre L, Rossy E, Pebay-Peyroula E, Cohen-Addad C, Covès J. J. Mol. Biol. 2004;339:161. doi: 10.1016/j.jmb.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 21.Schiering N, Kabsch W, Moore MJ, Distefano MD, Walsh CT, Pai EF. Nature. 1991;352:168. doi: 10.1038/352168a0. [DOI] [PubMed] [Google Scholar]
- 22.Johs A, Harwood IM, Parks JM, Nauss RE, Smith JC, Liang L, Miller SM. J. Mol. Biol. 2011;413:639. doi: 10.1016/j.jmb.2011.08.042. [DOI] [PubMed] [Google Scholar]
- 23.Wilson JR, Leang C, Morby AP, Hobman JL, Brown NL. FEBS Lett. 2000;472:78. doi: 10.1016/s0014-5793(00)01430-7. [DOI] [PubMed] [Google Scholar]
- 24.Howell SC, Mesleh MF, Opella SJ. Biochemistry. 2005;44:5196. doi: 10.1021/bi048095v. [DOI] [PubMed] [Google Scholar]
- 25.De Angelis AA, Howell SC, Nevzorov AA, Opella SJ. J. Am. Chem. Soc. 2006;128:12256. doi: 10.1021/ja063640w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pauli J, Baldus M, van Rossum B, de Groot H, Oschkinat H. ChemBioChem. 2001;2:272. doi: 10.1002/1439-7633(20010401)2:4<272::AID-CBIC272>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Franks W, Kloepper K, Wylie B, Rienstra C. J. Biomol. NMR. 2007;39:107. doi: 10.1007/s10858-007-9179-1. [DOI] [PubMed] [Google Scholar]
- 28.Pines A, Gibby MG, Waugh JS. J. Chem. Phys. 1973;59:569. [Google Scholar]
- 29.Levitt MH. Symmetry-Based Pulse Sequences in Magic-Angle Spinning Solid -State NMR. Chichester, UK: John Wiley & Sons; 2007. [Google Scholar]
- 30.Sahlman L, Wong W, Powlowski J. J. Biol. Chem. 1997;272:29518. doi: 10.1074/jbc.272.47.29518. [DOI] [PubMed] [Google Scholar]
- 31.Rulíšek Lr, Vondrášek J. J. Inorg. Biochem. 1998;71:115. doi: 10.1016/s0162-0134(98)10042-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.